Abstract
Phosphotyrosine hydrolysis by protein tyrosine phosphatases (PTPs) involves substrate binding by the PTP loop and closure over the active site by the WPD loop. The E loop, located immediately adjacent to the PTP and WPD loops, is conserved among human PTPs in both sequence and structure, yet the role of this loop in substrate binding/catalysis is comparatively unexplored. Hematopoietic tyrosine phosphatase (HePTP) is a member of the kinase interaction motif (KIM)-PTP family. Compared to the other PTPs, the KIM-PTPs have E loops that are unique in both sequence and structure. In order to understand the role of the E loop in the transition between the closed and open states of HePTP, we identified a novel crystal form of HePTP that allowed the closed-to-open state transition to be observed within a single crystal form. These structures, which include the first structure of the HePTP open state, show that the WPD loop adopts an ‘atypically open’ conformation and, importantly, that ligands can be exchanged at the active site, critical for HePTP inhibitor development. These structures also show that tetrahedral oxyanions bind at a novel, secondary site and function to coordinate the PTP, WPD and E loops. Finally, using both structural and kinetic data, we reveal a novel role for E loop residue Lys182 in enhancing HePTP catalytic activity through its interaction with Asp236 of the WPD loop, providing the first evidence for coordinated dynamics of the WPD and E loops in the catalytic cycle which, as we show, are relevant to multiple PTP families.
Keywords: Hematopoietic tyrosine phosphatase (HePTP), PTPN7, kinase interaction motif (KIM)-PTP, WPD loop, E loop
The past decade has seen an expanding interest in protein tyrosine phosphatases (PTPs) as potential therapeutic targets for the treatment of diseases ranging from diabetes to cancer.1-5 Atomic resolution structures of numerous classical class I human PTP catalytic domains have been solved, and comparison of these structures reveals that the overall structure of the catalytic domain is highly conserved.6 The architecture of the PTP active site is formed by several loop regions. The PTP loop, which contains the catalytic cysteine that is 100% conserved among classical human PTPs,7 forms the base of the active site, a highly positively charged pocket into which the phosphate group of the substrate phosphotyrosine binds. The WPD loop, which contains the catalytic aspartate that is 80% conserved among classical human PTPs,7 functions as a flexible ‘gate’ to the active site, and has been observed in the ‘open’ state (active site-accessible),8;9 the ‘closed’ state (active site-inaccessible),10;11 as well as in intermediate conformations.9 The roles of these loops in the PTP catalytic cycle involve: 1) opening of the WPD loop (‘open’ state), 2) binding of tyrosine-phosphorylated substrate at the active site, 3) closure of the WPD loop (‘closed’ state), 4) hydrolysis of the substrate phosphotyrosine, and 5) re-opening of the WPD loop and release of product phosphate and the dephosphorylated substrate.12-16
A third loop region, named the E loop after a glutamate residue present within the loop that is 100% conserved among classical human PTPs,7;17 is also located immediately adjacent to the active site. Primary sequence comparison of more than 20 classical class I human PTPs reveals that the E loop contains multiple highly conserved residues (Fig. 1a).7 A comparison of 74 different structures of PTPs (Table S1) also showed that, in more than 75% of these structures, the E loop adopts an identical β-hairpin conformation. The close proximity of the E loop to the active site and its conserved sequence/structure together suggest it may also play a role in substrate binding and catalysis.
Figure 1.
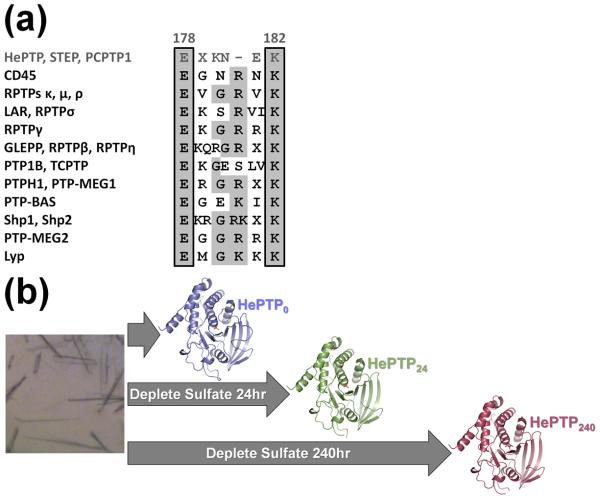
PTP E loop sequence alignment and schematic of soaking experiments with HePTP crystals. (a) Consensus sequence alignment of 12 human PTP subtypes. Conserved amino acids are shaded in light gray. Members of the KIM-phosphatase family, including HePTP, are in dark gray and the residue numbers above the alignment correspond to HePTP numbering. The 100% conserved E loop glutamate and 80% conserved E loop lysine are boxed. The sequences used for this analysis are described in Andersen et al. (2001).7 (b) Scheme for crystal soaking and data collection. HePTP crystals were grown in 1.7–1.9 M ammonium sulfate (left) and data collected immediately (HePTP0; blue), or soaked in 0.2 M ammonium tartrate for 24 hours (HePTP24; green) or 240 hours (HePTP240; red) prior to data collection. Protein models prepared using PyMOL.36
Hematopoietic tyrosine phosphatase (HePTP) is an immune cell-specific PTP that negatively regulates the activation and proliferation of the extracellular signal-regulated kinases 1/2 (Erk1/2) and p38. HePTP is one of three members of the kinase interaction motif (KIM)-PTP family, which also includes the striatum-enriched phosphatase (STEP) and STEP-like PTP (PTP-SL/PCPTP1). Interestingly, compared to the other human PTPs, the KIM-PTP family has a unique E loop. First, the KIM-PTPs have a truncated E loop, being shorter by one residue compared to the other human PTPs (Fig. 1a, top row). Second, in contrast to the β-hairpin structure formed in the other human PTPs, the KIM-PTP E loops form several unique structures. For instance, in STEP, the E loop has been observed to form a helical structure, to be partially disordered,9 or to be completely disordered (PDB ID: 2CJZ). Likewise, in every previously reported structure of HePTP,9;10;17 the E loop is also completely disordered. This differs from other PTPs in which the E loop is visible and highly ordered. The members of the KIM-PTP family also have an unusual WPD loop. For example, in previously reported structures of STEP,9 its WPD loop adopts an ‘atypically open’ conformation, even when bound to phosphotyrosine. Conversely, with the exception of CD45, HePTP is the only classical class I PTP in which the WPD loop has been observed exclusively in the closed conformation,9;10;17 in spite of extensive efforts to crystallize the HePTP open state (tens of 1000s of crystallization experiments; data not shown). This suggests that the dynamics of the HePTP WPD loop in the open state prohibits crystal formation.
Although visualizing conformational dynamics within individual protein crystals is not novel, it is rare, as it requires crystals in which the conformational changes of the protein do not disrupt the overall crystal lattice. For example, profilin:β-actin (PA) crystals grow in 1.8 M potassium phosphate (the ‘open’ state).18 Soaking these crystals in increasing concentrations of potassium phosphate or ammonium sulfate causes the actin to transform from the ‘open’ to the ‘closed’ state, in which large conformational changes between the actin subdomains cause them to collapse about the bound ATP, occluding it from solvent. Notably, the crystals do not lose their diffracting power, allowing the structures of both states to be determined to high resolution and providing novel structural insights into actin ATP hydrolysis and polymerization.18;19
In order to understand the unique role of the E loop and its interactions with the WPD loop in the transition between the closed and open states during the catalytic cycle, we identified a novel crystal form of HePTP that, when depleted of its active site oxyanion, maintained its diffraction quality, allowing the closed-to-open state transition to be observed within a single crystal form.
A rare crystal form provides the first HePTP crystals suitable for visualizing the closed-to-open state transition
HePTP residue Ser72 is an HePTP-specific phosphorylation target of Erk2.20 Recently, we generated the S72D mutant of HePTP (the aspartic acid mimics the size and charge distribution of phosphoserine21;22) in order to determine whether or not phosphorylation at this residue alters the structure of HePTP. Unexpectedly, HePTP S72D did not crystallize in the crystal form commonly observed for most apo HePTP wild-type (WT) and mutant structures (P61; PDB IDs: 1ZC0, 2GP0, 2HVL, 2QDC), but instead crystallized readily in a rare crystal form for apo HePTP (C2; PDB ID: 2QDM). The HePTP P61 crystal form has a high solvent content (70%) and the loops comprising the HePTP active site are located at a ternary crystal contact.10;17 Moreover, attempts to deplete phosphate from the active site by soaking these crystals in solutions lacking phosphate failed, as the crystals dissolved in these solutions. In contrast, the solvent content of the C2 crystal form is much lower (45%) and the HePTP active site is accessible and not located at crystal contacts. These characteristics of the C2 crystal form were of interest because they suggested that it might be amenable to crystal soaking experiments that would enable the structure of the open state and, more importantly, the structural changes that occur in the closed-to-open state transition of HePTP to be determined.
The HePTP S72D C2 crystals, which crystallized in 1.7–1.9 M ammonium sulfate pH 5.0, diffracted to 1.90 Å (hereafter referred to as HePTP0). In order to determine if these crystals would be suitable for depletion soaks, the HePTP0 crystals were transferred into a solution which did not contain sulfate (0.2 M ammonium tartrate pH 6.6, 20% (w/v) PEG 3,350; hereafter referred to as the depletion solution). This solution was selected because it had previously been shown to stabilize the HePTP C2 crystal form (HePTP:Erk2 peptide complexes crystallized in the C2 crystal form in this condition17). For 24 hour soaks, HePTP0 crystals were briefly washed through multiple drops of the depletion solution to remove any residual mother liquor, then transferred to a drop of fresh depletion solution and stored for 24 hours at 4°C. For 240 hour soaks, HePTP0 crystals were treated similarly, except transferred to drops of fresh depletion solution also on days 7, 9 and 10, for a total of eight transfers (the 24 hour- and 240 hour-soaked crystals are hereafter referred to as HePTP24 and HePTP240; Fig. 1b). Critically, both HePTP24 and HePTP240 crystals maintained their diffraction quality, diffracting to 2.6 Å and 2.25 Å, respectively, allowing their structures to be determined (Table 1; details of crystallization and structure determination are described in the table legend). Comparison of the HePTP0, HePTP24 and HePTP240 structures reveals that the closed-to-open state transition of HePTP was induced within a single crystal form. Unlike PTP1B,11 the closed-to-open transition does not involve a change in the water structure at the HePTP active site, but rather involves a novel, coordinated opening/disordering of the WPD and E loops.
Table 1.
Data Collection and Refinement Statistics.
| HePTP0e | HePTP24e | HePTP240e | |
|---|---|---|---|
| PDB ID | 3O4S | 3O4T | 3O4U |
| Data Collection | |||
| Space Group | C2 | C2 | C2 |
| Unit Cell | |||
| a, b, c (Å) | 117.8, 39.1, 83.4 | 118.0, 39.0, 83.7 | 119.0, 39.2, 84.0 |
| α, β, γ (°) | 90.0, 124.3, 90.0 | 90.0, 124.2, 90.0 | 90.0, 124.2, 90.0 |
| Wavelength (Å) | 1.0 | 1.54 | 1.1 |
| Resolution (Å) | 50.0-1.90 (1.93-1.90)a |
50.0-2.60 (2.64-2.60)a |
50.0-2.25 (2.29-2.25)a |
| No. protein molecules/ASU | 1 | 1 | 1 |
| Total/unique reflections | 92396/25241 | 20892/8972 | 64467/15443 |
| Redundancy | 3.7 (3.6)a | 2.3 (2.0)a | 4.2 (3.0)a |
| Completeness (%) | 99.7 (99.9)a | 90.3 (86.9)a | 99.0 (87.7)a |
| Rmerge (%)b | 9.2 (51.2)a | 8.8 (29.6)a | 11.3 (56.2)a |
| Mean I/σ(I) | 13.8 (3.5)a | 11.4 (3.7)a | 14.5 (2.6)a |
| Refinement | |||
| Resolution range | 20.00-1.90 | 20.00-2.60 | 20.00-2.25 |
| No. reflections (total) | 23923 | 8530 | 14619 |
| No. reflections (test) | 1287 | 440 | 772 |
| Rwork (%)c | 16.2 | 19.9 | 19.0 |
| Rfree (%)d | 21.2 | 25.3 | 24.3 |
| RMS deviations from ideal geometry | |||
| Bonds (Å) | 0.012 | 0.010 | 0.008 |
| Angles (°) | 1.31 | 1.10 | 1.08 |
| Ramachandran plot | |||
| Residues in allowed regions (%) | 99.7 | 99.6 | 99.6 |
| Residues in disallowed regions (%) | 0.3 | 0.4 | 0.4 |
| Mean B Value | |||
| Protein | |||
| Total | 21.3 | 24.2 | 32.0 |
| Active Sitee | 12.9 | 21.8 | 30.2 |
| Water | |||
| Active Site Sulfate | 16.6 | 25.4 | N/A |
| Active Site Tartrate | N/A | N/A | 44.8 |
| Glycerol Molecules | 44.3 | 44.6 | 43.1 |
| No. Atoms | |||
| Protein | 2340 | 2237 | 2189 |
| Water | 277 | 181 | 143 |
| Sulfate molecules | 6 | 1 | 0 |
| Tartrate molecules | 0 | 1 | 2 |
| Glycerol molecules | 5 | 2 | 2 |
Values in parentheses are for the highest resolution shell.
Rmerge = ∑|Ii−<Ii>|/∑|Ii| where Ii is the scaled intensity of the ith measurement, and <Ii> is the mean intensity for that reflection.
Rwork = ∑||Fobs|−|Fcalc||/∑|Fobs| where Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively.
Rfree = as for Rwork, but for 5.0% of the total reflections chosen at random and omitted from refinement.
Calculated for residues 270–276 of the HePTP PTP loop. HePTP (residues 44–339) containing the S72D mutation was subcloned into a derivative of the pET28a bacterial expression vector (Novagen) containing an N-terminal expression and hexahistidine purification tag (MGSDKIHHHHHH).30 Protein expression and purification was carried out using standard protocols.10;17 HePTP44–339 S72D initially formed clusters of small, one-dimensional ‘needle’ crystals in 1.8 M ammonium sulfate pH 5.0 using the sitting drop vapor diffusion method at 4°C. These initial crystals were used as seed for microseeding. This led to the formation larger, two-dimensional ‘plate’ crystals by microseeding into 1.7–1.9 M ammonium sulfate pH 5.0 using the sitting drop vapor diffusion method at 4°C. HePTP0: unsoaked HePTP44–339 S72D crystals (HePTP0) were cryoprotected in 1.28 M ammonium sulfate pH 5.0, 25% (v/v) glycerol for 30 seconds prior to diffraction screening and data collection. HePTP24: a subset of HePTP44–339 S72D crystals were transferred from the crystallization drop to 0.2 M ammonium tartrate pH 6.6, 20% (w/v) PEG 3,350 for 30 seconds at 4°C, then to a second drop of this solution for 30 seconds at 4°C, and subsequently to a third drop of this solution for 24 hours at 4°C, after which they were cryoprotected in 0.16 M ammonium tartrate pH 6.6, 16% (w/v) PEG 3,350, 20% (v/v) glycerol for 20 seconds prior to diffraction screening and data collection. HePTP240: another subset of HePTP44–339 S72D crystals were transferred from the crystallization drop through five drops of 0.2 M ammonium tartrate pH 6.6, 20% (w/v) PEG 3,350 for 30 seconds/drop at 4°C, then to a second drop of this solution for 142 hours at 4°C, subsequently to a third drop of this solution for 72 hours at 4°C, and finally a fourth drop of this solution for 26 hours at 4°C, after which they were cryoprotected in 0.15 M ammonium tartrate pH 6.6, 15% (w/v) PEG 3,350, 25% (v/v) glycerol for 20 seconds prior to diffraction screening and data collection. Crystallographic data for the HePTP0/HePTP24/HePTP240 crystals were collected at Brookhaven National Laboratory National Synchrotron Light Source (BNL-NSLS) Beamlines X6A and X25 at 100K using an ADSC QUANTUM 270 CCD detector or at Brown University at 100K using a Rigaku MicroMax-007 X-ray generator and R-AXIS IV++ imaging plate detector. All crystallographic data were indexed, scaled and merged using HKL2000 0.98.692i.31 The structures were solved by rigid body refinement using the program RefMac 5.2.001932 and the structure of HePTP44–339 D236A/C270S/Q314A (PDB ID: 2QDM) or HePTP0 as input models, after omitting solvent molecules, resulting in an initial Rfree = 31.2% and FOM = 0.75% for HePTP0, Rfree = 27.1% and FOM = 0.79 for HePTP24 and Rfree = 30.4% and FOM = 0.78 for HePTP240. All models were completed by cycles of manual building using the program Coot 6.0.233 coupled with structure refinement using RefMac 5.2.0019 against the datasets. The structure of HePTP0 was determined to 1.90 Å resolution and refined to Rwork = 16.2% and Rfree = 21.2%, and contains 1 molecule of HePTP, 277 water molecules, 6 sulfate molecules, and 5 glycerol molecules per asymmetric unit (HePTP residues 337–339 were not observed in the electron density map and so were not modeled). The structure of HePTP24 was determined to 2.60 Å resolution and refined to Rwork = 19.9% and Rfree = 25.3%, and contains 1 molecule of HePTP, 181 water molecules, 1 sulfate molecule, 1 tartrate molecule, and 2 glycerol molecules per asymmetric unit (HePTP residues 176–182, 235–239 and 337–339 were not observed in the electron density map and so were not modeled). The structure of HePTP240 was determined to 2.25 Å resolution and refined to Rwork = 19.0% and Rfree = 24.3%, and contains 1 molecule of HePTP, 143 water molecules, 2 tartrate molecules, and 2 glycerol molecules per asymmetric unit (HePTP residues 122–123, 175–183, 236–240 and 336–339 were not observed in the electron density map and so were not modeled). The stereochemical quality of the models was analyzed using MolProbity,34 which performs Ramachandran plot, Cβ deviation, and rotamer analyses. The agreement of the models to the diffraction data was analyzed using SFCheck 7.2.02.35 Atomic coordinates of the final models and experimental structure factors for the HePTP structures presented herein have been deposited with the Protein Data Bank (PDB) as entries 3O4S, 3O4T, 3O4U.
Calculated for ligand bound at the HePTP PTP loop.
An ordered HePTP E loop reveals a novel network of interactions between multiple catalytic site loops
The structure of HePTP0 is highly similar to the structure of WT HePTP (Fig. 2a), in spite of the fact that WT HePTP and HePTP0 crystallized in different space groups, P61 and C2, respectively, and that residue Ser72 does not mediate crystal contacts in either of these crystal forms. The structures of WT HePTP and HePTP0 superimpose with a root mean square difference (RMSD) of less than 0.5 Å, with differences localized primarily to peripheral loops located at crystal contacts (Fig. 2a). In addition, the S72D mutation does not induce any conformational changes in the loop containing Ser72, nor does it significantly alter the electrostatic surface in this region, showing that phosphorylation of Ser72 by Erk2 does not alter the overall charge or structure of HePTP. Notably, the conformation of the active site, and in particular the PTP and WPD loops, is essentially unchanged between the two structures.
Figure 2.
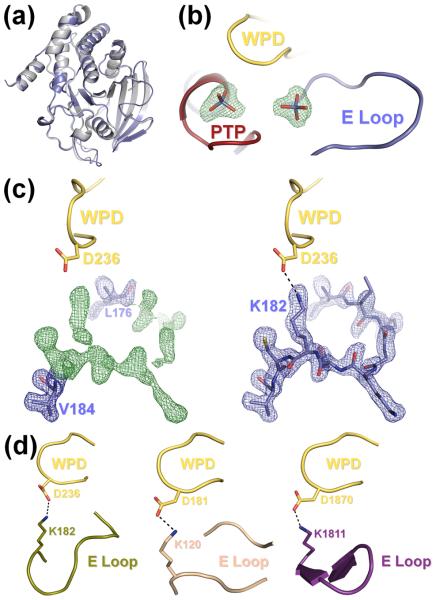
(a) The S72D mutation does not alter the overall structure of HePTP. Superposition of the structures of HePTP0 (blue) and WT HePTP (grey; PDB ID: 1ZC010). The structures are nearly identical (RMSD < 0.5 Å over all mainchain atoms). (b) HePTP0 contains sulfate bound at the active site and at a secondary binding site. mFO – DFC electron density map (green mesh; contoured at 3.0 σ to 1.90 Å) of the HePTP active site and interface of the PTP, WPD and E loops. Both sites contained well-defined, tetrahedral electron density (≥9.9 σ) from the early stages of refinement. (c) Visualization of the E loop in HePTP0. Left panel, 2mFO – DFC electron density map (blue mesh; contoured at 1.0 σ to 1.90 Å) and mFO – DFC electron density map (green mesh; contoured at 3.0 σ to 1.90 Å) of residues Leu176 and Val184 refined at an occupancy of 1.0 (blue, stick representation) with E loop residues 177–183 omitted from refinement. Right panel, 2mFO – DFC electron density map (blue mesh; contoured at 1.0 σ to 1.90 Å) of E loop residues 176–184 refined at an occupancy of 1.0 (blue). E loop residue Lys182 forms a hydrogen bond (illustrated by a dashed line) with the catalytic Asp236 of the WPD loop (yellow). Protein models prepared using PyMOL.36 (d) The WPD-E loop interaction is conserved across multiple, distinct PTP structures. Although the conformation of the E loop is different between HePTP0 (blue, right panel of (c)), HePTP C270S* (olive, left panel), PTP1B (beige, middle panel; PDB ID: 2B4S), and RPTPβ (purple, right panel: PDB ID: 2I4E), the catalytic aspartate of the WPD loop (yellow) in each of these structures forms a similar hydrogen bond with the conserved lysine of the E loop (illustrated by a dashed line).
From the early stages of refinement, a well-ordered, tetrahedron-shaped region of electron density (13.6σ in the initial mFO-DFC electron density map) was observed at the HePTP0 active site (Fig. 2b), which was modeled with a sulfate ion at an occupancy of 1. The WPD loop, which was closed about the active site, was also well-ordered and adopted the ‘closed’ conformation, with the catalytic acid/base Asp236 orientated towards the bound sulfate.
Unexpectedly, a second well-ordered tetrahedron-shaped region of electron density (9.9σ in the initial mFO-DFC electron density map) was also observed in a pocket immediately adjacent to the active site (Fig. 2b), hereafter referred to as the E loop oxyanion-binding pocket. This density was also modeled with a sulfate ion at an occupancy of 1. In HePTP0, the E loop sulfate binds via a bipartite hydrogen bonding interaction to PTP loop residue Arg276 (Fig. S1), which is 100% conserved among classical human PTPs. The E loop sulfate is also coordinated by E loop residue Lys182, the last residue in the E loop and which is also highly conserved among classical human PTPs (80% sequence identity, 100% sequence similarity; the only other residue at this position is either an arginine or a glutamine, both of which are capable of forming ionic and/or hydrogen bonding interactions with a bound oxyanion).
HePTP0 is also the first structure of HePTP with an ordered E loop, as this loop was disordered in all previously reported structures of HePTP (PDB ID: 1ZC0, 2A3K, 2GP0, 2HVL, 2QDM, 2QDC, 3D42, 3D449;10;17). This E loop flexibility is similar to what has been observed for other KIM-PTPs, in which the E loop has either adopted distinct conformations with high B-factors (STEP, PTP-SL; PDB ID: 2BV5, 2A8B) or is missing altogether (STEP; PDB ID: 2CJZ). Strikingly, from the initial stages of refinement, HePTP0 showed continuous, well-ordered electron density for the residues of the E loop (Fig. 2c). The E loop is stabilized in part through interactions at crystal contacts (Fig. S2; symmetry mate residues 122–125), but also through an intramolecular interaction with the WPD loop. Specifically, the catalytic acid/base Asp236 from the WPD loop hydrogen bonds with the conserved E loop Lys182 (Fig. 2c). Both the Asp236-Lys182 interaction (WPD and E loops) and the interaction between the E loop sulfate, Arg276 (PTP loop) and Lys182 (E loop) stabilize and order the E loop. Moreover, this interaction between the catalytic aspartate of the closed WPD loop and the conserved lysine of the E loop has been observed in structures of a number of other PTPs (Fig. 2d), including RPTPβ,23 RPTPρ,6 GLEPP1,24 PTP-BAS,25 and PTP1B,15;26;27 indicating that it is important for many PTPs.
Catalytic site dynamics within single crystals of HePTP reveal novel, coordinated motions of the WPD and E loops
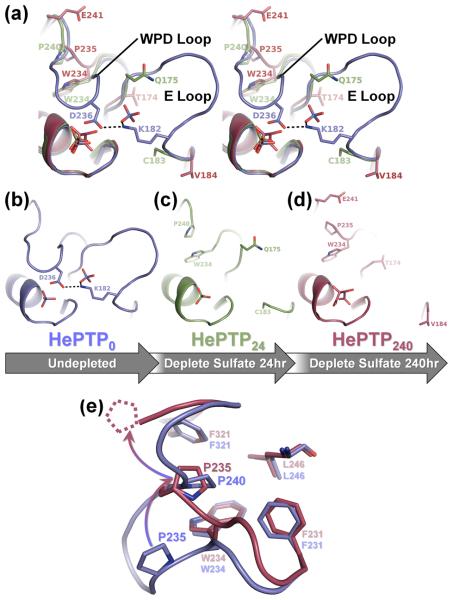
The transfer of HePTP0 crystals into solutions lacking sulfate resulted in substantial conformational changes at the active site, specifically in the WPD and E loops. In contrast to fresh HePTP0 crystals, soaking crystals in depletion solution for 24 hours (i.e. HePTP24) resulted in depletion of 50% of the sulfate ions from the HePTP active site and depletion of 100% of the sulfate ions from the E loop binding pocket. The depletion of oxyanions from these binding pockets was accompanied by a coordinated opening and disordering of both the WPD and E loops (Fig. 3a, 3d). In addition, HePTP24 residues 122–125 were also destabilized (Fig. S2), as evidenced by increased B-factors in the residues of this loop, consistent with the disruption of E loop interactions at crystal contacts with symmetry mate residues 122–125. In the structure of HePTP240, these changes were even more striking. Like HePTP24, the residues of the WPD and E loops were completely disordered (Fig. 3a, 3e) and not visible in the HePTP240 electron density map. However, in contrast to HePTP24, the active site oxyanion was completely depleted from the active site and instead replaced with tartrate (Fig. S3), a component of the depletion solution. The HePTP0, HePTP24 and HePTP240 structures show that the active site has a higher affinity for tetrahedral oxyanions than does the E loop binding pocket, since in HePTP24, the oxyanion was only 50% depleted from the active site, whereas the E loop binding pocket was fully depleted (Fig. 3d). A sulfate bound in the E loop binding pocket has also been observed in PTP1B (Fig. S4).26
Figure 3.
Depletion soaking induces coordinated movement of the HePTP WPD and E loops. (a) Stereo image of the superposition of the PTP, WPD and E loops of HePTP0 (blue), HePTP24 (green) and HePTP240 (red). In HePTP0 (blue), the WPD and E loops are well-ordered, and WPD loop residue Asp236 hydrogen bonds with E loop residue Lys182 (illustrated as a dashed line). In crystals depleted of sulfate for 24 hours (i.e. HePTP24; green) and 240 hours (i.e. HePTP240; red), the WPD and E loops simultaneously become disordered. Residues in HePTP24 and HePTP240 at ordered-to-disordered boundaries are shown in stick representation. The non-superposed PTP, WPD and E loops of HePTP0, HePTP24 and HePTP240 are shown in (b), (c) and (d), respectively. (e) Residue Pro235 adopts the ‘atypically open’ conformation in HePTP240. Superposition of the WPD loops from HePTP0 (blue) and HePTP240 (red). In the WPD-closed structure (i.e. HePTP0; blue), residue Pro240 packs into the hydrophobic pocket formed by Phe231, Trp234, Leu246, and Phe321. In the WPD-open structure (i.e. HePTP240; red), residue Pro240 is disordered (illustrated as dashed lines), and residue Pro235 is rotated upward by ~90° (illustrated as a gradated, curved arrow) and instead packs into the Phe231/Trp234/Leu246/Phe321 pocket. The conformation of the conserved WPD loop proline (i.e. Pro235 in HePTP) in HePTP240 is identical to that observed in ‘atypically open’ structures of STEP,9 Lyp29 and GLEPP16;24. Protein models prepared using PyMOL.36
The HePTPO structures also show an intramolecular interaction between the conserved residue Lys182 in the E loop and the catalytic residue Asp236 in the WPD loop (Fig. 2c). This interaction stabilizes the E loop in the closed conformation, as the WPD loop and E loop become increasingly disordered in both HePTP24 and HePTP240. An ordered E loop has only been observed in one other structure of HePTP, HePTP C270S*, which crystallized in P61 and not C2 (Table S2). Although the E loop adopts a different conformation in HePTP0 versus HePTP C270S*, in both structures the Lys182-Asp236 interaction is present and the WPD loop is in the closed conformation (Fig. 2c, 2d). This Lys182-Asp236 interaction may function to stabilize the WPD-closed conformation or alter WPD loop dynamics, possibly modulating HePTP catalytic activity. In order to test this possibility, we generated the K182A mutant of HePTP and measured its catalytic activity. Indeed, the catalytic activity of HePTP K182A (Fig. S5) is less than one third that of WT HePTP.17 Together, our structural and kinetic data reveal a novel role for Lys182 in enhancing HePTP catalytic activity. Notably, as described earlier, this interaction between the catalytic aspartate of the closed WPD loop and the conserved lysine of the E loop has been observed in other PTPs (Fig. 2d), including RPTPβ,23 RPTPρ,6 GLEPP1,24 PTP-BAS,25 and PTP1B.15;26;27 Moreover, for some of these PTPs, opening of the WPD loop and disruption of the WPD-E loop interaction has been accompanied by the E loop becoming disordered.23;24;28 Thus, the coordinated dynamics of the WPD and E loops is relevant not only to HePTP, but also to multiple other human PTP subtypes.
Finally, the differences in the structures of HePTP24 and HePTP240 show that the WPD loop adopts what has previously been referred to as an ‘atypically open’ conformation in HePTP240. Specifically, the conserved WPD loop Pro235 is rotated ~90° upward in HePTP240, displacing Pro240 in the hydrophobic pocket created by the conserved WPD loop Trp234 and other residues (Fig. 3e). Previously reported structures of STEP,9 Lyp29 and GLEPP16;24 contain a similar ‘atypically open’ WPD loop. In these structures, the C-terminal portion of the WPD loop forms a helical turn, thereby stabilizing the ‘atypically open’ conformation. These structures are also typified by the conserved WPD loop proline packing into the hydrophobic pocket formed by the conserved WPD loop tryptophan and other residues. Likewise, in the structure of HePTP240, the conserved WPD loop Pro235 also adopts the conformation observed in ‘atypically open’ PTP structures (Fig. 3e), although the C-terminal portion of the WPD loop is not visible in this structure. The ‘atypically open’ conformation of the WPD loop was previously proposed to represent a catalytically inactive conformation which could be targeted for the development inhibitors that stabilize this inactive state.6 The structure of HePTP240 provides the first evidence that HePTP also adopts an ‘atypically open’ WPD loop conformation.
Conclusion
The structures of HePTP0, HePTP24 and HePTP240, which were obtained from a single crystal form slowly depleted of bound oxyanion (i.e. sulfate), provide the first evidence for the coordinated dynamics of the WPD and E loops in the PTP catalytic cycle. Together, our structural and kinetic data reveal a novel role for Lys182 in enhancing HePTP catalytic activity. Because Lys182, like Asp236, is highly conserved among PTPs and because this WPD-E loop interaction has been observed in other PTPs, the coordinated dynamics between the WPD and E loops are likely relevant to multiple other human PTP subtypes. These structures also reveal that HePTP contains a secondary oxyanion binding pocket immediately adjacent to the active site, at the interface of the PTP, WPD and E loops, and that oxyanions bind at this site with lower affinity than at the active site. The structure of HePTP240 also provides the first evidence that HePTP adopts an atypically open conformation. Finally, these structures confirm that ligand bound at the active site in a preformed crystal of HePTP in the C2 crystal form can be replaced with exogenous ligand through soaking, providing a valuable tool for investigating the molecular basis for specificity of inhibitors that bind at the HePTP active site.
Supplementary Material
Acknowledgements
Crystallographic data was collected at beam lines X6A, funded by the National Institute of General Medical Sciences under agreement GM-0080, and X25. The National Synchrotron Light Source, Brookhaven National Laboratory is supported by the U.S. Department of Energy under contract number DE-AC02-98CH10886. This work was supported by an American Cancer Society Research Scholar Grant (RSG-08-067-01-LIB) to R.P. and NIH grants 1R21CA132121 and R03MH084230-01A1 to L.T.
Abbreviations
- PTP
protein tyrosine phosphatase
- HePTP
hematopoietic protein tyrosine phosphatase
- KIM-PTP
kinase interaction motif-protein tyrosine phosphatase
- STEP
striatum-enriched protein tyrosine phosphatase
- PTP-SL
STEP-like protein tyrosine phosphatase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
The coordinates and structure factors have been deposited in the PDB under accession numbers 3O4S, 3O4T and 3O4U for the HePTP0, HePTP24 and HePTP240 structures, respectively.
REFERENCES
- 1.Blaskovich MA. Drug discovery and protein tyrosine phosphatases. Curr Med Chem. 2009;16:2095–176. doi: 10.2174/092986709788612693. [DOI] [PubMed] [Google Scholar]
- 2.Jiang ZX, Zhang ZY. Targeting PTPs with small molecule inhibitors in cancer treatment. Cancer Metastasis Rev. 2008;27:263–72. doi: 10.1007/s10555-008-9113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ostman A, Hellberg C, Bohmer FD. Protein-tyrosine phosphatases and cancer. Nat Rev Cancer. 2006;6:307–20. doi: 10.1038/nrc1837. [DOI] [PubMed] [Google Scholar]
- 4.Tautz L, Pellecchia M, Mustelin T. Targeting the PTPome in human disease. Expert Opin Ther Targets. 2006;10:157–77. doi: 10.1517/14728222.10.1.157. [DOI] [PubMed] [Google Scholar]
- 5.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–46. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 6.Barr AJ, Ugochukwu E, Lee WH, King ON, Filippakopoulos P, Alfano I, Savitsky P, Burgess-Brown NA, Muller S, Knapp S. Large-scale structural analysis of the classical human protein tyrosine phosphatome. Cell. 2009;136:352–63. doi: 10.1016/j.cell.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersen JN, Mortensen OH, Peters GH, Drake PG, Iversen LF, Olsen OH, Jansen PG, Andersen HS, Tonks NK, Moller NP. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol Cell Biol. 2001;21:7117–36. doi: 10.1128/MCB.21.21.7117-7136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barford D, Flint AJ, Tonks NK. Crystal structure of human protein tyrosine phosphatase 1B. Science. 1994;263:1397–404. [PubMed] [Google Scholar]
- 9.Eswaran J, von Kries JP, Marsden B, Longman E, Debreczeni JE, Ugochukwu E, Turnbull A, Lee WH, Knapp S, Barr AJ. Crystal structures and inhibitor identification for PTPN5, PTPRR and PTPN7: a family of human MAPK-specific protein tyrosine phosphatases. Biochem J. 2006;395:483–91. doi: 10.1042/BJ20051931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mustelin T, Tautz L, Page R. Structure of the hematopoietic tyrosine phosphatase (HePTP) catalytic domain: structure of a KIM phosphatase with phosphate bound at the active site. J Mol Biol. 2005;354:150–63. doi: 10.1016/j.jmb.2005.09.049. [DOI] [PubMed] [Google Scholar]
- 11.Pedersen AK, Peters GG, Moller KB, Iversen LF, Kastrup JS. Water-molecule network and active-site flexibility of apo protein tyrosine phosphatase 1B. Acta Crystallogr D Biol Crystallogr. 2004;60:1527–34. doi: 10.1107/S0907444904015094. [DOI] [PubMed] [Google Scholar]
- 12.Stuckey JA, Schubert HL, Fauman EB, Zhang ZY, Dixon JE, Saper MA. Crystal structure of Yersinia protein tyrosine phosphatase at 2.5 A and the complex with tungstate. Nature. 1994;370:571–5. doi: 10.1038/370571a0. [DOI] [PubMed] [Google Scholar]
- 13.Zhang ZY, Wang Y, Dixon JE. Dissecting the catalytic mechanism of protein-tyrosine phosphatases. Proc Natl Acad Sci U S A. 1994;91:1624–7. doi: 10.1073/pnas.91.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang ZY, Wu L. The single sulfur to oxygen substitution in the active site nucleophile of the Yersinia protein-tyrosine phosphatase leads to substantial structural and functional perturbations. Biochemistry. 1997;36:1362–9. doi: 10.1021/bi9624043. [DOI] [PubMed] [Google Scholar]
- 15.Jia Z, Barford D, Flint AJ, Tonks NK. Structural basis for phosphotyrosine peptide recognition by protein tyrosine phosphatase 1B. Science. 1995;268:1754–8. doi: 10.1126/science.7540771. [DOI] [PubMed] [Google Scholar]
- 16.Neel BG, Tonks NK. Protein tyrosine phosphatases in signal transduction. Curr Opin Cell Biol. 1997;9:193–204. doi: 10.1016/s0955-0674(97)80063-4. [DOI] [PubMed] [Google Scholar]
- 17.Critton DA, Tortajada A, Stetson G, Peti W, Page R. Structural basis of substrate recognition by hematopoietic tyrosine phosphatase. Biochemistry. 2008;47:13336–45. doi: 10.1021/bi801724n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chik JK, Lindberg U, Schutt CE. The structure of an open state of beta-actin at 2.65 A resolution. J Mol Biol. 1996;263:607–23. doi: 10.1006/jmbi.1996.0602. [DOI] [PubMed] [Google Scholar]
- 19.Schutt CE, Myslik JC, Rozycki MD, Goonesekere NC, Lindberg U. The structure of crystalline profilin-beta-actin. Nature. 1993;365:810–6. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- 20.Saxena M, Williams S, Brockdorff J, Gilman J, Mustelin T. Inhibition of T cell signaling by mitogen-activated protein kinase-targeted hematopoietic tyrosine phosphatase (HePTP) J Biol Chem. 1999;274:11693–700. doi: 10.1074/jbc.274.17.11693. [DOI] [PubMed] [Google Scholar]
- 21.Robbins DJ, Zhen E, Owaki H, Vanderbilt CA, Ebert D, Geppert TD, Cobb MH. Regulation and properties of extracellular signal-regulated protein kinases 1 and 2 in vitro. J Biol Chem. 1993;268:5097–106. [PubMed] [Google Scholar]
- 22.Shen K, Hines AC, Schwarzer D, Pickin KA, Cole PA. Protein kinase structure and function analysis with chemical tools. Biochim Biophys Acta. 2005;1754:65–78. doi: 10.1016/j.bbapap.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 23.Evdokimov AG, Pokross M, Walter R, Mekel M, Cox B, Li C, Bechard R, Genbauffe F, Andrews R, Diven C, Howard B, Rastogi V, Gray J, Maier M, Peters KG. Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery. Acta Crystallogr D Biol Crystallogr. 2006;62:1435–45. doi: 10.1107/S0907444906037784. [DOI] [PubMed] [Google Scholar]
- 24.Almo SC, Bonanno JB, Sauder JM, Emtage S, Dilorenzo TP, Malashkevich V, Wasserman SR, Swaminathan S, Eswaramoorthy S, Agarwal R, Kumaran D, Madegowda M, Ragumani S, Patskovsky Y, Alvarado J, Ramagopal UA, Faber-Barata J, Chance MR, Sali A, Fiser A, Zhang ZY, Lawrence DS, Burley SK. Structural genomics of protein phosphatases. J Struct Funct Genomics. 2007;8:121–40. doi: 10.1007/s10969-007-9036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Villa F, Deak M, Bloomberg GB, Alessi DR, van Aalten DM. Crystal structure of the PTPL1/FAP-1 human tyrosine phosphatase mutated in colorectal cancer: evidence for a second phosphotyrosine substrate recognition pocket. J Biol Chem. 2005;280:8180–7. doi: 10.1074/jbc.M412211200. [DOI] [PubMed] [Google Scholar]
- 26.Li S, Depetris RS, Barford D, Chernoff J, Hubbard SR. Crystal structure of a complex between protein tyrosine phosphatase 1B and the insulin receptor tyrosine kinase. Structure. 2005;13:1643–51. doi: 10.1016/j.str.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 27.Salmeen A, Andersen JN, Myers MP, Tonks NK, Barford D. Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B. Mol Cell. 2000;6:1401–12. doi: 10.1016/s1097-2765(00)00137-4. [DOI] [PubMed] [Google Scholar]
- 28.Asante-Appiah E, Patel S, Desponts C, Taylor JM, Lau C, Dufresne C, Therien M, Friesen R, Becker JW, Leblanc Y, Kennedy BP, Scapin G. Conformation-assisted inhibition of protein-tyrosine phosphatase-1B elicits inhibitor selectivity over T-cell protein-tyrosine phosphatase. J Biol Chem. 2006;281:8010–5. doi: 10.1074/jbc.M511827200. [DOI] [PubMed] [Google Scholar]
- 29.Tsai SJ, Sen U, Zhao L, Greenleaf WB, Dasgupta J, Fiorillo E, Orru V, Bottini N, Chen XS. Crystal structure of the human lymphoid tyrosine phosphatase catalytic domain: insights into redox regulation. Biochemistry. 2009;48:4838–45. doi: 10.1021/bi900166y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peti W, Page R. Strategies to maximize heterologous protein expression in Escherichia coli with minimal cost. Protein Expr Purif. 2007;51:1–10. doi: 10.1016/j.pep.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 31.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. (Part A) 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 32.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–50. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 35.Vaguine AA, Richelle J, Wodak SJ. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr D Biol Crystallogr. 1999;55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 36.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.