Abstract
Aims
Anthracyclines such as daunorubicin (DNR) and doxorubicin are effective cancer chemotherapeutic agents, but can induce cardiotoxicity. GATA4 has been shown to serve as a survival factor of cardiac muscle cells, and anthracyclines promote apoptosis in part by down-regulating GATA4. The present study investigated the mechanism of anthracycline action to down-regulate GATA4.
Methods and results
DNR inhibited the transcriptional activity exhibited by the 250 bp conserved region immediately upstream from the transcriptional start site of the Gata4 gene. Mapping this region identified that the CCAAT-binding factor/nuclear factor-Y (CBF/NF-Y) binding to the CCAAT box was inhibited by DNR in HL-1 cardiac muscle cells and in perfused isolated mouse hearts. The DNR action on the Gata4 promoter was found to be dependent on p53, since DNR promoted nuclear binding of p53 to CBF/NF-Y and pifithrin-α (a p53 inhibitor) attenuated DNR down-regulation of GATA4.
Conclusion
Anthracycline down-regulation of GATA4 is mediated by the inhibition of Gata4 gene transcription via a novel mechanism that involves the p53-dependent inhibition of CBF/NF-Y binding to the CCAAT box within the Gata4 promoter.
Keywords: Anthracycline, Cardiotoxicity, GATA4, Heart, p53
1. Introduction
Anthracyclines including daunorubicin (DNR) and doxorubicin have been used as cancer chemotherapeutic agents. DNR is the first anthracycline developed and is effective against acute leukaemia. While these agents are effective in eliminating cancer cells, severe cardiotoxicity has been noted in >20% of patients treated with anthracyclines.1 Anthracyclines can cause myocarditis, myocardial infarction, cardiomyopathy, and heart failure. Apoptosis of cardiomyocytes may play an important role in anthracycline actions on the heart.2,3 Understanding the mechanisms of anthracycline actions should help in developing strategies for effective cancer therapy.
GATA4 is a member of the GATA family of zinc finger transcription factors. Six GATA family members have been identified and regulate transcription of target genes via binding to GATA elements including the consensus 5′-WGATAR-3′ sequence.4 While GATA-1, -2, and -3 play critical roles in the transcriptional regulation in blood cells and in endothelial cells, GATA-4, -5, and -6 are expressed in neonatal hearts. The GATA-binding activity in the adult heart is largely due to the activity of GATA4.5
We previously demonstrated that DNR can down-regulate GATA4 DNA-binding activity in cardiac muscle cells.5 The decrease in GATA4 activity is at least in part due to decreased GATA4 expression, as both protein and mRNA levels of GATA4 are down-regulated in response to DNR treatment. Since the restoration of GATA activity by overexpression of GATA4 using adenovirus-mediated gene transfer attenuated DNR-induced cardiac muscle cell apoptosis, we concluded that GATA4 down-regulation plays a role in the mechanism of DNR actions to induce apoptosis.5 Nemer and co-workers also reported that anthracyclines can down-regulate cardiac GATA4 expression in intact mice,6 and this may serve as a mechanism for anthracycline down-regulation of anti-apoptotic factors such as B-cell lymphoma 2 (Bcl-2) and Bcl-xL.7–9
In our previous studies, DNR was found to effectively down-regulate Gata4 mRNA levels, and experiments using actinomycin D to inhibit general gene transcription demonstrated that DNR does not alter Gata4 mRNA stability.5 Thus, we hypothesized that DNR inhibits Gata4 gene transcription. However, since information about the Gata4 promoter was not available, the mechanism of anthracycline actions on GATA4 gene expression had not been defined. More recently, others and we have identified the major transcriptional start site of the Gata4 gene in the mouse heart and cloned the immediately upstream region, which is conserved among species.10–12 In the present study, we tested the hypothesis that DNR affects Gata4 gene transcription. We found that DNR indeed suppressed Gata4 promoter-controlled gene transcription. Further, we identified a novel mechanism of DNR action, that is, to inhibit CCAAT-binding factor/nuclear factor-Y (CBF/NF-Y) binding to the CCAAT box in a p53-dependent fashion.
2. Methods
2.1. Culture of HL-1 cardiac muscle cells
HL-1 adult mouse cardiac muscle cells13 were maintained in Claycomb Medium (JRH Biosciences, Lenexa, KS, USA) supplemented with 10% foetal bovine serum (Invitrogen Corporation, Carlsbad, CA, USA), 100 µM norepinephrine, 100 units/mL penicillin, 100 µg/mL streptomycin, and 0.25 µg/mL amphotericin B (Sigma Chemical Company, St. Louis, MO, USA) in plastic dishes, coated with 12.5 µg/mL fibronectin and 0.02% gelatin, in a 5% CO2 atmosphere at 37°C.14 Nuclear extracts and cell lysates were prepared as previously described.5,7 Transfection and luciferase assay procedures are described in Supplementary material online.
2.2. Isolated mouse heart perfusion
Male C57BL/6 mice (20–25 g) were fed normal rat chaw and were used in accordance with approval by Georgetown University Animal Care and Use Committee and that by the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Hearts were rapidly removed from mice anaesthetized with inhalation of isoflurane. The aorta was cannulated with a cannula connected to the Langendorff apparatus.15 The Langendorff perfusion was initiated immediately after the heart excision with modified Krebs-Henseleit buffer, containing (in mM) 118.0 NaCl, 4.7 KCl, 1.7 CaCl2, 1.2 MgSO4, 1.2 KH2PO4, 25.0 NaHCO3, and 10.0 glucose. The buffer was continuously bubbled with 95% O2 and 5% CO2 (pH 7.4, 37°C) throughout the perfusion period. After 15 min of stabilization, the heart was perfused with 2 µM DNR for 120 min. Nuclear extracts were prepared from left and right heart ventricles as previously described.12
2.3. Electrophoretic mobility shift assays
Nuclear extracts from cells5 and tissues12 were prepared as described previously. Electrophoretic mobility shift assays (EMSA) were performed as described in Supplementary material online.
2.4. Reverse transcription-polymerase chain reaction
Total RNA (1 µg) extracted using TRIZOL (Invitrogen) was reverse-transcribed by oligo(dT) priming and reverse transcriptase (Applied Biosystems, Foster City, CA, USA). The resultant cDNA was amplified using Taq DNA polymerase (Invitrogen) and resolved on agarose gel containing ethidium bromide. See Supplementary material online for polymerase chain reaction (PCR) primer sequences and conditions.
2.5. Western blot analysis
Equal protein amounts of HL-1 cell nuclear extracts, whole cell lysates, or immunoprecipitates were electrophoresed through a reducing SDS polyacrylamide gel and electroblotted onto a nitrocellulose membrane. The membrane was blocked and incubated with antibodies for CBF-B, p53 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), or poly(ADP ribose; Trevigen, Gaithersburg, MD, USA), and the detection was made with the horseradish peroxidase-linked secondary antibody and Enhanced Chemiluminescence System (Amersham Life Science, Arlington Heights, IL, USA). Detection of protein carbonylation was performed as previously described.16 Immunoprecipitation was performed using GammaBind G-Sepharose (Amersham).
2.6. Statistical analysis
Means ± S.E. were calculated. Significant differences between all groups were computed by ANOVA with a Student-Newman–Keuls posthoc test. Significant differences between two groups were determined by the Student's t-test.
3. Results
3.1. DNR inhibits Gata4 promoter activity
We have previously shown that DNR reduced the Gata4 mRNA expression without altering the mRNA stability.5 We thus hypothesized that Gata4 gene transcription was inhibited by DNR. More recently, the transcriptional start site of the mouse Gata4 gene was identified.10–12 The 1000 bp region immediately upstream from the identified transcriptional start site is conserved among species. We cloned this 1000 bp region into a luciferase reporter vector and found that this region has transcriptional activity.10,12 Using this tool, we tested the hypothesis that DNR inhibits Gata4 gene transcription.
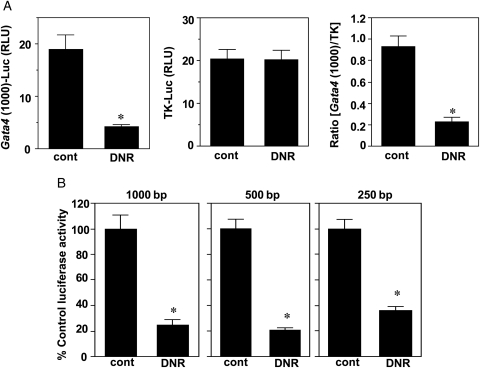
We found that DNR effectively inhibited this transcriptional activity of the 1000 bp Gata4 promoter region in HL-1 adult mouse cardiac muscle cells (Figure 1A; Supplementary material online, Figure SI-A). DNR did not have any effects on thymidine kinase (TK) promoter-controlled gene transcription (Figure 1A), indicating the specificity of DNR towards the Gata4 promoter. Our previous study has shown that another apoptotic inducer, tumour necrosis factor-α was ineffective in down-regulating GATA4.5 Consistently, tumour necrosis factor-α did not cause significant inhibition of the transcriptional activity of the 1000 bp Gata4 promoter region (Supplementary material online, Figure SI-B).
Figure 1.
Effects of DNR on gene transcription controlled by the proximal Gata4 promoter region. (A) HL-1 cells were co-transfected with firefly luciferase vector controlled by the proximal 1000 bp region of the mouse Gata4 promoter [Gata4 (1000)-Luc] and Renilla luciferase vector controlled by the TK promoter (TK-Luc). Seven hours after transfection, cells were treated with DNR (2 µM) for 17 h and then relative luciferase units (RLU) were measured. Values represent means ± S.E. (n = 9). (B) HL-1 cells were transfected with luciferase construct controlled by proximal 1000, 500, or 250 bp region of the Gata4 promoter then treated with DNR (2 µM) for 17 h. Values represent means ± S.E. (n = 9–12). (*) denotes values significantly different from the control value at P < 0.05.
To further identify the site(s) of the DNR action, we truncated the 1000 bp region to 500 and 250 bp proximal to the transcriptional start site. The truncation of the 1000 bp region to 500 bp or 250 bp did not significantly alter the basal transcriptional activity (Supplementary material online, Figure SI-C), suggesting that the 250 bp proximal region contains key regulatory elements for the basal expression.12 Further, this 250 bp region appears to contain the target site for DNR, as we observed dose-dependent effects of DNR on 250 bp Gata4 promoter-controlled luciferase expression, where 1 µM DNR caused significant inhibition (Supplementary material online, Figure SI-D). DNR equally inhibited the transcriptional activities controlled by the 250, 500, and 1000 bp regions (Figure 1B).
3.2. Identification of DNR targets
This 250 bp region that is immediately upstream from the transcriptional start site of the Gata4 gene contains putative binding sites for various transcription factors (Supplementary material online, Figure SII-A). Supershift analysis, using 32P-labelled double-stranded 250 bp region, demonstrated that early growth response factor 1, Sp1, upstream stimulatory factor 1, and upstream stimulatory factor 2 bind to this region.17 We performed similar supershift experiments using nuclear extracts from HL-1 cells treated with DNR to test the hypothesis that the DNA-binding activity of one or more of these transcription factors may be inhibited by the DNR treatment. However, DNR did not affect DNA-binding characteristics of any of these factors (Supplementary material online, Figure SII-B). Thus, these transcription factors do not appear to be the DNR targets.
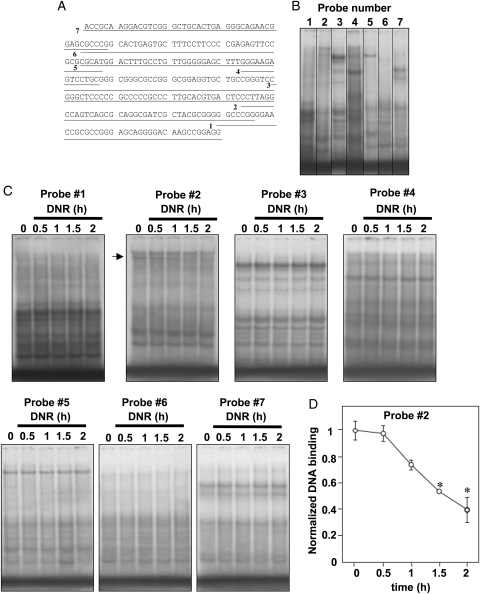
To identify DNR-sensitive transcriptional regulatory regions, we then used double-stranded EMSA probes that were constructed by truncating the 250 bp proximal Gata4 promoter region.12 Figure 2A shows the sequences of Probes #1–7. EMSA with untreated HL-1 cell nuclear extracts shows DNA-binding proteins, which interact with these probes (Figure 2B). Binding patterns of Probes #1, 3, 4, 5, 6, and 7, however, were not altered by treating cells with DNR (Figure 2C). Using Probe #2, which contains the CCAAT box where a transcription factor CBF/NF-Y binds,18 we identified a band that is reduced in cells treated with DNR (Figure 2C). Significant inhibition of the DNA-binding activity of CBF/NF-Y was observed by DNR treatment for 1.5 h (Figure 2D).
Figure 2.
Effects of DNR on DNA-binding activities towards various segments of proximal 250 bp Gata4 promoter. (A) Schematic of various probes containing sequences from the 250 bp Gata4 promoter region. (B) Basal DNA-binding activities of 32P-labelled Probes #1–7. (C) Effects of DNR (2 µM) on DNA-binding activities of nuclear extracts from control and DNR-treated HL-1 cells towards 32P-labelled Probes #1–7. The arrow indicates the band that was affected by DNR. (D) The line graph represents means ± S.E. of the intensity of the Probe #2 binding band affected by DNR as determined by densitometry (n = 4). (*) denotes values significantly different from the control value at P < 0.05.
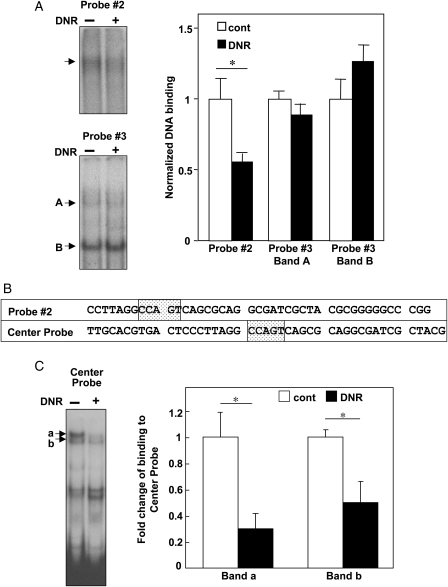
To confirm that observed events are not specific to HL-1 cells, we treated Langendorff-perfused, isolated mouse hearts with DNR. Similarly to results from HL-1 cells, treatment of isolated mouse hearts with DNR inhibited DNA-binding activity of CBF/NF-Y towards Probe #2 of the 250 bp Gata4 promoter without affecting Probe #3 (Figure 3A). Since Probe #2 contains the CCAAT box at the end of the probe, we also constructed an EMSA probe in which the CCAAT box is located in the centre of the probe, designated as Center Probe (Figure 3B). The use of this probe confirmed that factors which bind to the CCAAT box is influenced by DNR in the mouse heart (Figure 3C).
Figure 3.
Effects of DNR on perfused isolated mouse hearts. (A) Mouse hearts were subjected to Langendorff perfusion with or without 2 µM of DNR for 2 h. Nuclear extracts (5 µg) were subjected to EMSA using the 32P-labelled Probes #2 and #3 of the Gata4 promoter region. Values in the bar graph represent means ± S.E. (n = 4). (B) Sequences of Probe #2 and Center Probe. (C) Mouse heart nuclear extracts were subjected to EMSA using 32P-labelled Center Probe. Values in the bar graph represent means ± S.E. of fold change binding for Bands a and b. (*) denotes values significantly different from the untreated control at P < 0.05.
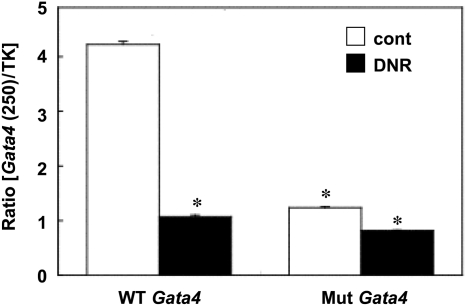
To determine if the CCAAT box plays functional roles in the regulation of gene transcription, a luciferase vector with a mutated CCAAT box site within the 250 bp Gata4 promoter18 was used. We found that degrees of the decreased transcriptional activity of the 250 bp Gata4 promoter region are comparable between mutating the CCAAT box and treating cells with DNR (Figure 4).
Figure 4.
Effects of DNR on Gata4 promoter activity. HL-1 cells were transfected with the luciferase construct controlled by the 250 bp proximal region of the wild-type (WT) Gata4 promoter, or mutant (Mut) Gata4 promoter at CCAAT box, then treated with DNR (2 µM) for 17 h. Values represent means ± S.E. of the ratio of 250 bp Gata4 promoter-controlled firefly luciferase activity to TK promoter-controlled Renilla luciferase activity (n = 6). (*) denotes values significantly different from the wild-type Gata4 promoter value at P < 0.05.
3.3. Role of p53
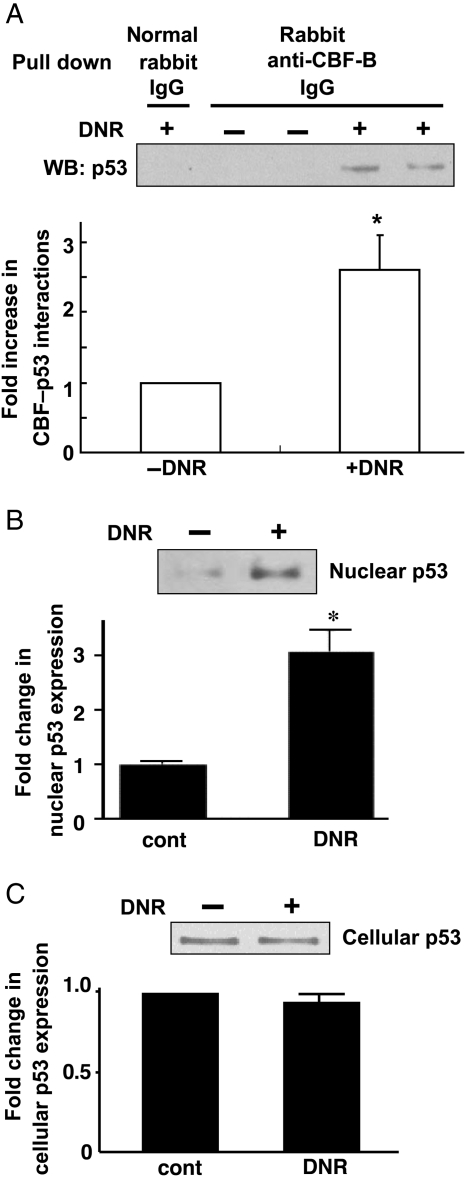
Anthracyclines have been shown to activate p53, and the disruption of p53 attenuates anthracycline-induced cardiotoxicity.19,20 We found that DNR promoted protein–protein interactions between CBF/NF-Y and p53 in the nucleus. As shown in Figure 5A, immunoprecipitation of nuclear extracts from DNR-treated HL-1 cells with rabbit anti-CBF-B IgG followed by immunoblotting with mouse anti-p53 IgG revealed the association between CBF-B and p53. In contrast, such interactions were absent in untreated cells. Control experiments using normal rabbit IgG in the immunoprecipitation reaction containing nuclear extracts from DNR-treated cells did not exhibit the band of interest.
Figure 5.
Effects of DNR on p53. (A) Nuclear extracts from HL-1 cells untreated or treated with DNR were immunoprecipitated with rabbit anti-CBF-B IgG or with normal rabbit IgG (as control), then immunoblotted with mouse anti-p53 IgG. Values in the bar graph represent means ± S.E. (n = 5). (B) HL-1 cells were treated with DNR (2 µM) for 4 h. p53 protein levels in nuclear extracts were measured by western blot. Values in the bar graph represent means ± S.E. (n = 4). (C) p53 protein levels in total cell lysates were measured by western blot. Values in the bar graph represent means ± S.E. (n = 4). (*) denotes values significantly different from the untreated control at P < 0.05.
Anthracyclines trigger poly(ADP-ribosyl)ation of p53, which in turn serves as a mechanism for p53 nuclear accumulation.21 Consistently, DNR promoted increased nuclear expression of p53 (Figure 5B). DNR did not change total cellular p53 levels (Figure 5C). Further, immunoprecipitation/immunoblotting experiments with antibodies for p53 and poly(ADP-ribosyl)ated proteins revealed that the p53 that is accumulated in the nucleus is poly(ADP-ribosyl)ated (Supplementary material online, Figure SIII-A). It has also been proposed that the nuclear p53 localization in response to anthracyclines is regulated through redox-dependent mechanisms.22 Immunoprecipitation/immunoblotting experiments with 2,4-dinitrophenylhydrazine-derivatized HL-1 cell nuclear extracts revealed that nuclear p53 is carbonylated (Supplementary material online, Figure SIII-B). Total carbonyl content was not significantly altered by DNR (Supplementary material online, Figure SIII-C). These results suggest that the nuclear accumulation of p53, perhaps in response to DNR-mediated p53 modifications through poly(ADP-ribosyl)ation and/or carbonylation, promotes p53–CBF/NF-Y interactions in the nucleus.
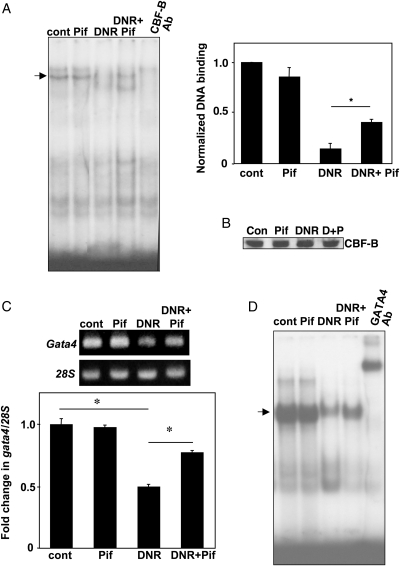
We hypothesized that DNR suppresses the CBF/NF-Y activity through p53 and in turn down-regulates Gata4 gene transcription. To test this hypothesis, a p53 inhibitor, pifithrin-α,23 was used. Pifithrin-α attenuated DNR-induced inhibition of CBF/NF-Y binding towards Probe #2 (Figure 6A), suggesting that the DNR action to influence CCAAT box binding is indeed p53 dependent. Reduced CBF/NF-Y binding is not due to down-regulation of CBF/NF-Y expression as western blotting of CBF-B showed no change in its expression in response to DNR treatment (Figure 6B). We also found that the inhibition of p53 by pifithrin-α attenuated DNR-induced down-regulation of Gata4 mRNA expression (Figure 6C) as well as the GATA4 DNA-binding activity (Figure 6D). These results provide evidence for the p53-dependent inhibition of CBF/NF-Y binding to the CCAAT box by DNR, which in turn suppresses Gata4 gene transcription and resultant GATA4 activity in cardiac muscle cells.
Figure 6.
Effects of p53 inhibitor. (A) HL-1 cells were treated with DNR (2 µM) and pifithrin-α (Pif; 10 µM) for 24 h. Nuclear extracts were subjected to EMSA using 32P-labelled Probe #2 of the Gata4 promoter. Bar graphs represent means ± S.E. (n = 4). (*) denotes values significantly different from each other at P < 0.05. CBF-B Ab indicates the supershift experiment with CBF-B antibody. (B) Nuclear extracts were subjected to western blotting with CBF-B antibody. D + P, DNR + Pif. (C) Total RNA was isolated, and levels of Gata4 mRNA and 28S rRNA were monitored by reverse transcription PCR. The bar graph represents means ± S.E. of the intensity of Gata4 mRNA levels determined by densitometry (n = 4). (*) denotes values significantly different from each other at P < 0.05. (D) Nuclear extracts were subjected to EMSA using the 32P-labelled double-stranded oligonucleotide containing GATA consensus elements. GATA4 Ab indicates supershifting with the GATA4 antibody.
4. Discussion
Anthracyclines such as DNR and doxorubicin have been shown to down-regulate Gata4 mRNA expression and in turn reduce GATA DNA-binding activity in cardiac muscle cells.5,6 Since the overexpression of GATA4 can attenuate the incidence of apoptosis induced by anthracyclines,5 GATA4 may serve as an anti-apoptotic factor in the heart. Indeed, GATA4 has been shown to regulate expression of anti-apoptotic proteins, Bcl-2 and Bcl-xL.6–8 Thus, GATA4 down-regulation may play an important role in anthracycline-induced cardiac muscle cell death and cardiotoxicity. However, the mechanism of GATA4 down-regulation by anthracyclines has not been defined. Here we report a p53-dependent mechanism for DNR action to down-regulate GATA4.
In HL-1 mouse cardiac muscle cells, whether DNR accelerates the Gata4 mRNA degradation was previously tested.5 Experiments with actinomycin D to inhibit general gene transcription suggested that DNR does not alter the Gata4 mRNA stability.5 Thus, we hypothesized that DNR may reduce the Gata4 mRNA expression by inhibiting Gata4 gene transcription. To test this hypothesis, we (i) first identified the transcriptional start site of mouse heart Gata4 mRNA, (ii) determined that the 1000 bp region immediately upstream from this identified transcriptional start site is conserved among species with putative transcriptional regulatory elements, and (iii) found that the cloned 1000 bp region exhibited strong transcriptional activity.10,12 Ohara et al.11 also studied mouse Gata4 promoter and reported that this region plays important roles in the promoter activity. Thus, we studied the effects of DNR on transcriptional activity controlled by this region of the Gata4 promoter.
DNR was found to effectively inhibit the luciferase activity controlled by the 1000 bp region, demonstrating that the mechanism of action to down-regulate GATA4 involves the inhibition of Gata4 gene transcription. To further identify the molecular mechanism of DNR on Gata4 gene transcription, the 1000 bp region was truncated to 500 and 250 bp upstream from the identified transcriptional start site. Such truncations did not reduce transcriptional activity, suggesting that the 250 bp region contains key transcriptional elements, which control basal gene transcription of Gata4.18 The present study has shown that DNR inhibits the gene transcription controlled by this 250 bp region, suggesting that the molecular target of DNR resides within this region.
Various fragments of the proximal 250 bp region of the Gata4 promoter were examined to identify factors that might be affected by DNR. We screened all of the fragments that span the 250 bp region and found that only one probe (designated Probe #2) was affected by DNR. This fragment contains the CCAAT box, and the truncation of the CCAAT box site or mutation of this region completely eliminated the DNA binding,18 suggesting that DNR targets CCAAT box-binding proteins.
While CCAAT/enhancer-binding protein-β has been shown to bind to the CCAAT box,24 our previous studies did not show evidence for the binding of this transcription factor to the Gata4 CCAAT box.18 A transcription factor CBF/NF-Y, a trimeric complex comprising of CBF-A, -B, and -C subunits, also binds to the CCAAT box sequence,25 and our laboratory showed that the CBF-B antibody interfered with CCAAT box-binding activity and pull-down with double-stranded oligonucleotide containing the Gata4 CCAAT box revealed DNA–protein interactions with CBF-B and the CCAAT box.18 The present study provided evidence to support that CBF/NF-Y, which has been shown to act as a cell survival factor,26 comprises the CCAAT box-binding complex that is affected by DNR. Since the mutation within the Gata4 CCAAT box sequence both suppresses protein binding by CBF/NF-Y and largely abolishes the Gata4 promoter activity, DNR-mediated interference of CBF/NF-Y binding to the CCAAT box should inhibit Gata4 gene transcription.
Our results further revealed that the mechanism of DNR-mediated inhibition of CBF/NF-Y binding to the CCAAT box involves p53. This is consistent with the previous reports that p53 inactivates CBF/NF-Y27 and that anthracyclines activate p53.19–21 One proposed mechanism of anthracycline action is that these compounds cause DNA damage and trigger modifications of nuclear proteins including p53, which in turn serve as mechanisms for inhibiting the nuclear export and the degradation of p53, thereby increasing its nuclear accumulation.21 We indeed found that DNR triggers increased nuclear p53, which is associated with the promotion of poly(ADP-ribosyl)ation as well as oxidative modification. Our results demonstrating that an inhibitor of p53 attenuated the DNR action on CBF/NF-Y support the role of p53. While the mechanism of p53 inactivation of CBF/NF-Y has not yet been previously determined, the present study provides evidence for protein–protein interactions between CBF/NF-Y and p53. These results suggest that DNR via p53 inactivates CBF/NF-Y through a mechanism involving protein–protein interactions and in turn suppresses Gata4 gene transcription (Supplementary material online, Figure SIV).
Anthracyclines are effective cancer chemotherapeutic agents, however, severe cardiotoxicity has been noted in patients treated with anthracyclines.1 Anthracyclines cause myocarditis, myocardial infarction, cardiomyopathy, and heart failure, and apoptosis of cardiac muscle cells may play important roles.2,3 Our laboratory and others have demonstrated that GATA4 is a cardiac muscle cell survival factor.5–8 More recent studies using genetically modified mice further shown that knock-down of GATA4 promotes cardiac muscle cell apoptosis and heart failure.28,29 Results from the present study revealed a novel molecular mechanism of the action of DNR to inhibit a cardiac muscle cell survival factor, GATA4, through the involvement of interactions between p53 and CBF/NF-Y. In addition to the role of this mechanism in anthracycline cardiotoxicity, p53 regulation of GATA4 down-regulation may be involved in heart failure subsequent to pressure overload-induced cardiac hypertrophy, as the importance of p53 in this clinically important event has been documented.30
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Funding
This work was supported in part by grants from National Institutes of Health (grant numbers: R01HL67340, R01HL72844 and R01HL97514 to Y.J.S.) and American Heart Association Mid-Atlantic Affiliate (grant number: Grant-in-Aid 0855337E to Y.J.S.).
Acknowledgements
We thank Karen Pitlyk for excellent technical assistance.
Conflict of interest: none declared.
References
- 1.Singal PK, Iliskovic N. Doxorubicin-induced cardiomyopathy. New Engl J Med. 1998;339:900–905. doi: 10.1056/NEJM199809243391307. doi:10.1056/NEJM199809243391307. [DOI] [PubMed] [Google Scholar]
- 2.Arola OJ, Saraste A, Pulkki K, Kallajoki M, Parvinen M, Voipio-Pulkki LM. Acute doxorubicin cardiotoxicity involves cardiomyocyte apoptosis. Cancer Res. 2000;60:1789–1792. [PubMed] [Google Scholar]
- 3.Kang YJ, Zhou ZX, Wang GW, Buridi A, Klein JB. Suppression by metallothionein of doxorubicin-induced cardiomyocyte apoptosis through inhibition of p38 mitogen-activated protein kinases. J Biol Chem. 2000;275:13690–13698. doi: 10.1074/jbc.275.18.13690. doi:10.1074/jbc.275.18.13690. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki YJ, Evans T. Regulation of cardiac myocyte apoptosis by the GATA-4 transcription factor. Life Sci. 2004;74:1829–1838. doi: 10.1016/j.lfs.2003.10.002. doi:10.1016/j.lfs.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Kim Y, Ma AG, Kitta K, Fitch SN, Ikeda T, Ihara Y, et al. Anthracycline-induced suppression of GATA-4 transcription factor: implication in the regulation of cardiac myocyte apoptosis. Mol Pharmacol. 2003;63:368–377. doi: 10.1124/mol.63.2.368. doi:10.1124/mol.63.2.368. [DOI] [PubMed] [Google Scholar]
- 6.Aries A, Paradis P, Lefebvre C, Schwartz RJ, Nemer M. Essential role of GATA-4 in cell survival and drug-induced cardiotoxicity. Proc Natl Acad Sci USA. 2004;101:6975–6980. doi: 10.1073/pnas.0401833101. doi:10.1073/pnas.0401833101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kitta K, Day RM, Kim Y, Torregroza I, Evans T, Suzuki YJ. Hepatocyte growth factor induces GATA-4 phosphorylation and cell survival in cardiac muscle cells. J Biol Chem. 2003;278:4705–4712. doi: 10.1074/jbc.M211616200. doi:10.1074/jbc.M211616200. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi S, Lackey T, Huang Y, Bisping E, Pu WT, Boxer LM, et al. Transcription factor GATA4 regulates cardiac BCL2 gene expression in vitro and in vivo. FASEB J. 2006;20:800–802. doi: 10.1096/fj.05-5426fje. [DOI] [PubMed] [Google Scholar]
- 9.Negoro S, Oh H, Tone E, Kunisada K, Fujio Y, Walsh K, et al. Glycoprotein 130 regulates cardiac myocyte survival in doxorubicin-induced apoptosis through phosphatidylinositol 3-kinase/Akt phosphorylation and Bcl-xL/caspase-3 interaction. Circulation. 2001;103:555–651. doi: 10.1161/01.cir.103.4.555. [DOI] [PubMed] [Google Scholar]
- 10.Nagase H, Vinod Kumar S, Szwergold NR, Day RM, Suzuki YJ. Characterizations of the cardiac GATA-4 promoter. Proc Am Thorac Soc. 2006;3:A686. (abstract) [Google Scholar]
- 11.Ohara Y, Atarashi T, Ishibashi T, Ohashi-Kobayashi A, Maeda M. GATA-4 gene organization and analysis of its promoter. Biol Pharm Bull. 2006;29:410–419. doi: 10.1248/bpb.29.410. doi:10.1248/bpb.29.410. [DOI] [PubMed] [Google Scholar]
- 12.Park AM, Nagase H, Vinod Kumar S, Suzuki YJ. Acute intermittent hypoxia activates myocardial cell survival signaling. Am J Physiol. 2007;292:H751–H757. doi: 10.1152/ajpheart.01016.2006. [DOI] [PubMed] [Google Scholar]
- 13.Claycomb WC, Lanson NA, Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, et al. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci USA. 1998;95:2979–2984. doi: 10.1073/pnas.95.6.2979. doi:10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitta K, Clément SA, Remeika J, Blumberg JB, Suzuki YJ. Endothelin-1 induces phosphorylation of GATA-4 transcription factor in HL-1 atrial-muscle cell line. Biochem J. 2001;359:375–380. doi: 10.1042/0264-6021:3590375. doi:10.1042/0264-6021:3590375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park AM, Suzuki YJ. Effects of intermittent hypoxia on oxidative stress-induced myocardial damage in mice. J Appl Physiol. 2007;102:1806–1814. doi: 10.1152/japplphysiol.01291.2006. doi:10.1152/japplphysiol.01291.2006. [DOI] [PubMed] [Google Scholar]
- 16.Wong CM, Cheema AK, Zhang L, Suzuki YJ. Protein carbonylation as a novel mechanism in redox signaling. Circ Res. 2008;102:310–318. doi: 10.1161/CIRCRESAHA.107.159814. doi:10.1161/CIRCRESAHA.107.159814. [DOI] [PubMed] [Google Scholar]
- 17.Park AM, Nagase H, Vinod Kumar SV, Suzuki YJ. Effects of intermittent hypoxia on the heart. Antioxid Redox Signal. 2007;9:723–729. doi: 10.1089/ars.2007.1460. doi:10.1089/ars.2007.1460. [DOI] [PubMed] [Google Scholar]
- 18.Park AM, Wong CM, Jelinkova L, Liu L, Nagase H, Suzuki YJ. Pulmonary hypertension-induced GATA4 activation in the right ventricle. Hypertension. 2010;56:1145–1151. doi: 10.1161/HYPERTENSIONAHA.110.160515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.L'Ecuyer T, Sanjeev S, Thomas R, Novak R, Das L, Campbell W, et al. DNA damage is an early event in doxorubicin-induced cardiac myocyte death. Am J Physiol. 2006;291:H1273–H1280. doi: 10.1152/ajpheart.00738.2005. [DOI] [PubMed] [Google Scholar]
- 20.Shizukuda Y, Matoba S, Mian OY, Nguyen T, Hwang PM. Targeted disruption of p53 attenuates doxorubicin-induced cardiac toxicity in mice. Mol Cell Biochem. 2005;273:25–32. doi: 10.1007/s11010-005-5905-8. doi:10.1007/s11010-005-5905-8. [DOI] [PubMed] [Google Scholar]
- 21.Kanai M, Hanashiro K, Kim SH, Hanai S, Boulares AH, Miwa M, et al. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nature Cell Biol. 2007;9:1175–1183. doi: 10.1038/ncb1638. doi:10.1038/ncb1638. [DOI] [PubMed] [Google Scholar]
- 22.Nithipongvanitch R, Ittarat W, Cole MP, Tangpong J, St. Clair DK, Oberley TD. Mitochondrial and nuclear p53 localization in cardiomyocytes: redox modulation by doxorubicin (adriamycin)? Free Radic Biol Med. 2007;9:1001–1008. doi: 10.1089/ars.2007.1632. [DOI] [PubMed] [Google Scholar]
- 23.Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, et al. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285:1733–1737. doi: 10.1126/science.285.5434.1733. doi:10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- 24.Nerlov C. The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 2007;17:318–324. doi: 10.1016/j.tcb.2007.07.004. doi:10.1016/j.tcb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. doi:10.1016/S0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 26.Bhattacharya A, Deng JM, Zhang Z, Behringer R, de Crombrugghe B, Maity SN. The B subunit of the CCAAT box binding transcription factor complex (CBF/NF-Y) is essential for early mouse development and cell proliferation. Cancer Res. 2003;63:8167–8172. [PubMed] [Google Scholar]
- 27.Jung MS, Yun J, Chae HD, Kim JM, Kim SC, Choi TS, et al. p53 and its homologues, p63 and p73, induce a replicative senescence through inactivation of NF-Y transcription factor. Oncogene. 2001;20:5818–5825. doi: 10.1038/sj.onc.1204748. doi:10.1038/sj.onc.1204748. [DOI] [PubMed] [Google Scholar]
- 28.Bisping E, Ikeda S, Kong SW, Tarnavski O, Bodyak N, McMullen JR, et al. Gata4 is required for maintenance of postnatal cardiac function and protection from pressure overload-induced heart failure. Proc Natl Acad Sci USA. 2006;103:14471–14476. doi: 10.1073/pnas.0602543103. doi:10.1073/pnas.0602543103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oka T, Maillet M, Watt AJ, Schwartz RJ, Aronow BJ, Duncan SA, et al. Cardiac-specific deletion of Gata4 reveals its requirement for hypertrophy, compensation, and myocyte viability. Circ Res. 2006;98:837–845. doi: 10.1161/01.RES.0000215985.18538.c4. doi:10.1161/01.RES.0000215985.18538.c4. [DOI] [PubMed] [Google Scholar]
- 30.Sano M, Minamino T, Toko H, Miyauchi H, Orimo M, Qin Y, et al. p53-induced inhibition of Hif-1 causes cardiac dysfunction during pressure overload. Nature. 2007;446:444–448. doi: 10.1038/nature05602. doi:10.1038/nature05602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.