Abstract
Background
Dextran sodium sulfate (DSS) is used to induce murine colitis. Although the exact mechanism by which DSS administration causes disease is unknown, evidence suggests that the resident bacteria play a role in the development of murine DSS colitis, analogous to their role in human inflammatory bowel diseases.
Methods
C57BL/6 mice received 5% DSS in the drinking water, and were euthanized three days and 14 days after the initiation of DSS treatment. Culture-independent methods were used to follow changes in the community structure of the gut’s microbiota following DSS treatment. Histologic evidence of disease and changes in host gene expression were assessed.
Results
Histologic colitis was minimal in DSS-treated animals at three days, but severe after 14 days. Analysis of 16S rRNA-encoding gene clone libraries demonstrated that the microbial communities in the ceca of DSS-treated mice were distinct from those in control mice. The microbiota in the cecum of DSS-treated animals was characterized by an overall decrease in microbial richness, an increase in members of the phylum Verrucomicrobia, and decrease in Tenericutes. Changes in the host’s inflammatory response and microbial communities occurred before the histologic appearance of severe disease in the colon, but were seen concurrently in the cecum.
Conclusion
DSS administration is associated with reproducible changes in the gut microbial diversity of mice. Microbial and immunological changes appeared before the development of severe inflammation in the colon. This indicates that these changes in microbial community may play role in the potentiation of the abnormal inflammatory response seen in DSS-treated animals.
Keywords: Microbiota, DSS, 16S rRNA-encoding gene, ecology, colitis
Introduction
Microbes are essential to the health and well-being of their hosts (1, 2). However, the presence of bacteria alone is not enough to impart these benefits, rather the composition and relative abundance of specific microbes have an important role in maintaining health (3). Alterations of the community structure of the indigenous microbiota have also been implicated in the development of disease. One such condition in which the resident bacteria are thought to play a critical role in pathogenesis is inflammatory bowel disease (IBD) (3, 4). Dysbiosis, defects in immunoregulation and defects in the barrier function are all thought to contribute to the onset of disease (5–7). Evidence of a key role for the microbiota in pathogenesis is provided by studies that demonstrate that antibiotics can reduce or prevent inflammation both in patients and in murine models of disease (8, 9). In IL10−/ − mice, which develop colitis after infection with certain bacteria or in the setting of certain housing conditions, treatment with antibiotics is associated with alterations in the microbial gut community (10), and the prevention or amelioration of disease (11).
Of the multiple murine models of IBD, one commonly used system involves the administration of dextran sodium sulfate (DSS) which induces disease very similar to human ulcerative colitis (12). Antibiotic administration has been shown to ameliorate DSS-induced colitis (13), and cathelicidin, an antimicrobial peptide, was also found to have protective effects in this model of colitis (14), indicating that the microbiota plays a role in this disease model system.
Multiple hypotheses have been postulated for the mechanism by which DSS-triggers gut mucosal inflammation but the exact pathogenesis remains unclear (14, 15). Given that human IBD is associated with an altered microbial diversity (5, 16) and evidence for a role of the microbiota in DSS-induced colitis (14, 17), we hypothesize that this model is also associated with an altered diversity of the intestinal microbiota. To test whether DSS treatment can alter the microbial community diversity we employed molecular techniques targeting the 16S rRNA-encoding gene to follow the community structure of the gut bacteria in animals receiving DSS. We correlated changes in the community structure with development of disease and host responses.
Materials and Methods
Animals
C57BL/6 mice from a breeding colony initially established with breeding stock Jackson laboratories (Bar Harbor, ME) were used for experiments. The animal studies were conducted at Michigan State University and were approved by the Michigan State University Ethics Committee. Mice were housed with autoclaved bedding, given sterile food and water ad libitum, and exposed to 12:12 h light:dark cycles. The mice, between 12 to 16 wks old, were assigned to cages according to gender. Dextran sodium sulfate (DSS) (36 000– 50 000 MW, ICN Biochemicals Inc., CA.) was administered as a 5% solution in the drinking water (8).
Three groups of mice were used in the experiment: ten control mice that were maintained on sterile drinking water; ten mice that were placed on 5% DSS in sterile drinking water for 3 days before being euthanized; and eleven mice that were given 5% DSS for 14 days before euthanasia (18).
Necropsy and Histology
The mice were euthanized by CO2 asphyxiation and the cecal tissue harvested as described previously (19). The tissues were gently washed with 1X phosphate-buffered saline (PBS) to remove the fecal contents, cut into sections and snap-frozen in liquid nitrogen. One of these sections was used for terminal restriction fragment length polymorphism (T-RFLP) analyses and clone library analysis, and another for RNA extraction.
The remainder of the ceca was processed for histology as follows: The luminal contents were removed, washed with PBS, placed in tissue cassettes and submerged in 10% formalin for 24 hours. The tissue cassettes were transferred to a 60% ethanol solution and then processed for paraffin embedding and staining with haematoxylin and eosin (H&E).
Scoring was completed using the colitisindex histological scoring system used by Berndt et al. (20) and adapted from Rachmilewitz et al. (21). Briefly, the sections assessed on inflammation, transmural infiltration, cell wall thickening and bleeding; and scored on a scale ranging from 0 to 40.
RNA extraction and PCR array analysis
Total RNA from cecal tissue was isolated using TRIzolR (Invitrogen, Carlsbad, CA) as directed by the manufacturer’s protocol. The High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) was used to convert the total RNA to cDNA. Changes in host gene expression were measured using an array of gene-specific primers from SuperarrayR (Frederick, MD), designed for the following targets (22): Jun, Cd80, Cd209a, Il12b, Irak3, Smad3, Arg1, Cd86, Mapk3, Il17a, Mapk8, Tgfb1, Cd274, Creb1, Mapk1, Il1b, Tlr2, Tlr5, Vtcn1, Cx3cr1, Foxp3, Il2, Tlr4, Tlr9, Ccr2, Cxcl1, Tnfrsf18, Il23a, H2-DMb1, Tnf, Ccr7, Cxcl10, Infg, Il4, Nfkb1, Vegfa, Itgax, Cxcl5, Il10, Il6, Mapk14, Gapdh, Cd40, Cxcl2, Il12a, Irak4, Pik3r1, ActB. The neutrophil marker, Ly-6G, was also quantified using the primer set from Sasmono et al. (Ly-6G forward primer 5'-TGGACTCTCACAGAAGCAAAG-3' and reverse primer 5'-GCAGAGGTCTTCCTTCCAACA-3') as well as the Gapdh primer set from Cui et al. (forward primer 5'-ACCACAGTCCATGCCATCAC-3' and reverse primer 5'-TCCACCACCCTGTTGCTGTA-3). Quantization was performed using LightCyclerR 480 SYBR Green I Master and analyzed on a LightCyclerR 480 system (Roche Diagnostics, Indianapolis, IN) as directed by manufacture’s protocol, with the following cycling conditions: 95°C activation for 10 min; 40 cycles of 95°C denaturation for 15s, and 60°C annealing for 1 min. Resulting threshold values were analyzed by calculating the 2−Δ ΔCt values, using GAPDH as the reference, to find fold regulation compared to the no-DSS control (23–25).
DNA extraction
Genomic DNA was extracted from tissue using the Qiagen DNeasyR Blood & Tissue kit (Cat. No.69504, Valencia, CA). Briefly, the tissue was incubated overnight in lysis buffer and proteinase K at 56 °C. The following day, the enzyme was denatured at 95 °C and DNA was purified using ethanol through filter columns, as directed by the manufacturer’s protocol, and eluted in 30μl of elution buffer. This genomic DNA was then used in preparation of the clone libraries and T-RFLP.
Clone library construction
For clone library construction, reaction was set up as described previously (26), with illustra PuReTaqTM Ready To Go PCRTM beads (GE Healthcare, Piscataway, NJ). Briefly, amplification by polymerase chain reaction was performed using broad-ranged primers, (8F, 5'-AGAGTTTGATCCTGGCTCAG-3'; 1492R, 5'-GGTTACCTTGTTACGACTT-3'). Amplicon purification was done using a commercial kit (GFX, GE Healthcare, Piscataway, NJ) as directed by the manufacturer. Products were ligated into the TOPO 4R vector (Invitrogen K4575-01, Carlsbad, CA) according to manufacturer’s specifications, and transformed into Escherichia coli. Colonies were picked into Luria Broth (LB) with carbenicillin (50 μg/ml)) and grown overnight at 37 °C. Vector specific primers (M13F, 5'-CAGTCACGACGTTGTAAAACGACGGC-3'; and M13R, 5'-CAGGAAACAGCTATGACCATG-3') were used to screen these colonies for bacterial clones containing the appropriate 1.5 KB amplicon insert. Partial 16S sequences were determined by a single sequencing run using the 8F primer, at the Genomic Core at Michigan State University. Raw sequence data were processes through an automated “information pipeline” available through the Ribosomal Database Project (RDP) Web site (http://rdp.cme.msu.edu/). Data were screened using the Chimera Check Program before uploading into the RDP. Following alignment of the sequences via myRDP (27) distance matrices representing each of the libraries were downloaded and then taxonomic assignments designated (80% confidence cut-off) using Classifier through the RDP website. These distance matrices were also input into mothur (28) to be grouped into operational taxonomic units (OTUs). Analyses were done using a 97% sequence similarity to denote species level. An input table was also generated for the EstimateS program via the RDP Pipeline. EstimateS (29) was used to calculate ecological diversity indices from the aligned sequences. Dendrograms based on the Bray-Curtis similarity index were constructed using the Mega3 program (30).
16S small subunit rRNA gene sequences obtained by clone library analysis were subjected to in silico terminal restriction fragment length polymorphism (T-RFLP) analysis using the TRF-cut program in the ARB suite of programs (31). The MspI enzyme, a four base cutter, was used to calculate predicted terminal restriction fragments (TRFs) from the clone sequences. Histograms were then constructed displaying the relative abundance of these in silco generated TRFs and compared to the actual traces retrieved from the T-RFLP analysis described below. The fragment sizes obtained by T-RFLP anlaysis were compared to the sizes of the TRFs generated in silico from the cloned 16S sequences. These fragments were identified within two basepairs (±2 bp) of the predicted TRFs (32).
T-RFLP analysis
T-RFLP was performed as described elsewhere (33) using primers 1492R and FAM labeled 8F. In summary, genomic DNA was amplified using the aforementioned primers with PCR conditions: 94 °C denaturation for 2 min; 30 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 45 s, and extension at 72 °C for 90 s; and final extension at 72 °C for 4 min. The 1.5 kb PCR product was verified and purified with GFX columns (GFX, GE Healthcare, Piscataway, NJ), then subjected to digestion with the MspI restriction enzyme for two hours. The digested DNA was submitted for analysis to the Genome Technology Support Facility (GTSF) at Michigan State University. Traces were visualized using the program Genescan® (Applied Biosystems, Foster City, CA).
The T-RFLP Stats collection of programs (http://styx.ibest.uidaho.edu/ibest/research.html) (34) were used to analyze the traces from the T-RFLP output data. Briefly, the peaks of each trace were binned into operational taxonomic units (OTU) to produce data categorized by both OTUs and abundance. This was then used as input for the EstimateS (http://viceroyeebuconnedu/estimates) (29) program where the Bray-Curtis values were calculated and used to compare the diversity among the communities. The Mega3 program (30) was used to construct dendrograms showing the relationship between community structures using the Bray-Curtis values. The significance differences between the community structures were calculated using the parsimony test from the mothur suite of programs.
qPCR analysis of microbial communities
The quantity of 16S rRNA operons in the samples relative to a single-copy host gene was measured using a primer/probe set that targets a broad range of rRNA-encoding gene sequences (5’-TCCTACGGGAGGCAGCAGT-3 ’), the reverse primer (5’-GGACTACCAGGGTATCTAATCCTGTT-3’), and the probe (5’-[6-FAM]-203CGTATTACCGCGGCTGCTGGCAC-[TAMRA]-3’) (35). A primer/probe set targeting a 264-bp portion of the TNF alpha gene was used as a reference using 200 nanomoles of the forward (TNFα _mu_se; 5’-GGCTTTCCGAATTCACTGGAG-3’) and reverse primers (TNFα _mu_as; 5’-CCCCGGCCTTCCAAATAAA-3’), and 100 nanomoles of the probe (TNFα _mu_probe; 5’-[Cy5]-ATGTCCATTCCTGAGTTCTGCAAAGGGA-[Iowa Black RQ ]-3’) 213 (36). The reaction mix consists of LightCycler® 480 Probes Master reaction mix (Roche) at 1X concentration, and appropriate primer/probe pair.
Amplification of each gene was done under separate run conditions: cycling conditions for the 16S target involved an activation step of 50 °C for 2 min followed by 95 °C for 10 min. Forty-five cycles was done at 95 °C for 15 s and 60 °C for 1 min before holding. For the TNF reference gene cycling conditions included an activation step of 50 °C for 2 min followed by 95 °C for 10 min and forty-fives cycles of 95 °C for 20 s and 64 °C for 30 s. Calculations of 2−Δ ΔCt were made to compare changes in the amount of 16S from samples between treatment groups (21,22).
Statistical Analysis
The non-parametric Kruskal-Wallis test was used to analyze histological scores. Differences in the abundances of specific operational taxonomic units among the treatment groups were analyzed by ANOVA. Statistical differences in the dendrograms comparing distances between microbial communities were calculated using the parsimony test function in mothur (28). Probability values less than 0.05 were considered significantly different.
Results
DSS treatment leads to inflammation of the murine intestinal tract
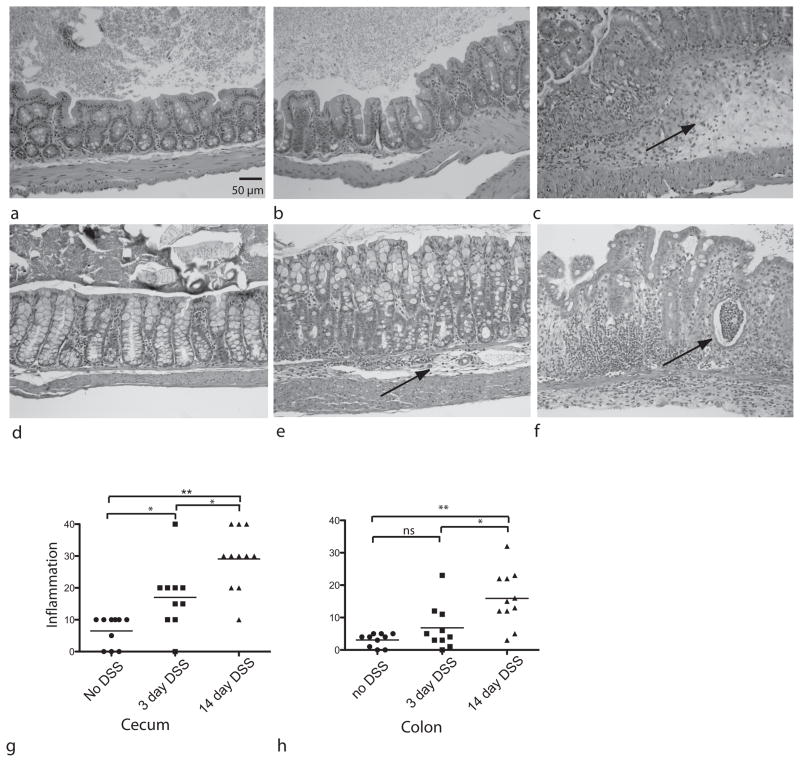
Wildtype mice were treated with 5% DSS in drinking water and the gastrointestinal tissue examined histologically after 3 and 14 days of treatment. All animals survived the DSS treatment and no significant clinical signs were noted. A mild inflammatory infiltrate developed in the cecum 3 days after DSS administration (figure 1b), with notable progression of disease after 14 days characterized the development of a massive inflammatory cell infiltrate and severe edema (figure 1c). These changes were significantly different from the control state (p<0.05), as well as between the 3 day and 14 day treatment outcomes (figure 1g).
Figure 1.
Histopathology in DSS-treated mice. Hemotoxylin and eosin (H&E) stained sections were prepared from the cecum of (a) untreated control mice (b) mice after 3 days of DSS treatment and (c) after 14 days of DSS. Arrow indicates submucosl edema. H&E sections were also prepared from colon samples of (d) untreated controls (e) mice after 3 days of DSS treatment (arrow points to inflammation) and (f) animals after 14 days of DSS (arrow shows abscess). Histopathologic scores were calculated for sections from all 31 animals for (g) cecum and (h) colon sections. Statistical analysis done was Kruskal-Wallis test. *p< 0.05 **p<0.001. Initial magnification 40X
Similarly in the colon, a few animals developed morphological and inflammatory changes after 3 days of DSS treatment, but as a group, these changes were not significant when compared to controls. However, following 14 days of DSS administration colonic tissue showed evidence of severe inflammation and edema which was significantly greater than controls of animals after 3 days of treatment (figure 1h).
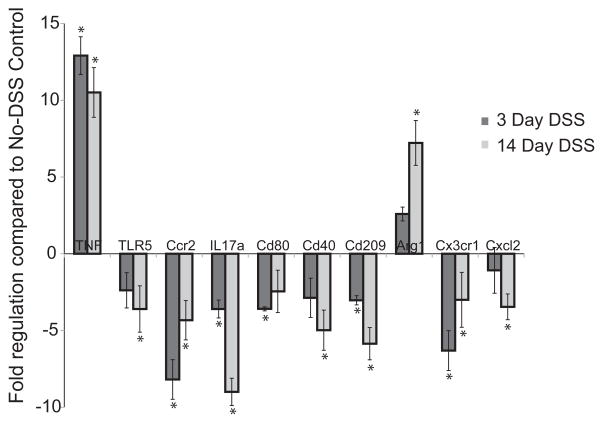
Changes in host gene expression following DSS treatment
Using a quantitative PCR array, we found significant changes in the expression of ten host genes following DSS treatment (figure 2, supplemental figure 1). The expression of arginase 1 began to increase after 3 days and was significantly up-regulated ~seven fold (p<0.05) after 14 days of DSS administration. A significant increase in tumor necrosis factor-alpha (TNFα ) was also seen after 3 days of DSS treatment, with a 12.91 fold up-regulation (p<0.05), and remained significantly up-regulated at the 14-day time point.
Figure 2.
Changes in host gene expression after 3-days (dark bars) or 14-days (grey bars) ± standard deviation of DSS administration. Expression levels were compared to expression in tissue from animals not exposed to DSS. Statistical analysis done was Student’s t-test. Statistically significant differences (p<0.05) are denoted by asterisks (*).
The other eight genes were down-regulated, compared to untreated controls. They were TLR 5, IL17, Ccr2, Cx3cr1, Cxcl2, Cd40, Cd80 and Cd209a. TLR 5 had significant down-regulation after 14-days of DSS exposure, as did Cd40 and Cxcl2. Cd80 only showed significant change after 3 days of DSS and only marginal change after 14 days. Ccr2, Cx3cr1, cd209a and IL17a showed significant down-regulation both at the 3 day and 14 day time points.
The presence of neutrophils, as indicated by measuring the Ly-6G marker, increased significantly with DSS treatment with a 7.7 ± 1.2 fold increase after 3 days of DSS administration, and 14.0 ± 1.8 fold increase after 14 days of DSS (supplemental figure 1).
Shifts in gut microbial diversity following DSS treatment
To obtain an overview of the status of the microbial community present in the gastrointestinal tract of each of the animals in this study, 16S terminal restriction fragment length polymorphism (T-RFLP) analysis was performed on DNA extracted from cecal tissue. Analysis of the microbial communities in the cecum of all 31 animals in the study revealed that the structure of the microbial communities from DSS treated mice were significantly different from the no-DSS controls (p<0.001) (supplemental figures 2).
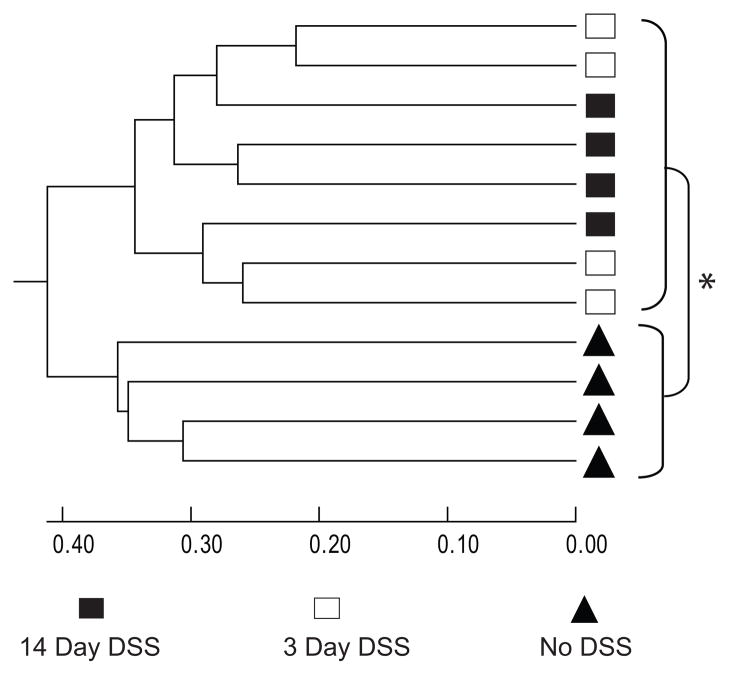
Since T-RFLP provides a broad overview of the community structure without information regarding changes in specific organisms, 16S clone libraries were constructed to further investigate the microbial community in four representative mice from each treatment group. A total of 328 partial 16S rRNA-encoding gene sequences were retrieved from control animals (71, 84, 89 and 84 per animal), 364 sequences from animals 3 days after the initiation of DSS treatment (88, 92, 93 and 91) and 370 from the animals treated with 14 days of DSS (92, 94, 89 and 95). Clone library analysis confirmed the T-RFLP findings, indicating that the community compositions of the microbiota in DSS treated animals were more similar to each other than they were to those of the control mice (figure 3, supplemental 4). The T-RFLP traces were compared to in silico TRFs of the clone libraries to verify that the OTUs matched the traces (supplemental figures 5–8). The communities in all DSS-treated animals were significantly different from those in the no-DSS controls (p<0.001 by the parsimony test implemented in the analysis suite mothur (28)). Rarefaction analysis of the clone libraries revealed that DSS treatment resulted in a decrease in phylotype richness (supplemental 4). This finding was also supported by a decrease in the Simpson diversity index (1/D) following DSS treatment from 43.0 ± 12.1 to 21.3 ± 8.3 (3 days) and 19.7 ± 7.0 (14 days) (p<0.05). However, quantitative PCR of the 16S gene from each treatment group showed that there were no significant changes in overall bacterial biomass relative to the no-DSS control (fold change for 3 day DSS= 0.68 ± 0.92 SD, 14 day DSS= 0.97 ± 0.82 SD).
Figure 3.
Comparison of the cecal community in control animals (black triangles) and in animals following 3 days of DSS treatment (white squares) and 14 days of treatment (black squares). Using an OTU definition of 97% similarity, the Bray-Curtis similarity metric was calculated for each pair-wise comparison and then the results displayed in denrogram format. Analysis by the parsimony test indicates that there is a statistically significant (*p<0.001) difference between the communities in control animals and in both groups of DSS-treated animals.
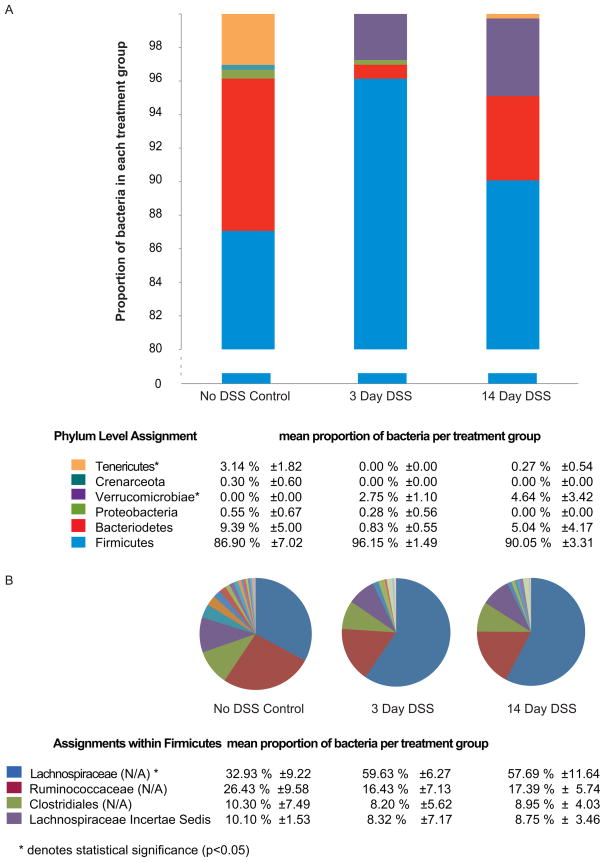
We analyzed changes in the relative abundance of specific phylotypes by classification of the 16S rRNA-encoding gene sequences. DSS treatment resulted in a decrease in members of the phyla Bacteriodetes and Tenericutes. Alterations in the microbial community were also apparent at lower taxonomic levels. For example, there was an increase in the unclassified genera under the family Lachnospiraceae (figure 4, supplemental 3). The majority of classified phylotypes in animals from all three experimental groups were members of the phylum Firmicutes, which include the classes Clostridia, Mollicutes and Bacilli. The Bacteriodetes was most abundant in the control animals and represented to a lesser extent in the DSS treated groups (figure 4A, supplemental 3). A striking observation was that members of the phylum Verrucomicrobiae were detected only in the DSS treated mice and not in the control mice (figure 4A).
Figure 4.
Changes in the cecal gut microbial community following DSS administration (A) at the phylum level (B) within the phylum Firmicutes. Analysis was done by ANOVA and statistical significance (p<0.05) is denoted by asterisks (*).
Among members of the phylum Bacteriodetes the richness of the population did not change, indicating that the changes were mainly due to shift in abundances of different bacteria within this phylum. Among the Firmicutes, the unclassified genera among the families Ruminoccocaceae and Lachnospiraceae were most prevalent. These groups showed treatment related shifts as seen in the pie charts of figure 4B. When analysis was done using 97% sequence similarity, there was high richness among the Lachnospiraceae, compared to other major groups within the Firmicutes (table 1), indicating increased diversity within this category (p<0.05). These groups also showed treatment related shifts (table 1).
Table 1.
showing distribution of OTU’s across treatments from major groups within the Firmicutes.
| Clostridiales N=17 (%) | Ruminococcus N=36 (%) | Lachnospiracea N=65 (%) | |
|---|---|---|---|
| Shared OTUs among all groups | 6 (35) | 16 (44) | 15 (23) |
| OTUs in control group only | 4 (24) | 8 (22) | 13 (20) |
| OTUs in 3 day DSS group only | 2 (12) | 4 (11) | 8 (12) |
| OTUs in 14 day DSS group only | 2 (12) | 5 (14) | 12 (18) |
| Shared OTU in DSS treated groups | 3 (18) | 3 (8) | 17 (26) |
Discussion
The indigenous gut microbiota are felt to play a key role in the pathogenesis of inflammatory bowel disease. Much of the evidence for the involvement of intestinal bacteria in IBD comes from studies with murine models of disease (37, 38). Many of these models have specific alterations in host defenses ranging from altered epithelial cell function to altered innate or adaptive immunity (39–42). These alterations in host defense result in abnormal interactions between the host and the indigenous microbiota that lead to disease (43). Evidence for this comes from the fact that antibiotic administration reduces the severity of disease in these models and rederivation of these mice to the germ-free state prevents initiation of disease (13, 44).
Administration of DSS consistently triggers intestinal inflammation in rodents. Similar to other models, antibiotic administration has a beneficial effect in the system again pointing to a role for the indigenous microbiota in the disease process (45). The effects of DSS on germfree mice have been variable. In some strains of mice more severe colitis is encountered while early death, without the development of significant inflammation, is observed in others (46, 47). Germ-free IQI/Jic mice have a high mortality rate, compared to conventional mice, when given a concentration of 5% DSS in drinking water (45). These mice died secondary to hemorrhage within three days after DSS treatment, indicating a protective function of the microbiota against the toxic effects of DSS on the gut epithelium. These results are consistent with the finding that antibiotic administration or genetic defects in TLR signaling also increased mortality in DSS-treated conventional mice (48, 49). Differences in these two studies employing germ-free mice likely reflects the differential effects of the two doses of DSS on the epithelium, but may also reflects differences in the composition of the microbiota of the conventional control animals. Since specific subsets of the “normal” microbiota have differing ability to initiate or sustain experimental colitis (45, 50) these results emphasize the importance of characterizing the changes in the gut community associated with DSS-induced colitis across time.
Although it was originally suggested that IBD resulted from infection with an unknown bacterial pathogen (51), the more contemporary view is that entire shifts in the microbial community structure may be the trigger for disease (5). An altered gut microbial community has been associated with disease as shown in human studies and animal models (52, 53), suggesting that there is a change in abundance of bacteria that are necessary for maintaining homeostasis (54, 55). After administering DSS to mice, we show that treatment results in changes in microbial community structure in the gut, also noted by others (49, 56). Additionally, we found that these changes occur early (within 3 days) and are characterized by reduced overall diversity while bacterial load remains unchanged. Some community members are below the limits of detection after DSS treatment, while others increase in abundance demonstrating a selective effect of DSS treatment on specific members of the community. It has been shown that certain bacteria can de-polymerization DSS and thus can grow in a DSS-rich environment better than others (57). This may explain the bloom of Verrucomicrobia that we observed in DSS treated mice. This group of microbes, which has been found in mammalian intestine at very low abundances (58), metabolize sulfur and degrade mucin (59). Therefore, perturbations of the community can cause previously underrepresented members of the community to become dominant (60), resulting in shifts in community structure, as seen in the phylum Bacteriodetes and families Ruminococcaceae and Lachnospiraceae.
Another study has profiled the effects of DSS treatment on the microbial community (49). Similar to our study, it has been noted that shifts in the relative abundances of specific phylotypes were observed following DSS treatment, although the specific changes were different from what we observed. One obvious reason for these differences could be explained by the fact that different anatomic locations were sampled (cecal mucosa versus distal colonic contents), as well as length and concentrations of DSS administration. Additionally, it is likely that the baseline microbial communities were quite different between the animals in this study and the animals we used. Recent reports have demonstrated that the baseline microbiota can differ in genetically identical animals obtained from different sources and these differences can have a profound effect on host immune responses (61, 62).
Disease severity in the cecum increased after 3 days and 14 days post DSS administration, concurrent with the changes observed in the microbial community, indicating a link between the changes in diversity of the microbiota and disease. Since shifts in the community structure can reduce beneficial members of the indigenous microbiota that act to maintain epithelial health, such as the production of short-chain fatty acids and stimulation of mucin production (55, 63–66), it is likely that such changes can affect the host’s inflammatory response (43, 67).
Others have suggested that DSS-induced colitis occurs due to breaches in the epithelial barrier with exposure to antigens produced by the luminal microbes (5, 68–74). However, there is evidence that the detection of an altered microbial community is an important factor in the host response (75, 76). Our data indicate that DSS induces changes in the microbiota, and together with a compromised barrier, appears to facilitate disease initiation and progression. It has been shown that the host responds to microbial-associated molecular patterns (MAMPS) via toll-like receptors (TLRs) in the development of colitis (17, 38, 77). We found significant change in TLR 5, the expression of which was decreased as colitis progressed. It has been previously demonstrated that TLR 5 is down-regulated in patients with severe ulcerative colitis (78), in concurrence with our data. However, the role of flagella in DSS colitis is not dependent on TLR5 (38, 79), suggesting that the down-regulation is perhaps a response from the over-stimulation from microbes through the deteriorating epithelial layer.
It is noteworthy that we saw an increase in arginase 1 after 14 days of DSS treatment. This could be due to the increase of neutrophils after DSS administration, as indicated by increased expression of the neutrophil marker Ly6-G. Arginase 1 also has a protective role in the Citrobacter rodentium model of colitis where enzyme inhibition aggravated disease (80). It has been seen that arginase expression increases with TNFα /lipopolysaccharides (LPS) stimulation in human intestinal microvascular endothelial cells (81). With breaches in the epithelial layer that allows increased stimulation by LPS, together with an increase in TNFα as seen here and in other studies with DSS (71, 82), it follows that arginase expression is likely to be up-regulated. Interestingly, although TNFα appeared to be down-regulated at 14 days compared to 3 days, arginase was up-regulated at 14 days compared to 3 days, consistent with the presumed role for this gene product in the epithelial repair response. To our knowledge, this study is the first to document the significant increase of this enzyme in DSS-induced colitis.
Although others have noted changes in the gut microbiota following DSS treatment (49) after development of disease, this is the first study to monitor DSS-induced changes in the intestinal microbiota over time. In this study, we not only corroborate that changes in the gut microbial community are associated with DSS-induced disease, but we also show that diversity of the microbiota changed as early as after 3 days and continued to 14 days of DSS administration. Early shifts in the microbial community were characterized with increases in abundances of microbes previously undetectable by the methods used. These altered communities are present at the onset of inflammation, and maintained as inflammation severity increased, reinforcing the idea that disease occurs, and persists, in the presence of an altered intestinal community. These early changes in the microbiota support the view that disturbance of the resident bacterial community structure may have a very important role in the onset of disease, such as IBD.
Supplementary Material
Supplemental figure 2. Comparison of the cecal community in control animals (black triangles), and animals following 3 days of DSS treatment (white squares) and days of treatment (black squares). Using T-RFLP, the Bray-Curtis similarity metric was calculated for each pair-wise comparison and then the results displayed in dendrogram format. Communities that were further analyzed, by construction of clone libraries, are indicated by arrows. Analysis by the parsimony test indicates that there is a statistically significant (*p<0.05 and **p<0.001).
Supplemental figure 4. The rarefaction curves at 97% sequence similarity comparing the microbial communities in mice after 3-days of DSS treatment (pink), 14-days of DSS treatment (blue) and the controls (yellow).
Supplemental figure 5. Comparisons between T-RFLP traces and histograms of the terminal restriction fragments (TRFs) of in silico digest of the clone library constructed from the control communities.
Supplemental figure 6. Comparisons between T-RFLP traces and histograms of the terminal restriction fragments (TRFs) of in silico digest of the clone library constructed from the 3-day DSS administered communities.
Supplemental figure 7. Comparisons between T-RFLP traces and histograms of the terminal restriction fragments (TRFs) of in silico digest of the clone library constructed from the 14-day DSS administered communities.
Acknowledgments
I would like to extend thanks to Jason Pratt for technical assistance, and Dr. Courtney Robinson, Dr. Grace Chen and Dr. Gary Huffnagle for helpful discussions and review of the manuscript. I am especially grateful for the funding provided by the Center for Pathogenesis, MSU and the NIH. I would also like to acknowledge the Genomics Core at Michigan State University where all the sequencing and T-RFLP data were generated, as well as the RDP staff for facilitating the work.
Center for Microbial Pathogenesis, Michigan State University. NIH grant DK070875 (VBY), Crohn’s and Colitis Foundation of America Senior Investigator Award (VBY).
References
- 1.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human – microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley RE, Peterson DA, Gordon JI. Ecological and Evolutionary Forces Shaping Microbial Diversity in the Human Intestine. Cell. 2006;24:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 3.Round J, Mazmanian S. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Subramanian S, Campbell BJ, Rhodes JM. Bacteria in the pathogenesis of inflammatory bowel disease. Curr Opin Infect Dis. 2006:475–484. doi: 10.1097/01.qco.0000244054.69253.f3. [DOI] [PubMed] [Google Scholar]
- 5.Packey C, Sartor R. Interplay of commensal and pathogenic bacteria, genetic mutations, and immunoregulatory defects in the pathogenesis of inflammatory bowel diseases. J Intern Med. 2008;263:597–606. doi: 10.1111/j.1365-2796.2008.01962.x. [DOI] [PubMed] [Google Scholar]
- 6.Swidsinski A, Ladhoff A, Pernthaler A, et al. Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002;122:44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- 7.Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 8.Kang SS, Bloom SM, Norian LA, et al. An antibiotic-responsive mouse model of fulminant ulcerative colitis. PLoS Medicine. 2008;5:e41. doi: 10.1371/journal.pmed.0050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swidsinski A, Weber J, Loening-Baucke V, et al. Spatial Organization and Composition of the Mucosal Flora in Patients with Inflammatory Bowel Disease. J Clin Microbiol. 2005;43:3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sellon RK, Tonkonogy S, Schultz M, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–5231. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madsen KL, Doyle JS, Tavernini MM, et al. Antibiotic therapy attenuates colitis in interleukin 10 gene-deficient mice. Gastroenterology. 2000;118:1094–1105. doi: 10.1016/s0016-5085(00)70362-3. [DOI] [PubMed] [Google Scholar]
- 12.Okayasu I, Hatakeyama S, Yamada M, et al. A Novel Method in the Induction of reliable Experimental Acute and Chronic Ulcerative Colitis in Mice. Gastroenterology. 1990;98:694–702. doi: 10.1016/0016-5085(90)90290-h. [DOI] [PubMed] [Google Scholar]
- 13.Rath H, Schultz M, Freitag R, et al. Different Subsets of Enteric Bacteria Induce and Perpetuate Experimental Colitis in Rats and MIce. Infect Immun. 2001;69:2277–2285. doi: 10.1128/IAI.69.4.2277-2285.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tai EKK, Wu WKK, Wong HPS, et al. A New Role for Cathelicidin in Ulcerative Colitis in Mice. Exp Biol Med (Maywood) 2007;232:799–808. [PubMed] [Google Scholar]
- 15.Kitajima S, Takuma S, Morimoto M. Changes in Colonic Mucosal Permeability in Mouse Colitis Induced with Dextran Sulfate Sodium. Exp Anim. 1999;48:137–143. doi: 10.1538/expanim.48.137. [DOI] [PubMed] [Google Scholar]
- 16.Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. PNAS. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, et al. Recognition of Commensal Microflora by Toll–Like Receptors Is Required for Intestinal Homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Vowinkel T, Kalogeris T, Mori M, et al. Impact of Dextran Sulfate Sodium Load on the Severity of Inflammation in Experimental Colitis. Dig Dis Sci. 2004;49:556–564. doi: 10.1023/b:ddas.0000026298.72088.f7. [DOI] [PubMed] [Google Scholar]
- 19.Young VB, Knox KA, Pratt JS, et al. In Vitro and In Vivo Characterization of Helicobacter hepaticus Cytolethal Distending Toxin Mutants. Infect Immun. 2004;72:2521–2527. doi: 10.1128/IAI.72.5.2521-2527.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berndt BE, Zhang M, Chen G-H, et al. The Role of Dendritic Cells in the Development of Acute Dextran Sulfate Sodium Colitis. J Immunol. 2007;179:6255–6262. doi: 10.4049/jimmunol.179.9.6255. [DOI] [PubMed] [Google Scholar]
- 21.Rachmilewitz D, Karmeli F, Takabayashi K, et al. Immunostimulatory DNA ameliorates experimental and spontaneous murine colitis. Gastroenterology. 2002;122:1428–1441. doi: 10.1053/gast.2002.32994. [DOI] [PubMed] [Google Scholar]
- 22.Lees CW, Zacharias WJ, Tremelling M, et al. Analysis of Germline GLI1 Variation Implicates Hedgehog Signalling in the Regulation of Intestinal Inflammatory Pathways. PLoS Med. 2008;5:1761–1775. doi: 10.1371/journal.pmed.0050239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfaffl MW. A new mathematical model for relative quantification in real-time RT- PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guide to Performing Relative Quantitation of Gene Expression Using Real-Time Quantitative PCR. Applied Biosystems. 2004 [Google Scholar]
- 25.Schmittgen TD, KJL Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 26.Young VB, Schimdt TM. Antibiotic-Associated Diarrhea Accompanied by Large-Scale Alterations in the Composition of the Fecal Microbiota. J Clin Microbiol. 2004;42:1203–1206. doi: 10.1128/JCM.42.3.1203-1206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cole JR, Chai B, Farris RJ, et al. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nuclic Acids Res. 2007;35:D169–172. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schloss P, Westcott S, Ryabin T, et al. Introducing mothur: Open Source, Platform-independent, Community-supported Software for Describing and Comparing Microbial Communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colwell RK. EstimateS: Statistical estimation of species richness and shared species from samples. User's Guide and application. 2006 published at: http://purloclcorg/estimates.
- 30.Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 31.Ludwig W, Strunk O, Westram R, et al. ARB: a software environment for sequence data. Nucleic Acid Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu WT, Marsh TL, Cheng H, et al. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keuhl CJ, Wood HD, Marsh TL, et al. Colonization of the Cecal Mucosa by Helicobacter hepaticus Impacts the Diversity of the Indigenous Microbiota. Infect Immun. 2005;73:6952–6961. doi: 10.1128/IAI.73.10.6852-6961.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdo Z, Schuette UME, Bent SJ, et al. Statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environ Microbiol. 2006;8:929–938. doi: 10.1111/j.1462-2920.2005.00959.x. [DOI] [PubMed] [Google Scholar]
- 35.Martin FE, Nadkarni MA, Jacques NA, et al. Quantitative Microbiological Study of Human Carious Dentine by Culture and Real-Time PCR: Association of Anaerobes with Histopathological Changes in Chronic Pulpitis. J Clin Microbiol. 2002;40:1698–1704. doi: 10.1128/JCM.40.5.1698-1704.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nitsche A, Becker M, Junghahn I, et al. Quantification of human cells in NOD/SCID mice by duplex real-time polymerase-chain reaction. Haematologica. 2001;86:693–699. [PubMed] [Google Scholar]
- 37.Wirtz S, Neurath MF. Mouse models of inflammatory bowel disease. Adv Drug Deliv Rev. 2007;59:1073–1083. doi: 10.1016/j.addr.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 38.Erridge CDS, Bereswill S, Heimesaat MM. The Induction of Colitis and Ileitis in Mice Is Associated with Marked Increases in Intestinal Concentrations of Stimulants of TLRs 2, 4, and 5. PLoS. 2010;5:e9125. doi: 10.1371/journal.pone.0009125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang Y, Clayburgh DR, Mittal N, et al. Epithelial NF-{kappa}B Enhances Transmucosal Fluid Movement by Altering Tight Junction Protein Composition after T Cell Activation. Am J Pathol. 176:158–167. doi: 10.2353/ajpath.2010.090548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shea-Donohue T, Thomas K, Cody MJ, et al. Mice deficient in the CXCR2 ligand, CXCL1 (KC/GRO-{alpha}), exhibit increased susceptibility to dextran sodium sulfate (DSS)-induced colitis. Innate Immun. 2008;14:117–124. doi: 10.1177/1753425908088724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blumberg RS, Saubermann LJ, Strober W. Animal models of mucosal inflammation and their relation to human inflammatory bowel disease. Curr Opin Immunol. 1999;11:648–656. doi: 10.1016/s0952-7915(99)00032-1. [DOI] [PubMed] [Google Scholar]
- 42.Belardelli F, Ferrantini M. Cytokines as a link between innate and adaptive antitumor immunity. Trends Immunol. 2002;23:201–208. doi: 10.1016/s1471-4906(02)02195-6. [DOI] [PubMed] [Google Scholar]
- 43.Abt MC, Artis D. The intestinal microbiota in health and disease: the influence of microbial products on immune cell homeostasis. Curr Opin Gastroenterol. 2009;25:496–502. doi: 10.1097/MOG.0b013e328331b6b4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 45.Rath HC, Schultz M, Freitag R, et al. Different subsets of enteric bacteria induce and perpetuate experimental colitis in rats and mice. Infect Immun. 2001;69:2277–2285. doi: 10.1128/IAI.69.4.2277-2285.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hudcovic T, Stepankova R, Cebra J, et al. The Role of Microflora in the Development of Intestinal Inflammation: Acute and Chronic Colitis Induced by Dextran Sulfate in Germ-Free and Conventionally Reared Immunocompetent and Immunodeficient Mice. Folia Microbiol (Praha) 2001;46:565–572. doi: 10.1007/BF02818004. [DOI] [PubMed] [Google Scholar]
- 47.Kitajima S, Morimoto M, Sagara E, et al. Dextran sodium sulfate-induced colitis in germ-free IQI/Jic mice. Exp Anim. 2001;50:387–395. doi: 10.1538/expanim.50.387. [DOI] [PubMed] [Google Scholar]
- 48.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, et al. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 49.Heimesaat M, Fischer A, Siegmund B, et al. Shift towards pro-inflammatory intestinal bacteria aggravates acute murine colitis via Toll-like receptors 2 and 4. PLoS One. 2007;25:e662. doi: 10.1371/journal.pone.0000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoentjen F, Harmsen HJ, Braat H, et al. Antibiotics with a selective aerobic or anaerobic spectrum have different therapeutic activities in various regions of the colon in interleukin 10 gene deficient mice. Gut. 2003;52:1721–1727. doi: 10.1136/gut.52.12.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hansen R, Thomson JM, El-Omar EM, Hold GL. The role of infection in the aetiology of inflammatory bowel disease. J Gastroenterol. 2010;45:266–76. doi: 10.1007/s00535-009-0191-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alexander S, Vera L-B, Mario V, et al. Active Crohn's disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm Bowel Dis. 2008;14:147–161. doi: 10.1002/ibd.20330. [DOI] [PubMed] [Google Scholar]
- 53.Suchodolski JS, Xenoulis P, Paddock CG, et al. Molecular analysis of the bacterial microbiota in duodenal biopsies from dogs with idiopathic inflammatory bowel disease. Vet Microbiol. 2010;142:394–400. doi: 10.1016/j.vetmic.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 54.Maukonen J, Satokari R, Matto J, et al. Prevalence and temporal stability of selected clostridial groups in irritable bowel syndrome in relation to predominant faecal bacteria. J Med Microbiol. 2006;55:625–633. doi: 10.1099/jmm.0.46134-0. [DOI] [PubMed] [Google Scholar]
- 55.Cummings JH, Macfarlane GT. Role of intestinal bacteria in nutrient metabolism. JPEN J Parenter Enteral Nutr. 1997;21:357–365. doi: 10.1177/0148607197021006357. [DOI] [PubMed] [Google Scholar]
- 56.Deplancke B, Finster K, Graham WV, et al. Gastrointestinal and Microbial Responses to Sulphate-Supplemented Drinking Water in Mice. Exp Biol Med. 2003;228:424–433. doi: 10.1177/153537020322800413. [DOI] [PubMed] [Google Scholar]
- 57.Araki YMK, Sugihara H, Fujiyama Y, Hattori T. Proteus mirabilis sp. intestinal microflora grow in a dextran sulfate sodium-rich environment. Int J Mol Med. 2010;25:203–208. [PubMed] [Google Scholar]
- 58.Wang M, Ahrne S, Jeppsson B, et al. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol Ecol. 2005:219–231. doi: 10.1016/j.femsec.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 59.Derrien M, Vaughan EE, Plugge CM, et al. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 60.Antonopoulos D, Huse S, Morrison H, et al. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ivanov II, Frutos RL, Manel N, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ahmad MS, Krishnan S, Ramakrishna BS, et al. Butyrate and glucose metabolism by colonocytes in experimental colitis in mice. Gut. 2000;46:493–499. doi: 10.1136/gut.46.4.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barcenilla A, Pryde SE, Martin JC, et al. Phylogenetic Relationships of Butyrate-Producing Bacteria from the Human Gut. Appl Environ Microbiol. 2000;66:1654–1661. doi: 10.1128/aem.66.4.1654-1661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arijs I, De Hertogh G, Lemaire K, et al. Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PLoS One. 2009;24:e7984. doi: 10.1371/journal.pone.0007984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wostmann B. The germfree animal in nutritional studies. Annu Rev Nutr. 1981:1. doi: 10.1146/annurev.nu.01.070181.001353. [DOI] [PubMed] [Google Scholar]
- 67.Clarke TB, Davis KM, Lysenko ES, et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:160–161. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johansson MEV, Phillipson M, Petersson J, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. PNAS. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Joerg DS, Svenja P, Maren A, et al. Epithelial Tight Junctions in Intestinal Inflammation. Ann NY Acad Sci. 2009;1165:294–300. doi: 10.1111/j.1749-6632.2009.04062.x. [DOI] [PubMed] [Google Scholar]
- 70.Nenci A, Becker C, Wullaert A, et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446:557. doi: 10.1038/nature05698. [DOI] [PubMed] [Google Scholar]
- 71.te Velde A, de Kort F, Sterrenburg E, et al. Comparative analysis of colonic gene expression of three experimental colitis models mimicking inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:325–330. doi: 10.1002/ibd.20079. [DOI] [PubMed] [Google Scholar]
- 72.Podolsky D, Gerken G, Eyking A, et al. Colitis-associated variant of TLR2 causes impaired mucosal repair because of TFF3 deficiency. Gastroenterology. 2009;137:209–220. doi: 10.1053/j.gastro.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yan Y, Kolachala V, Dalmasso G, et al. Temporal and spatial analysis of clinical and molecular parameters in dextran sodium sulfate induced colitis. PLoS One. 2009;4:e6073. doi: 10.1371/journal.pone.0006073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ito R, Kita M, Shin-Ya M, et al. Involvement of IL-17A in the pathogenesis of DSS- induced colitis in mice. Biochem Biophys Res Commun. 2008;377:12–16. doi: 10.1016/j.bbrc.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 75.Stefan L, Dick SD, Eva J, et al. The role of the Lps gene in experimental ulcerative colitis in mice. APMIS. 1996;104:823–833. doi: 10.1111/j.1699-0463.1996.tb04948.x. [DOI] [PubMed] [Google Scholar]
- 76.Salzman NH, Bevins CL. Negative interactions with the microbiota: IBD. Adv Exp Med Biol. 2008;635:67–78. doi: 10.1007/978-0-387-09550-9_6. [DOI] [PubMed] [Google Scholar]
- 77.Rakoff-Nahoum S, Hao L, Medzhitov R. Role of Toll–like Receptors in Spontaneous Commensal-Dependent Colitis. Immunity. 2006;25:319–329. doi: 10.1016/j.immuni.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 78.Stanislawowski M, Wierzbicki PM, Golab A, et al. Decreased Toll-like receptor-5 (TLR-5) expression in the mucosa of ulcerative colitis patients. J Physiol Pharmacol. 2009;60:71–75. [PubMed] [Google Scholar]
- 79.Sabine MI, Megan EH, Gijs H, et al. TLR5 is not required for flagellin-mediated exacerbation of DSS colitis. Inflamm Bowel Dis. 16:401–409. doi: 10.1002/ibd.21097. [DOI] [PubMed] [Google Scholar]
- 80.Gobert AP, Cheng Y, Akhtar M, et al. Protective Role of Arginase in a Mouse Model of Colitis. J Immunol. 2004;173:2109–2117. doi: 10.4049/jimmunol.173.3.2109. [DOI] [PubMed] [Google Scholar]
- 81.Horowitz S, Binion DG, Nelson VM, et al. Increased arginase activity and endothelial dysfunction in human inflammatory bowel disease. Am J Physiol Gastroint Liver Physiol. 2007;292:G1323–1336. doi: 10.1152/ajpgi.00499.2006. [DOI] [PubMed] [Google Scholar]
- 82.Philip A, Nicholas CZ, Thuan N, et al. Distinct cytokine patterns identified from multiplex profiles of murine DSS and TNBS-induced colitis. Inflamm Bowel Dis. 2009;15:341–352. doi: 10.1002/ibd.20753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figure 2. Comparison of the cecal community in control animals (black triangles), and animals following 3 days of DSS treatment (white squares) and days of treatment (black squares). Using T-RFLP, the Bray-Curtis similarity metric was calculated for each pair-wise comparison and then the results displayed in dendrogram format. Communities that were further analyzed, by construction of clone libraries, are indicated by arrows. Analysis by the parsimony test indicates that there is a statistically significant (*p<0.05 and **p<0.001).
Supplemental figure 4. The rarefaction curves at 97% sequence similarity comparing the microbial communities in mice after 3-days of DSS treatment (pink), 14-days of DSS treatment (blue) and the controls (yellow).
Supplemental figure 5. Comparisons between T-RFLP traces and histograms of the terminal restriction fragments (TRFs) of in silico digest of the clone library constructed from the control communities.
Supplemental figure 6. Comparisons between T-RFLP traces and histograms of the terminal restriction fragments (TRFs) of in silico digest of the clone library constructed from the 3-day DSS administered communities.
Supplemental figure 7. Comparisons between T-RFLP traces and histograms of the terminal restriction fragments (TRFs) of in silico digest of the clone library constructed from the 14-day DSS administered communities.