Abstract
With the advent of transcriptome data, it has become clear that mRNA-like noncoding RNAs (mlncRNAs) are widespread in eukaryotes. Although their functions are poorly understood, these transcripts may play an important role in development and could thus be involved in determining developmental complexity and phenotypic diversification. However, few studies have assessed their potential roles in the divergence of closely related species. Here, we identify and study patterns of sequence and expression divergence in ten novel candidate mlncRNAs from Drosophila pseudoobscura and its close relative D. persimilis. The candidate mlncRNAs were identified by randomly sequencing a group of 734 cDNA clones from a microarray that showed either no difference in expression (187 clones) or differential expression (547 clones) in comparisons between D. pseudoobscura and D. persimilis and between these two species and their F1 hybrids. Candidate mlncRNAs are overrepresented among differentially expressed transcripts between males of D. pseudoobscura and D. persimilis, and although they have high sequence conservation between these two species, seven of them have no putative homologs in any of the other ten Drosophila species whose genomes have been sequenced. Expression of eight of the ten candidate mlncRNAs was detected either in whole bodies (adults) or testes using a custom-designed oligonucleotide microarray. Three of the ten candidate mlncRNAs are highly expressed (in the top 4% of the male transcriptome), differentially expressed between species, and show extreme levels of sex-bias, with one transcript having the highest level of male bias in the whole transcriptome. Proteomic data from testes show no traces of any predicted peptides from the candidate mlncRNAs. Our results suggest that these mlncRNAs may be important in male-specific processes related to sexual dimorphism and species divergence in this species group.
Keywords: mlncRNA, noncoding RNA, Drosophila pseudoobscura, species divergence, sex-bias
Introduction
Understanding the genetic basis of phenotypic diversity is a major challenge for biologists. Protein-coding genes traditionally have been considered the most important group of genomic elements underlying developmental complexity, and changes either in their amino acid sequences or in their patterns of expression are considered the exclusive mechanisms underlying phenotypic diversification (King and Wilson 1975; Carroll 2005; Hoekstra and Coyne 2007). However, two general findings in the last decade suggest that protein-coding genes are not the only set of genomic elements that play an important role in determining developmental complexity (Taft et al. 2007). First, in eukaryotes, only small parts of the genome code for proteins (e.g., less than 5% in humans) , whereas most of the genome is noncoding (the so-called “junk DNA”) (IHGS 2004; Frith et al. 2005). Second, in eukaryotes, the number of protein-coding genes does not increase comparably with either genome size or organismal/developmental complexity. For example, the human genome is about 30 times larger than that of the nematode Caenorhabditis elegans and the fruit fly D. melanogaster. However, despite having more physiological and developmental complexity, the human genome contains only a few thousand additional protein-coding genes (21,257) than the genomes of C. elegans (20,224) or D. melanogaster (13,781) (Ensembl release 59, August 2010).
Recent whole-genome tiling microarray studies, large-scale sequencing of cDNA libraries, next-generation RNA sequencing, and other experimental work (Manak et al. 2006; Birney et al. 2007; Nagalakshmi et al. 2008) suggest that a large fraction of the eukaryotic genome is transcribed (e.g., ∼85% in Drosophila) and that an enormous number of RNA transcripts in eukaryotes do not code for proteins (Okazaki et al. 2002; Carninci et al. 2005; Tupy et al. 2005; Kapranov et al. 2007). Such transcripts are called noncoding RNAs (ncRNAs) and include all RNAs that are not translated into functional protein products. The size of ncRNAs varies considerably, from small microRNAs (miRNAs) of 18 nucleotides up to very large RNAs of several thousand nucleotides like the ∼17 kb human Xist RNA (Brockdorff et al. 1992). Most ncRNAs have between 20 and 500 nucleotides and are thus shorter than the majority of mRNAs. Some authors have suggested that ncRNAs may be particularly important for explaining the genetic basis of biological complexity and thus may be fundamental in explaining the seeming lack of association among organismal complexity, genome size, and the number of protein-coding genes (Mattick 2003; Kapranov et al. 2007; Mattick 2007; Mattick 2009; Mercer et al. 2009; Wilusz et al. 2009).
mRNA-like ncRNAs (mlncRNAs) are a major group of ncRNAs that possess many of the properties of mRNAs (e.g., presence of introns and 3′ polyadenylation) but have limited protein-coding ability (Erdmann et al. 2000; Prasanth and Spector 2007; Rymarquis et al. 2008; Ponting et al. 2009). Identification of mlncRNAs is difficult because they do not have structural or sequence features that facilitate their identification in silico (Tupy et al. 2005; Hiller et al. 2009). Some mlncRNAs function as long RNAs (Nakamura et al. 1996), whereas others are precursors of small RNAs (Riccardo et al. 2007; Carlile et al. 2008). mlncRNAs are likely transcribed by RNA polymerase II, which transcribes mRNAs, small RNAs, and miRNAs (Mattick and Makunin 2006). Most mlncRNAs evolve faster than transfer RNAs and ribosomal RNAs (rRNAs), which have strong secondary or tertiary structures, thus making de novo computational prediction of mlncRNAs much harder than for classic non-coding RNAs such as rRNA (Mattick and Makunin 2006).
In Drosophila, the first mlncRNA was discovered two decades ago (Lipshitz et al. 1987), but additional mlncRNA transcripts have been identified only recently through tiling array experiments and large-scale screening of cDNA libraries (Stolc et al. 2004; Inagaki et al. 2005; Tupy et al. 2005; Manak et al. 2006; Hiller et al. 2009; Li et al. 2009). So far, 169 mlncRNA genes have been identified in D. melanogaster (version 5.29) and just two in D. pseudoobscura (version 2.12). Although the function and expression profiles of only a handful of Drosophila mlncRNAs have been investigated (Rajendra et al. 2001; Hardiman et al. 2002; Stuckenholz et al. 2003; Martinho et al. 2004; Inagaki et al. 2005; Tupy et al. 2005; Sanchez-Elsner et al. 2006), some have been shown to be vital for normal development and cell function. For instance, loss of the bereft RNA in the peripheral nervous system of D. melanogaster causes aberrant development of both extrasensory organs and interommatidial bristles of the eye (Hardiman et al. 2002). Furthermore, in Drosophila, the roX1 and roX2 RNAs are essential for dosage compensation (Stuckenholz et al. 2003), and the Pgc RNA is expressed only in primordial germ cells, regulates transcriptional repression during early embryonic development, and is required for maintenance of germ cell fate (Nakamura et al. 1996; Martinho et al. 2004).
Despite the potential importance of ncRNAs in explaining organismal complexity and phenotypic diversity, no studies have investigated their possible role in the divergence of closely related species. Here, we identify ten putative mlncRNAs from D. pseudoobscura and its close relative, D. persimilis, and describe their patterns of sequence and transcriptional divergence between these two recently diverged (<1 Ma) species that constitute an important model system for studying the genetic basis of speciation and the process of species divergence (Dobzhansky 1933; Dobzhansky and Epling 1944; Orr 1987; Wang et al. 1997; Noor, Grams, Bertucci, and Reiland 2001; Noor, Grams, Bertucci, et al. 2001; Machado et al. 2002; Machado and Hey 2003; Machado et al. 2007; Noor et al. 2007; Kulathinal et al. 2009).
Materials and Methods
cDNA Library Preparation, Clone Selection, and Sequencing
We constructed a normalized cDNA library of D. pseudoobscura to build a cDNA microarray and 734 cDNA clones from that library were sequenced after selecting them based on results from a microarray experiment (see details below). Detailed information about the cDNA library and microarray construction, array hybridization, and data analyses can be found in the supplementary materials (Supplementary Material online). Briefly, we pooled total RNA from all life stages (embryo to seven-day-old adult) of four inbred lines of D. pseudoobscura: Mather17, Mather48, Abajo36, and AFC2 (Machado et al. 2002). The RNA was used to construct a D. pseudoobscura cDNA library using the SMART cDNA library construction kit (Clontech). We normalized the cDNA library (Ali et al. 2000) to reduce the overall frequency of highly expressed genes. Close to 10,000 clones were picked and stored individually in 96 well plates, and cDNA inserts were amplified for half of the clones using primers from the SMART kit. Clones that showed a clean band in polymerase chain reaction (PCR) amplification products (4,416 total) were printed on nylon filters (Hybond N+) with a GeneMachines Omnigrid Arrayer. cDNA probes were synthesized from total RNA with α33P-dCTP by oligo dT-primed polymerization following standard protocols (Becker et al. 2003). RNA samples from each sex were obtained separately from virgin seven-day-old (sexually mature) adults, from three lines of each species (D. pseudoobscura: Mather10, Mather32, Flagstaff18; D. persimilis: MatherG, MSH3, MSH42), and from F1 hybrids of the two species. Hybrids were created by mass mating virgin female or male individuals from a mixture of 13 inbred lines of D. pseudoobscura (MSH10, AF2, AFC7, AFC3, Abajo36, Easton54, MSH42, Mather17, Flagstaff16, MSH9, MSH21, MSH32, and MSH24) with virgin individuals of the opposite sex from a mixture of ten inbred lines of D. persimilis (MSH25, Mather6, Mather10, MSH1, MSH7, Mather40, Mather37, MSH42, MSH7, and Mather39). Array hybridizations were conducted using standard methods (Becker et al. 2003), and the ImaGene software (Biodiscovery) was used to grid the phosphor image, record the pixel density of each spot, and perform background subtractions.
The normalized data sets were log-transformed and analyzed in SAS v.9.0 (SAS Institute, Inc.) using a mixed-model analysis of variance (ANOVA). Signal intensity across membranes was normalized using this ANOVA model: yi = μ + Mi + ϵi, where yj is the log2-transformed score for each spot, μ is the overall mean intensity for genes across the membranes, species, sexes, and spots, Mi is the effect of the ith membrane, and ϵi is the random error. The residuals from this model were fitted to clone-specific mixed-model ANOVAs of the form yijkl = μ + Mi + Xj + Gk + Sl + (G × S)kl + ϵijkl, where Xj is the jth spot in each membrane (each clone was printed twice in each membrane), Gk is the kth species (D. pseudoobscura, D. persimilis, F1 ♀ D. pseudoobscura × ♂ D. persimilis, F1 ♀ D. persimilis × ♂ D. pseudoobscura), Sl is the lth sex (male or female), (G × S)kl is the species by sex interaction, and ϵijkl is the residual. The gene-specific models were fitted using membrane (Mi) and spot (Xj) as random effects. A total of 734 cDNAs were selected for sequencing: 208 clones that were differentially expressed between females of D. pseudoobscura and D persimilis or between females of these two parental species and their F1 hybrids, 339 clones that were differentially expressed between males of the same comparisons, and 187 clones that showed no significant expression differences in any of those comparisons. Clones were sequenced unidirectionally from the 5′ end using the TriplEx5′LD primer (5′-CTCGGGAAGCGCGCCATTGTGTTGGT-3′) (Clontech).
Identification of Putative mlncRNAs
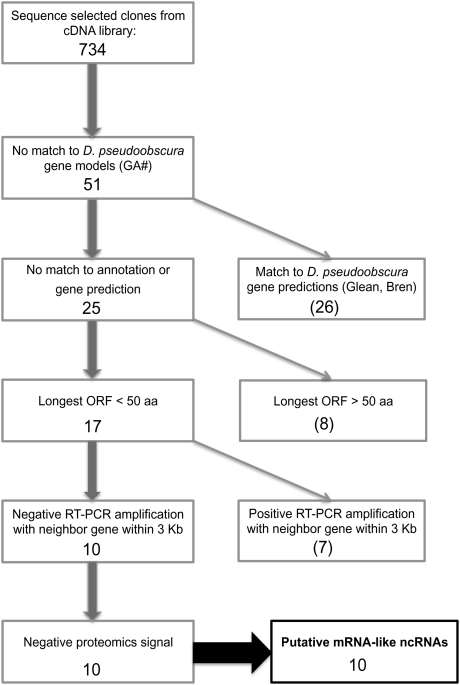
The sequences of the 734 selected clones were blasted against the D. melanogaster and D. pseudoobscura annotated genomes. We found that 71 clones (9.6% of all sequenced clones), corresponding to 51 unique transcripts, did not match the D. melanogaster annotation (version 5.29), the D. pseudoobscura annotation or its intron set (version 2.12) (supplementary table S1, Supplementary Material online). “Full-length” cDNAs among these 51 unique transcripts were identified by manually checking against the chromatogram for possible sequencing errors and for the existence of a poly-A tail. We aligned the full-length cDNAs to the D. pseudoobscura genome sequence with BLAT (Kent 2002) to determine genome location and identify putative introns (highest score; identity >95%). Because the genome assembly available for BLAT is outdated (version: November 2004 Flybase 1.03/dp3), we translated the coordinates to the current version of the genome assembly (release 2.12). For these candidate mlncRNAs, we wrote PERL scripts that identified open reading frame (ORF) sequences and calculated ORF length. We also compared the longest Met-initiated ORF with the longest non-Met–initiated ORF to rule out the possibility that the start codon of the current cDNA sequence is missing. When there was a difference in length, we kept the longer ORF for further analyses.
We further removed transcripts that partially intersected predictions generated by additional algorithms because they are likely to be protein-coding genes that are not yet included in the genome annotation. We ran a local blast of the 51 unique transcripts to predicted D. pseudoobscura genes (release 2.12) and observed matches (E < 10−10) in 26 transcripts to predictions from the following prediction programs: twinscan, genscan, BREN_N-Scan, SNAP, GleanR, PACH_genemapper, RGUI_geneid, and DGIL_snap (supplementary table S2, Supplementary Material online). This filtering step left a total of 25 unique transcripts that we considered candidate mlncRNAs (fig. 1). Because of the difficulty in differentiating small peptide–coding genes from noncoding genes, we set a stringent size criterion and only included sequences with ORFs that were less than 50 codons in our set of candidates. This is very conservative because only 0.5% of the annotated protein-coding genes (71 of 13,752) in the entire D. melanogaster genome (release 5.29) have such small ORF lengths, and most previous ncRNA studies have identified putative ncRNA sequences using an ORF size cutoff up to 100 codons (Inagaki et al. 2005; Tupy et al. 2005; Dinger et al. 2008). This criterion resulted in the removal of eight transcripts rendering 17 candidate mlncRNAs (supplementary table S3, Supplementary Material online).
FIG. 1.
Workflow of the filtering process used to identify candidate mRNA-like ncRNA genes. Numbers are the remaining candidate ncRNAs after each filtering step, and numbers in parenthesis are the number of candidate genes eliminated after each filtering step.
Nine of the 17 putative mlncRNAs are located within 3 kb of annotated protein-coding genes and in the same strand (supplementary table S3, Supplementary Material online). Those transcripts could be unannotated exons, 5′ untranslated regions (UTRs), or 3′ UTRs that are part of those genes. To determine if their expression was independent of the neighboring genes, we conducted reverse transcriptase PCR (RT-PCR) reactions with RNA from all life stages using a primer in the candidate mlncRNA and a primer from the nearest exon of the coding gene (see supplementary Methods, Supplementary Material online). In two of the nine cases no RT-PCR products were observed (clones 2731 and 2338, supplementary table S3, Supplementary Material online), suggesting that expression of those putative mlncRNAs is independent of the expression of neighboring annotated protein-coding genes. The positive RT-PCR reactions for the remaining seven cases suggest that gene models need to be revised for those genes (supplementary table S3 and fig. S1, Supplementary Material online). This filtering step left us with a final list of ten candidate mlncRNAs (table 1).
Table 1.
Location and Sequence Divergence Patterns of 10 Candidate mlncRNAs from Drosophila pseudoobscura.
| Clone ID | Chromosome Scaffold (Strand) | Length (bp) | ORF Length (codons) | Intron | Distance to Closest Gene 5′ End Scaffold (strand)a | Distance to Closest Gene 3′ End Scaffold (strand)a | Percent Sequence Similarity to D. persimilis | Ka/Ksb |
| 991 | 2 (−) | 260 | 38 | N | 6,622 bp 5′ end GA15970 (−) | 1,607 bp 5′ end GA11876 (+) | 98.5 | NC |
| 2731 | 2 (+) | 206 | 47 | N | 891 bp 3′ end GA27083 (+) | 19,940 bp 5′ end GA27083 (+) | 97.3 | NC |
| 1383 | 2 (−) | 260 | 36 | N | 4,558 bp 5′ end GA22066 (−) | 3,009 bp 3′ end GA22058 (−) | 99.4 | NC |
| 354 | 3 (−) | 270 | 28 | N | 398 bp 3′ end GA19331 (+) | 2,202 bp 5′ end GA24816 (+) | 99.3 | NC |
| 233 | 3 (−) | 330 | 22 | N | 19,098 bp 5′ end GA12588 (−) | 19,714 bp 5′ end GA24784 (+) | 97.2 | NC |
| 97 | 4_group1 (−) | 358 | 35 | Y | 22,774 bp 3′ end GA25317 (+) | 8,793 bp 5′ end GA16361 (+) | 98.1 | 1.20 |
| 3982 | 4_group3 (−) | 290 | 36 | Y | 6,537 bp 5′ end GA19649 (−) | 732 bp 5′ end GA10584 (+) | 98.7 | NC |
| 1108 | 4_group4 (−) | 149 | 9 | Y | 1,286 bp 3′ end GA21432 (+) | 3,833 bp 3′ end GA16495 (−) | 98.2 | Indel |
| 2090 | XL_group1e (+) | 316 | 36 | N | 23,962 bp 5′ end GA15495 (−) | 430 bp 3′ end GA25756 (−) | 96.0 | 3.52 |
| 2338 | XR_group8 (+) | 236 | 31 | N | 20 bp 3′ end GA28512 (+) | 23,39 bp 5′ end GA12013 (+) | 98.3 | Stop |
Distances are approximate, based on the range of the Blat hit to the genome.
Estimated between the aligned ORF sequences from the D. pseudoobscura and D. persimilis genomes. NC, no change. Stop and indel denotes the presence of stop codons or indels in either of the two genome sequences.
To identify orthologous sequences of these genes in the genomes of other sequenced Drosophila species, we used the BlastN tool in Flybase using a cutoff E value of 10−5. To determine if the sequences were homologous to identified ncRNAs, we searched the Rfam database (Gardner et al. 2009) using WU-Blast with an E value threshold of 1. To determine if the sequences were possible miRNA precursors, we searched the miRBase database (Griffiths-Jones et al. 2008) using BlastN with an E value threshold of 0.1. Using the ORFs, we also calculated pairwise Ka/Ks between D. pseudoobscura and the ortholog sequences in D. persimilis. The longest ortholog ORF sequences were aligned using ClustalW (Thompson et al. 1994), and the alignments were passed to PAML for estimation of Ks and Ka using the codeml program with the pairwise distance estimation option (runmode = −2) (Yang 1997). The sequences of the unique transcripts, including the putative ncRNAs considered here, were deposited in Genbank (accession numbers GW774481–GW774563).
Proteomics
To further confirm that candidate clones are mlncRNAs, we searched against a testis proteomics data set prepared from D. pseudoobscura adults of the genome sequence line MV2-25 (Richards et al. 2005). A detailed description of protocols applied to acquire the proteomics data can be found in the supplementary material (Supplementary Material online). Briefly, proteins were extracted from 50 seven-day old virgin adult testes, digested with trypsin, and fractionated into 24 fractions by isoelectric focusing. Each fraction was injected into a Pepmap C18 trapping cartridge for liquid chromatography–mass spectrometry analysis. Data files from the 24 fractions were merged into a single file and searched against a peptide database consisting of the D. pseudoobscura protein database (version 2.12) plus the longest predicted peptides from the original 51 candidate ncRNA transcripts.
Expression of Putative mlncRNAs
To investigate expression divergence of these mlncRNAs in D. pseudoobscura and D. persimilis, we designed 60-mer oligonucleotide probes for the putative mlncRNAs and included them in an Agilent custom-designed oligonucleotide microarray (Jiang and Machado 2009). Probes were designed to be identical in sequence between D. pseudoobscura and D. persimilis to reduce potential hybridization differences due to sequence mismatches. A detailed description of methods and statistical analyses of the Agilent microarray data have been described elsewhere (Jiang and Machado 2009). We corrected for multiple statistical tests in the context of the whole genome by estimating the false discovery rate with the Q-value software (Storey and Tibshirani 2003). Significance cutoff was set at q < 10−6, which is slightly more stringent than the Bonferroni correction (P = 2.65 × 10−6). The microarray data are available in the Gene Expression Omnibus Depository (Accession GSE17192).
In addition, we validated microarray results for two candidate mlncRNAs using quantitative real-time PCR (for protocols and primer sequences, see supplementary material, Supplementary Material online). Transcription levels were measured in three lines of D. pseudoobscura (MV2-25, MSH21, and Mather 10) and two lines of D. persimilis (Mather G and MSH3), using RNA collected from whole bodies. Furthermore, as these transcripts have extremely male-biased patterns of expression, we measured expression in testes and in male bodies without testes to determine if expression is predominantly testes-specific.
Results
Identification of Putative mlncRNAs
We found 51 unique transcripts that did not match the current D. pseudoobscura genome annotation (version 2.12) (supplementary table S1, Supplementary Material online). All transcripts are single-copy except transcript (4048), which has two identical copies on the same chromosome but on opposite strands. Among those 51 transcripts, our stringent set of filtering criteria (fig. 1) identified ten transcripts that are likely to be mRNA-like ncRNAs (table 1). Most of the putative mlncRNAs are located in intergenic regions, several kilobases away from the closest annotated protein-coding gene (table 1). RT-PCR results show that the expression of transcripts that are within 3 kb of gene models on the same strand is independent of the neighboring annotated gene (supplementary table S3 and fig. S1, Supplementary Material online).
None of these putative mlncRNAs have annotated homologs in D. melanogaster or in the Rfam (version 9.1) (Gardner et al. 2009) and miRBase (Griffiths-Jones et al. 2008) databases (E < 10−6). Furthermore, none of the transcripts match any of the annotated miRNAs from D. pseudoobscura (release 2.12; 152 miRNAs). More importantly, no peptides from the ten putative mlncRNAs were detected in the proteomics data, whereas peptides from six of the 41 transcripts that failed to pass our filtering pipeline were found under the same search conditions (supplementary table S4, Supplementary Material online).
The lack of conservation of ORFs and observation of Ka/Ks > 1 are helpful criteria for distinguishing putative mlncRNAs from protein-coding genes (Dinger et al. 2008). We found that the longest ORFs of two of the ten putative mlncRNAs have either an indel or a stop codon in the homologous region of the genome of D. persimilis (strain MSH3; Clark et al. 2007) (table 1), supporting their identification as putative mlncRNAs. We calculated pairwise Ka/Ks between the predicted translations of the longest ORF of the eight remaining cDNA sequences and the putative orthologous sequences in the D. persimilis genome (table 1). The longest ORFs in two of the eight putative mlncRNAs have Ka/Ks > 1, also consistent with their classification as putative mlncRNAs. However, the remaining six evaluated ORFs have identical sequences in the two species.
Sequence Divergence
To assess evolutionary conservation and rate of sequence divergence of these putative mlncRNAs, we first Blasted the transcripts against the genome sequence of D. persimilis, the closest relative of D. pseudoobscura among the 12 genome-sequenced Drosophila species (Richards et al. 2005; Clark et al. 2007). The average sequence divergence among these putative mlncRNAs is not significantly different from the average sequence divergence among 12,973 orthologous protein-coding genes (<20% aa divergence) shared between the two species (P = 0.06, Wilcoxon rank-sum test).
Despite the observed sequence conservation of the ten candidate mlncRNAs between D. pseudoobscura and D. persimilis, partial putative homolog sequences in other sequenced Drosophila species (BlastN E < 10−5) were found in only three of the ten transcripts (354, 2731, 1383; supplementary table S5, Supplementary Material online). In each case, the significant match covered less than 26% of the transcript length. Therefore, at least seven of the ten putative mlncRNAs described here appear to be newly evolved transcripts that could have originated during the evolution of the obscura group of Drosophila.
Source of the ncRNA Sequences
Five of the ten putative mlncRNAs (five clones in total) came from the group of 187 clones (2.7%, 5/187) that showed no differences in expression in the cDNA array between D. pseudoobscura and D. persimilis and between the F1 hybrids and the pure species (table 2). The other five putative mlncRNAs (11 clones in total) came from the group of differentially expressed clones between males of D. pseudoobscura and D. persimilis (10.9%, 8/73) or between F1 male hybrids and pure species males (1.1%, 3/266). Interestingly, none of the putative mlncRNAs came from the 208 clones differentially expressed between females of both species or between F1 female hybrids and pure-species females. Thus, the putative mlncRNAs identified here are significantly more likely to be differentially expressed in males than in females (χ2 = 6.95, df = 1, P = 0.008) and make up a sizable fraction (10.9%) of clones differentially expressed between males of both species.
Table 2.
Source of the 10 Putative mRNA-Like ncRNAs from Drosophila pseudoobscura (D.ps) and D. persimilis (D.per).
| Category | Sequenced Clones | mRNA-Like ncRNAs | Clones Identified as ncRNAs (%) |
| Not differentially expressed in any of the comparisons | 187 | 5 | 5 (2.7) |
| Differentially expressed between D.ps and D.per males | 73 | 3 | 8 (10.9) |
| Differentially expressed between pure species and F1 hybrid males | 266 | 2 | 3 (1.1) |
| Differentially expressed between D.ps and D.per females | 49 | 0 | 0 |
| Differentially expressed between pure species and F1 hybrid females | 159 | 0 | 0 |
| Total | 734 | 10 | 16 (2.2) |
Patterns of Expression and Expression Divergence
To investigate patterns of expression of these transcripts in adult D. pseudoobscura and D. persimilis, the ten putative mlncRNAs were printed in a whole-genome oligonucleotide array designed for D. pseudoobscura (Jiang and Machado 2009). Six of the ten putative mlncRNAs show significant above-background levels of expression in whole bodies of virgin seven-day-old flies, and six transcripts are also expressed in testes (table 3). We could not detect expression in whole adult bodies of four of the putative mlncRNAs printed in the oligonucleotide array (table 3). The most likely explanation for the latter result is the fact that although the cDNA library was constructed using RNA from all life stages, we only surveyed expression in adults (see supplementary material, Supplementary Material online).
Table 3.
Microarray Expression Results for the 10 Putative mlncRNAs.
| Clone ID | Normalized Microarray Expression Intensity |
Sex-Bias Ratio (♂/♀)c | Sex-Bias Patternd | q Value Expression Divergence (sex, ↑species)e | ||||
| D.ps ♀a | D.ps ♂a | D.per ♀a | D.per ♂a | Testesb | ||||
| 233 | 0.1264 | 5.5171 | 0.7067 | 4.9454 | 5.2426 | 43.66 | MB | 8.7 × 10−8 (♂ ps), 8.8 × 10−9 (♀ pe) |
| 991 | 0.1279 | 3.2753 | 0.1554 | 2.3997 | 3.6826 | 25.62 | MB | 2.3 × 10−13 (♂ ps) |
| 97 | 0.1254 | 2.3763 | 0.2112 | 2.9429 | 3.7234 | 18.96 | MB | 3.2 × 10−8 (♂ pe) |
| 354 | 0.2904 | 0.6492 | 0.3292 | 0.5868 | N/Af | 2.24 | MB | NS |
| 2731 | 1.8406 | 1.3989 | 1.7612 | 1.5144 | 1.0216 | 0.76 | FB in D.ps | NS |
| 1383 | 0.1348 | 0.1327 | 0.1419 | 0.1642 | N/Af | 0.98 | NSB | NS |
| 1108 | N/Af | N/Af | N/Af | N/Af | 0.3153 | — | — | — |
| 3982 | N/Af | N/Af | N/Af | N/Af | N/Af | — | — | — |
| 2090 | N/Af | N/Af | N/Af | N/Af | N/Af | — | — | — |
| 2338 | N/Af | N/Af | N/Af | N/Af | 0.0526 | — | — | — |
Average normalized log-transformed expression values in whole adult bodies (Jiang and Machado 2009). 12,507 expressed genes of Median in D. pseudoobscura males: 0.416; 2.5th percentile: 0.154; 97.5th percentile: 2.765.
Average normalized log-transformed expression values for testes (Jiang Z-F and Machado CA, in preparation). 12,272 expressed genes. Median in D. pseudoobscura: 0.325; 2.5th percentile: 0.049; 97.5th percentile: 2.705.
In D. pseudoobscura.
MB: male-biased in both species; FB: female biased (in D. pseudoobscura); NSB: non-sex-biased in both species.
Only significant values shown (q < 10−6). In parenthesis: sex in which there is a significant difference and species in which transcript is expressed at higher level. NS, no significant difference between species.
N/A: no expression detected in the oligonucleotide micro array.
The expression levels of three of the ten putative mlncRNAs (233, 991, and 97) fall in the top 4% of the adult male transcriptome. Furthermore, putative mlncRNA 233 is one of the top ten transcripts of the adult male transcriptome of D. pseudoobscura, with levels of expression similar to that of several mitochondrial genes (see table S1 in Jiang and Machado 2009). The same three top transcripts detected in adult whole bodies (233, 991, and 97) are in the top 1% of the most highly expressed transcripts in testes, with putative mlncRNA 233 being also one of the top ten transcripts in this tissue (Jiang Z-F and Machado CA, in preparation). The fact that predicted peptides from these highly expressed transcripts were not observed in the testes proteomics data set provides additional strong support to their identification as ncRNAs.
Four of the ten putative mlncRNAs have male-biased expression in both species, one has female-biased expression only in D. pseudoobscura, and one has non-sex-biased expression in both species (table 3). Three male-biased mlncRNAs (233, 991, and 97) show greater than 10-fold differences in expression between sexes in D. pseudoobscura and two of them in D. persimilis (991 and 97). Three of the six expressed putative mlncRNAs are differentially expressed between species in at least one sex under a stringent significance cutoff (q < 10−6) (table 3). Most expression differences are less than 2-fold, consistent with previous observations that in the transcriptome, sex differences are greater than species differences (Ranz et al. 2003; Jiang and Machado 2009). Interestingly, transcript 233 is the most extremely male-biased transcript in the entire transcriptome of D. pseudoobscura showing greater than a 40-fold difference in expression between males and females. This enrichment of highly sex-biased transcripts in our small sample (3/10; 30%) is quite remarkable considering that only 4.1% of the D. pseudoobscura transcriptome shows greater than 10-fold sex-biased expression (Jiang and Machado 2009) (χ2 = 11.046, df = 1, P = 0.0009).
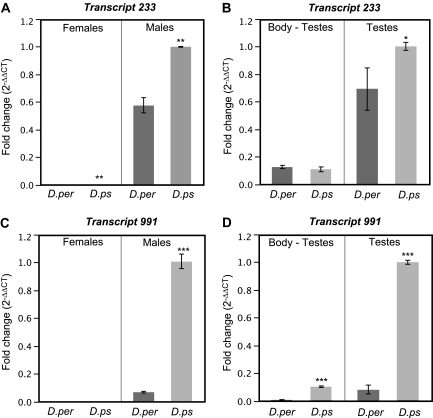
To validate the microarray results from the Agilent oligonucleotide, we conducted quantitative real-time PCR for two putative mlncRNAs (233 and 991) (fig. 2). The results from real-time PCR are consistent with the results from Agilent oligonucleotide microarray: the transcripts are male biased and differentially expressed between males of the two species (233: ♂D.ps > ♂D.per [P < 0.001]; 991: ♂D.ps > ♂D.per [P < 0.0001]) (fig. 2A and C). Furthermore, the two putative mlncRNAs are expressed predominantly in testes in both species (testes > body-testes: 233 D.ps [P < 0.0001], 233 D.per [P = 0.004], 991 D.ps [P < 0.0001], 991 D.ps [P = 0.04]) (fig. 2B and D).
FIG. 2.
Real-time PCR measurements of relative expression in two putative mlncRNAs from Drosophila pseudoobscura (D.ps) and D. persimilis (D.per). Panels on the left (A, C) show expression differences between sexes and species for whole-body samples; values are shown relative to expression in D. pseudoobscura males. Panels on the right (B, D) show expression differences in males between tissues and species for testes and whole-body/no testes (Body–Testes); values are shown relative to expression in D. pseudoobscura testes. These two transcripts are extremely male-biased, differentially expressed between species (except for 991 in females), and predominantly expressed in testes. ***P < 0.0001, **P < 0.001, *P < 0.01.
Discussion
One of the most exciting recent advances in genome biology has been the discovery of a large number of ncRNAs expressed in diverse organisms (Mattick 2003; Prasanth and Spector 2007; Ponting et al. 2009). Although the functions of most ncRNAs are unknown, some case studies suggest that they play important roles in a broad spectrum of developmental and physiological phenomena, such as dosage compensation and the expansion of human brain function (reviewed in Mattick 2009). The discovery of ncRNAs and the realization of their biological importance are providing new and potentially fundamental insights on the genetic basis of biological complexity, as these findings shift explanations away from traditional views that have emphasized the roles of protein-coding genes. To date, most ncRNA studies have simply cataloged and characterized new genes in model species. However, very few evolutionary studies of ncRNA sequence and expression divergence in closely related species have been conducted (Pollard et al. 2006; Yang et al. 2007; Lu et al. 2008), and thus the potential role of ncRNAs in the process of species divergence remains largely unexplored.
In this study, we have identified ten novel putative mlncRNAs in D. pseudoobscura, an important increase in the number of identified mlncRNAs in this species considering that there are only two currently annotated ncRNAs in the D. pseudoobscura genome (version 2.12). Owing to the difficulty in differentiating small peptide–coding genes from mlncRNAs with some coding potential (Dinger et al. 2008), we conservatively considered only loci with ORFs smaller than 50 codons in length to be candidate mlncRNAs. However, we cannot completely reject the possibility that some of these putative mlncRNAs are small peptide–coding genes (e.g., the “polished rice” locus in D. melanogaster; Kondo et al. 2007; Kondo et al. 2010). Similarly, some additional transcripts that were not among the final list of putative mlncRNAs (supplementary tables S1–S3, Supplementary Material online) may actually be ncRNAs despite having ORFs longer than 50 codons. The fact that none of the candidate transcripts were detected in a proteomics data set of D. pseudoobscura testes provides further support to their identification as candidate mlncRNAs. That evidence is particularly important for the three highly expressed, and likely testes-specific, transcripts (233, 991, and 97) that are among the most highly expressed transcripts in males. Interestingly, none of the male-biased putative mlncRNAs are located on the X chromosome, consistent with results showing a demasculinization of the X chromosome in several Drosophila species (Parisi et al. 2003; Ranz et al. 2003; Sturgill et al. 2007; Jiang and Machado 2009).
Although only 2.2% of the total number of sequenced cDNA clones (16/734) were identified as mlncRNAs, close to 11% of the clones (8/73) differentially expressed between males of D. pseudoobscura and D. persimilis were mlncRNAs (table 2). In contrast, none of the 208 clones differentially expressed cDNAs between females of both species or in female hybrids were mlncRNAs. Furthermore, a large proportion (4/6) of putative mlncRNAs expressed in sexually mature adults are male-biased, and the most highly expressed (233, 991, 97) are likely testes-specific (table 3, fig. 2). In contrast to previous efforts that focus on the rapid evolution of protein-coding genes as the underlying reason behind the general observation of faster evolution of male-specific characters in the divergence of animals (Civetta and Singh 1998; Swanson and Vacquier 2002), our data suggest the possibility that mlncRNAs could also be a major factor underlying the rapid evolution of the male transcriptome. However, this conclusion will need to be substantiated by data from a comprehensive transcriptome survey.
Seven of the ten candidate mlncRNAs have no homologs in any of the other ten sequenced Drosophila species suggesting that they are novel transcripts. This observation is consistent with previous studies showing that most mlncRNAs known in D. melanogaster have no homologs in D. pseudoobscura (Inagaki et al. 2005; Tupy et al. 2005). It is, however, not clear if the patterns of sequence divergence in these new mlncRNAs, conserved between closely related species but not over longer evolutionary distances, imply lack of function or newly evolved functions. Although standard molecular evolution theory suggests that there should be a positive correlation between the level of sequence conservation and the functional importance of a gene, recent work has shown that this may not hold, at least for noncoding regions. For example, the Xist RNA gene is responsible for the fundamental process of dosage compensation in eutherians but shows very large sequence divergence over short evolutionary divergences (Pang et al. 2006). Furthermore, experimental studies in mice show that removal of ultraconserved regions do not have detrimental effects in development (Nobrega et al. 2004; Ahituv et al. 2007).
Although sequence divergence may not be the best indicator of function, expression is an important indicator of functionality. Eight of the ten putative mlncRNAs have above-background levels of expression either in whole bodies or in testes. Moreover, data from a time course development study show that the expression of these transcripts is different in larval and pupal stages than in adults (Jiang Z-F and Machado CA, in preparation), suggesting modulation during development. Three of the transcripts (233, 991, and 97), together with mitochondrial genes involved in metabolic activity, are among the most highly expressed genes in the transcriptome, further suggesting that they may be involved in important functions. mlncRNAs that were not detected by the microarray expression assay may also have important functions. Expression of important genes may have been too low for detection, in part because of our conservative criteria for inclusion in the set of expressed genes (Jiang and Machado 2009). Previous studies suggest that genes expressed at low levels in certain tissues during specific life stages are critical for normal development (Ponting et al. 2009).
The advent of RNA-Seq (Nagalakshmi et al. 2008; Sultan et al. 2008), a method that allows complete characterization of transcriptomes at the sequence level, will permit conducting a thorough identification and characterization of all coding and noncoding elements of a genome (e.g., the modENCODE project in D. melanogaster and C. elegans; Celniker et al. 2009). Results from comprehensive transcriptome divergence surveys using RNA-Seq will provide the opportunity to conduct comparative genome-wide studies of ncRNA evolution in closely related species and will allow testing our suggestion, based on this partial survey of cDNAs, that mlncRNAs are important in male-specific processes related to sexual dimorphism and species divergence in Drosophila.
Supplementary Material
Supplementary tables S1–S3, fig S1, and Methods are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
Thanks to three anonymous reviewers and Willie Swanson for providing useful comments. We thank Bento Soares and Douglas L. Crawford for technical advice with the cDNA library normalization; Khew-Voon Chin for help with the cDNA microarray construction; Laura Rascón for conducting the RT-PCR and real-time PCR experiments. Author contributions: C.A.M and J.H. designed research; Z.J., Y.W., and C.A.M. performed research; C.A.M. and J.H. contributed reagents; C.A.M., Z.J., and D.A.C. wrote the paper. The proteomics core facility at the University of Maryland is supported by the College of Chemical and Life Sciences. Purchasing of the LTQ Orbitrap XL mass spectrometer was supported by National Institutes of Health (NIH) RR 023383 awarded to C. Fenselau. Support for D.A.C. was provided by the Center for Insect Science, University of Arizona, through NIH Training Grant # 1K12 GM000708. National Science Foundation Grant DEB-0520535/0941217 and startup funds to C.A.M. supported this work.
References
- Ahituv N, Zhu Y, Visel A, Holt A, Afzal V, Pennacchio LA, Rubin EM. Deletion of ultraconserved elements yields viable mice. PLoS Biol. 2007;5:e234. doi: 10.1371/journal.pbio.0050234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S, Holloway B, Taylor WC. Normalisation of cereal endosperm EST libraries for structural and functional genomic analysis. Plant Mol Biol Rep. 2000;18:123–132. [Google Scholar]
- Becker KG, Wood WH, III, Cheadle C. Membrane-based spotted cDNA arrays. In: Botwell D, Sambrook J, editors. DNA microarrays. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2003. pp. 289–306. [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- Carlile M, Nalbant P, Preston-Fayers K, McHaffie GS, Werner A. Processing of naturally occurring sense/antisense transcripts of the vertebrate Slc34a gene into short RNAs. Physiol Genomics. 2008;34:95–100. doi: 10.1152/physiolgenomics.00004.2008. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Carroll SB. Evolution at two levels: on genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Dillon LA, Gerstein MB, et al. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civetta A, Singh RS. Sex-related genes, directional sexual selection, and speciation. Mol Biol Evol. 1998;15:901–909. doi: 10.1093/oxfordjournals.molbev.a025994. [DOI] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Mattick JS. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PloS Comput Bio. 2008;4:e1000176. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. On the sterility of the interracial hybrids in Drosophila pseudoobscura. Proc Natl Acad Sci U S A. 1933;19:397–403. doi: 10.1073/pnas.19.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T, Epling T. Contributions to the genetics, taxonomy, and ecology of Drosophila pseudoobscura and its relatives. Washington (DC): Carnegie Institute of Washington; 1944. [Google Scholar]
- Erdmann VA, Szymanski M, Hochberg A, Groot N, Barciszewski J. Non-coding, mRNA-like RNAs database Y2K. Nucleic Acids Res. 2000;28:197–200. doi: 10.1093/nar/28.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Pheasant M, Mattick JS. The amazing complexity of the human transcriptome. Eur J Hum Genet. 2005;13:894–897. doi: 10.1038/sj.ejhg.5201459. [DOI] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate JG, et al. (11 co-authors) Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–D140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardiman KE, Brewster R, Khan SM, Deo M, Bodmer R. The bereft gene, a potential target of the neural selector gene cut, contributes to bristle morphogenesis. Genetics. 2002;161:231–247. doi: 10.1093/genetics/161.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, Findeiss S, Lein S, et al. (11 co-authors) Conserved introns reveal novel transcripts in Drosophila melanogaster. Genome Res. 2009;19:1289–1300. doi: 10.1101/gr.090050.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. The locus of evolution: evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- IHGSC. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Inagaki S, Numata K, Kondo T, Tomita M, Yasuda K, Kanai A, Kageyama Y. Identification and expression analysis of putative mRNA-like non-coding RNA in Drosophila. Genes Cells. 2005;10:1163–1173. doi: 10.1111/j.1365-2443.2005.00910.x. [DOI] [PubMed] [Google Scholar]
- Jiang Z-F, Machado CA. Evolution of sex-dependent gene expression in three recently diverged species of Drosophila. Genetics. 2009;183:1175–1185. doi: 10.1534/genetics.109.105775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, et al. (22 co-authors) RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S, Kageyama Y. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol. 2007;9:660–665. doi: 10.1038/ncb1595. [DOI] [PubMed] [Google Scholar]
- Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F, Kageyama Y. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science. 2010;329:336–339. doi: 10.1126/science.1188158. [DOI] [PubMed] [Google Scholar]
- Kulathinal RJ, Stevison LS, Noor MA. The genomics of speciation in Drosophila: diversity, divergence, and introgression estimated using low-coverage genome sequencing. PLoS Genet. 2009;5:e1000550. doi: 10.1371/journal.pgen.1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Liu M, Zhang L, Zhang WX, Gao G, Zhu ZY, Wei LP, Fan QC, Long MY. Detection of intergenic non-coding RNAs expressed in the main developmental stages in Drosophila melanogaster. Nucleic Acids Res. 2009;37:4308–4314. doi: 10.1093/nar/gkp334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipshitz HD, Peattie DA, Hogness DS. Novel transcripts from the Ultrabithorax domain of the bithorax complex. Genes Dev. 1987;1:307–322. doi: 10.1101/gad.1.3.307. [DOI] [PubMed] [Google Scholar]
- Lu J, Shen Y, Wu Q, Kumar S, He B, Shi S, Carthew RW, Wang SM, Wu CI. The birth and death of microRNA genes in Drosophila. Nat Genet. 2008;40:351–355. doi: 10.1038/ng.73. [DOI] [PubMed] [Google Scholar]
- Machado CA, Haselkorn TS, Noor MA. Evaluation of the genomic extent of effects of fixed inversion differences on intraspecific variation and interspecific introgression in Drosophila pseudoobscura and D. persimilis. Genetics. 2007;175:1289–1306. doi: 10.1534/genetics.106.064758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado CA, Hey J. The causes of phylogenetic conflict in a classic Drosophila species group. Proc R Soc Lond B Biol Sci. 2003;270:1193–1202. doi: 10.1098/rspb.2003.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado CA, Kliman RM, Markert JA, Hey J. Inferring the history of speciation using multilocus DNA sequence data: the case of Drosophila pseudoobscura and close relatives. Mol Biol Evol. 2002;19:472–488. doi: 10.1093/oxfordjournals.molbev.a004103. [DOI] [PubMed] [Google Scholar]
- Manak JR, Dike S, Sementchenko V, et al. (11 co-authors) Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat Genet. 2006;38:1151–1158. doi: 10.1038/ng1875. [DOI] [PubMed] [Google Scholar]
- Martinho RG, Kunwar PS, Casanova J, Lehmann R. A noncoding RNA is required for the repression of RNApolII-dependent transcription in primordial germ cells. Curr Biol. 2004;14:159–165. doi: 10.1016/j.cub.2003.12.036. [DOI] [PubMed] [Google Scholar]
- Mattick JS. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays. 2003;25:930–939. doi: 10.1002/bies.10332. [DOI] [PubMed] [Google Scholar]
- Mattick JS. A new paradigm for developmental biology. J Exp Biol. 2007;210:1526–1547. doi: 10.1242/jeb.005017. [DOI] [PubMed] [Google Scholar]
- Mattick JS. The genetic signatures of noncoding RNAs. PloS Genetics. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Suppl 1):R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Amikura R, Mukai M, Kobayashi S, Lasko PF. Requirement for a noncoding RNA in Drosophila polar granules for germ cell establishment. Science. 1996;274:2075–2079. doi: 10.1126/science.274.5295.2075. [DOI] [PubMed] [Google Scholar]
- Nobrega MA, Zhu Y, Plajzer-Frick I, Afzal V, Rubin EM. Megabase deletions of gene deserts result in viable mice. Nature. 2004;431:988–993. doi: 10.1038/nature03022. [DOI] [PubMed] [Google Scholar]
- Noor MAF, Garfield DA, Schaeffer SW, Machado CA. Divergence between the Drosophila pseudoobscura and D. persimilis genome sequences in relation to chromosomal inversions. Genetics. 2007;177:1417–1428. doi: 10.1534/genetics.107.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MAF, Grams KL, Bertucci LA, Almendarez Y, Reiland J, Smith KR. The genetics of reproductive isolation and the potential for gene exchange between Drosophila pseudoobscura and D. persimilis via backcross hybrid males. Evolution. 2001;55:512–521. doi: 10.1554/0014-3820(2001)055[0512:tgoria]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Noor MAF, Grams KL, Bertucci LA, Reiland J. Chromosomal inversions and the reproductive isolation of species. Proc Natl Acad Sci U S A. 2001;98:12084–12088. doi: 10.1073/pnas.221274498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y, Furuno M, Kasukawa T, et al. (139 co-authors) Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Orr HA. Genetics of male and female sterility in hybrids of Drosophila pseudoobscura and D. persimilis. Genetics. 1987;116:555–563. doi: 10.1093/genetics/116.4.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Parisi M, Nuttall R, Naiman D, Bouffard G, Malley J, Andrews J, Eastman S, Oliver B. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299:697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, et al. (16 co-authors) An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- Rajendra TK, Prasanth KV, Lakhotia SC. Male sterility associated with overexpression of the noncoding hsromega gene in cyst cells of testis of Drosophila melanogaster. J Genet. 2001;80:97–110. doi: 10.1007/BF02728335. [DOI] [PubMed] [Google Scholar]
- Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- Riccardo S, Tortoriello G, Giordano E, Turano M, Furia M. The coding/non-coding overlapping architecture of the gene encoding the Drosophila pseudouridine synthase. BMC Mol Biol. 2007;8:15. doi: 10.1186/1471-2199-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Liu Y, Bettencourt BR, et al. (52 co-authors) Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene and cis-element evolution. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymarquis LA, Kastenmayer JP, Huttenhofer AG, Green PJ. Diamonds in the rough: mRNA-like non-coding RNAs. Trends Plant Sci. 2008;13:329–334. doi: 10.1016/j.tplants.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Sanchez-Elsner T, Gou D, Kremmer E, Sauer F. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311:1118–1123. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- Stolc V, Gauhar Z, Mason C, et al. (12 co-authors) A gene expression map for the euchromatic genome of Drosophila melanogaster. Science. 2004;306:655–660. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckenholz C, Meller VH, Kuroda MI. Functional redundancy within roX1, a noncoding RNA involved in dosage compensation in Drosophila melanogaster. Genetics. 2003;164:1003–1014. doi: 10.1093/genetics/164.3.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill D, Zhang Y, Parisi M, Oliver B. Demasculinization of X chromosomes in the Drosophila genus. Nature. 2007;450:238–241. doi: 10.1038/nature06330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan M, Schulz MH, Richard H, et al. (16 co-authors) A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays. 2007;29:288–299. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tupy JL, Bailey AM, Dailey G, Evans-Holm M, Siebel CW, Misra S, Celniker SE, Rubin GM. Identification of putative noncoding polyadenylated transcripts in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2005;102:5495–5500. doi: 10.1073/pnas.0501422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RL, Wakeley J, Hey J. Gene flow and natural selection in the origin of Drosophila pseudoobscura and close relatives. Genetics. 1997;147:1091–1106. doi: 10.1093/genetics/147.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YF, Li Z, Fan QC, Long MY, Zhang WX. Significant divergence of sex-related non-coding RNA expression patterns among closely related species in Drosophila. Chi Sci Bull. 2007;52:748–754. [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analyses by maximum likelihood. CABIOS. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.