Abstract
Aerobic fermentation has evolved independently in two yeast lineages, the Saccharomyces cerevisiae and the Schizosaccharomyces pombe lineages. In the S. cerevisiae lineage, the evolution of aerobic fermentation was shown to be associated with transcriptional reprogramming of the genes involved in respiration and was recently suggested to be linked to changes in nucleosome occupancy pattern in the promoter regions of respiration-related genes. In contrast, little is known about the genetic basis for the evolution of aerobic fermentation in the Sch. pombe lineage. In particular, it is not known whether respiration-related genes in Sch. pombe have undergone a transcriptional reprogramming or changes in nucleosome occupancy pattern in their promoter regions. In this study, we compared genome-wide gene expression profiles of Sch. pombe with those of S. cerevisiae and the aerobic respiration yeast Candida albicans. We found that the expression profile of respiration-related genes in Sch. pombe is similar to that of S. cerevisiae, but different from that of C. albicans, suggesting that their transcriptional regulation has been reprogrammed during the evolution of aerobic fermentation. However, we found no significant nucleosome organization change in the promoter of respiration-related gene in Sch. pombe.
Keywords: aerobic fermentation, nucleosome organization, gene expression, Schizosaccharomyces pombe
Introduction
The evolution of aerobic fermentation in yeasts is a good example of phenotypic evolution. In eukaryotes, glucose is mainly assimilated through the respiration pathway in mitochondria to produce CO2 and H2O for maximum energy yield in the presence of oxygen. However, Crabtree-positive yeasts such as Saccharomyces cerevisiae undergo aerobic fermentation in which glucose is predominantly fermented to ethanol even in the presence of oxygen (Merico et al. 2007). It was proposed that the evolution of aerobic fermentation in yeasts was an adaptation to glucose-rich environments for rapid consumption of glucose (De Deken 1966). Recent studies indicated that the evolution of aerobic fermentation in the S. cerevisiae lineage was associated with regulatory reprogramming of genes involved in respiration and mitochondrial functions (Ihmels et al. 2005; Field et al. 2009). The loss of a specific cis-regulatory element in many genes coding for mitochondrial proteins in the S. cerevisiae lineage was speculated to have contributed to the transcriptional reprogramming process (Ihmels et al. 2005).
Recently, the regulatory evolution of respiratory genes in the S. cerevisiae lineage was linked to chromatin structure change in their promoter regions (Field et al. 2009; Tsankov et al. 2010). In eukaryotes, DNA is repetitively wrapped around nucleosomes, which form barriers to direct interaction between transcription factors and their binding sites. Several studies have found that genes with different expression profiles are associated with distinct nucleosome occupancy patterns in the promoter regions (Tirosh and Barkai 2008; Jiang and Pugh 2009). The promoters of constantly expressed genes usually contain a nucleosome-depleted region where most transcription factor–binding sites are located (Yuan et al. 2005; Lee et al. 2007). In contrast, conditionally expressed genes, such as stress-response genes, are associated with nucleosome-occupied promoters (Tirosh and Barkai 2008). By comparing the nucleosome organization patterns, Field et al. (2009) found that the promoters of respiration-related genes tend to be more depleted of nucleosomes in the aerobic respiration yeasts than that in aerobic fermentation species in the hemiascomycete lineage. They concluded that in the aerobic fermentation yeasts, respiration-related gene promoters have evolved from the nucleosome-depleted type to the nucleosome-occupied type and that this change has contributed to the evolution of aerobic fermentation in hemiascomycete yeasts (Field et al. 2009).
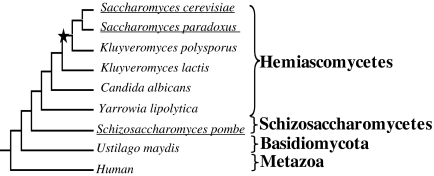
The fission yeast Schizosaccharomyces pombe separated from the hemiascomycete lineage at least 300 Ma (Sipiczki 2000) but, like S. cerevisiae, is capable of aerobic fermentation in the presence of excess sugars (Alexander and Jeffries 1990). Therefore, aerobic fermentation appears to have evolved at least twice during yeast evolution (fig. 1). However, the underlying genetic basis for the evolution of aerobic fermentation in the Sch. pombe lineage has been little explored. This study has two purposes. First, we investigated whether in Sch. pombe regulatory reprogramming of respiration-related genes was also associated with the evolution of aerobic fermentation. Second, we studied whether nucleosome organization change in the respiration-related gene promoters was associated with this process, using the newly available nucleosome occupancy data of Sch. pombe (Lantermann et al. 2010).
FIG. 1.
Schematical illustration of independent evolution of aerobic fermentation in two distantly related yeast lineages. The names of species with aerobic fermentation were underlined. The star indicates the occurrence of whole-genome duplication. The tree shows the evolutionary relationships of representative species; the branch lengths are not scaled.
Materials and Methods
Data Sources
We downloaded the large collections of microarray data of S. cerevisiae (1,011 expression profiles) and Candida albicans (198 expression profiles) from Ihmels et al. (2005) and 1,161 expression profiles in Sch. pombe from Pancaldi et al. (2010). These microarray data were obtained under a large variety of growth conditions, stress conditions, cell cycle stages, or genetic backgrounds. We retrieved 3,113 orthologous groups that contain at least one gene in each of the three species from the yeast orthology maps (Wapinski et al. 2007). We compiled Gene Ontology (GO) hierarchy data from gene ontology association data of C. albicans (Revision: 1.587), S. cerevisiae (Revision: 1.149), Sch. pombe (Revision: 1.144), respectively (Ashburner et al. 2000).
Computing Expression Correlations between Gene Sets
We computed the Pearson correlation coefficient (ρ) between every pair of genes in each species using the collections of microarray data. Then, we normalized the Pearson correlations by subtracting them from their mean and by dividing each value by the standard deviation to correct the potential biases that may arise due to different sample sizes of expression data among the three species (Field et al. 2009). For each GO gene set, we calculated the average of normalized correlations between all pairs of genes to determine if the genes in a GO group have similar expression profiles.
Because cytosol ribosomal protein (CRP)–encoding genes are consistently expressed under different conditions and show a strong correlation with cell growth (Mager and Planta 1991; Gasch et al. 2000), we used these genes as a reference to characterize the expression profiles for each GO gene group. We obtained 132, 133, and 78 CRP genes for S. cerevisiae, Sch. pombe, and C. albicans, respectively, based on their genomic annotation. The expression profile of a GO group was measured by the average of the normalized correlations between the expression of every gene in the GO group and the expression of every CRP gene.
Calculating the Average Nucleosome Occupancy of a Gene Set
The in vivo nucleosome occupancy data of S. cerevisiae and C. albicans were downloaded from Field et al. (2009). We obtained the in vivo nucleosome occupancy data of Sch. pombe from Lantermann et al. (2010). These nucleosome data were obtained from cells during the mid log phase growth in rich media. The nucleosome occupancy of promoters of a gene set was calculated as the average nucleosome occupancy of all genes for the region between 400 bp upstream and 100 bp downstream of the translation start site.
Results
Sch. pombe Respiration-Related Genes Have Undergone a Regulatory Reprogramming
To investigate if the evolution of aerobic fermentation in Sch. pombe was associated with gene transcription remodeling, we compared its genome-wide gene expression profiles with those of the aerobic fermentation yeast S. cerevisiae and the aerobic respiratory yeast C. albicans, using large collections of gene expression data and the gene sets from Gene Ontology (see Materials and Methods). To avoid small sample sizes, we only selected the GO sets with at least ten orthologous genes in each species, and we obtained 1,791, 1,547, and 1,486 GO sets for S. cerevisiae, Sch. pombe, and C. albicans, respectively. We only used the GO groups whose genes have coherent coexpression patterns (i.e., having an average normalized correlation >0.5) for further analysis. There are 698, 352, and 253 GO sets above the threshold for S. cerevisiae, Sch. pombe, and C. albicans, respectively. The expression profiles of each GO group are measured by its expression correlation with the CRP genes (see Method and Materials).
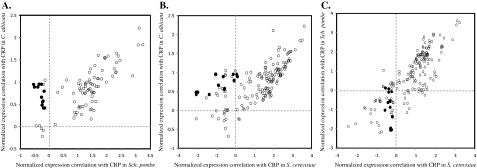
We compared the expression profiles of all selected GO groups between each pair of the three species (fig. 2A–C). Among the selected GO groups, 218 of them are shared by Sch. pombe and S. cerevisiae, and the expression profiles of these GO groups have a strong positive correlation between the two species (Pearson correlation ρ = 0.83). In contrast, the ρ value between Sch. pombe and C. albicans and that between S. cerevisiae and C. albicans are both 0.63. Thus, although S. cerevisiae is evolutionarily closer to C. albicans, its genome-wide gene expression profile is more similar to that of another aerobic fermentation species Sch. pombe. As shown in figure 2A, there are 20 GO groups whose gene expressions are positively correlated with expressions of CRP genes in C. albicans but negatively correlated with expressions of CRP genes in Sch. pombe. After removing these 20 GO groups, the ρ value increases from 0.63 to 0.82, suggesting that these GO groups are the outliers that lower the correlation of gene expressions between the two species. Eighteen of the 20 GO groups are coherent in all three species, and all of these 18 GO groups are negatively or not correlated with CRP genes in S. cerevisiae but have a much higher correlation with CRP genes in C. albicans (fig. 2B). In contrast, the 18 GO gene groups are negatively or not correlated with CRP genes in both Sch. pombe and S. cerevisiae (fig. 2C). Therefore, these 18 GO groups tend to be actively expressed under typical growth conditions in C. albicans, but they are likely to be inactive or lowly expressed during normal growth in Sch. pombe and S. cerevisiae. As shown in table 1, all the 18 GO group genes are involved in respiration and mitochondrial functions, suggesting that a regulatory reprogramming of these genes was associated with the evolution of aerobic fermentation in the Sch. pombe lineage. Therefore, we called the genes in these 18 GO groups as respiration-related genes (R genes; see the gene list in supplementary table 1, Supplementary Material online). Because the two aerobic fermentation lineages evolved from ancestral aerobic respiration species and because one would expect that the R genes of a typical aerobic respiration species are actively transcribed during normal growth, similar to that of C. albicans, it is reasonable to conclude that the transcription of the R genes has been independently reprogrammed during evolution of aerobic fermentation in both the Sch. pombe and the S. cerevisiae lineages. Therefore, as in the S. cerevisiae lineage, the evolution of aerobic fermentation in the Sch. pombe lineage was apparently also associated with a regulatory reprogramming of respiration-related genes.
FIG. 2.
Changes in the transcriptional programs of respiration-related genes in the two aerobic fermentation species. Each spot represents the average of normalized Pearson correlations between the expression of genes of a GO group and the expression of the CRP genes. (A) Schizosaccharomyces pombe versus Candida albicans. A total of 116 GO sets were used. The 18 GO sets that show different correlation patterns are shown as solid black dots; the genes in these 18 GO sets are called the R genes. These 18 GO gene sets are also shown in black in B and C. (B) Saccharomyces cerevisiae versus C. albicans. A total of 194 GO sets were used. (C) Sch. pombe versus S. cerevisiae. A total of 218 GO sets were used.
Table 1.
A list of the 18 GO groups and the numbers of genes in each GO set in Schizosaccharomyces pombe, Saccharomyces cerevisiae and Candida albicans.
| GO Accession | GO Definition | Sch. pombe | S. cerevisiae | C. albicans |
| GO:0005746 | Mitochondrial respiratory chain | 24 | 22 | 22 |
| GO:0022904 | Respiratory electron transport chain | 24 | 22 | 19 |
| GO:0042773 | ATP synthesis coupled electron transport | 18 | 22 | 19 |
| GO:0042775 | Mitochondrial ATP synthesis coupled electron transport | 18 | 22 | 19 |
| GO:0070469 | Respiratory chain | 24 | 26 | 22 |
| GO:0022900 | Electron transport chain | 25 | 55 | 19 |
| GO:0005753 | Mitochondrial proton-transporting ATP synthase complex | 16 | 15 | 14 |
| GO:0045259 | Proton-transporting ATP synthase complex | 16 | 15 | 14 |
| GO:0006119 | Oxidative phosphorylation | 41 | 49 | 33 |
| GO:0046933 | Hydrogen ion–transporting ATP synthase activity, rotational mechanism | 16 | 15 | 12 |
| GO:0006818 | Hydrogen transport | 55 | 55 | 42 |
| GO:0015992 | Proton transport | 55 | 55 | 42 |
| GO:0015078 | Hydrogen ion transmembrane transporter activity | 55 | 51 | 37 |
| GO:0006754 | ATP biosynthetic process | 20 | 42 | 14 |
| GO:0016469 | Proton-transporting two-sector ATPase complex | 30 | 32 | 16 |
| GO:0015985 | Energy-coupled proton transport, down electrochemical gradient | 20 | 27 | 14 |
| GO:0015986 | ATP synthesis coupled proton transport | 20 | 27 | 14 |
| GO:0033177 | Proton-transporting two-sector ATPase complex, proton-transporting domain | 14 | 15 | 10 |
| Total unique genes | 68 | 111 | 47 |
Nucleosome Organization in the Promoters of Respiratory Genes Differs between Sch. pombe and S. cerevisiae
To determine if change in nucleosome occupancy in the promoters of R genes was associated with their expression change during the evolution of aerobic fermentation in the Sch. Pombe lineage, we compared their promoter nucleosome organization with the category I (CI) and II (CII) genes defined by Field et al. (2009), which was based on a comparative study of gene expression data between S. cerevisiae and C. albicans. The CI genes are mainly involved in cellular growth, amino acid biosynthesis, and RNA processing and are actively expressed during cell growth, whereas the CII genes are enriched in specific cellular states, in response to environmental stresses. In contrast to the CI genes, the CII genes are mainly expressed under specific conditions and are inactive under the normal growth condition (Field et al. 2009). Accordingly, the promoter regions of CI genes were found to be more depleted of nucleosome than that of CII in both C. albicans and S. cerevisiae (Field et al. 2009).
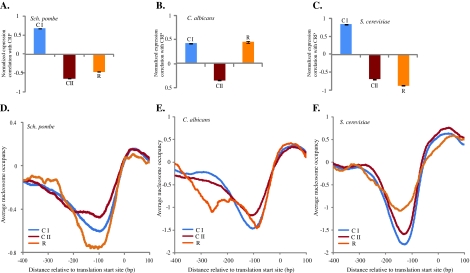
To determine if Sch. pombe CI and CII genes have similar expression patterns as their orthologous genes in the other two species, we calculated the averages of normalized correlations between CRP genes and the experimentally defined CI and CII genes for each species. In Sch. pombe, the averages of normalized correlations are ρ = 0.67 ± 0.005 between expressions of CI and CRP genes, and ρ = −0.64 ± 0.006 between expression of CII and CRP genes (fig. 3A). Therefore, in general, the expression of Sch. pombe CI genes tends to be positively correlated with that of CRP genes, but the CII gene expression shows a negative correlation with that of CRP genes, consistent with the observations in S. cerevisiae and C. albicans (fig. 3A–C). Moreover, in Sch. pombe, the average nucleosome occupancy in CI gene promoters is lower than that in CII gene promoters, a pattern that is shared by all three species (fig. 3D–F). We also calculated the average normalized correlations of expression profiles between the R genes and CRP genes. As shown in figure 3A–C, the expression of R genes is positively correlated with that of CRP genes in C. albicans (ρ = 0.45 ± 0.02) but is negatively correlated with that of CRP genes in S. cerevisiae (ρ = −0.87 ± 0.012) and in Sch. pombe (ρ = −0.46 ± 0.01). Interestingly, the nucleosome organization of R gene promoters of Sch. pombe is more similar to that of C. albicans R genes than to that of S. cerevisiae R. genes. In contrast to the nucleosome occupancy of R gene promoters in S. cerevisiae, which is higher than that of the CII genes, the nucleosome occupancy of R gene promoters in Sch. pombe is lower than that of CII genes, which is similar to the situation in C. albicans. We also used two other sets of respiration-related genes identified in Ferea et al. (1999) and Ihmels et al. (2002) and obtained the same results (supplementary figs. 1 and 2, Supplementary Material online). Thus, although the expression regulation of the R genes in Sch. pombe has been reprogrammed during the evolution of aerobic fermentation, their promoter nucleosome organization remains depleted as in aerobic respiration species. Therefore, regulatory reprogramming of respiration-related genes during evolution of aerobic fermentation in Sch. pombe was not coupled with change in nucleosome occupancy.
FIG. 3.
Comparisons of gene expression profiles and nucleosome occupancy patterns in Schizosaccharomyces pombe, Candida albicans, and Saccharomyces cerevisiae. (A–C) Averages of normalized correlations between the CRP genes and CI, CII, and R genes in Sch. pombe, C. albicans, and S. cerevisiae, respectively. Error bars indicate the standard errors of means. The expression profiles of CI and CII genes are conserved among the three species, but the expression profiles of R genes in Sch. pombe and S. cerevisiae have been independently reprogrammed during the evolution of aerobic fermentation. (D–F) Average nucleosome occupancies of the CI, CII, and R gene promoters in Sch. pombe, Candida albicans, and S. cerevisiae, respectively. The promoters of R genes in S. cerevisiae have changed from nucleosome depleted to nucleosome occupied. In contrast, the promoters of R genes in Sch. pombe remained nucleosome depleted.
Discussion
We have shown here that gene expression divergence was not coupled with change in nucleosome organization during the evolution of aerobic fermentation in the Sch. pombe lineage, contrary to the observation in the S. cerevisiae lineage (Field et al. 2009). The presence of nucleosome may hinder the direct interaction between a transcription factor and its binding sites and may therefore obstruct the transcriptional initiation of a gene. Because S. cerevisiae assimilates glucose predominantly through the fermentation pathway and because in S. cerevisiae the transcription of respiration-related genes is repressed under the normal growth condition (Carlson 1999), the presence of nucleosome-occupied promoters in these genes might be favored by natural selection. As nucleosome positioning appears to be determined by the intrinsic property of nearby DNA sequences (Kaplan et al. 2009; Tirosh et al. 2010), change in nucleosome occupancy in the promoter regions may be largely explained by accumulation of mutations. Therefore, one might expect that the switch from the nucleosome-depleted to nucleosome-occupied promoters in respiration-related genes has contributed to their transcriptional change and the evolution of aerobic fermentation in the S. cerevisiae lineage (Field et al. 2009).
If the above hypothesis is correct, how does one explain the observation that no significant change of promoter nucleosome organization was associated with transcriptional changes of respiratory-related genes in Sch. pombe during the evolution of aerobic fermentation? To answer this question, factors that might prevent the change of promoter nucleosome organization should be considered. Although both S. cerevisiae and Sch. pombe undergo aerobic fermentation in the presence of excess sugars, there are physiological differences between them. In particular, they have distinct requirements for oxygen. Like many other eukaryotes, Sch. pombe cannot survive without the presence of oxygen, whereas S. cerevisiae is able to grow under anaerobic conditions (Visser et al. 1990). Another important difference is that the two species have different requirements of mitochondrial functions. Pyrimidines and purines are two of the building blocks of nucleic acids. The fourth step of de novo pyrimidine biosynthesis in Sch. pombe and most other eukaryotes is catalyzed by the dihydroorotate dehydrogenase (DHODase), which is localized in mitochondria, and the enzymatic activity of DHODase is dependent on the function of the respiratory chain (Andreasen and Stier 1953; Nagy et al. 1992; Chabes et al. 2000). However, during the evolution of S. cerevisiae, the pyrimidine biosynthesis pathway has been modified because the S. cerevisiae DHODase, which is localized in cytosol, does not require a functional respiratory chain and thus pyrimidine synthesis in S. cerevisiae is independent of oxygen (Nagy et al. 1992). This change is probably because the S. cerevisiae DHODase gene was acquired from bacteria by horizontal gene transfer after its divergence from the C. albicans lineage (Gojkovic et al. 2004). Unlike S. cerevisiae, the respiration-related genes in Sch. pombe cannot be completely repressed under normal growth condition due to the absolute requirement of mitochondrial functions. This difference is consistent with our observation that the expression of S. cerevisiae respiratory chain genes (GO:0070469) is much more negatively correlated with the CRP genes than that in Sch. pombe (normalized Pearson correlation: −1.97 vs. −0.25). Therefore, a nucleosome-depleted promoter would be favored for Sch. pombe respiration-related genes, which might at least partly explain the difference of nucleosome organization in the R gene promoters between S. cerevisiae and Sch. pombe.
To test the above hypothesis, we compared the gene expression levels of the R, CI, and CII groups during the exponential growth phase in Sch. pombe (Lantermann et al. 2010), S. cerevisiae (Nagalakshmi et al. 2008), and C. albicans (Bruno et al. 2010). As expected, the R genes in C. albicans are highly expressed and their expression level is even significantly higher than the growth-related CI genes (supplementary fig. 3, Supplementary Material online; P value <4.07 × 10−12, two-sided Student's t-test). Interestingly, the R genes in neither Sch. pombe nor S. cerevisiae were strictly repressed during fermentative growth. Although the mean expression levels of R genes in the two species are not significantly different from that of CI genes (P value = 0.14 and 0.06, respectively), they are significantly higher than that of CII genes (P value<2 × 10−10). To confirm these observations, we repeated this analysis using the mRNA level data of C. albicans and S. cerevisiae from Tsankov et al. (2010). We observed the similar pattern (supplementary fig. 4, Supplementary Material online), further supporting that the R genes in fermentative yeasts are not expressed as high as that in respiratory yeasts but they are not completely repressed. DeRisi et al. (1997) monitored the temporal gene expression change during the diauxic shift in S. cerevisiae and revealed that the expression of a large number of respiration-related genes were substantially induced when S. cerevisiae cells were forced to use nonfermentable ethanol as the carbon source because of the depletion of glucose in media. Using this diauxic shift expression data, we found that the expression levels of R genes increased >6-fold after S. cerevisiae cells switch from fermentation to the respiration mode (supplementary fig. 5, Supplementary Material online). These observations indicate that the transcription of R genes in either Sch. pombe or S. cerevisiae is not completely repressed, and they are transcribed at relatively low level in the fermentative mode than in the respiratory mode. Because expression levels of R genes change significantly under different growth conditions, it can explain why we observed a low or anticorrelation with the expression of CRP genes in Sch. pombe and S. cerevisiae. Therefore, the relative repression of R gene expression in presence of glucose was associated with the evolution of aerobic fermentation in both Sch. pombe and S. cerevisiae lineages. As suggested by previous studies, the nucleosome occupancy change might have facilitated this repression process in the S. cerevisiae lineage (Field et al. 2009; Tsankov et al. 2010). However, our results indicated that a different mechanism may underlie the same process in the Sch. pombe lineage.
In summary, our study revealed that in Sch. pombe, the proximal promoter regions of CI (growth related) genes have much lower nucleosome occupancy than those of CII (stress related) genes. The dichotomy between promoter packaging patterns of growth- and stress-related genes in Sch. pombe is generally consistent with the dichotomy observed in multiple hemiascomycete yeasts (Field et al. 2008; Tirosh et al. 2010; Tsankov et al. 2010). However, some discrepancies about the role of nucleosome change in gene regulatory evolution have been also reported. A recent study of the contribution of nucleosome organization to the gene expression divergence between S. cerevisiae and Saccharomyces paradoxus revealed that nucleosome occupancy change has very limited effects on the gene expression divergence between the two closely related species (Tirosh et al. 2010). In addition, based on a study of 12 hemiascomycete yeasts, Tsankov et al. also found that the genes involved in glycolysis and gluconeogenesis, which are highly expressed in all species, lack deep nucleosome free region (NFR) in their promoter regions. In the present study, we showed that although the respiration-related genes have been reprogrammed in Sch. pombe, the nucleosome occupancy pattern of these genes remains similar to that in the aerobic respiratory yeasts. This observation does not reject the view that change in nucleosome organization in respiration-related genes played a significant role in the evolution of aerobic fermentation in the S. cerevisiae lineage. However, it suggests that change of nucleosome occupancy is not a major contributor to the evolution of aerobic fermentation or for transcriptional reprogramming in Sch. pombe. It would be worthwhile to study if the modification of cis-elements in the promoters of Sch. pombe R genes was associated with their regulatory reprogramming.
Supplementary Material
Supplementary table S1 and figures S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We greatly appreciate Dr Philipp Korber and Alexandra Lantermann for providing the nucleosome occupancy data of Sch. pombe and Dr Jürg Bähler and Dr Vera Pancaldi for providing the compiled microarray data of Sch. pombe. We thank Dr Yong Woo for his assistance and helpful advice on data analysis and Chris Hittinger for valuable comments on the manuscript. We also gratefully thank Dr Ken Wolfe and two anonymous reviewers for their valuable critical and helpful comments. This study was funded by National Institutes of Health grant GM30998.
References
- Alexander MA, Jeffries TW. Respiratory efficiency and metabolite partitioning as regulatory phenomena in yeasts. Enzyme Microb Technol. 1990;12:2–19. [Google Scholar]
- Andreasen AA, Stier TJ. Anaerobic nutrition of Saccharomyces cerevisiae. I. Ergosterol requirement for growth in a defined medium. J Cell Physiol. 1953;41:23–36. doi: 10.1002/jcp.1030410103. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, et al. (20 co-authors). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno VM, Wang Z, Marjani SL, Euskirchen GM, Martin J, Sherlock G, Snyder M. Comprehensive annotation of the transcriptome of the human fungal pathogen Candida albicans using RNA-seq. Genome Res. 2010;20:1451–1458. doi: 10.1101/gr.109553.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M. Glucose repression in yeast. Curr Opin Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- Chabes A, Domkin V, Larsson G, Liu A, Graslund A, Wijmenga S, Thelander L. Yeast ribonucleotide reductase has a heterodimeric iron-radical-containing subunit. Proc Natl Acad Sci U S A. 2000;97:2474–2479. doi: 10.1073/pnas.97.6.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Deken RH. The Crabtree effect: a regulatory system in yeast. J Gen Microbiol. 1966;44:149–156. doi: 10.1099/00221287-44-2-149. [DOI] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Ferea TL, Botstein D, Brown PO, Rosenzweig RF. Systematic changes in gene expression patterns following adaptive evolution in yeast. Proc Natl Acad Sci U S A. 1999;96:9721–9726. doi: 10.1073/pnas.96.17.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y, Fondufe-Mittendorf Y, Moore IK, Mieczkowski P, Kaplan N, Lubling Y, Lieb JD, Widom J, Segal E. Gene expression divergence in yeast is coupled to evolution of DNA-encoded nucleosome organization. Nat Genet. 2009;41:438–445. doi: 10.1038/ng.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojkovic Z, Knecht W, Zameitat E, Warneboldt J, Coutelis JB, Pynyaha Y, Neuveglise C, Moller K, Loffler M, Piskur J. Horizontal gene transfer promoted evolution of the ability to propagate under anaerobic conditions in yeasts. Mol Genet Genomics. 2004;271:387–393. doi: 10.1007/s00438-004-0995-7. [DOI] [PubMed] [Google Scholar]
- Ihmels J, Bergmann S, Gerami-Nejad M, Yanai I, McClellan M, Berman J, Barkai N. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science. 2005;309:938–940. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- Ihmels J, Friedlander G, Bergmann S, Sarig O, Ziv Y, Barkai N. Revealing modular organization in the yeast transcriptional network. Nat Genet. 2002;31:370–377. doi: 10.1038/ng941. [DOI] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, et al. (11 co-authors). The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantermann AB, Straub T, Stralfors A, Yuan GC, Ekwall K, Korber P. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat Struct Mol Biol. 2010;17:251–257. doi: 10.1038/nsmb.1741. [DOI] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- Mager WH, Planta RJ. Coordinate expression of ribosomal protein genes in yeast as a function of cellular growth rate. Mol Cell Biochem. 1991;104:181–187. doi: 10.1007/BF00229818. [DOI] [PubMed] [Google Scholar]
- Merico A, Sulo P, Piskur J, Compagno C. Fermentative lifestyle in yeasts belonging to the Saccharomyces complex. Febs J. 2007;274:976–989. doi: 10.1111/j.1742-4658.2007.05645.x. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy M, Lacroute F, Thomas D. Divergent evolution of pyrimidine biosynthesis between anaerobic and aerobic yeasts. Proc Natl Acad Sci U S A. 1992;89:8966–8970. doi: 10.1073/pnas.89.19.8966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancaldi V, Schubert F, Bahler J. Meta-analysis of genome regulation and expression variability across hundreds of environmental and genetic perturbations in fission yeast. Mol Biosyst. 2010;6:543–552. doi: 10.1039/b913876p. [DOI] [PubMed] [Google Scholar]
- Sipiczki M. Where does fission yeast sit on the tree of life? Genome Biol. 2000;1:REVIEWS1011. doi: 10.1186/gb-2000-1-2-reviews1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Sigal N, Barkai N. Divergence of nucleosome positioning between two closely related yeast species: genetic basis and functional consequences. Mol Syst Biol. 2010;6:365. doi: 10.1038/msb.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankov AM, Thompson DA, Socha A, Regev A, Rando OJ. The role of nucleosome positioning in the evolution of gene regulation. PLoS Biol. 2010;8:e1000414. doi: 10.1371/journal.pbio.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser W, Scheffers WA, Batenburg-van der Vegte WH, van Dijken JP. Oxygen requirements of yeasts. Appl Environ Microbiol. 1990;56:3785–3792. doi: 10.1128/aem.56.12.3785-3792.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.