Abstract
Mammals have ten voltage-dependent sodium (Nav) channel genes. Nav channels are expressed in different cell types with different subcellular distributions and are critical for many aspects of neuronal processing. The last common ancestor of teleosts and tetrapods had four Nav channel genes, presumably on four different chromosomes. In the lineage leading to mammals, a series of tandem duplications on two of these chromosomes more than doubled the number of Nav channel genes. It is unknown when these duplications occurred and whether they occurred against a backdrop of duplication of flanking genes on their chromosomes or as an expansion of ion channel genes in general. We estimated key dates of the Nav channel gene family expansion by phylogenetic analysis using teleost, elasmobranch, lungfish, amphibian, avian, lizard, and mammalian Nav channel sequences, as well as chromosomal synteny for tetrapod genes. We tested, and exclude, the null hypothesis that Nav channel genes reside in regions of chromosomes prone to duplication by demonstrating the lack of duplication or duplicate retention of surrounding genes. We also find no comparable expansion in other voltage-dependent ion channel gene families of tetrapods following the teleost–tetrapod divergence. We posit a specific expansion of the Nav channel gene family in the Devonian and Carboniferous periods when tetrapods evolved, diversified, and invaded the terrestrial habitat. During this time, the amniote forebrain evolved greater anatomical complexity and novel tactile sensory receptors appeared. The duplication of Nav channel genes allowed for greater regional specialization in Nav channel expression, variation in subcellular localization, and enhanced processing of somatosensory input.
Keywords: sodium channel, tetrapods, amniotes, terrestriality, gene duplication, brain
Introduction
Voltage-dependent sodium (Nav) channels are critical for electrical excitability and neuronal computation. Mammals have ten Nav channels with distinct biophysical properties, types of modulation by neurotransmitters, and tissue and subcellular distributions (Angelino and Brenner 2007). For example, a distinct Nav channel (Nav1.4) is predominantly expressed in skeletal muscle and another Nav channel (Nav1.5) predominantly in cardiac muscle. Different Nav channels are expressed in unmyelinated axons (Nav1.2) and at the nodes of Ranvier in myelinated axons (Nav1.6) (Westenbroek et al. 1989; Caldwell et al. 2000). Specific Nav channels (Nav1.7, 1.8, and 1.9) are highly expressed in nociceptors (Akopian et al. 1996; Cummins et al. 1999; Dib-Hajj et al. 2002) or may be upregulated specifically in neurons in the nociceptive pathway following injury (Nav1.3) (Hains et al. 2003). Some cell types, such as fast-firing parvalbumin-positive inhibitory neurons, mainly express one type of Nav channel (Nav1.1), whereas another Nav channel is expressed in neighboring pyramidal neurons (Nav1.6) (Ogiwara et al. 2007; Lorincz and Nusser 2010). Different Nav channels may even be expressed in different subcellular domains in neurons: distinct Nav channels are responsible for initiating the action potential at the axon initial segment (Nav1.6) and for backpropagation of the action potential into the soma (Nav1.2), a critical function for activity-dependent synaptic plasticity (Hu et al. 2009).
Recent studies have clarified the evolutionary relationships among and timing of the origin of vertebrate Nav channels (Okamura et al. 1994; Plummer and Meisler 1999; Lopreato et al. 2001; Goldin 2002; Piontkivska and Hughes 2003; Novak et al. 2006; Hill et al. 2008). Early in vertebrate evolution, a single Nav channel gene of early chordates (Okamura et al. 1994) duplicated twice, presumably during two consecutive whole genome duplication (WGD) events, giving rise to four Nav channel genes, each presumed on a different chromosome (Plummer and Meisler 1999; Lopreato et al. 2001; Novak et al. 2006). In teleosts, this number jumped to eight Nav channel genes via a third teleost-specific WGD (Lopreato et al. 2001; Novak et al. 2006), whereas a series of tandem duplications on two of these chromosomes at unknown times in the lineage leading to mammals resulted in a total of ten Nav channel genes in rodents and humans and presumably other mammals (Plummer and Meisler 1999). A major goal of this study was to determine the timing and significance of these tandem duplications for tetrapod evolution. Additionally, we wished to investigate whether the duplication and retention was unique to Nav channel genes and, therefore, possibly adaptive or merely the result of passive factors such as chromosomal “hotspots” for duplication. Finally, we asked whether the expansion of the Nav channel gene family was part of a general expansion of other ion channel gene families or a unique event.
Materials and Methods
Genomic Sequences
We obtained the whole complement of Nav channel amino acid sequences from human (Homo sapiens) and rat (Rattus norvegicus) from GenBank. Using a BLAT search with human and rodent Nav channel genes, we derived and translated nucleotide sequences from the Ensemble genome databases for western clawed frog (Xenopus tropicalis, v4.1, August 2005), green anole lizard (Anolis carolinensis, AnoCar1, assembly 2007), platypus (Ornithorhynchus anatinus, v5.0, assembly January 2007), gray short-tailed opossum (Monodelphis domestica; MonDom5, October 2006), chicken (Gallus gallus, v2.1, May 2006), and elephant shark (Callorhinchus milii; v1.0, 2007). About half of the Nav channel genes from these species had already been deposited in GenBank, but the other half had not yet been annotated. Additional sequences from lamprey (Petromyzon marinus), newt (Cynops pyrrhogaster), and the African clawed frog (Xenopus laevis) were also used for phylogeny estimation (supplementary table I, Supplementary Material online).
Because Xenopus is extensively used as a developmental biological model, very good Xenopus expressed sequence tag (EST) databases are available from the TIGR database (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/Blast/index.cgi). We utilized this EST database to confirm Xenopus genomic sequences (supplementary fig. 1, Supplementary Material online).
Nav channels comprise four repeating domains (DI–DIV), interconnecting extra- and intracellular loops and N and C termini. Sequences from all mammals, Xenopus, Anolis, and Gallus, were mostly full length (N to C termini) with the occasional small region of missing sequence due to gene assembly problems. Due to the low coverage (1.4×) of the Callorhinchus genome, contigs were short and typically contained only one or a few exons.
Sequences Derived by Reverse transcription Polymerase Chain Reaction
We cloned additional Nav channel sequences by Reverse transcription (RT) polymerase chain reaction (PCR) from various tissues of a few key species for which sequenced genomes were unavailable. Lungfish (Dipnoi) and coelacanths are the most basal living sarcopterygii and are basal to the tetrapods, so we cloned Nav channel transcripts from heart, muscle, brain, and spinal cord of the South American lungfish (Lepidosiren paradoxa). Additionally, we cloned Nav channel genes from genomic DNA of lungfish to attempt to capture any genes that might be expressed in low levels or not expressed in the tissues from which we extracted RNA.
The Chondrichthyes (e.g., sharks, rays, skates, and chimeras) diverged from the lineage leading to tetrapods ∼525 Ma in the mid-Cambrian period (Hedges 2009), presumably following the second vertebrate WGD (Kuraku 2008). We were able to obtain muscle and heart from a horn shark (Heterodontus francisci) and the brain of an Atlantic stingray (Dasyatis sabina) and cloned Nav channel transcripts from them to estimate the number of Nav channel genes in vertebrates well before the emergence of tetrapods.
We used nested primer sets developed in our laboratory for cloning Nav channel genes from a range of vertebrate species. These primarily targeted domains I, II, and/or III and variable interconnecting intracellular loops. The RT reaction either utilized the lower primer of step one to attempt to target Nav channel genes or a polyTTT primer to ensure that we had not missed any transcripts. For lungfish, the primers were as follows: first PCR (upper: CCRTGGAAYTGKCTKGATTT and lower: RTRAARRADGABCCRAADRTGATG) and second PCR (upper: ATGRCGTAYVYYACVGAGTT and lower: TACATDATNYCCATCCANCCTTT). We used the following primer sets for: —1) shark heart: first PCR (upper: TGYGGYGARTGGATYGARAC and lower: RTRAARRADGABCCRAADRTGATG) and second PCR (upper: ATGTGGGAYTGYATGGARGT and lower: TACATDATNYCCATCCANCCTTT) and 2) shark muscle: first PCR (upper: TCYMGAGGBTTCTGYDTTGG and lower: RTRAARRADGABCCRAADRTGATG) and second PCR (upper: CCRTGGAAYTGKCTKGATTT and lower: TACATDATNYCCATCCANCCTTT). For skate brain, we used a similar primer set as in lungfish muscle. Temperatures and times were 53 °C for annealing and 94 °C for denaturing steps (30–45 s), and 74 °C for extension steps (extension time dependent on the length of predicted PCR products, 1 min/1,000 bp) for a total of 35 cycles.
Nav Channel Phylogeny
Nav channel amino acid sequences were aligned in ClustalX using default parameters, and poorly aligned regions, mainly long intracellular loops, were removed. Final alignments were output into NEXUS files. To reconstruct Nav channel gene phylogeny, two independent Bayesian analyses (Mr Bayes, version 3) were conducted, each with five MCMC chains and run for 200,000 generations, assuming six substitution types and a gamma distribution of rate variation among sites. For each analysis, posterior probabilities were calculated using majority-rule consensus of all trees saved after the log-likelihood asymptote (burn-in). Lamprey Nav channel gene sequences were specified for rooting.
Analysis of Non-Nav Channel Genes
We wished to determine whether duplications of non-Nav channel genes occurred within the early tetrapod lineage following the teleost–tetrapod divergence. Human nucleotide sequences were initially used to search the National Center for Biotechnology Information (NCBI) (nr/nt) database for orthologous or paralogous sequences from chicken, frog, and teleosts (supplementary table II, Supplementary Material online). Then, all the above nucleotide sequences were used for BLAT searches of Ensemble genome databases of zebrafish and chicken to assure that no unannotated genes were missed. No attempt was made to reconstruct the history of these genes within teleosts, so extensive searches were not made of teleost genomes. However, if no teleostean ortholog was initially recovered from GenBank or from the zebrafish genome, further searches were made in the genomes of stickleback (Gasterosteus aculeatus), medaka (Oryzias latipes), and pufferfish (Takifugu rubripes and Tetraodon nigroviridis) in case a putative teleostean ortholog might have been lost in zebrafish but retained in other teleost lineages. Finally, the teleostean sequences were used for BLAT searches of the NCBI and Ensemble databases to ensure that all tetrapod orthologs had been uncovered. If a gene had duplicated in tetrapods, each duplicate would be equally likely to be identified by its teleostean ortholog. Analysis of teleostean and tetrapod non-Nav channel amino acid trees was done using the Neighbor-Joining algorithm with default values in ClustalX with 1,000 bootstrap replications.
Determination of Synteny
Chromosomal location of human and chicken Nav channel and flanking genes was available from NCBI. Synteny was determined for lizard and frog Nav and flanking genes by manually assembling genes from scaffolds from Ensemble, determining gene identity by BLAT with the NCBI database and by their locations in comparison with human and chicken chromosomes.
Results
Description of Data
From BLAT searches on archived genomes, we recovered six Nav channel genes from Xenopus, nine from lizard, six from platypus (plus two smaller fragments that were not used for analysis), eight from opossum, and nine from chicken. Where possible, predicted X. tropicalis gene sequences were confirmed by ESTs, and in two cases (xt236 and xt464b), incomplete genomic sequences were filled in by overlap with ESTs (supplementary fig. 1, Supplementary Material online).
We derived pieces of Nav channel genes from elephant shark. Due to the low coverage, each contig had one or at most a few exons, and contigs could not be unambiguously connected. However, we identified 3–4 distinct pieces corresponding to most exons of the Nav channel gene that suggested a total of four Nav channel genes. We recovered four contigs with all or part of the most 3′ exon (the longest exon in all vertebrate Nav channel genes); one contig included two other exons. This gave us sequences of from ∼200 to 500 amino acids.
By RT-PCR, we cloned three Nav channel genes from lungfish, one each from brain and spinal cord, muscle, and heart tissue; two from horn shark, one from muscle and one from heart; and one from skate brain. These were not complete sequences but covered domains II–III (lungfish muscle and skate brain) or domains I–III (shark and lungfish heart, lungfish brain and lungfish muscle). Given the limitations of our tissue samples and the RT-PCR method, we do not claim that this provides the full complement of Nav channel genes of these species.
As we show below, we have a strong case for homologizing the amniote Nav channel genes; therefore, we use the mammalian gene nomenclature (SCNxA, where x = a number) for Nav channel genes from mammals, birds, and lizards (protein designation = Nav; gene designation = SCN). But because not all the nonamniote genes are orthologs of amniote genes, we did not use the mammalian nomenclature for these.
Expansion of Nav Channel Genes Occurred in the Devonian and Carboniferous Periods
We generated phylogenetic trees without (alignment1: fig. 1) and with (alignment2: supplementary fig. 2, Supplementary Material online) the four elephant shark sequences. Resulting tree topologies of alignment1 were identical for the two runs with minor variation in the posterior probabilities for a few branches. Alignment2 was run only once due to the low support values in the parts of the tree including the elephant shark sequences (supplementary fig. 2, Supplementary Material online). We will focus on alignment1.
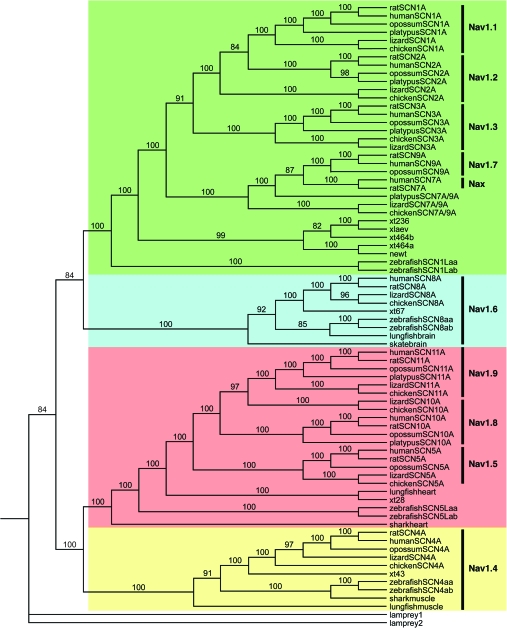
FIG. 1.
Phylogenetic tree for amino acid translation of tetrapod Nav channel genes determined by Bayesian analysis. Amniote Nav channel genes are homologized with and named according to human gene nomenclature. Genes from the frog, Xenopus tropicalis, are named after their scaffold location and genes from lungfish, skate, and shark according to the tissue from which they were amplified. Names of the mammalian Nav channel proteins are given on the right side of the figure. Human Nav channel genes that are on the same chromosome and their homologs from other species are within a block of color.
SCN8A has a simple history with no duplications tracing back to an ancestral gene that is also represented in elasmobranchs (skatebrain), lungfish (lungfishbrain), and amphibians (xt67) (fig. 1: shaded in light blue). There are two duplicates in zebrafish due to a teleost-specific WGD. As in humans, SCN8A orthologs of frog, chicken, and lizard reside alone on a chromosome (supplementary table I, Supplementary Material online).
SCN4A shows a similar history with amniote orthologs grouping with genes from frog (xt43), lungfish (lungfishmuscle), and shark (sharkmuscle) and a pair of duplicate genes in teleosts (fig. 1: shaded in yellow). The orthologs of SCN4A also reside singly on a chromosome (supplementary table I, Supplementary Material online).
Orthologs of the three Nav channel genes on human chromosome 3 (SCN5A, SCN10A, and SCN11A) are found in other mammals, chicken, and lizard, and these three genes have shared synteny (fig. 1: shaded in light red). They derive from a single gene represented in our sample by orthologs in shark (sharkheart), zebrafish (SCN5Laa and SCN5Lab), lungfish (lungfishheart), and frog (xt28). The single frog gene is syntenically related to the amniote genes (fig. 2). The presence of a single gene at the amphibian–amniote split and of three genes before the synapsid (mammals)–diapsid (reptiles and birds) divergence means that two duplications of the ancestral gene occurred in a 30-My window at the origin of amniotes in the lower to mid-Carboniferous periods.
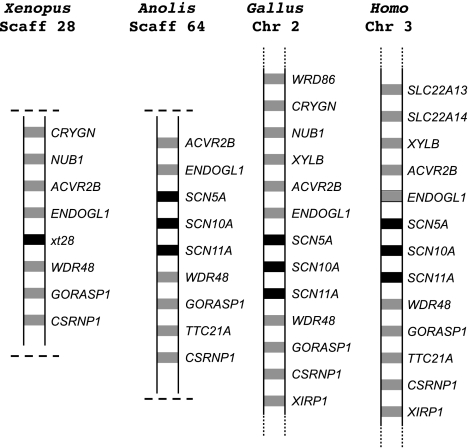
FIG. 2.
Synteny for tetrapod Nav channel genes referenced to Nav channel genes on human chromosome 3. Black boxes represent Nav channel genes, and gray boxes represent other genes.
The history of the Nav channel genes on human chromosome 2 is more complex. These genes derive from a fourth ancestral gene (Novak et al. 2006), although we were unable to detect an ortholog of this gene in lungfish tissues or skate brain despite extensive attempts to amplify it from RNA and genomic DNA. As expected, there are two gene duplicates in zebrafish (SCN1Laa and SCN1Lab). The gene tree suggests that a single ancestral gene underwent independent duplications in amphibians and amniotes (fig. 1: shaded in green). In this scenario, multiple duplications of a putative ancestral Nav channel gene would have generated SCN1A, SCN2A, SCN3A, and SCN7A/SCN9A (the precursor to separate SCNA7a and SCN9A genes). Because these all have orthologs in mammals, lizards, and chicken, these duplications would have occurred within the same 30-My window as the triplicated genes on human chromosome 3 (fig. 1). The final duplication of SCN7A/SCN9A into SCN7A and SCN9A likely occurred after the divergence of monotreme and therian mammals (220 Ma) preceding the marsupial–placental split (175 Ma). However, given the low values of the posterior probabilities in our trees, there is some uncertainty about the timing of this duplication.
On the other hand, the most parsimonious interpretation of the synteny (fig. 3) is that two duplications had already occurred in the common ancestor of amphibians and amniotes. This is because the Nav channel and other genes in this region of the amphibian and amniote chromosomes have the same syntenic relationships. Additionally, one Nav channel gene in each lineage and in the same relative chromosomal position (amphibian xt464b and amniote SCN2A) is oppositely oriented on the chromosome to all the other Nav channel genes (arrows, fig. 3). This suggests that most of the duplications occurred at a slower rate over 130 My.
FIG. 3.
Synteny for tetrapod Nav channel genes referenced to Nav channel genes on human chromosome 2. Black boxes represent Nav channel genes, and gray boxes represent other genes. Arrows represent Nav channel gene whose chromosomal orientation is opposite to all the other Nav channel genes.
What might account for the discrepancy in interpretation between the gene tree and synteny data? It is unlikely to be due to Xenopus Nav channel genes that were missed or misassembled in genome sequencing. First, all the ESTs that we uncovered uniquely matched a specific gene (in most cases multiple ESTs were mapped to each gene), and all Xenopus Nav channel genes were represented in the EST database (supplementary fig. 1, Supplementary Material online). Second, the few additional amphibian Nav channel genes available from GenBank of sufficient length to align (e.g., newt and xlaev1.2) appeared to be orthologs of genes that we had already uncovered in X. tropicalis. Finally, the Xenopus scaffolds were assembled from overlapping reads of shotgun sequence de novo so that the apparent synteny is not an artifact (Hellsten et al. 2010). It is possible, but seems unlikely, that independent duplications in amphibians and amniotes could have resulted in identical patterns of synteny. If the duplications had occurred in the common ancestor of amphibians and amniotes as suggested by synteny, then the nonoverlapping clustering of amphibian and amniote genes in the tree might be explained by some amount of gene conversion within the amniote and/or amphibian lineages, as sometimes occurs following gene duplications (Kellis et al. 2004).
Alignment2 included the fragments from elephant shark (supplementary fig. 2, Supplementary Material online). As expected, the inclusion of these short sequences resulted in low posterior probabilities for them and their neighboring branches; these were too low to trust their exact positioning in the tree. However, each of the 4 sequences grouped with 1 of the 4 clades of Nav channel genes with extremely strong (posterior probabilities = 100) support. Furthermore, a BLAT search of GenBank with the nucleotide sequences of each of the four 3′ exons had top matches with sequences in one specific clade. The inclusion of these sequences supports our contention (Lopreato et al. 2001; Novak et al. 2006) that the ancestor of teleosts and tetrapods had four Nav channel genes and gives an indication of the “missing” ancestor of the fourth Nav channel gene clade.
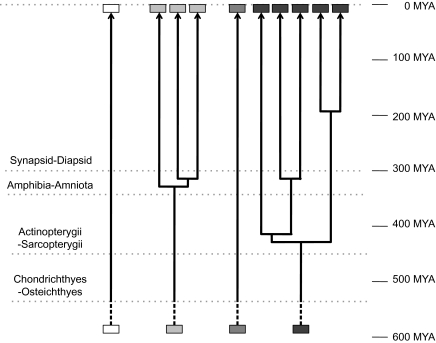
In sum, we suggest that after the second WGD (estimated at ∼550 Ma, Meyer and Schartl 1999; Dehal and Boore 2005; Panopoulou and Poustka 2005; Blomme et al. 2006) brought the number of Nav channel genes to four, there was a period of stasis. Then a series of tandem Nav channel gene duplications occurred in a 30- to 130-My period during early tetrapod evolution, following which the Nav channel gene family remained largely stable for another 300 My (fig. 5).
FIG. 5.
Schematic timeline for Nav channel gene duplications. Each set of boxes represents the lineage of four ancestral genes. Timing of the duplication of the orthologs of human chromosomes 2 (the darkest boxes) is conservatively estimated according to the most parsimonious interpretation offered by synteny. Four Nav channel genes were present in the last common ancestor of teleosts and tetrapods (actinopterygian–sarcopterygian divergence, ∼450 Ma) and likely also in the common ancestor of chondrichthyes and osteichthyes (∼525 Ma). The vertical dotted lines imply that these four genes resulted from the second of two vertebrate WGD events estimated at ∼550 Ma. Divergence times from Hedges (2009), Madsen (2009), and Shedlock and Edwards (2009).
Genes Flanking the Nav Channel Genes Did Not Duplicate or Were Not Retained
We posit that the retention of the Nav channel gene paralogs was due to selection. The null hypothesis is that the Nav channel gene expansion was simply a consequence of instability in the regions of these two chromosomes in which the Nav channel genes reside that led to duplication and retention of all the genes in this region. We tested the null hypothesis by examining whether flanking genes show a similar history of duplication and retention. We sampled only those flanking genes that were located on chromosomes or scaffolds of the species for which we had synteny information. In our sample (figs. 2 and 3) 14 of 15 genes showed no tandem duplications on these chromosomes, with SLC22A13 and SLC22A14 being a duplicate pair. Given that the Nav channel genes on these two chromosomes have both duplicated (2 out of 2: this analysis ignores the fact that the Nav channel genes underwent multiple duplications), this is significantly greater than expected given the number of duplications of the flanking non-Nav channel genes on these two chromosomes (1 out of 14) (P < 0.0001, two-tailed chi-square).
However, it is also possible that these flanking genes duplicated but were then dispersed throughout the genome, whereas the Nav channel genes were retained where they duplicated. We derived sequences for the 16 flanking genes in figures 2 and 3 from GenBank or searches from genome databases and constructed trees for them. We found that most showed indications of the initial two rounds of WGD but no duplications in the 450 My following the actinopterygian–sarcopterygian split (fig. 4A). The only exception, mentioned above, was the duplicate pair of SLC22A members13 and 14. This gene family has a history of extensive duplication (>25 members) with the “center of gravity” of duplication elsewhere in the genome.
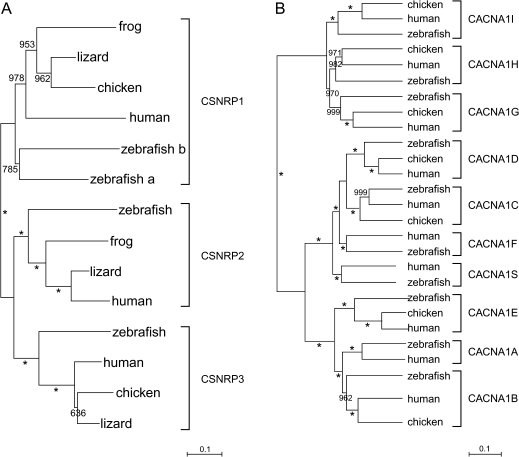
FIG. 4.
Representative non-Nav channel gene trees. (A) CSNRP, a gene flanking the Nav channel genes, which was studied to test for duplication and retention of neighboring non-Nav genes. (B) CACNA1, the calcium channel gene family, an example of another ion channel family. No attempt was made to systematically sample teleostean orthologs; therefore, these trees do not represent detailed phylogenies of teleostean genes. Teleost orthologs were only used to establish the number of duplications that occurred in tetrapods following the teleost–tetrapod divergence. Asterisks, bootstrap values of 100.
We conclude that the genes flanking the Nav channel genes either did not duplicate or, if they did so, were not retained. This supports the hypothesis that the Nav channel gene duplications were retained as a result of selection.
Other Ion Channel Gene Families Did Not Duplicate or Were Not Retained
Voltage-dependent ion channels are fundamental to the electrical activity of the brain. We next asked whether there was a general expansion of other six transmembrane (6TM) voltage-dependent ion channel gene families during tetrapod evolution or whether this expansion was specific to the Nav channel gene family. We addressed this question using both published literature (Saito and Shingai 2006; Hoegg and Meyer 2007; Jackson et al. 2007) and gene trees that we constructed with sequences from teleost, human, frog, and chicken databases. The channel that we investigated included the major depolarizing (Ca2+, transient receptor potential [TRP], and hyperpolarization-activated cyclic nucleotide-gated ion channel [HCN]) and hyperpolarizing (Kv, ether-a-go-go related [ERG], and slo) channels. We sampled all 17 members of the voltage-dependent K+ channel (Kv), 4 members of the HCN), 7 members of the canonical TRP, 6 members of the TRP, vanilloid sensitive (TRPV), 4 members of the TRP, melastatin (TRPM), 1 member of the TRP ankyrin repeat, 8 members of the ERG, 3 members of the large-conductance calcium-activated K+ family (slo), and 10 members of the voltage-dependent calcium (CACNA1 or Cav) gene families (fig. 4B).
Most (54 out of 60) of the ion channel genes in our sample showed no duplications following the teleost–tetrapod divergence. There is a mammalian-specific duplication in the slo gene family (slo3). A number of TRP channels do not have teleost orthologs (TRPV1/TRPV2, TRPV3, TRPV5/TRPV6, TRPM6, and TRPM8). However, the absence of TRPM8 and TRPV3 in teleosts is likely due to losses in teleosts rather than duplications in amniotes (Saito and Shingai 2006). TRPV5/TRPV6 are a pair that clearly duplicated in amniotes (Saito and Shingai 2006). The timing of the TRPV1/TRPV2 duplication is not resolvable (Saito and Shingai 2006), but it may represent a tetrapod-specific duplication. Even assuming that all apparent duplications in tetrapods are real rather than reflecting losses of teleost genes, Nav channel genes duplicated (2 out of 4: again, this analysis ignores repeated duplications of each ancestral paralog) significantly more than other ion channel genes (6 out of 60) (P < 0.007, two-tailed chi-square).
Discussion
Relationship to Previous Nav Channel Phylogenies
Based on chromosomal location and phylogeny of the Nav channel family in mammals, Plummer and Meisler (1999) proposed that the ten mammalian Nav channels resulted from a single ancestral chordate Nav channel gene that underwent two rounds of genome duplication early in vertebrate evolution. These duplications ended in a single Nav channel gene on each of four chromosomes followed by a series of tandem duplications on two of those chromosomes. Inclusion of teleosten Nav channel sequences in subsequent analyses supported this idea and further demonstrated that the initial two rounds of duplications preceded the teleost–tetrapod divergence, teleosts have eight Nav channel genes likely as the result of a WGD, and the duplications in teleosts and tetrapods occurred independently (Lopreato et al. 2001; Novak et al. 2006). Novak et al. (2006), which also included chicken sequences, did not attempt to resolve the timing of the tandem duplications within tetrapods. The trees from that study would suggest that many of the Nav channel genes from chicken and mammals duplicated independently. However, the sequences then available from the chicken were likely misassembled (and some have been discontinued by NCBI), thereby confusing the relationship of mammalian and avian genes. In the current study, we addressed this issue by determining the chicken sequences manually from a newer Ensemble release of the chicken genome (version 2.1). Furthermore, we added sequence from key taxa such as amphibians, lizard, and monotreme and marsupial mammals as well as including sequences cloned in our laboratory from lungfish and elasmobranchs. A strong conclusion from the current study is that, with the exception of a single mammalian-specific duplication, all mammalian Nav channel genes were present in an early amniote ancestor (fig. 5).
In addition, the current phylogeny supports our previous conclusion that four Nav channel genes existed in the common ancestor of teleosts and tetrapods. A novel result in the current study is that we identified orthologs of these four genes in lungfish and elasmobranchs. Orthologs to 3 of these 4 were easily cloned from lungfish and elasmobranch tissues. Despite numerous attempts, we were unable to amplify the fourth gene, the ancestor of the SCNA1, SCN2A, etc. complex (which are mainly neural-expressing genes), from lungfish brain, spinal cord, skate brain, or lungfish genomic DNA. However, we identified a likely ortholog in the elephant shark genome. We suggest that this gene is not expressed, or at least not at high levels, in the central nervous system of elasmobranchs and lungfish.
Nav Channel Gene Expression and Tetrapod Evolution
The Nav channel gene family expansion occurred in two chromosomal regions. It was accompanied neither by duplication and/or retention of flanking genes nor by widespread expansion of other ion channel genes; it was specific and presumably advantageous. The Nav channel gene tandem duplications were concurrent with the origin of tetrapods and their invasion of the terrestrial habitat. It largely preceded the diversification of amniotes into synapsid (mammal) and diapsid (reptiles/birds) lineages (Hedges 2009; Shedlock and Edwards 2009).
As tetrapods took to land, they evolved new modes of locomotion, coped with loss of buoyancy, confronted a novel sensory environment, and exploited new food resources (Glenner et al. 2006; Clack 2007). Meeting these challenges was facilitated by the evolution of new sensory receptors in their skin and muscles. For example, early tetrapods evolved muscle spindles (Maeda et al. 1983; Ross et al. 2007), and different lineages of tetrapods later evolved other kinds of somatosensory receptors (e.g., lamellated Pacinian/Herbst corpuscles in amniotes, dome pressure receptors in crocodilians, etc.) (Hunt 1961; Proske 1969; Dorward and Macintyre 1971; von Düring and Miller 1979; Soares 2002). This was accompanied by greater anatomical and physiological complexity in the dorsal root ganglion system (Sneddon 2002; Sneddon et al. 2003). Little is known about the expression of Nav channel genes in the dorsal root ganglia of nonmammalian tetrapods. In mammals, a number of different Nav channels are expressed in dorsal root ganglion neurons and axons—Nav1.6, Nav1.3, Nav1.8, Nav1.9, and Nav1.7—and these contribute to the systematic variation in conduction velocity and other biophysical properties of different classes of dorsal root ganglion neurons (Herzog et al. 2003). Of these channels, all but Nav1.6 derive from the tetrapod-specific duplications. Thus, the duplications of Nav channel genes enabled the diversity of Nav channel types in dorsal root ganglion cells.
Besides the obvious advantages of possessing a repertoire of Nav channels that can be called upon for performing various coding “jobs,” matching Nav channel biophysical properties with signaling requirements also results in metabolic efficiency (Hasenstaub et al. 2010; Schmidt-Hieber and Bischofberger 2010). A more expansive repertoire of Nav channel genes might also have been selected on the basis of energy savings.
The Nav channel gene duplications also occurred at a time when the amniote brain, especially the forebrain, was robustly expanding and adding new anatomical regions (Northcutt 2002). We do not believe that the increase in number of Nav channel genes was causal to the increase in forebrain complexity. Rather, these likely happened in parallel, both as manifestations of increasing brain complexity. Also, because we have only examined 6TM ion channel gene families, we cannot comment on the possible role of other brain-expressing genes, such as neurotransmitters and their receptors, etc. in the evolution of the amniote brain.
Parallel Expansion of Nav Channel Gene Family in Teleosts
Global cataclysmic events in the late Devonian initiated a period of mass extinction for most vertebrate lineages (Clack 2007; Sallan and Coates 2010). The main survivors of this event were the tetrapods (the other sarcopterygian taxa mostly went extinct and are represented today solely by lungfishes and coelacanths), on land, and the actinopterygii (and chondrichthyes), in water. Similar to the evolutionary success of tetrapods, one group of actinopterygii, the teleosts, eventually came to dominate the marine and aquatic habitats.
Interestingly, the Nav channel gene family independently expanded in teleosts to almost the same number of genes (eight Nav channel genes) as in amniotes (Lopreato et al. 2001; Novak et al. 2006). In contrast to tetrapods, this duplication arose suddenly as a result of a third teleost-specific WGD (Meyer and Schartl 1999; Hurley et al. 2007) and no tandem duplications occurred over the next ∼250–300 My. As in tetrapods, all the Nav channel gene duplicates have been retained. It will be interesting to reconstruct the detailed histories of other ion channel gene families in teleosts and determine if these are all retained as the Nav channel genes have been or whether there has been greater loss of other ion channel genes back to a “baseline” pre-WGD number. In other words, is there a relative increase in Nav over other ion channel genes in teleosts as in tetrapods? In a further parallel to the amniotes, the teleostean forebrain also increased in complexity compared with that of nonteleost actinopterygian fishes (e.g., bowfin, gar, and sturgeon) (Northcutt 2002).
Recent Nav Channel Gene Duplicates Are Differentially Expressed
It has been proposed that recently duplicated genes show more restricted expression or greatest sequence divergence than those that duplicated in the distant past (Farré and Alba 2009; Milinkovitch et al. 2010). SCN4A, with no history of duplication since the last WGD, is predominantly expressed in mammalian muscle; in lungfish, its ortholog was only expressed in muscle. SCN8A, also with no history of duplication since the WGD, is expressed in brain (and not heart or muscle) in lungfish and is expressed at uniform levels throughout the mammalian brain (Whitaker et al. 2000, 2001) (no data from reptiles and birds; although SCN8A is also expressed in mammalian heart, Maier et al. 2004). On the other hand, two of the complex of triplicated genes represented by human chromosome 3 and one from the complex localized to human chromosome 2 are expressed in neurons of the peripheral somatosensory system, and some have “unusual” biophysical properties (Akopian et al. 1996; Cummins et al. 1999, 2001; Dib-Hajj et al. 2002).
The complex of genes that show the greatest duplication (human chromosome 2) are mainly expressed in brain and show the greatest variation in regional patterns of expression (telencephalon vs. brain stem) or in subcellular distribution (axons vs. somata) in the mammalian brain (Westenbroek et al. 1989; Akopian et al. 1996; Cummins et al. 1999, 2001; Caldwell et al. 2000; Whitaker et al. 2000, 2001; Dib-Hajj et al. 2002; Jarnot and Corbett 2006; Ogiwara et al. 2007; van Wart et al. 2007; Duflocq et al. 2008; Hu et al. 2009). The final gene duplication that occurred before the origin of therian mammals gave rise to a unique Nav channel with a highly derived sequence that has lost its voltage sensitivity but is still permeable to Na+ ions (Nax) (Watanabe et al. 2006). This channel is more involved in Na+ ion regulation than neural computation.
Conclusion
The Nav channel gene family of tetrapods underwent a series of duplications 300–450 Ma largely during their early evolution (fig. 5). This wave of duplications did not involve the duplication or retention of flanking genes or other ion channel genes. We speculate that the rapid expansion of the Nav channel gene family accommodated greater complexity in neural processing and was a seminal event in the evolution of the amniote brain.
Supplementary Material
Supplementary figures 1 and 2 and tables I and II are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors gratefully acknowledge Marianna Grenadier for artwork, Dr Robert Dores, University of Denver, for supplying tissues, and Dr Hans Hofmann for commenting on the manuscript.
National Science Foundation (IBN 0236147 to H.H.Z. and M.C.J.); National Institutes of Health (R01GM084879 to H.H.Z.).
References
- Akopian A, Sivilotti L, Wood J. A tetrodotoxin-resistant voltage-gated sodium channel expressed by sensory neurons. Nature. 1996;379:257–262. doi: 10.1038/379257a0. [DOI] [PubMed] [Google Scholar]
- Angelino E, Brenner M. Excitability constraints on voltage-gated sodium channels. PLoS Comput Biol. 2007;3:e177. doi: 10.1371/journal.pcbi.0030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomme T, Vandepoele K, De Bodt S, Simillion C, Maere S, Van de Peer Y. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 2006;7:R43. doi: 10.1186/gb-2006-7-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell J, Schaller K, Lasher R, Peles E, Levinson S. Sodium channel Nav1.6 is localized at nodes of Ranvier, dendrites, and synapses. Proc Natl Acad Sci U S A. 2000;97:5616–5620. doi: 10.1073/pnas.090034797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack J. Devonian climate change, breathing, and the origin of the tetrapod stem group. Integr Comp Biol. 2007;47:510–523. doi: 10.1093/icb/icm055. [DOI] [PubMed] [Google Scholar]
- Cummins T, Aglieco F, Renganathan M, Herzog R, Dib-Hajj S, Waxman S. Nav1.3 sodium channels: rapid repriming and slow closed-state inactivation display quantitative differences after expression in a mammalian cell line and in spinal sensory neurons. J Neurosci. 2001;21:5952–5961. doi: 10.1523/JNEUROSCI.21-16-05952.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins T, Dib-Hajj S, Black J, Akopian A, Wood J, Waxman S. A novel persistent tetrodotoxin-resistant sodium current in SNS-null and wild-type small primary sensory neurons. J Neurosci. 1999;19:1–6. doi: 10.1523/JNEUROSCI.19-24-j0001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Boore J. Two rounds of genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib-Hajj S, Black J, Cummins T, Waxman S. NaN/Nav1.9: a sodium channel with unique properties. Trends Neurosci. 2002;25:253–259. doi: 10.1016/s0166-2236(02)02150-1. [DOI] [PubMed] [Google Scholar]
- Dorward P, Macintyre A. Responses from vibration-sensitive receptors in the interosseous region of the duck's hind limb. J Physiol. 1971;219:77–87. doi: 10.1113/jphysiol.1971.sp009650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duflocq A, Le Bras B, Bullier E, Couraud F, Davenne M. Nav1.1 is predominantly expressed in nodes of Ranvier and axon initial segments. Mol Cell Neurosci. 2008;39:180–192. doi: 10.1016/j.mcn.2008.06.008. [DOI] [PubMed] [Google Scholar]
- Farré D, Alba M. Heterogenous patterns of gene-expression diversification in mammalian gene duplicates. Mol Biol Evol. 2009;27:325–335. doi: 10.1093/molbev/msp242. [DOI] [PubMed] [Google Scholar]
- Glenner H, Thomsen P, Hebsgaard M, Sørensen M, Willwerslev E. The origin of insects. Science. 2006;314:1883–1884. doi: 10.1126/science.1129844. [DOI] [PubMed] [Google Scholar]
- Goldin A. Evolution of voltage-gated Na(+) channels. J Exp Biol. 2002;205:575–584. doi: 10.1242/jeb.205.5.575. [DOI] [PubMed] [Google Scholar]
- Hains B, Klein J, Saab C, Craner M, Black J, Waxman S. Upregulation of sodium channel Nav1.3 and functional involvement in neuronal hyperexcitability associated with central neuropathic pain after spinal cord injury. J Neurosci. 2003;23:8881–8892. doi: 10.1523/JNEUROSCI.23-26-08881.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenstaub A, Otte S, Callaway E, Sejnowski TJ. Metbolic cost as a unifying principle governing neuronal biophysics. Proc Natl Acad Sci U S A. 2010;107:12329–12334. doi: 10.1073/pnas.0914886107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges S. Vertebrates (Vertebrata) New York: Oxford University Press; 2009. pp. 309–314. [Google Scholar]
- Hellsten U, Harland R, Gilchrist M, et al. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog R, Cummins T, Ghassemi F, Dib-Hajj S, Waxman S. Distinct repriming and closed-state inactivation kinetics of Nav1.6 and Nav1.7 sodium channels in mouse spinal sensory neurons. J Physiol. 2003;551:741–750. doi: 10.1113/jphysiol.2003.047357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A, Nishino A, Nakajo K, Zhang G, Fineman J, Selzer M, Okamura Y, Cooper E. Ion channel clustering at the axon initial segment and node of Ranvier evolved sequentially in early chordates. PLoS Genet. 2008;4:e1000317. doi: 10.1371/journal.pgen.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegg S, Meyer A. Phylogenomic analyses of KCNA gene clusters in vertebrates: why do gene clusters stay intact? BMC Evol Biol. 2007;7:139. doi: 10.1186/1471-2148-7-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Tian C, Li T, Yang M, Hou H, Shu Y. Distinct contributions of Nav1.6 and Nav1.2 in action potential initiation and backpropagation. Nat Neurosci. 2009;12:996–1002. doi: 10.1038/nn.2359. [DOI] [PubMed] [Google Scholar]
- Hunt C. On the nature of vibration receptors in the hind limb of the cat. J Physiol. 1961;155:175–186. doi: 10.1113/jphysiol.1961.sp006621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley I, Mueller R, Dunn K, et al. A new time-scale for ray-finned fish evolution. Proc R Soc Lond B Biol Sci. 2007;274:489–498. doi: 10.1098/rspb.2006.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson H, Marshall C, Accill E. Evolution and structural diversification of hyperpolarization-activated cyclic nucleotide-gated channel genes. Physiol Genomics. 2007;29:231–245. doi: 10.1152/physiolgenomics.00142.2006. [DOI] [PubMed] [Google Scholar]
- Jarnot M, Corbett A. Immunolocalization of NaV1.2 channel subtypes in rat and cat brain and spinal cord with high affinity antibodies. Brain Res. 2006;1107:1–12. doi: 10.1016/j.brainres.2006.05.090. [DOI] [PubMed] [Google Scholar]
- Kellis M, Birren B, Lander E. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;28:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- Kuraku S. Insights into cyclostome phylogenomics: pre-2R or post-2R. Zool Sci. 2008;25:960–968. doi: 10.2108/zsj.25.960. [DOI] [PubMed] [Google Scholar]
- Lopreato G, Lu Y, Southwell A, Atkinson A, Hillis D, Wilcox T, Zakon H. Evolution and divergence of sodium channel genes in vertebrates. Proc Natl Acad Sci U S A. 2001;98:7588–7592. doi: 10.1073/pnas.131171798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz A, Nusser Z. Molecular identity of dendritic voltage-gated sodium channels. Science. 2010;328:906–909. doi: 10.1126/science.1187958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen O. Mammals (Mammalia) New York: Oxford University Press; 2009. pp. 459–461. [Google Scholar]
- Maeda N, Miyoshi S, Toh H. First observation of a muscle spindle in fish. Nature. 1983;302:61–62. doi: 10.1038/302061a0. [DOI] [PubMed] [Google Scholar]
- Maier SKG, Westenbroek RE, McCormick KA, Curtis R, Scheuer T, Catterall WA. Distinct subcellular localization of different sodium channel α and β subunits in single ventricular myocytes from mouse heart. Circulation. 2004;109:1421–1427. doi: 10.1161/01.CIR.0000121421.61896.24. [DOI] [PubMed] [Google Scholar]
- Meyer A, Schartl M. Gene and genome duplications in vertebrates: the one-to-four (-to eight in fish) rule and the evolution of novel gene function. Curr Opin Cell Biol. 1999;11:699–704. doi: 10.1016/s0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- Milinkovitch M, Helaers R, Tzika A. Historical constraints on vertebrate genome evolution. Genome Biol Evol. 2010;2:13–18. doi: 10.1093/gbe/evp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcutt R. Understanding brain evolution. Integr Comp Biol. 2002;42:743–756. doi: 10.1093/icb/42.4.743. [DOI] [PubMed] [Google Scholar]
- Novak A, Jost M, Lu Y, Taylor A, Zakon H, Ribera A. Gene duplications and evolution of vertebrate voltage-gated sodium channels. J Mol Evol. 2006;63:208–221. doi: 10.1007/s00239-005-0287-9. [DOI] [PubMed] [Google Scholar]
- Ogiwara I, Miyamoto H, Morita N, et al. Nav1.1 localizes to axons of parvalbumin-positive inhibitory interneurons: a circuit basis for epileptic seizures in mice carrying an Scn1a gene mutation. J Neurosci. 2007;27:5903–5914. doi: 10.1523/JNEUROSCI.5270-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura Y, Ono F, Okagaki R, Chong J, Mandel G. Neural expression of a sodium channel gene requires cell-specific interactions. Neuron. 1994;13:937–948. doi: 10.1016/0896-6273(94)90259-3. [DOI] [PubMed] [Google Scholar]
- Panopoulou G, Poustka A. Timing and mechanisms of ancient vertebrate genome duplications—the adventure of a hypothesis. Trends Genet. 2005;121:559–567. doi: 10.1016/j.tig.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Piontkivska H, Hughes A. Evolution of vertebrate voltage-gated ion channel alpha chains by sequential gene duplication. J Mol Evol. 2003;56:277–285. doi: 10.1007/s00239-002-2399-9. [DOI] [PubMed] [Google Scholar]
- Plummer N, Meisler M. Evolution and diversity of mammalian sodium channel genes. Genomics. 1999;57:323–331. doi: 10.1006/geno.1998.5735. [DOI] [PubMed] [Google Scholar]
- Proske U. Vibration sensitive mechanoreceptors in snake skin. Exp Neurol. 1969;23:187–194. doi: 10.1016/0014-4886(69)90055-7. [DOI] [PubMed] [Google Scholar]
- Ross C, Eckhardt A, Herrel A, Hylander W, Metzger K, Shaerlaeken V, Washington R, Williams S. Modulation of intra-oral processing in mammals and lepidosaurs. Integr Comp Biol. 2007;47:118–136. doi: 10.1093/icb/icm044. [DOI] [PubMed] [Google Scholar]
- Saito S, Shingai R. Evolution of thermo TRP ion channel homologs in vertebrates. Physiol Genomics. 2006;27:219–230. doi: 10.1152/physiolgenomics.00322.2005. [DOI] [PubMed] [Google Scholar]
- Sallan L, Coates M. End-Devonian extinction and a bottleneck in the early evolution of modern jawed vertebrates. Proc Natl Acad Sci U S A. 2010;107:10131–10135. doi: 10.1073/pnas.0914000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Hieber C, Bischofberger J. Fast sodium channel gating supports localized and efficient axonal action potential initiation. J Neurosci. 2010;30:10233–10242. doi: 10.1523/JNEUROSCI.6335-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedlock A, Edwards S. Amniotes (Amniota) New York: Oxford University Press; 2009. pp. 375–379. [Google Scholar]
- Sneddon L. Anatomical and electrophysiological analysis of the trigeminal nerve in a teleost fish, Oncorhynchus mykiss. Neurosci Lett. 2002;319:167–171. doi: 10.1016/s0304-3940(01)02584-8. [DOI] [PubMed] [Google Scholar]
- Sneddon L, Braithwaite V, Gentle M. Do fishes have nociceptors? Evidence for the evolution of a vertebrate sensory system. Proc R Soc Lond B Biol Sci. 2003;270:1115–1121. doi: 10.1098/rspb.2003.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares D. Neurology: an ancient sensory organ in crocodilians. Nature. 2002;417:241–242. doi: 10.1038/417241a. [DOI] [PubMed] [Google Scholar]
- van Wart A, Trimmer J, Matthews G. Polarized distribution of ion channels within microdomains of the axon initial segment. J Comp Neurol. 2007;500:339–352. doi: 10.1002/cne.21173. [DOI] [PubMed] [Google Scholar]
- von Düring M, Miller M. Sensory nerve endings in the skin and deep structures. In: Gans C, Northcutt R, Ulinski P, editors. Biology of the Reptilia. New York: Academic Press; 1979. pp. 407–442. [Google Scholar]
- Watanabe E, Hiyama T, Shimizu H, Kodama R, Hayashi N, Miyata S, Yanagawa Y, Obara K, Noda M. Sodium-level-sensitive sodium channel Na(x) is expressed in glial laminate processes in the sensory circumventricular organs. Am J Physiol Regul Integr Comp Physiol. 2006;290:R568–R576. doi: 10.1152/ajpregu.00618.2005. [DOI] [PubMed] [Google Scholar]
- Westenbroek R, Merrick D, Catterall W. Differential subcellular localization of the RI and RII Na+ channel subtypes in central neurons. Neuron. 1989;3:695–704. doi: 10.1016/0896-6273(89)90238-9. [DOI] [PubMed] [Google Scholar]
- Whitaker W, Clare J, Powell A, Chen Y-H, Faull R, Emson P. Distribution of voltage-gated sodium channel alpha-subunit and beta-subunit mRNAs in human hippocampal formation, cortex, and cerebellum. J Comp Neurol. 2000;422:123–139. doi: 10.1002/(sici)1096-9861(20000619)422:1<123::aid-cne8>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Whitaker WR, Faull RL, Waldvogel HJ, Plumpton CJ, Emson PC, Clare JJ. Comparative distribution of voltage-gated sodium channel proteins in human brain. Brain Res Mol Brain Res. 2001;88:37–53. doi: 10.1016/s0169-328x(00)00289-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.