Abstract
The Solute Carriers (SLCs) are membrane proteins that regulate transport of many types of substances over the cell membrane. The SLCs are found in at least 46 gene families in the human genome. Here, we performed the first evolutionary analysis of the entire SLC family based on whole genome sequences. We systematically mined and analyzed the genomes of 17 species to identify SLC genes. In all, we identified 4,813 SLC sequences in these genomes, and we delineated the evolutionary history of each of the subgroups. Moreover, we also identified ten new human sequences not previously classified as SLCs, which most likely belong to the SLC family. We found that 43 of the 46 SLC families found in Homo sapiens were also found in Caenorhabditis elegans, whereas 42 of them were also found in insects. Mammals have a higher number of SLC genes in most families, perhaps reflecting important roles for these in central nervous system functions. This study provides a systematic analysis of the evolutionary history of the SLC families in Eukaryotes showing that the SLC superfamily is ancient with multiple branches that were present before early divergence of Bilateria. The results provide foundation for overall classification of SLC genes and are valuable for annotation and prediction of substrates for the many SLCs that have not been tested in experimental transport assays.
Keywords: Pfam, SLC, solute carrier, transporter
Introduction
Genes coding for membrane proteins constitute approximately 27% of all human genes (Lander et al. 2001; Almen et al. 2009). The largest family of phylogenetically related membrane proteins are the G protein–coupled receptors with about 800 genes in the human genome coding for proteins from this family (Fredriksson et al. 2003). The solute carriers (SLCs) are the second largest family of membrane proteins with at least 384 proteins in human (Fredriksson, Hagglunde, et al. 2008). The SLCs mediate the flow of various substances such as sugars, amino acids, nucleotides, inorganic ions, and drugs over the cell membrane. The SLC families include genes that encode passive transporters, ion transporters, and exchangers. The different SLC families are functionally related in that they all, with a few exceptions, rely on an ion gradient over the cell membrane as the driving force for transportation. The SLC family does not, however, include the primary active transporters, such as ABC transporters, nor ion channels or aquaporins. There are currently 46 distinct families of SLCs recognized in humans (Fredriksson, Nordstrom, et al. 2008). Some of these are known to be phylogenetically linked into larger clusters such as SLC32, SLC36, and SLC38 (Sundberg et al. 2008). The HUGO Gene Nomenclature Committee (HGNC) (http://www.genenames.org) provides a list of transporter families of the SLC gene series. In this, a transporter is assigned to a given SLC family if it has at least 20–25% amino acid sequence identity to another member of the family (Hediger et al. 2004) but not by strict phylogenetic criteria.
One method of classifying the entire proteome of an organism is provided by the Protein family (Pfam) database (http://pfam.sanger.ac.uk). Pfam is a comprehensive collection of protein domains and protein families based on the concept of clans. A clan is a compilation of Pfam entries that are judged likely to be homologous. Clans are built manually based on a wide array of information including literature, known structures, and different databases. The Pfam-domain thresholds are curated in a conservative manner to ensure high specificity at the expense of some sensitivity. In the Pfam release version 22.0, there are 283 clans, containing 1,808 of the 9,318 Pfam families. This shows that many families in the Pfam classification are related, although many of the families have yet to be assigned to clans. The clan classification is expected to grow further because many automatically detected relationships still need to be manually verified (Finn et al. 2007). The SLC families populate three clans. The major facilitator superfamily (MFS) clan holds SLCs and other groups of membrane proteins. The MFS clan is one of largest clans of membrane transporters found in humans (Venter et al. 2001). The clan has diverse functions and it includes members that can function by solute uniport, solute/cation symport, solute/cation antiport, and/or solute/solute antiport with inwardly and/or outwardly directed polarity and are known to transport a large variety of different substrates (Pao et al. 1998). The second clan containing SLCs is the amino acid-–polyamine-–organocation (APC) superfamily that is known to primarily transport amino acids. Jack et al. (2000) suggested that the relatively high degree of sequence similarity among the proteins within the APC superfamily could not have arisen either by chance or by evolutionary sequence convergence and that the APC proteins are most likely homologous and thus constitute a single superfamily. Moreover, the monovalent cation:proton antiporter (CPA)/anion transporter (AT) clan contains transporter proteins from the SLC family. These belong to the CPA superfamily and the AT superfamily. Although their presence is established, the overall evolutionary history of these clans and their relationship with other SLC families has not been investigated in detail. Genomic sequence information from a large number of species is currently available, although the quality of the genomes varies greatly. The availability of genome information from different evolutionary lineages provides a unique opportunity for evolutionary studies of large gene families like the SLCs.
In this paper, we systematically mined the genomes of 17 species aiming to identify most of the SLC genes in these genomes that have resemblance to the human SLCs. The results were analyzed using Hmmpfam of the HMMER 2.3.2 package. This provided the first systematic analysis of the evolutionary history of the human SLC families in Eukaryotes. In all, we identified 4,813 sequences and we delineated the evolutionary history of each of the subgroups.
Materials and Methods
Description of the Original Data Sets
The “pep all” and “ab initio” data sets for Homo sapiens (Lander et al. 2001; Venter et al. 2001), Mus musculus (Waterston et al. 2002), Drosophila melanogaster (Adams et al. 2000), Anophelessgambiae (Holt et al. 2002), Caenorhabditiselegans (1998; Gupta and Sternberg 2003), and Saccharomycescerevisiae (Goffeau et al. 1996) were downloaded from the ensemble ftp site (ftp://ftp.ensembl.org/) (Flicek et al. 2007). The best (filtered) models for Laccaria bicolor (Martin et al. 2008), Monosiga brevicollis (King et al. 2008), Ostreococcus tauri (Palenik et al. 2007), Trichoplax adhaerens (Srivastava et al. 2008), and Nematostella vectensis (Putnam et al. 2007) were downloaded from the Joint Genome Institute (http://www.jgi.doe.gov/). In addition, the protein data sets for Schizosaccharomyces pombe (Wood et al. 2002) and Arabidopsis thaliana (2000) were downloaded from Sanger (ftp://ftp.sanger.ac.uk/) and the Arabidopsis Information Resource ftp site (ftp://ftp.arabidopsis.org/), respectively. Protein data set for Cyanidioschyzon merolae (Nozaki et al. 2007) was downloaded from http://merolae.biol.s.u-tokyo.ac.jp/, for Dictyostelium discoideum (Eichinger et al. 2005) from http://www.sanger.ac.uk/Projects/D_discoideum/, for Paramecium tetraurelia (Arnaiz et al. 2007) from http://paramecium.cgm.cnrs-gif.fr/, and for Trypanozoma cruzi (El-Sayed et al. 2005) from http://tritrypdb.org/tritrypdb/. Unmasked genomic DNA sequences were downloaded from the same resources as the protein data sets.
Construction of Hidden Markov Models
The sequences for all known human SLC proteins were downloaded from the NCBI (http://www.ncbi.nlm.nih.gov/) database, using the Entrez data retrieval tool based on the accession numbers obtained from the SLC database (http://www.bioparadigms.org/slc/menu.asp). This resulted in a data set with 321 sequences, grouped into 46 families. The sequences from each family were aligned using ClustalW 1.83 (Thompson et al. 1994) using default settings. From the alignments, sequence hidden Markov models (HMMs) were constructed using the HMMER 2.3.2 package (Eddy 1998). The models were constructed using HMMbuild with default settings and calibrated using HMMcalibrate.
Constructing a Reference Database
A reference database was constructed from the RefSeq data set version 35 (May 2005 release) (ftp://ftp.ncbi.nih.gov/refseq/H_sapiens/) by identifying all SLC proteins in the RefSeq database with BlastP (The Standalone BLAST 2.2.14 package) searches using the 384 SLC sequences previously presented (Fredriksson, Nordstrom, et al. 2008). The FASTA names of all identified SLCs were modified with a tag (_SLC_) to allow for reliable and easy identification. This modified RefSeq data file contained 29,061 protein sequences.
Identification of SLC Sequences
SLCs were identified in the protein data sets from the 17 species included in the study using HMMsearch from HMMER 2.3.2 (Eddy 1998) with a cutoff at E = 10. All hits were searched against our reference data set to remove non-SLC sequences and for each species, the top five hits were manually inspected and had to match either criteria A or B below or otherwise they were excluded from further analysis. Criteria A: a minimum of four of five top hits had to be an SLC for inclusion in the particular data set. Criteria B: If an SLC family had fewer than five members, it was enough that all the family members were listed among the top five hits. All protein sequences obtained from the previous step were aligned to the corresponding genome sequence using BLAT 32x1 (Kent 2002), and all protein sequences with their best alignment at the same position in the genome were considered duplicates and only the longest protein sequence was kept.
Subdivision of SLCs
We searched the remaining proteins in each of our 17 analyzed species against our reference data set with BlastP with an E value of 1e–3. For each protein, the top five hits were manually inspected, and a minimum of four of these had to be from a given SLC family (or if a family had fewer than four members, all members had to be represented among the top five hits, see criteria A or B above) to be classified as a member of that family. Proteins with a significant hit in the HMM searches (see above, “Identification of SLC Sequences”), but not matching these criteria were called unclassified SLCs. Proteins that only had hits above the 1e–3 threshold were excluded from further analysis. The unclassified group was further analyzed with blastclust (The Standalone BLAST 2.2.14 package) to identify novel groups. All sequences can be found in supplementary data 1, Supplementary Material online. This is a FASTA file where each sequence name is preceded by a tag indicating the species for that sequence with hs for H. sapiens, mm for M musculus, and the equivalent for all other species.
Validation of our Predicted SLC Gene Analysis Model
We compared the number of SLC sequences from different SLC families from our previous H. sapiens data (Fredriksson, Nordstrom, et al. 2008) and the HGNC online database (http://www.genenames.org) with the results of our analysis model. The results of our M. musculus genome analysis were compared with the MGI database (Mouse Genome Informatics, http://www.informatics.jax.org/). The results of the comparisons can be found in supplementary table 1, Supplementary Material online.
Pfam Analysis of SLC
The Pfam database is a comprehensive collection of protein domains and families. We downloaded the Pfam MySQL database (ftp://ftp.sanger.ac.uk/pub/databases/Pfam/database_files/) and built it locally using default values. The SLC proteins in our reference database were searched against the database with Hmmpfam of the HMMER 2.3.2 package using a script (http://www.sanger.ac.uk/Users/sgj/code/pfam/scripts/search/pfam_scan.pl) with default values, with the exception of not using the less- sensitive BLAST preprocessing option. The result was analyzed using the Pfam-A family, which is a family of curated set of families based on profile HMMs and clans (Finn et al. 2007). More information on clans and Pfam-A were obtained from http://pfam.sanger.ac.uk/.
Cluster Analysis
All unclassified proteins were collected and processed with blastclust of the BLAST package with default settings. Additional analyses with blastclust of the unclassified proteins together with the NCBI RefSeq and our own SLC database were also performed.
Results
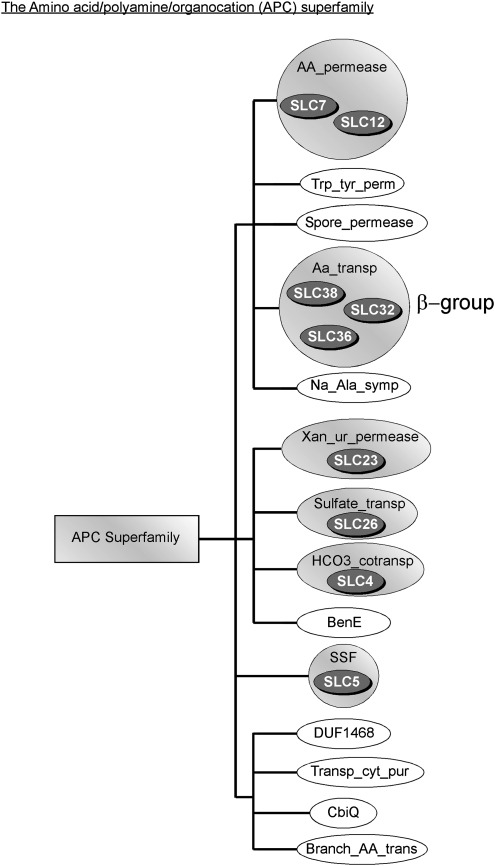
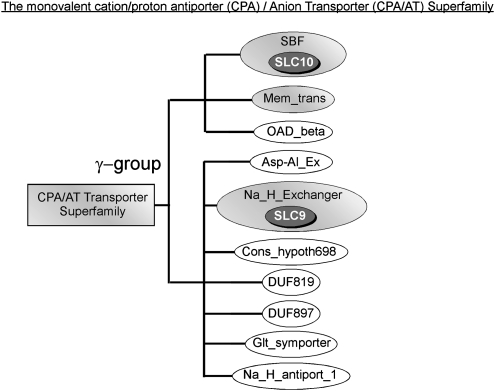
The SLC proteins in our reference database were searched with Hmmpfam of the HMMER 2.3.2 package and the resulting sequences can be found in supplementary data 1, Supplementary Material online. The Pfam analysis of our reference protein data set reveals that 26 of the 46 human SLC families belong to eight different Pfam clans, where a Pfam clan is a collection of related Pfam families (Finn et al. 2006, 2007). The other 20 SLC families belong to Pfam families that do not belong to any clan. Among these, there is in general only one SLC family associated to each Pfam family. The only exception is that the SLC8 and SLC24 families both belong to the Pfam Sodium/calcium exchanger protein (Na_Ca_Ex) family (table 1). Three clans, the APC superfamily, the CPA/AT superfamily, and the MFS, contained more than one established SLC family (see figs. 2–4). In the figures, the rectangles indicate the clans and the ellipses indicate the Pfam families. Shaded ellipses indicate Pfam families with proteins present in human and white ellipses families without human members. Presence of SLCs within a Pfam family is indicated with a dark gray ellipse.
Table 1.
Data Table Generated by Searching the Proteins in our Human Reference Database with Hmmpfam of the HMMER 2.3.2 Package. The Columns of the Table List the Pfam Clan, the Pfam Accession Number, and the SLC Families Belonging to Each clan.
| Pfam Clan of Family | Pfam Clan Accession Number | Pfam Family Accession Number | SLC Families |
| APC superfamily | CL0062 | SLC4, 5, 7, 12, 23, 26, 32, 36, 38 | |
| CPA/AT transporter superfamily; Ion channel (VIC) superfamily | CL0064; CL0030 | SLC9, 10 | |
| Drug/metabolite transporter superfamily; EF-hand like superfamily | CL0184; CL0220 | SLC35 | |
| Ion transporter superfamily | CL0182 | SLC13 | |
| MFS | CL0015 | SLC2, 15, 16, 17, 18, 19, 21, 22, 29, 33, 37, 43, 45, 46 | |
| Tim barrel glycosyl hydrolase superfamily | CL0058 | SLC3 | |
| SDF superfamily | No clan | PF00375 | SLC1 |
| SNF superfamily | No clan | PF00209 | SLC6 |
| Na_Ca_Ex superfamily | No clan | PF01699 | SLC8, SLC24 |
| Nramp superfamily | No clan | PF01566 | SLC11 |
| UT superfamily | No clan | PF03253 | SLC14 |
| PHO4 superfamily | No clan | PF01384 | SLC20 |
| Mito_carr superfamily | No clan | PF00153 | SLC25 |
| LuxE superfamily | No clan | PF04443 | SLC27 |
| Nucleos_tra2_C superfamily | No clan | PF07662 | SLC28 |
| Cation_efflux superfamily | No clan | PF01545 | SLC30 |
| Ctr superfamily | No clan | PF04145 | SLC31 |
| Na_Pi_cotrans superfamily | No clan | PF02690 | SLC34 |
| Zip superfamily | No clan | PF02535 | SLC39 |
| FPN1 superfamily | No clan | PF06963 | SLC40 |
| MgtE superfamily | No clan | PF01769 | SLC41 |
| Ammonium_transp superfamily | No clan | PF00909 | SLC42 |
| DUF580 superfamily | No clan | PF04515 | SLC44 |
FIG. 2.
Presentation of the SLC representation in the APC superfamily. The squared boxes indicate the clans, the ellipses indicate a Pfam Family, and the gray ellipses indicate the presence of a certain SLC family. The amino acid–polyamine–organoCation (APC) superfamily with 14 Pfam families; HCO3-transporter (HCO3_cotransp) family, sodium:solute symporter (SSF) family, amino acid permease (AA_Permease) family, permease (Xan_ur_permease) family, the sulfate transporter (Sulfate_transp) family, the transmembrane amino acid transporter protein (Aa_transp) family, benzoate membrane transport protein (BenE) family, branched-chain amino acid transport protein (Branch_AA_trans) family, cobalt transport protein (CbiQ) family, domains of unknown function 1468 (DUF1468) family, sodium:alanine symporter (Na_Ala_symp) family, spore germination protein (Spore_permease) family, permease for cytosine/purines, uracil, thiamine, allantoin (Transp_cyt_pur) family, and Tryptophan/tyrosine permease (Trp_tyr_perm) family.
FIG. 3.
Presentation of the SLC representation in the CPA/AT superfamily. The squared boxes indicate the clans, the ellipses indicate a Pfam Family, and the gray ellipses indicate the presence of a certain SLC family. The CPA/AT (CPA superfamily and AT superfamily) transporter superfamily with ten Pfam families; sodium/hydrogen exchanger (Na_H_Exchanger) family, the sodium bile acid symporter (SBF) family, predicted permease membrane region (Asp-Al_Ex) family, Cons_hypoth698 (Cons_hypoth698) family, domain of unknown function 819 (DUF819) family, domain of unknown function 897 (DUF897), sodium/glutamate symporter (Glt_symporter) family, membrane transport protein (Mem_trans), Na+/H+ antiporter 1 (Na_H_antiport_1) family, and the Na+-transporting methylmalonyl-CoA/oxaloacetate decarboxylase, beta subunit (OAD_beta) family.
FIG. 4.
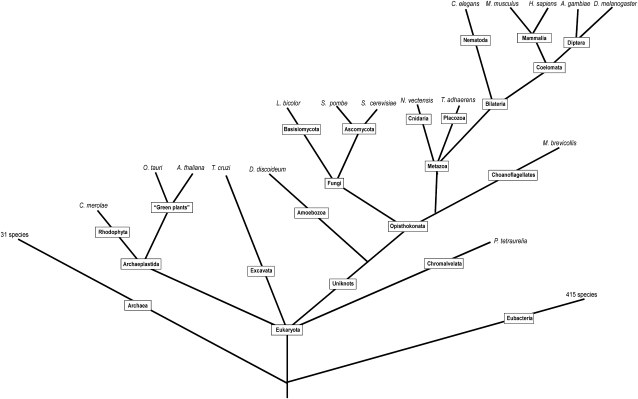
Schematic phylogenetic tree showing the relationship of the species in our analysis. The figure is constructed using the NCBI taxonomy browser (http://www.ncbi.nlm.nih.gov/Taxonomy); data and references from the tree of life Web project (http://tolweb.org/tree/phylogeny.html) and from Hampl et al. (2009). The rectangular-shaped boxes show that the evolutionary lineages and species are denoted at the leaves. Branch lengths are not drawn to represent actual evolutionary distances.
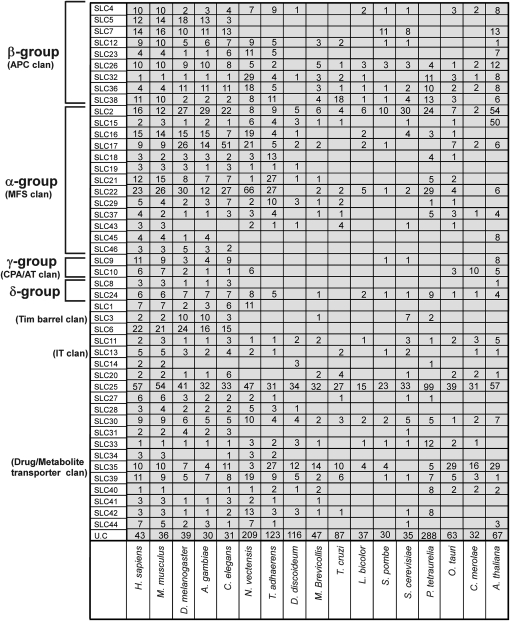
We identified, annotated, and analyzed proteins from 17 eukaryotic species with our HMM, Blast, and BLAT approach, see Materials and Methods for details of species selection, and supplementary fig. 1, Supplementary Material online, for an overview of the methodology. A schematic description of the evolutionary interrelationship between these species can be seen in figure 4. In figure 5, we present the results of this analysis where we show that two of the SLC families (SLC25 and SLC30) were found in all species. Four families were found in all species except one (SLC26, SLC36, SLC35, and SLC39) and six families were found in 15 of the 17 species investigated (SLC32, SLC38, SLC22, SLC24, SLC11, and SLC33). The families found in most species can be considered ancestral and the species that are missing members are likely to have undergone lineage- or species-specific reductions. In figure 5, we also denote how each of these groups are connected to specific SLC families (see Fredriksson, Nordstrom, et al. 2008) and pfam clans (Finn et al. 2006, 2007). We found that representatives from two (α and β) of the four major phylogenetic groups are present in all lineages and were thus most likely present in the early eukaryotic ancestor. Also, members from the δ group, which codes for Na+/Ca2+ exchangers, were present in all species except D. discoideum and T. cruzi. The lack of members from this group in D. discoideum is most likely due to a specific loss in this species as these are present in all Opisthokonta as well as in Archaeplastida and Chromalveolata. The lack of representatives from the δ group in T. cruzi is possibly a result of reductions in T. cruzi because the δ group is found in all the other main lineages. The fact that T. cruzi is a parasite could explain why these, and possibly also many other genes found in other eukaryotes, lack orthologous genes in this species. This also compliments the notion that T. cruzi has a reduced genome, first estimated to have about 12,500 genes (El-Sayed et al. 2005), with current assembly predictions around 10,500 genes.
FIG. 5.
Tabulated representation of the number of SLCs in each of the 46 families for 17 species; Homo sapiens, Mus musculus, Drosophila melanogaster, Anopheles gambiae, Caenorhabditis elegans, Nematostella vectensis, Trichoplax adhaerens, Dictyostelium discoideum, Monosiga brevicollis, Trypanozoma cruzi, Laccaria bicolour, Schizosaccharomyces pombe, Saccharomyces cerevisiae, Paramecium tetraurelia, Cyanidioschyzon merolae, Ostreococcus tauri, and Arabidopsis thaliana. The main SLC groups α–δ (Fredriksson et al. 2008b), as well as Pfam (Finn et al. 2007) clans not included in this classification, is noted in the left margin of the figure.
Our HMM models performed well when validated against the HGNC database and against our in-house SLC data set. Our method yielded 400 human genes compared with the HGNC data set (360) and our previous analysis (384, Fredriksson, Nordstrom, et al. 2008), see supplementary table 1, Supplementary Material online, for details. We used both sequence data sets based on experimental data and sequence data sets based solely on computer predictions. The method also found genes not previously identified, which were similar to SLCs. Our initial analysis with HMM searches and the high E value of 10 yields very high sensitivity. Due to the use of different data sets, our initial analysis gives us a high number of false positives, very few false negatives, and many duplicate entries of the same genes. We removed the false positives using a semiautomatic approach with Blast and kept the proteins matching our criteria. Next, we used BLAT, an in-house Java program, and manual inspection to remove all duplicates. This resulted in the identification of ten new human genes and four new mouse genes, not previously annotated as SLCs.
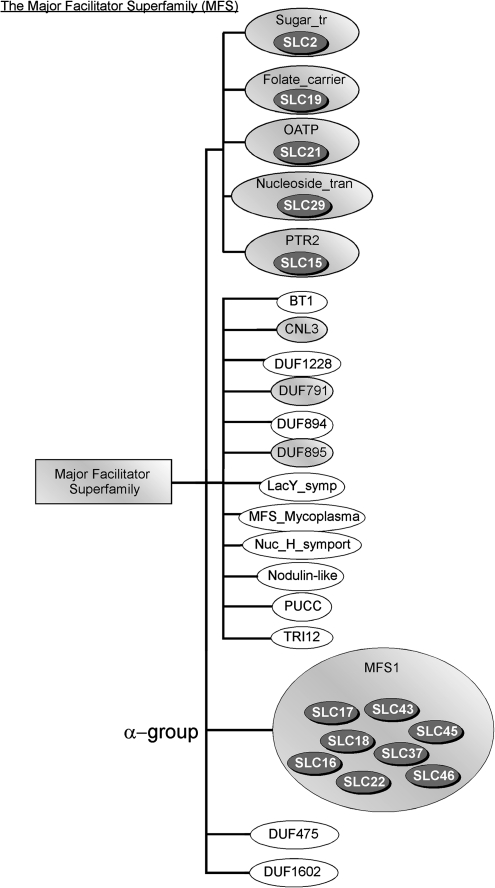
We obtained information on the hierarchical grouping of Pfam families into clans from the Pfam database (http://www.pfam.org/). Furthermore, we added information from our analysis regarding the Pfam family to which each of the SLC families belonged. In figure 1, we present the largest of the Pfam clans containing SLC families, the MFS. A subbranch of MFS, denoted as MFS_1 in the Pfam system, is equivalent to the α family of SLCs (Fredriksson, Nordstrom, et al. 2008). In addition to the SLC α family, MFS contained four main subbranches (as defined in the Pfam database), one of which has human genes annotated as SLCs. Moreover, three other families from one subbranch contained human proteins, although these are currently not annotated as SLCs. The proteins in the MFS clan are the largest cluster of phylogenetically related SLCs so far described. Of the 13 SLC families, SLC2 (facilitated hexose and polyol transporters) and SLC22 (organic cation, zwitterions/cation, and organic AT) are present in all (SLC2) or most (SLC22) eukaryotic genomes investigated. SLC18 (vesicular amine transporter family), SLC19 (folate/thiamine transporter family), SLC21/OATP/SLC0 (organic AT), SLC29 (nucleoside transporter family), and SLC46 (heme transporter) are mainly present in animals, with SLC46 present only in Bilateria. Some of the α members, the SLC15 (proton oligopeptide cotransporter), SLC16 (transport a variety of substrates), SLC17 (organic AT), and SLC37 (sugar-phosphate/phosphate exchanger) are prevalent also outside the Bilaterian phyla. SLC43 (large neutral amino acids transporter) is only found in low numbers in each species but in many eukaryotic lineages.
FIG. 1.
Presentation of the SLC representation in the MFS clan. The squared boxes indicate the clans, the ellipses indicate a Pfam family, and the gray ellipses indicate the presence of a certain SLC family. The MFS with 20 Pfam families; sugar (and other) transporter (Sugar_tr) family, proton-dependent oligopeptide transport), proton-dependent oligopeptide transport (PTR2) family, MFS_1 family, reduced folate carrier (Folate_carrier) family, organic anion transporter polypeptide (OATP) family, nucleoside transporter (Nucleoside_tran), biopterin transport 1 (BT1) family, CLN3 gene (CLN3) family, domain of unknown function 1228 (DUF1228) family, domain of unknown function 1602 (DUF1602) family, domain of unknown function 475 (DUF475) family, domain of unknown function 791 (DUF791) family, domain of unknown function 894 (DUF894) family, domain of unknown function (DUF895) family, LacY proton/sugar symporter (LacY_symp) family, Mycoplasma MFS transporter (MFS_Mycoplasma) family, nodulin-like (nodulin-like) family, nucleoside H+ symporter (Nuc_H_symport) family, PUCC protein (PUCC) family, and the fungal trichothecene efflux pump (TRI12) family.
Within the APC superfamily (see fig. 2), we identified nine known SLC families, making the APC superfamily the second largest cluster of human SLCs. In total, the APC superfamily contains 14 Pfam families grouped into four clusters. The SLC32, SLC36, and SLC38 have previously been annotated as the β family of SLCs (Fredriksson, Nordstrom, et al. 2008) and these constitute one of the clusters in the APC superfamily. Of the 14 Pfam families, 6 contain human SLCs, while the other 8 families did not contain any human members. Of the known human SLCs found in the APC superfamily, SLC4 (sodium bicarbonate and AT family), SLC12 (cation-coupled Cl− cotransport family), SLC23 (sodium-dependent ascorbic acid transporter), SLC26 (anion exchanger family), SLC32 (synaptic uptake and release of inhibitory amino acids), SLC36 (proton-coupled amino acid transporter), and SLC38 (sodium-coupled neutral amino acid transporter) were present in all Bilaterian species investigated here as well as in N. vectensis and T. adhaerens (see fig. 5). These are probably the β family members found in most animals; losses of these in animal species are most likely the result of reductions. SLC7 (the cation and heterodimeric amino acid transporter) and SLC5 (sodium glucose cotransporter family) are mainly found in Bilateria species, although SLC7 is found in A. thaliana as well as in the unicellular yeast species, suggesting that this family could have been present before animals and that it was lost in several species in this study.
The CPA and the AT families constitute the CPA/AT superfamily (fig. 3). This clan contains in total, ten families of which two have proteins annotated as SLCs. The NA_H_Exchanger and SBF family contain proteins from the SLC9 and SLC10 families, respectively. These two families have previously been grouped into the γ family (Fredriksson et al. 2008). Evolutionary, SLC9 (Na+/H+ exchanger) and SLC10 (sodium-dependent bile acid cotransporter) were present in all Bilateria species and in A. thaliana, whereas they were found only in a few other species investigated (fig. 5). In addition, one more of the families in the CPA/AT superfamily, the Mem_trans family, contained human members, although these have not been annotated as SLCs.
The δ family is the smallest cluster of known SLC families, containing proteins from the SLC8 and SLC24 families (Fredriksson, Nordstrom, et al. 2008). In the Pfam nomenclature, they both belong to the Na_Ca_Ex family, which is not a member of any Pfam clan. SLC8 (Na+/Ca2+) existed in Bilateria and Viridiplantae but neither in Archaea nor in Eubacteria, whereas SLC24 (Na+/Ca2+-K+ exchanger) was present in Eubacteria and Archaea as well as in most Eukaryotic species analyzed (fig. 5). The sodium/calcium exchanger family contained the SLC8 and SLC24 family, but no other human families or nonclassified sequences.
Of the SLC families not belonging to any larger phylogenetic clusters (fig. 5), SLC13 (dicarboxylate and sulfate transporter), SLC25 (mitochondrial transporter), SLC11 (H+-coupled metal ion transporters), SLC30 (zinc transporter), SLC35 (nucleoside-sugar transporter), SLC39 (metal- ion transporters) were present in Eubacteria and Archaea as well as most Eukaryotic species we analyzed, where SLC13 seems to have undergone a few more losses than the other families. It is likely therefore that these families were present in the last common ancestor of these three lineages. SLC1 (neutral amino acid transporter), SLC6 (neurotransmitter transporter), SLC27 (fatty acid transporter), and SLC28 (nucleoside transporter family) were present predominantly in Bilateria and SLC41 (Mg2+ transporter) and were found in most animal species. SLC6 has earlier been shown to be present in Bilateria, but not in Viridiplantae or Ascomycota (Hoglund et al. 2005). Also the other SLC families, SLC3 (heteromeric amino acid transporters), SLC40 (ion transporter), SLC42 (Rhesus ammonium transporter), SLC44 (choline transporter), SLC20 (sodium-phosphate cotransporter), SLC14 (urea transporters), SLC31 (copper transporter), and SLC34 (phosphate cotransporter) were found in Bilateria, although one or a few sequences from these families were found in one or a few or several genomes outside Bilateria. These families, which are found in a more scattered pattern in the species in this study, could be a result of misclassification in our method. This could be due to new families that have appeared in specific lineages evolving over time to differ significantly from their parent family. As our classification method is based on the human SLC families and these lack orthologous families in humans, they would be classified to the most similar human SLC family. In addition, these patterns could be the result of multiple local losses or parallel evolutionary events with multiple local gains. Another explanation is that these patterns could be due to horizontal transfers. This explanation is more likely for some species, like T. cruzi, because horizontal transfers are more common in protists, whereas very rare in multicellular organisms. It is likely that some of these families are more recent and their presence in nonanimal species could be due to secondary events. It is also possible that our selection of genomes had some impact on the identification of missing families. This is especially pronounced with lineages represented by only one species, such as the Amoebozoa, which are represented by D. discoideum. Therefore, data regarding missing families in these species should be interpreted with some caution, especially with SLC families that have few members, as one or few independent gene losses would erase an entire family.
The SLCs not placed into any of the 46 known SLC families according to our criteria were placed in a category designated unclassified (fig. 5). Some of the genomes we have investigated (D. discoideum, L. bicolor, M. brevicollis, N. vectensis, O. tauri, P. tetraurelia, T. cruziesmaraldo, and T. adhaerens) are in a relatively early stage of assembly and the gene prediction pipelines are not tailored for these genomes. The gene models from these species can therefore be considered highly preliminary. This could be one underlying factor for a higher relative number of unclassifiable proteins from these genomes. It is known from the progression of the annotation of mammalian genomes that the number of predicted proteins with low similarity to other sequences (i.e., unclassifiable) decreases as the gene prediction pipelines are refined (Collins et al. 2003). It is therefore likely that the number of proteins in these classes will be reduced with time.
We investigated eight of the more mature genomes in our study further regarding the unclassified sequences. We found 43 unclassified SLCs in H. sapiens, 36 in M. musculus, 39 in D. melanogaster, 30 in A. gambiae, 31 in C. elegans, 30 in S. pombe, 35 in S. cerevisiae, and 67 in A. thaliana. Although the unclassified SLCs could not be placed into any of the known SLC families using our criteria, some could still be placed into the Pfam clans. Of these, 59 belong to the APC superfamily, 76 to CPA/AT, 6 to IT (ion transporter) superfamily, 150 to MFS, whereas 73 did not belong to a clan. We analyzed these 364 nonclassified SLCs further using blastclust to identify possible novel groupings and found 2 clusters with three proteins and 18 clusters with two. Of these clusters, 13 consisted of mouse and human proteins, 6 consisted solely of A. thaliana proteins and 1 of solely S. cerevisiae proteins. About 322 unclassified SLCs did not cluster and our hypothesis is that these are rapidly evolving SLCs, as neither orthologous nor paralogous genes could be identified.
Discussion
We provide the first evolutionary study of all 46 known human SLC families from a range of fully sequenced genomes. We investigated 17 eukaryotic species, covering four basal eukaryotic branches (fig. 4), and found that a large number of the SLC families in H. sapiens are highly evolutionary conserved in Bilaterian species (fig. 5). We found that 43 of the 46 (93%) of the human SLC families are also found in C. elegans, whereas 42 of the 46 human families were also found in insects. All SLC families except SLC14, SLC34, SLC40, SLC43, and SLC45 are present in all Bilaterian species investigated and we observe that most families are also present in N. vectensis. In contrast, we see a marked reduction in the family representation in the other species. There are three SLC families that are notable as represented in all eukaryotes in the study, the mitochondrial transporters (SLC25), the nucleotide-sugar transporters (SLC35), and the sugar transporters (SLC2) families. In addition, it appears that all species have members from at least one of the three closely related amino acid transporters from the SLC32, SLC36, and SLC38 families. We have previously shown that these families are closely related and we suggested that they have a common evolutionary origin (Sundberg et al. 2008). It is possible that members of these families, in species distantly related to humans, have proteins representing unique evolutionary branches of a common ancestor, which are classified arbitrarily into SLC32, SLC36, or SLC38. These three families should perhaps be considered one family in evolutionary terms. It is tempting to speculate that all eukaryotes (and possibly the eukaryotic ancestor) have these transport functions, whereas the other SLC functions are more recent amalgamations.
SLCs were originally defined as ATP-independent transporters and this superfamily includes ion-coupled transporters, exchangers, and passive transporters. However, the degree of structural and primary sequence similarity between different SLC families varies greatly. The current practice has been that new sequences are assigned to a SLC family if they have a sequence identity more than 20–25% to any other family of SLCs (Hediger et al. 2004). However, it is important to note that many of the 46 SLC families lack any significant sequence identity to some other SLC families, which means that they cannot be aligned in pairwise alignments. This differentiates the SLC family from other large super families of membrane proteins such as voltage-gated ion channels (Yu and Catterall 2004), G protein–coupled receptors (Fredriksson et al. 2003), and tyrosine kinase receptors (Manning et al. 2002), which have a clearer relationship at the primary sequence level. We have previously shown using phylogenetic analysis based on multiple sequence alignments, that 15 of the SLC families found in humans could be grouped into four main clusters (Fredriksson, Nordstrom, et al. 2008). In the current study, we used sequence HMMs, which in general has a higher sensitivity than multiple alignment-based methods (Madera and Gough 2002). We extend this grouping here, showing that, as many as 26 of the human SLC families belong to the four major groups (see fig. 1–3), termed α, β, γ, and δ groups. The largest of these is the α group. We have previously shown that seven SLC families, namely SLC2, SLC16, SLC17, SLC18, and SLC22 (including the related family SVOP), SLC37, and SLC46 could be phylogenetically placed into the α group (Fredriksson, Hagglund, et al. 2008). Here, we extend this group to also include additional six families, SLC15, SLC19, SLC21, SLC29, SLC43, and SLC45, resulting in a cluster of 13 human SLC families. Pfam has a large supergroup called MFS (Saier et al. 1999; Lemieux 2007; Law et al. 2008). This system includes a high number of groups that have not been classified as SLCs and many of these groups are not found in humans. Here, we have put the recognized SLC families into the perspective of the large Pfam MFS superfamily system of membrane-bound proteins mentioned in the introduction (fig. 1). The MFS system currently contains 20 families as illustrated in figure 1. The α group of SLCs (Fredriksson, Nordstrom, et al. 2008) forms a separate cluster within the MFS system and these genes are all members of the MFS_1 family, although the MFS_1 family also contains other sequences. Within the α group, we see that SLC22, SLC37, and SLC45 are well represented in A. thaliana, suggesting that these families have ancient origin. Interestingly, the α group families have very diverse substrates as the SLC22 transport a wide range of organic cations (including sugar), SLC37 and possibly SLC45 also transports sugars, SLC17 and SLC18 are vesicular neurotransmitter transporters, SLC16 and SLC43 transports amino acids, and SLC46 is a family of heme transporters. Some proteins from these families have been characterized in distant eukaryotes. For example, the ZFS1 transporter from A. thaliana, which we show here to be most similar to the SLC22 family, has been associated with the transportation of organic cations, with a preference for carbohydrates (Haydon and Cobbett 2007). This indicates that the substrate preference for the SLC22 family has been conserved over a long evolutionary time. In general, we see that some individual families of the MFS transporter group have a high degree of similarity, yet as a group, the superfamily contains an enormous diversity of substrates as well as high degree of sequence diversity, which has previously been indicated (Law et al. 2008).
It has previously been shown that the SLC32, SLC36, and SLC38 families are phylogenetically related (Sundberg et al. 2008) and are forming the foundation of the β group. Here, we show that SLC7 and SLC12 also belong to the β cluster (fig. 2). The β group shares functional features as they are all postulated to have 11 TM regions and they have relatively long N termini (Sundberg et al. 2008). The β group belongs to the APC clan. This clan also contains a number of other nonmammalian groups that have not been considered to belong to SLCs. In total, nine human SLC families belong to the APC clan (see fig. 1). The substrates of several APC members have been carefully studied and the indication is that these transporters are mainly amino acid transporters. Some of these transporters have exceptionally broad specificity, whereas others are restricted to just one or a few amino acids or related compounds (Jack et al. 2000). Interestingly, most of the members of the β group are amino acid transporters. However, the other SLC families that belong to the APC clan, but are not members of the β group, are not amino acid transporters as SLC26 transports sulfate ions, whereas SLC4 transports bicarbonates. The SLC families in the APC clan are very well represented in A. thaliana with several members from all the nine families except for the glucose-transporting SLC5 family. As glucose is clearly taken up by A. thaliana cells (Horemans et al. 2008), this function is most likely mediated by proteins from other SLC families.
In our previous work, we have shown that the third main family is the γ group. This groups belongs, according to Pfam, to the CPA/AT clan, which stands for the monovalent CPA superfamily and the AT superfamily. This clan contains ten families, three of these families have human members (Na_H_Exchanger, SBF, and Mem_trans), and of these, Na_H_Exchanger and SBF have known human SLCs families, the SLC9 and SLC10, respectively. Both these γ group SLC families have 12 TM regions, one large fourth extracellular loop and both the N and C termini toward the cellular lumen but their substrates differ as SLC10 transports amino acids, whereas SLC9 transports hydrogen and sodium ions. Moreover, both these families are well represented in vertebrate genomes and in A. thaliana, but we are not aware of reports that specify what they transport in species distantly related to human.
Our present study shows that some sequences do not fulfill our criteria for classification into specific SLC families but clearly belong to the SLC superfamily. We investigated these unclassified sequences from eight genomes more closely. Of these, 43 are found in H. sapiens, 39 are found in D. melanogaster, and 67 are found in A. thaliana. Further analysis using pairwise sequence alignments suggested that 24 of the 43 unclassified human sequences could be classified to specific SLC families. Of the remaining 19 sequences, 5 sequences are likely to belong to the SLC22-related group SV2 (Jacobsson et al. 2007) and 4 are annotated sequences not previously classified as SLCs (spinster homolog 1, spinster homolog 2, and P protein (melanocyte-specific transporter protein) and transmembrane protein 104). The remaining ten are other novel genes not yet annotated.
We also analyzed these unclassified proteins with blastclust to examine similarity to any previously existing SLC or other human RefSeq sequence. The search yielded two clusters with 3 and 18 clusters with two sequences; all other sequences did not cluster with any human RefSeq sequence. Of these clusters, 13 consist only mouse and human sequences, 6 consist solely A. thaliana sequences, and 1 consist only S. cerevisiae sequences. There are 290 proteins in the eight species where we investigated the unclassified sequences that do not cluster with any known human RefSeq sequence or with any of the SLCs in this study. About 30 genes are from the A. gambiae, 70 in A. thaliana, 34 in C. elegans, 41 in D. melanogaster, 13 in H. sapiens, 15 in M. musculus, 47 in S. cerevisiae, and 40 in S. pombe. This shows that there are many proteins similar to SLC that are lineage- or species specific, which seem to evolve rapidly, as there are neither paralogous nor orthologous sequences of these found in our data set. It is also possible that some of these represent families that were either lost on the evolutionary path leading to humans or they have appeared in only some lineages. This is a limitation of the current analysis as some SLC families might not exist in humans but are still present in other eukaryotic species. However, the fact that we are unable to group most of these unclassified sequences into clusters suggests that majority of them are not members of larger families that are lineages specific or families lost in humans. These results suggest that beyond several of the main families of SLCs that have very long evolutionary history, there are also a large number of SLC genes showing a high rate of divergent evolution. However, further analysis is needed to classify and establish relationships among these proteins. There are possibly proteins that could be classified as new SLC families as functional tests come along. An example of one such divergent and more recently established family is the multidrug and toxin-extrusion family, now termed SLC47 (Terada and Inui 2008).
In conclusion, we have performed the first evolutionary analysis of the human SLC families based on whole genome sequences. Using sequence HMMs, we found that 26 of the 46 SLC families belong to four evolutionary clusters, whereas the others have no or low sequence similarity. We also show that three of the SLC families are present in all the eukaryotes we investigated. Ten other families are missing in only one or two species, which is possibly a result of lineage- or species-specific losses. Therefore, we suggest that the ancestral eukaryotic repertoire of the SLC families found in human could be represented by these 13 families. It is also clear that SLCs were present in the common ancestor of Eukaryotes, Eubacteria, and Archaea. Overall, 59% of the human SLC families are found in prokaryotes and 51% in Archaea, suggesting that the SLC family have in general the richest evolutionary history among the main families of membrane-bound proteins. We also identify ten new human sequences not previously classified as SLCs, which most likely belong to the SLC family, and show that there are a high number of unclassified SLC-like sequences in a number of other genomes. The results are valuable for annotation and prediction of substrates for the many SLCs that have not been tested in experimental transport assays.
Supplementary Material
Supplementary data 1, figure S1, and table S1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This study was supported by the Swedish Research Council, the Åhlens Foundation, The Novo Nordisk Foundation, The Göran Gustafsson Foundation, Engkvist foundation, the Magnus Bergvall Foundation and Carl Tryggers foundation.
References
- Arabidopsis Genome Initiative, Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Adams MDSE, Celniker RA, Holt CA, et al. (193 co-authors) The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Almen MS, Nordstrom KJ, Fredriksson R, Schioth HB. Mapping the human membrane proteome: a majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biol. 2009;7:50. doi: 10.1186/1741-7007-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaiz O, Cain S, Cohen J, Sperling L. ParameciumDB: a community resource that integrates the Paramecium tetraurelia genome sequence with genetic data. Nucleic Acids Res. 2007;35:D439–D444. doi: 10.1093/nar/gkl777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caenorhabditis elegans Sequencing Consortium, Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Collins JE, Goward ME, Cole CG, Smink LJ, Huckle EJ, Knowles S, Bye JM, Beare DM, Dunham I. Reevaluating human gene annotation: a second-generation analysis of chromosome 22. Genome Res. 2003;13:27–36. doi: 10.1101/gr.695703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Eichinger L, Pachebat JA, Glockner G, et al. (97 co-authors) The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed NM, Myler PJ, Bartholomeu DC, et al. (82 co-authors) The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, et al. Pfam: clans, web tools and services. Nucl Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, et al. The Pfam protein families database. Nucl Acids Res. 2008;36:D281–D248. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Aken BL, Beal K, et al. (59 co-authors) Ensembl 2008. Nucl Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson R, Hagglund M, Olszewski PK, Stephansson O, Jacobsson JA, Olszewska AM, Levine AS, Lindblom J, Schioth HB. The obesity gene, FTO, is of ancient origin, up-regulated during food deprivation and expressed in neurons of feeding-related nuclei of the brain. Endocrinology. 2008;149:2062–2071. doi: 10.1210/en.2007-1457. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Nordstrom KJ, Stephansson O, Hagglund MG, Schioth HB. The solute carrier (SLC) complement of the human genome: phylogenetic classification reveals four major families. FEBS Lett. 2008;582:3811–3816. doi: 10.1016/j.febslet.2008.10.016. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, et al. (16 co-authors) Life with 6000 genes. Science. 1996;274:546, 563–547. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Gupta BP, Sternberg PW. The draft genome sequence of the nematode Caenorhabditis briggsae, a companion to C. elegans. Genome Biol. 2003;4:238. doi: 10.1186/gb-2003-4-12-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampl V, Hug L, Leigh JW, Dacks JB, Lang BF, Simpson AG, Roger AJ. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc Natl Acad Sci U S A. 2009;106:3859–3864. doi: 10.1073/pnas.0807880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon MJ, Cobbett CS. A novel major facilitator superfamily protein at the tonoplast influences zinc tolerance and accumulation in Arabidopsis. Plant Physiol. 2007;143:1705–1719. doi: 10.1104/pp.106.092015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger MA, Romero MF, Peng JB, Rolfs A, Takanaga H, Bruford EA. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteinsIntroduction. Pflugers Arch. 2004;447:465–468. doi: 10.1007/s00424-003-1192-y. [DOI] [PubMed] [Google Scholar]
- Hoglund PJ, Adzic D, Scicluna SJ, Lindblom J, Fredriksson R. The repertoire of solute carriers of family 6: identification of new human and rodent genes. Biochem Biophys Res Commun. 2005;336:175–189. doi: 10.1016/j.bbrc.2005.08.048. [DOI] [PubMed] [Google Scholar]
- Holt RAGM, Subramanian A, Halpern GG, et al. (123 co-authors) The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Horemans N, Szarka A, De Bock M, Raeymaekers T, Potters G, Levine M, Banhegyi G, Guisez Y. Dehydroascorbate and glucose are taken up into Arabidopsis thaliana cell cultures by two distinct mechanisms. FEBS Lett. 2008;582:2714–2718. doi: 10.1016/j.febslet.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack DL, Paulsen IT, Saier MH. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology. 2000;146(Pt 8):1797–1814. doi: 10.1099/00221287-146-8-1797. [DOI] [PubMed] [Google Scholar]
- Jacobsson JA, Haitina T, Lindblom J, Fredriksson R. Identification of six putative human transporters with structural similarity to the drug transporter SLC22 family. Genomics. 2007;90:595–609. doi: 10.1016/j.ygeno.2007.03.017. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, et al. (36 co-authors) The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ESLM, Linton B, Birren C, et al. (254 co-authors) Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Law CJ, Maloney PC, Wang DN. Ins and outs of major facilitator superfamily antiporters. Annu Rev Microbiol. 2008;62:289–305. doi: 10.1146/annurev.micro.61.080706.093329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux MJ. Eukaryotic major facilitator superfamily transporter modeling based on the prokaryotic GlpT crystal structure. Mol Membr Biol. 2007;24:333–341. doi: 10.1080/09687680701496507. [DOI] [PubMed] [Google Scholar]
- Madera M, Gough J. A comparison of profile hidden Markov model procedures for remote homology detection. Nucleic Acids Res. 2002;30:4321–4328. doi: 10.1093/nar/gkf544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Martin F, Aerts A, Ahren D, et al. (68 co-authors) The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452:88–92. doi: 10.1038/nature06556. [DOI] [PubMed] [Google Scholar]
- Nozaki H, Takano H, Misumi O, et al. (18 co-authors) A 100%-complete sequence reveals unusually simple genomic features in the hot-spring red alga Cyanidioschyzon merolae. BMC Biol. 2007;5:28. doi: 10.1186/1741-7007-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palenik B, Grimwood J, Aerts A, et al. (38 co-authors) The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc Natl Acad Sci U S A. 2007;104:7705–7710. doi: 10.1073/pnas.0611046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao SS, Paulsen IT, Saier MH., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, et al. (19 co-authors) Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Saier MH, Jr, Beatty JT, Goffeau A, et al. (14 co-authors) The major facilitator superfamily. J Mol Microbiol Biotechnol. 1999;1:257–279. [PubMed] [Google Scholar]
- Srivastava M, Begovic E, Chapman J, et al. (21 co-authors) The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- Sundberg BE, Waag E, Jacobsson JA, et al. (11 co-authors) The evolutionary history and tissue mapping of amino acid transporters belonging to solute carrier families SLC32, SLC36, and SLC38. J Mol Neurosci. 2008;35:179–193. doi: 10.1007/s12031-008-9046-x. [DOI] [PubMed] [Google Scholar]
- Terada T, Inui K. Physiological and pharmacokinetic roles of H+/organic cation antiporters (MATE/SLC47A) Biochem Pharmacol. 2008;75:1689–1696. doi: 10.1016/j.bcp.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JCMD, Adams EW, Myers PW, et al. (275 co-authors) The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Waterston RHK, Lindblad-Toh E, Birney J, et al. (220 co-authors) Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wood VR, Gwilliam MA, Rajandream M, et al. (134 co-authors) The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Yu FH, Catterall WA. The VGL-chanome: a protein superfamily specialized for electrical signaling and ionic homeostasis. Sci STKE. 2004;re15:1–17. doi: 10.1126/stke.2532004re15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.