Abstract
Sets of SNARE proteins mediate membrane fusion by assembling into core complexes. Multiple SNAREs are thought to function in different intracellular trafficking steps but it is often unclear which of the SNAREs cooperate in individual fusion reactions. We report that syntaxin 7, syntaxin 8, vti1b and endobrevin/VAMP-8 form a complex that functions in the fusion of late endosomes. Antibodies specific for each protein coprecipitate the complex, inhibit homotypic fusion of late endosomes in vitro and retard delivery of endocytosed epidermal growth factor to lysosomes. The purified proteins form core complexes with biochemical and biophysical properties remarkably similar to the neuronal core complex, although each of the four proteins carries a transmembrane domain and three have independently folded N-terminal domains. Substitution experiments, sequence and structural comparisons revealed that each protein occupies a unique position in the complex, with syntaxin 7 corresponding to syntaxin 1, and vti1b and syntaxin 8 corresponding to the N- and C-terminal domains of SNAP-25, respectively. We conclude that the structure of core complexes and their molecular mechanism in membrane fusion is highly conserved between distant SNAREs.
Keywords: endocytosis/endosomes/membrane fusion/SNARE proteins
Introduction
SNARE proteins are thought to play a key role in intracellular membrane fusion events (Rothman, 1994). They comprise a superfamily (Weimbs et al., 1998) of small and mostly membrane-associated proteins that are differentially distributed among intracellular membranes (for review see Bock and Scheller, 1997; Jahn and Südhof, 1999). Best characterized are the SNAREs functioning in the exocytosis of synaptic vesicles. They include the vesicle protein synaptobrevin 2 (also referred to as VAMP-2), and the plasma membrane proteins syntaxin 1 and SNAP-25. These proteins form a stable ternary complex that can be reversibly disassembled by the chaperone ATPase NSF in conjunction with α-SNAP (Söllner et al., 1993). It is currently thought that assembly of the complex drives the fusion reaction by means of forming a tight connection between the membranes (Hanson et al., 1997).
Syntaxin 1 and synaptobrevin 2 each possess a single transmembrane domain at their C-terminal ends. SNAP-25 has a cysteine-rich stretch in the middle of the molecule that is palmitoylated and probably serves as membrane anchor. Like all other SNAREs, these proteins contain conserved stretches of ∼60 amino acids that are related to each other and are referred to as SNARE motifs (Jahn and Südhof, 1999). Syntaxin 1 and synaptobrevin 2 each possess a single SNARE motif directly adjacent to the transmembrane domain. SNAP-25 contains two SNARE motifs that are separated by the cysteine-rich region. Unlike synaptobrevin and SNAP-25, syntaxin 1 has an additional domain at the N-terminus that is independently folded (Fernandez et al., 1998).
Assembly of the neuronal SNARE complex is mediated by the SNARE motifs. While monomeric SNARE motifs are largely unstructured, they assemble into a protease-resistant core complex (Fasshauer et al., 1997, 1998b; Poirier et al., 1998). Its crystal structure represents a bundle of four intertwined α-helices that are aligned in parallel (Sutton et al., 1998). Syntaxin 1 and synaptobrevin 2 each contribute one helix whereas SNAP-25 contributes two helices. In the center of the bundle, the helices form 16 layers of interacting amino acid side chains that are mostly hydrophobic and that are perpendicular to the axis of the helix bundle. In the middle of the bundle, an unusual hydrophilic layer was discovered that consists of three glutamines (Q), contributed by syntaxin 1 and SNAP-25, and one arginine (R), contributed by synaptobrevin 2 (Sutton et al., 1998). Sequence comparison of a large number of SNARE proteins showed that these amino acids are highly conserved throughout the entire SNARE superfamily (Weimbs et al., 1998), resulting in a classification as Q-SNAREs and R-SNAREs (Fasshauer et al., 1998b). The amino acids contributing to the hydrophobic layers are also conserved whereas the amino acids on the surface of the bundle are more variable.
While the SNAREs functioning in exocytosis of yeast resemble those of neurons (Ferro-Novick and Jahn, 1994), the composition and stoichiometry of SNARE complexes operating in other intracellular fusion steps are less well characterized. The reasons for the difficulties in identifying functional SNARE complexes are several fold. First, some SNAREs are involved in more than one fusion step and interact with different sets of SNARE partners (Fischer von Mollard et al., 1997; Holthuis et al., 1998; Spang and Schekman, 1998). Secondly, stable complexes form in vitro between SNAREs that do not interact in vivo (Fasshauer et al., 1999; Yang et al., 1999). Thirdly, it cannot be excluded that more than one type of SNARE complex operates in parallel in an individual fusion reaction, resulting in functional redundancy.
The question then arises as to what extent the characteristics of the neuronal SNARE complex are valid for distantly related SNARE complexes. To address this issue, we set out to identify and characterize a SNARE complex operating in the fusion of endosomes. As intermediates of the endocytic pathway, endosomes can be labeled by extracellular probes and their fusion can be monitored using both cell-free assays and intact cells. Delivery of endocytosed material to lysosomes involves sequential transport via early and late endosomes (Mellman, 1996). At least two different homotypic fusion steps can be distinguished, involving early and late endosomes, respectively (Gruenberg and Howell, 1989). Recently, several SNAREs have been implicated in these fusion reactions. These include the R-SNAREs VAMP-7 (Advani et al., 1999; Ward et al., 2000) and endobrevin/VAMP-8 (Antonin et al., 2000a), and the Q-SNAREs syntaxin 7 (Prekeris et al., 1999; Mullock et al., 2000; Nakamura et al., 2000; Ward et al., 2000), syntaxin 8 (Prekeris et al., 1999) and syntaxin 13 (McBride et al., 1999).
Here we report that the R-SNARE endobrevin forms a SNARE complex with the Q-SNAREs syntaxin 7, syntaxin 8 and vti1b, which functions in the homotypic fusion of late endosomes. Despite significant differences in the overall structure of the proteins, the core of the complex exhibits properties remarkably similar to those of the neuronal complex, indicating that the molecular mechanism of SNARE action in membrane fusion is preserved between distant family members.
Results
Identification of a novel SNARE complex
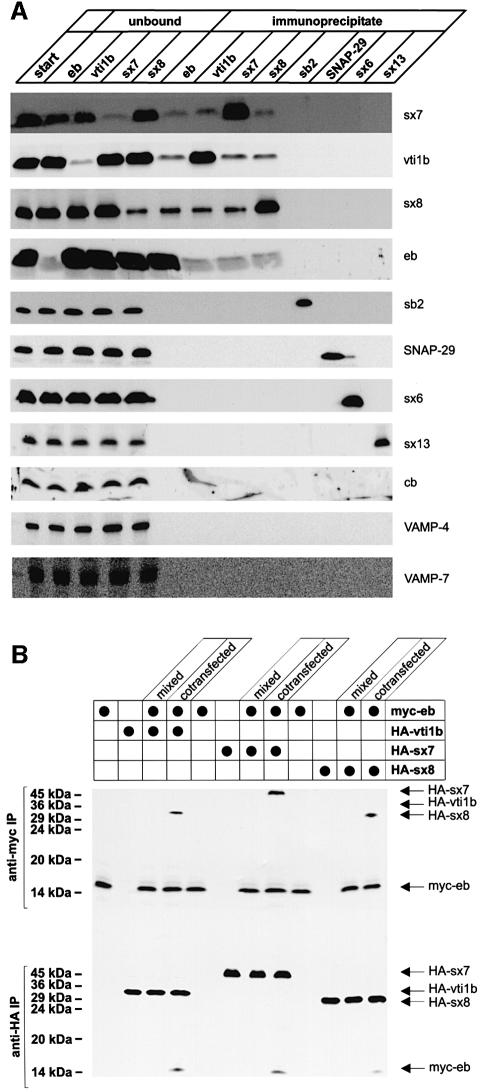
We have previously shown that the R-SNARE endobrevin is involved in the homotypic fusion of both early and late endosomes (Antonin et al., 2000a). To identify SNAREs interacting with endobrevin, immunoprecipitations were performed using detergent extracts of rat liver membranes. The precipitates were analyzed by immunoblotting for the presence of additional SNAREs. Of a total of 11 SNAREs examined, three were found to coprecipitate with endobrevin, including syntaxin 7, syntaxin 8 and vti1b. To determine whether these SNAREs are all in the same complex, immunoprecipitations were performed with antibodies specific for syntaxin 7, syntaxin 8 and vti1b. As shown in Figure 1A, the same set of proteins coprecipitated irrespective of the antibody used. For control, the blots were also analyzed for synaptobrevin 2, SNAP-29, syntaxin 6, syntaxin 13, cellubrevin, VAMP-4 and VAMP-7. None of these proteins was detectable in the immunoprecipitates. Furthermore, immunoprecipitations were also performed with antibodies specific for synaptobrevin 2, SNAP-29, syntaxin 6 and syntaxin 13. While each of these antibodies precipitated its respective antigen, no other SNAREs were detected in the precipitates.
Fig. 1. Endobrevin, syntaxin 7, syntaxin 8 and vti1b form a complex in native membranes. (A) A complex of endobrevin, syntaxin 7, syntaxin 8 and vti1b co-immunoprecipitates from membrane extracts of rat liver. Triton X-100 solubilized membrane extracts were used for immunoprecipitations with antibodies specific for endobrevin (eb), vti1b, syntaxin 7 (sx7), syntaxin 8 (sx8), synaptobrevin 2 (sb2), SNAP-29, syntaxin 6 (sx6) and syntaxin 13 (sx13). The figure shows an immunoblot analysis of the starting material (start), unbound material obtained after precipitation (only for endobrevin, vti1b, syntaxin 7 and syntaxin 8) and immunoprecipitates. In each case, equal proportions of starting material, unbound material and immunoprecipitates were analyzed using antibodies specific for the indicated proteins. Cb, cellubrevin. (B) The complex of endobrevin, syntaxin 7, syntaxin 8 and vti1b exists in membranes before detergent solubilization. NRK cells were transfected in order to express myc-endobrevin, HA-vti1b, HA-syntaxin 7 and HA-syntaxin 8 either individually in different sets of cells, or to co-express HA-vti1b, HA-syntaxin 7 and HA-syntaxin 8 each together with myc-endobrevin (cotransfected). Membrane fractions prepared from each set of cells were extracted with Triton X-100 and used for immunoprecipitation with antibodies specific for the myc epitope (upper part) or the HA epitope (lower part). To check for complex formation after solubilization, aliquots of membranes obtained from individually transfected cells were mixed before detergent extraction (mixed). Note that coprecipitation of myc- and HA-tagged protein is only observed upon cotransfection.
A potential complication of SNARE co-immunoprecipitations may arise from complexes that only form after detergent solubilization and thus do not reflect the assembly status in the intact membrane (Otto et al., 1997). To exclude this possibility, myc-tagged endobrevin and HA-tagged vti1b, syntaxin 7 and syntaxin 8 were transfected in various combinations in NRK cells. In each case, the expressed proteins were distributed very similarly to the endogenous proteins, indicating correct sorting (not shown). Pair-wise transfections of endobrevin with each of the other SNAREs were performed in two ways. First, both proteins were cotransfected, resulting in their expression within the same cells, followed by membrane extraction and immunoprecipitation using either myc- or HA-specific antibodies. Secondly, separate batches of cells were transfected with either protein (resulting in their expression in different cells), and were then homogenized and mixed before membrane extraction and immunoprecipitation. As shown in Figure 1B, each of the HA-tagged SNAREs co-immunoprecipitated with myc-endobrevin only when they were cotransfected and not when they were expressed in different cells, and the membranes were mixed before detergent extraction. The same results were obtained when HA-specific antibodies were used for immunoprecipitation. Together, these data exclude the possibility that coprecipitation of the SNAREs is based on a detergent-induced artefact.
The novel SNARE complex mediates fusion of late endosomes
Next we investigated in which of the fusion steps involving endobrevin the SNARE complex is operating. Two complementary approaches were used. In the first set of experiments, fusion of both early and late endosomes was studied in vitro using established content-mixing assays (Gruenberg and Howell, 1989; Holroyd et al., 1999; Antonin et al., 2000a). Batches of PC12 cells were labeled with two different sets of markers, either for 5 min (fusion of early endosomes) or for 20 min, followed by a chase of 60 min (fusion of late endosomes). The cells were then homogenized and post-nuclear supernatants were prepared from each batch. To examine which of the SNAREs are involved in fusion, Fab fragments were prepared from affinity-purified antibodies and added to the post-nuclear supernatants. Fusion was initiated by combining the post-nuclear supernatants.
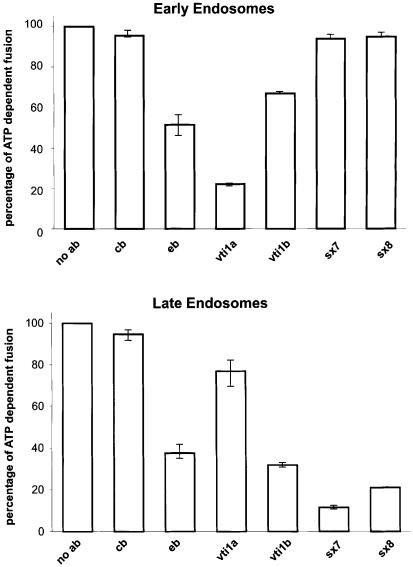
Fusion of late endosomes was strongly inhibited by Fab fragments specific for endobrevin, as published previously (Antonin et al., 2000a), vti1b, syntaxin 7 and syntaxin 8 (Figure 2). In contrast, fusion of early endosomes was unaffected by Fab fragments specific for syntaxin 7 and syntaxin 8, and only slightly inhibited by vti1b antibodies. To assess further the specificity of the Fab effects, we used Fab fragments specific for cellubrevin, an R-SNARE operating in the exocytic limb of the pathway (Galli et al., 1994), and vti1a (also referred to as Vti1-rp2), which has been implicated in intra-Golgi traffic (Xu et al., 1998) although the fusion step it mediates is unknown. Cellubrevin antibodies did not affect either of the fusion reactions. In contrast, Fab fragments specific for vti1a strongly inhibited the fusion of early endosomes but were largely ineffective in inhibiting the fusion of late endosomes. These data indicate that the newly identified SNARE complex mediates the fusion of late but not early endosomes.
Fig. 2. Fab fragments specific for endobrevin, vti1b, syntaxin 7 and syntaxin 8 inhibit homotypic fusion of late endosomes. Homotypic fusion of early and late endosomes derived from PC12 cells was monitored in vitro using a content-mixing assay. Fab fragments (1.2 µM) specific for cellubrevin, endobrevin, vti1a, vti1b, syntaxin 7 and syntaxin 8 were used for pre-incubation of post-nuclear supernatants. ATP-dependent fusion activity in the absence of Fab fragments is defined as 100%. Values are given as means of three independent experiments, bars indicate the range. Fusion was initiated by mixing donor and acceptor fractions while simultaneously adding ATP and cytosol, followed by 30 min of incubation. See legend to Figure 1 for abbreviations.
As an independent approach, endocytic transport was analyzed in intact HeLa cells following microinjection of Fab fragments. To monitor transport, the cells were incubated with fluorescently labeled epidermal growth factor (EGF). EGF is known to be endocytosed, together with its receptor, by clathrin-coated vesicles and to be transported to lysosomes for degradation. Interference with transport to the lysosomes results in retardation of EGF breakdown (Yoshimori et al., 1991, 2000). The DNA dye 4′,6-diamidine-2-phenylindole (DAPI) was co-injected to identify injected cells, and cells were fixed and analyzed 1 and 3 h after initiating endocytosis (see Materials and methods for details). As shown in Figure 3, injection of Fab fragments for each individual component of the novel SNARE complex had profound effects on EGF processing. After 1 h, non-injected cells displayed large fluorescent puncta, as expected for late endosomes and lysosomes. In contrast, labeling was disperse in antibody-injected cells, probably reflecting its distribution in small transport vesicles typical for early endosomal compartments. After 3 h, non-injected cells had largely lost their label due to proteolytic degradation of EGF in lysosomes. Injected cells contained brightly fluorescent puncta, indicating that EGF has finally reached the lysosomes but is not yet degraded. Cellubrevin antibodies injected as control had no effect on transport and processing of EGF.
Fig. 3. Endocytic transport and lysosomal degradation of EGF is retarded by Fab fragments specific for endobrevin, vti1b, syntaxin 7 and syntaxin 8. HeLa cells were microinjected with Fab fragments specific for cellubrevin, endobrevin, vti1b, syntaxin 7 and syntaxin 8. EGF labeled with the fluorescent dye Texas Red was then added. After binding of EGF, endocytotic transport was allowed to proceed for 1 (left) or 3 h (right). At the end of the incubation, the cells were fixed and analyzed by fluorescence microscopy. Microinjected cells were identified by the nuclear staining by co-injected DAPI. Bar, 10 µm.
Biochemical characterization of the late endosomal SNARE complex
The SNARE complex identified above for the fusion of late endosomes differs in several important aspects from its counterpart functioning in neuronal exocytosis. First, it consists of four rather then three different SNAREs, three of which possess extended N-terminal regions beyond the SNARE motif. Secondly, all four components possess C-terminal transmembrane domains. Thirdly, while the overall composition of three Q-SNARE motifs and one R-SNARE motif is preserved, the presence of two syntaxins is surprising. For these reasons, we used recombinant proteins lacking the transmembrane domains in order to study the properties of the complex in vitro.
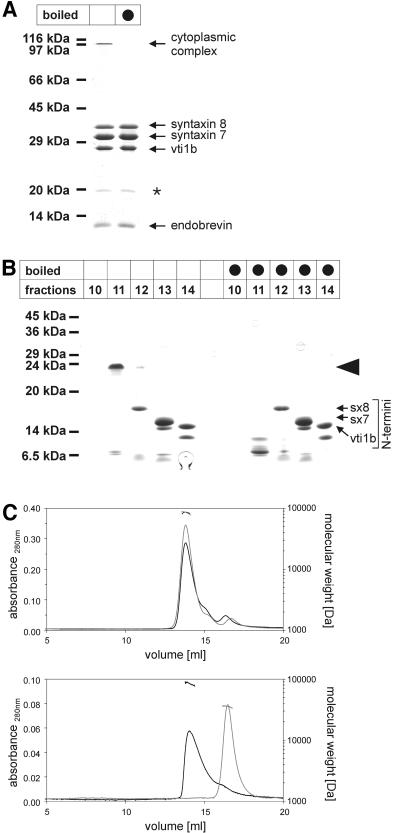
When the four recombinant proteins were mixed at equal stoichiometry, a quaternary complex formed that was separated from the monomers by ion exchange chromatography (not shown). Analysis of the purified complex by SDS–PAGE shows a band with an apparent molecular weight of ∼100 000 Da, in addition to the bands corresponding to the monomers (Figure 4A). The 100 kDa band disappears upon preheating of the sample, indicating that the complex is partially resistant to SDS at room temperature. In the purified complex (referred to as cytoplasmic complex since it lacks transmembrane domains), the monomers were present at equal stoichiometry as determined by comparison with standardized monomer solutions (not shown).
Fig. 4. Biochemical characterization of the endosomal SNARE complex. (A) Recombinant endobrevin, syntaxin 7, syntaxin 8 and vti1b (each lacking their transmembrane domain) form a partially SDS-resistant complex. After assembly, the complex was purified using ion exchange chromatography and analyzed by SDS–PAGE, with or without boiling in SDS sample buffer. The asterisk denotes a contaminating breakdown product of syntaxin 8. (B) Limited proteolysis of the cytoplasmic complex gives rise to a partially SDS-resistant core complex. The complex shown in (A) was treated with trypsin for 30 min. The reaction mixture was separated by size-exclusion chromatography, and the protein-containing fractions were analyzed by SDS–PAGE with or without boiling in SDS sample buffer. The arrowhead denotes the position of the protease-resistant core complex in the non-boiled sample. All N-termini were identified by sequencing. Note that a similar pattern was obtained when proteinase K or chymotrypsin (not shown) was used. (C) Cytoplasmic and core complexes are defined particles with a 1:1:1:1 stoichiometry. Cytoplasmic complexes (upper panel) and core complexes (lower panel) were analyzed by size-exclusion chromatography. Absorption was measured at 280 nm (continuous lines, left axis), and molecular masses were determined by multi-angle laser light scattering (broken lines, right axis). Separations were carried out in either low salt buffer (20 mM Tris pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM DTT; black lines) or high salt buffer (1 M NaCl instead of 150 mM; grey lines). Protein concentrations of the injected samples were 10 µM.
Next, the purified complex was subjected to limited proteolysis in order to determine whether it contains protease-resistant domains. In order to achieve an improved resolution of the fragments, the reaction mixture was separated by size-exclusion chromatography prior to analysis by SDS–PAGE and Coomassie Blue staining. As shown in Figure 4B, a protease-resistant core complex eluted first that contained the SNARE motifs of all four constituents and that was partially resistant to SDS. In addition, smaller protease-resistant fragments eluted subsequently. Sequencing revealed that they correspond to N-terminal regions of syntaxin 7, syntaxin 8 and vti1b, suggesting that each of these proteins contains independently folded N-terminal domains.
For further characterization, we determined the molecular weights of the single proteins and of the assembled complexes using size-exclusion chromatography in combination with multi-angle laser light scattering. In this and all subsequent experiments, the core complex was formed directly from the purified SNARE motifs, which were defined by alignments with the neuronal SNARE motifs (see Materials and methods for details). As shown in Figure 4C (upper panel), the cytoplasmic complex migrates as a single molecular species with a molecular weight of 84.8 ± 0.5 kDa, as predicted for a complex with a 1:1:1:1 stoichiometry (calculated Mr 89.900). In contrast, the core complex exhibited a molecular weight of 83.4 ± 0.6 kDa, suggesting oligomerization (Figure 4C, lower panel). However, increasing the NaCl concentration to 1 M resulted in a shift to a molecular weight of 35.2 ± 0.5 kDa, i.e. the mass predicted for a particle with a 1:1:1:1 stoichiometry (calculated Mr 38 100). A similar salt-sensitive oligomerization was observed for the neuronal complex (Fasshauer et al., 1997). The single proteins migrated as monomers (not shown).
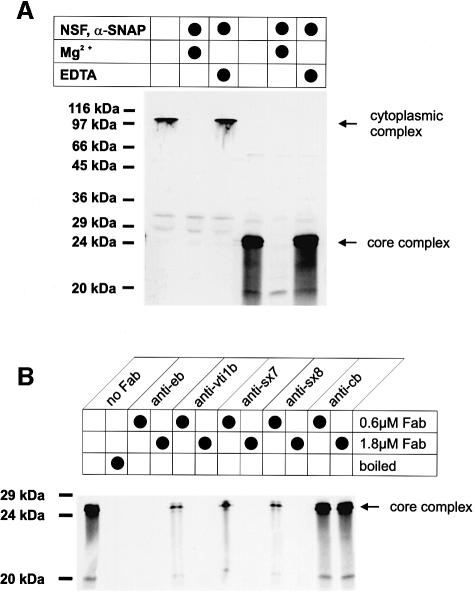
In the next experiment we tested whether the endosomal complexes are sensitive to disassembly by NSF and α-SNAP. This was examined using SDS resistance as a measure of the presence of complexes. As shown in Figure 5A, both the cytoplasmic and the core complexes disappeared when ATP cleavage by NSF was permitted.
Fig. 5. Disassembly and assembly of endosomal SNARE complexes. (A) Disassembly by NSF and α-SNAP of the cytoplasmic complex and the core complex. Purified cytoplasmic complexes and core complexes were incubated as indicated in the presence of ATP. NSF-driven disassembly was monitored by the disappearance of the SDS-resistant complexes using an endobrevin-specific antiserum specific for their detection after immunoblotting. (B) Fab fragments specific for the endosomal SNARE proteins inhibit assembly of the core complex. SNARE motifs of endobrevin, vti1b, syntaxin 7 and syntaxin 8 were each incubated with two different concentrations of the respective Fab fragments. The other three SNARE motifs were then added to allow formation of core complexes that were determined as in (A). For control, endobrevin was incubated with Fab fragments specific for cellubrevin. SDS-resistant core complexes are dissociated by boiling (left).
The data described so far suggest that the late endosomal SNARE complex (hereafter referred to as endosomal complex) contains a protease-resistant core that is partially resistant to SDS and that is responsible for assembly and NSF-driven disassembly. Since assembly of core complexes is thought to be a key step in membrane fusion, we tested whether the antibodies (Fab fragments) used above for the inhibition of endosome fusion and of endocytic transport of EGF inhibit the formation of this core complex. As shown in Figure 5B, pre-incubation of SNARE motifs with the respective Fab fragments, using a concentration range similar to that in the fusion experiments, prevented complex formation.
Formation of the endosomal SNARE complex increases α-helicity
In the following experiments, we used circular dichroism (CD) spectroscopy to investigate whether the endosomal SNAREs undergo structural changes upon assembly that are similar to those of the neuronal SNAREs. Previous studies have shown that the SNARE motifs of both synaptobrevin 2 and SNAP-25 are largely unstructured as monomers but assume an α-helical conformation upon assembly with syntaxin 1 (Fasshauer et al., 1997). The assembled core complex is extraordinarily resistant to heat denaturation and starts melting only above 80°C (Fasshauer et al., 1998a). Furthermore, an increase in α-helical content is observed when syntaxin 1 is mixed with any two of the three SNARE motifs of the neuronal complex. Such ‘partial’ complexes may be of functional relevance, e.g. complexes between syntaxin 1 and SNAP-25 on the plasma membrane prior to vesicle docking.
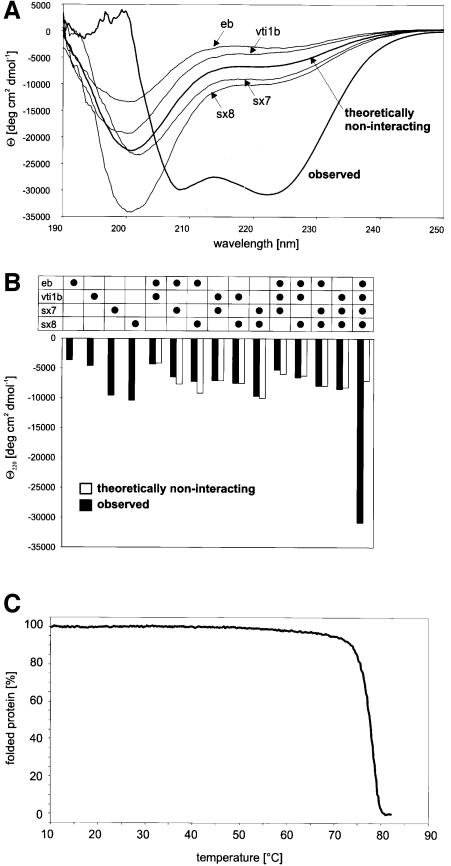
Figure 6A shows the CD spectra for each of the purified individual SNARE motifs and a stoichiometric mix of all four components. Similar to the neuronal SNAREs, none of the individual SNARE motifs exhibited significant α-helical content. In contrast, the mix showed a strongly α-helical spectrum (minima at 208 and 222 nm), indicating that an α-helical core complex forms from largely unstructured monomer precursors. Unlike the neuronal SNAREs, however, combination of any two or three of the individual SNARE motifs failed to induce α-helices (Figure 6B), suggesting that no partial complexes are formed.
Fig. 6. Structural characterization of endosomal SNARE motifs and their interactions. (A) CD spectra of purified SNARE motifs of endobrevin, vti1b, syntaxin 7 and syntaxin 8, and of an equimolar mixture of all four. CD was recorded in 40 mM sodium phosphate at 25°C at a protein concentration of 15 µM. All samples were incubated overnight at 4°C prior to spectroscopy. (B) Changes in the mean residue ellipticity (Θ) at 220 nm induced by interaction between SNARE motifs of endobrevin, vti1b, syntaxin 7 and syntaxin 8. CD was recorded in buffer containing 40 mM sodium phosphate at 25°C at a protein concentration of 15 µM. White columns represent theoretically non-interacting mean residue ellipticity at 220 nm calculated from the observed CD spectra of the individual proteins (columns 1–4). To obtain the observed CD spectra (black columns) of the various combinations, the proteins were incubated overnight at 4°C using equimolar ratios. (C) Percentage of α-helical structure of the purified core complex as a function of temperature. Change in the ellipticity at 220 nm was monitored and the α-helical content was calculated.
For further characterization, we monitored heat denaturation by CD spectroscopy of the purified core complex. As shown in Figure 6C, the complex remained stable up to 70°C and melted with a T1/2 of 78°C, only slightly lower than the neuronal complex. We also recorded CD spectra from the intact soluble domains of the Q-SNAREs and from the cytoplasmic complex (not shown). In each case, the α-helical content was significantly higher than that of the SNARE motifs, ranging from 32 (syntaxin 8) to 55% (syntaxin 7 and vti1b). These observations support the view that the N-terminal regions represent independently folded domains.
Vti1b and syntaxin 8 occupy the position of SNAP-25 in the endosomal SNARE complex
In the final set of experiments, we investigated which of the endosomal Q-SNAREs corresponds to syntaxin 1, to the N-terminal (SN1) and to the C-terminal (SN2) SNARE motif of SNAP-25, respectively. It has been shown previously that in vitro corresponding SNAREs can be substituted for each other in a wide variety of SNARE complexes without compromising the structure and stability of the resulting core complexes (Fasshauer et al., 1999; Yang et al., 1999). Although such non-cognate complexes probably have no physiological relevance, they provide a convenient tool for the assignment of equivalent SNARE motifs.
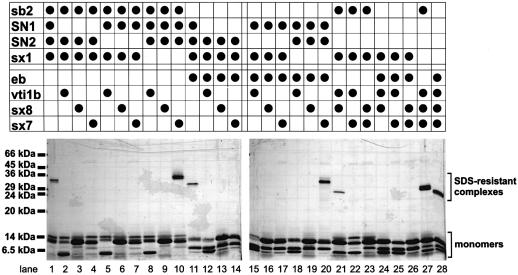
Purified SNARE motifs of both the endosomal and the neuronal complex were combined in vitro in such a way that all possible combinations of three Q-SNAREs were tested for the formation of SDS-resistant complexes with either endobrevin or synaptobrevin 2. As shown in Figure 7, non-cognate SDS-resistant complexes were only formed under the following conditions. First, the exchange of endobrevin and synaptobrevin 2 (lanes 11 and 27). This is not surprising since they represent the R-SNAREs of their respective complexes. Secondly, syntaxin 7 can substitute for syntaxin 1 in the neuronal complex (lane 10), suggesting that syntaxin 7 represents the ‘true’ syntaxin of the endosomal complex. Thirdly, SDS-resistant complexes are seen with endobrevin–SN1– SN2–syntaxin 7 (lane 20) and synaptobrevin 2–vti1b– syntaxin 8–syntaxin 1 (lane 21). Thus, it appears that vti1b and syntaxin 8 together substitute for the two SNARE motifs of SNAP-25, whereas none of the individual components is able to substitute.
Fig. 7. SDS-resistant complexes between neuronal and endosomal SNAREs form only when equivalent SNARE motifs are exchanged. Combinations of purified SNARE motifs were mixed as indicated in approximately equimolar ratios, incubated overnight, and analyzed by SDS–PAGE without boiling and staining with Coomassie Blue.
Several conclusions can be drawn from these experiments. Syntaxin 7 is equivalent to syntaxin 1, whereas syntaxin 8 and vti1b together represent the ‘SNAP-25’ of the complex. This assignment is supported by sequence comparisons (Figure 8). Both syntaxin 1 and syntaxin 7 share a Phe residue in the –3 layer position with many other syntaxins, whereas syntaxin 8 (and some other syntaxins, e.g. syntaxin 6) has a small amino acid in this position. The crystal structure of the neuronal complex shows that this layer is highly asymmetric, with two large hydrophobic residues (Phe of syntaxin 1 and Met of the R-SNARE synaptobrevin 2) occupying most of the space, leaving room for only small side chains (Gly and Ala) contributed by the two SNAP-25 helices (Fasshauer et al., 1998b). In the endosomal complex, this layer is composed of Phe (syntaxin 7), Met (endobrevin) and two Gly residues (vti1b and syntaxin 8). Another asymmetric interaction involves arginine 198 of syntaxin 1, i.e. a position corresponding to ‘layer’ –8. Although synaptobrevin 2 is no longer participating in layer formation at this position, the arginine is interacting with a Ser (SN1) and a Met residue (SN2). Again, the arginine is conserved among syntaxins (matching the conservation of Phe in layer –3) but is lacking in syntaxin 8 (Gln) and vti1b (Ala). With respect to syntaxin 8 and vti1b, it appears that syntaxin 8 exhibits more similarities with SN2, and vti1b with SN1, respectively (see Figure 8). In addition to the previous example (Ala corresponding to Ser, and Met corresponding to Gln), sequence comparison reveals the following overall similarities of the SNARE motifs: vti1b/SN1, 24.6% (vti1b/SN2, 14.0%), and syntaxin 8/SN2, 28.1% (syntaxin 8/SN1, 17.5%).
Fig. 8. Sequence alignment of the SNARE motifs of different Q-SNAREs. Shading denotes the position of residues that in the crystal structure of the neuronal SNARE complex form interacting layers of the four associating α-helices [numbered according to Fasshauer et al. (1998b)]. Side chains that differentiate between syntaxin, SNAP-25/N-terminal domain (SN1) and SNAP-25/C-terminal domain (SN2) subfamily members are boxed. DDBJ/EMBL/GenBank accession Nos are: sx1, P3285; sx2, L20823; sx3, Q08849; sx4, Q08850; sx5, Q08851; sx7, AF031430; sx13, AF031430; sx6, U56815; sx8, AF033109; SNAP-23, AF052596; SNAP-25, AF245227; SNAP-29, AF260577; vti1a, AF035209; and vti1b, AF035208.
Discussion
In the present study we have shown that the three Q-SNAREs syntaxin 7, syntaxin 8 and vti1b, form a SNARE complex with the R-SNARE endobrevin, which functions in late endosome fusion. The core of the complex represents a four-helix bundle with properties very similar to the neuronal core complex, although the SNARE components are only distantly related. We conclude that the features of the neuronal core complex are paradigmatic for all SNARE complexes and, thus, are essential for SNARE function in membrane fusion.
Since SNAREs are known to assemble promiscuously in vitro (Fasshauer et al., 1999; Yang et al., 1999), it is important to define criteria that differentiate between functional and non-physiological complexes. We believe that the endosomal complex characterized here is genuine for the following reasons. First, antibodies for each of the SNAREs coprecipitated comparable amounts of the other three proteins, indicating that we are dealing with a single entity of defined stoichiometry. Secondly, the complex is present in intact membranes and does not form after detergent solubilization, a problem previously encountered with the neuronal complex (Otto et al., 1997). Thirdly, we found no evidence for additional SNAREs in the immunoprecipitates. Conversely, antibodies for several other SNAREs including SNAP-29, synaptobrevin 2, syntaxin 6 and syntaxin 13, did not precipitate detectable amounts of syntaxin 7, syntaxin 8, vti1b and endobrevin. The possibility cannot be excluded, however, that some of the SNAREs participate in additional complexes that are too low in abundance for detection. For instance, endobrevin is likely to function in at least one additional SNARE complex that operates in the fusion of early endosomes (Antonin et al., 2000a). Fourthly, Fab fragments derived from antibodies specific for each of the SNAREs all inhibit the same fusion step with comparable efficiency.
Recent studies by other laboratories have also invoked syntaxin 7 and syntaxin 8 in endosomal trafficking. For syntaxin 7, our data confirm previous reports, which, using approaches similar to those here, allow the conclusion that the protein is involved in the fusion of late endosomes and of late endosomes with lysosomes, but not in the homotypic fusion of early endosomes and lysosomes (Mullock et al., 2000; Nakamura et al., 2000; Ward et al., 2000). Furthermore, antibodies specific for syntaxin 8 have been shown to retard lysosomal trafficking of endocytosed EGF, again in good agreement with our findings (Prekeris et al., 1999). It should be pointed out that, due to the relatively long chase time (1 h), a contribution of lysosomes to the in vitro fusion reaction cannot be excluded in our assay. In contrast, the involvement of vti1b was surprising since the protein was previously localized to the Golgi apparatus (Advani et al., 1998). However, this may be partly due to overexpression since the native protein is also detectable in endosomes (our unpublished observations). Moreover, in accordance with our data it has recently been reported that vti1b coprecipitates with syntaxin 8 (Subramaniam et al., 2000) and that endobrevin coprecipitates with syntaxin 7 (Mullock et al., 2000).
Several additional SNAREs, including VAMP-4 (Steegmaier et al., 1999), VAMP-7 (Advani et al., 1999), syntaxin 6 (Simonsen et al., 1999) and syntaxin 13 (Prekeris et al., 1998; Tang et al., 1998), have been localized to endocytic compartments, but functional data are only available for syntaxin 13 and VAMP-7. Syntaxin 13 appears to be involved in the fusion of early endosomes and in the recycling of plasma membrane receptors (Prekeris et al., 1998; McBride et al., 1999). In contrast, VAMP-7 (also referred to as Ti-VAMP) was assigned to a late fusion step of the endocytic pathway (Advani et al., 1999; Ward et al., 2000), although in other studies the protein was assigned to apical membrane delivery in epithelial cells and to neurite outgrowth (Galli et al., 1998; Martinez-Arca et al., 2000).
Our findings concerning vti1a and vti1b shed new light on the evolution of SNARE proteins. Yeast contains only a single vti homolog, Vti1p, which functions in multiple intracellular fusion steps involving distinct sets of SNARE partners. These include vesicular transport from the trans-Golgi network to the prevacuolar/endosomal compartment (Fischer von Mollard et al., 1997) and all trafficking pathways to the vacuole/lysosome (Ungermann et al., 1999; Fischer von Mollard and Stevens, 1999). As in mammalian species, two homologs were identified in Arabidopsis (AtVTI1a and AtVTI1b) (Zheng et al., 1999). If expressed in yeast, AtVTI1a substitutes for yeast Vti1p in the trafficking step to the prevacuolar compartment whereas AtVTI1b substitutes in the trafficking to the vacuole, but not vice versa. While it is not yet known which fusions are catalyzed by AtVTI1a and AtVTI1b, our data show clearly that vti1a and vti1b function in different fusion reactions of mammalian cells. Thus it appears that SNAREs originate from common ancestors that were involved in multiple trafficking steps. During evolution, the proteins diversified and specialized progressively, which resulted in multiple homologs specialized in a few (such as endobrevin) or single (such as the neuronal SNAREs) fusion events.
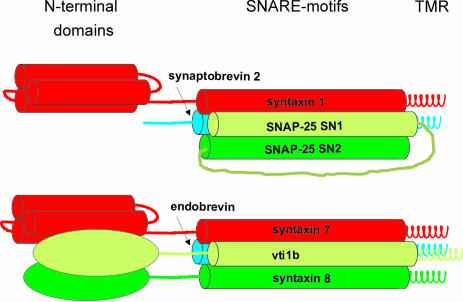
The neuronal and endosomal SNAREs exhibit only a low degree of homology and differ in their overall structure. Unlike the neuronal complex, the endosomal complex contains four distinct proteins, each of which has a single C-terminal transmembrane domain (see Figure 9). Furthermore, three of the four SNAREs contain independently folded N-terminal domains, whereas in the neuronal complex this is true only for syntaxin 1. Moreover, the fusion reactions mediated by the endosomal and the neuronal complex are very different. Fusion of late endosomes constitutes a basic and constitutive trafficking event in the endocytic pathway of every eukaryotic cell, whereas exocytosis of synaptic vesicles is a highly regulated and very rapid fusion reaction of specialized cells.
Fig. 9. Cartoon showing a comparison of the neuronal SNARE complex and the endosomal SNARE complex. TMR, transmembrane region. See text for details. Note that the connection between the N-terminal domains and the helical SNARE motifs may be structured and that the N-terminal domains may fold back and interact with the helical bundle in the endosomal complex. Also, note that it is not known whether the N-terminus of syntaxin 7 forms a trihelical bundle like that of syntaxin 1.
The comparison between these different complexes allows us to deduce general features of SNARE complexes. Most importantly, our findings strongly support the view (Fasshauer et al., 1998b) that each SNARE complex contains a core that is composed of four different helices contributed by the SNARE motifs. Three of the helices are derived from Q-SNAREs whereas one is derived from an R-SNARE. Moreover, the positions of the three Q-SNAREs in the complex are not equivalent. Using a combination of substitutions between neuronal and endosomal Q-SNARE, and sequence comparisons based on the crystal structure of the neuronal core complex, it was possible to assign each of the endosomal Q-SNAREs to one of the neuronal helices. The common features of equivalent helices may permit the development of criteria for a correct assignment of corresponding SNAREs in other SNARE complexes. For instance, the SNARE motif of syntaxin 6 has more similarities with that of syntaxin 8 and the C-terminal motif of SNAP-25 than with ‘genuine’ syntaxins. Likewise, vti1a is most similar to the N-terminal SNARE motif of SNAP-25. In fact, both syntaxin 6 and vti1a may be part of the same SNARE complex (as suggested by co-immunoprecipitations; Xu et al., 1998) where we would predict that they occupy helical positions corresponding to those of SNAP-25.
The structural similarities between the neuronal and endosomal core complexes are indeed remarkable. First, both are resistant to protease digestion and, as the sequence analysis of the resulting fragments shows, probably have a similar length. When the protease-resistant fragment of vti1b is aligned with the N-terminal part of SNAP-25, it corresponds almost precisely to the SNAP-25 fragment used for crystallization (i.e. 17 residues upstream of the –8 ‘layer’ position), and similar results were also obtained for endobrevin, which remained uncleaved. Secondly, assembly of both core complexes proceeds from largely unstructured monomers to a highly helical assembly, i.e. it involves major conformational changes. Interestingly, however, we did not find any evidence for the formation of partial complexes between endosomal SNAREs. Thirdly, both core complexes are highly resistant to heat denaturation, with similar melting temperatures. Fourthly, the endosomal complex is partially resistant to denaturation by SDS, although this feature is less pronounced than in the neuronal complex. Finally, both complexes are disassembled by NSF and α-SNAP, as expected for SNARE complexes.
Although a high-resolution structure is available only for the neuronal complex (Sutton et al., 1998), these similarities strengthen the view that all core complexes consist of parallel four-helix bundles with a layer structure that matches that of the neuronal complex. Of course, SNAREs are known to form assemblies with other proteins in which the structure of the SNAREs may be different, as documented by two recent examples (Hanson and Stevens, 2000; Misura et al., 2000). Also, it is possible that partial complexes form that are of physiological importance. However, the requirement for core complex formation during membrane fusion lends strong support to the view that the assembly of core complexes is the decisive step in bringing the fusing membranes together and initiating the fusion reaction.
Materials and methods
Reagents were used from the following sources: enzymes for DNA manipulations from New England Biolabs (Beverly, MA); GSH–Sepharose 4B and CNBr–Sepharose 4B from Pharmacia (Uppsala, Sweden); Ni–NTA Agarose from Qiagen (Hilden, Germany). All other reagents were purchased from Sigma (Deisenhofen, Germany). Plasmid manipulations were performed in the Escherichia coli strain XL1Blue.
NSF and α-SNAP in pQE-9 plasmids encoding His6-tagged fusion proteins were kindly provided by S.Whiteheart and J.E.Rothman (Sloan-Kettering Center, New York). The following expression constructs were described previously: thrombin-cleavable GST–endobrevin (residues 1–74) (Fasshauer et al., 1999); GST–SNAP-29 (Pabst et al., 2000); expression constructs for the purification of the core fragments of the neuronal complex including sx1 (residues 180–262), SN2 (residues 120–206), SN1 (residues 1–83) and sb2 (residues 1–96) (Fasshauer et al., 1998a); and an expression construct for the cytosolic part of vti1b (residues 1–207) with an N-terminal His6 tag (Antonin et al., 2000b). The cDNA for rat syntaxin 1A was kindly provided by R.H.Scheller (Stanford, CA). cDNA for cellubrevin and SNAP-25a was provided by T.C.Südhof (Dallas, TX).
Molecular cloning
Rat syntaxin 7 was amplified using primers (cgcggatccatgtcttacactccggggat and cgcggtacctcagcctttcagaccccatac) based on the published sequence (DDBJ/EMBL/GenBank accession No. AF031430) from rat kidney and rat cerebellum cDNA lambda ZAPII libraries (Stratagene, La Jolla, CA). The PCR products were subcloned into pBS vector (Stratagene). The sequences of all clones derived from the two tissues were identical. However, comparison with the published syntaxin 7 sequence revealed one nucleotide discrepancy (c416a) resulting in an amino acid exchange (A139E). Both the human (DDBJ/EMBL/GenBank accession No. HSU77942) and mouse (DDBJ/EMBL/GenBank accession No. AF056323) syntaxin 7 possess a glutamate in this position.
Rat syntaxin 12/13 was amplified using primers (gctctagatgtcctacggtcccttagac and gcggtacctcacttagaagcaacccagataac) based on the published sequence (DDBJ/EMBL/GenBank accession No. AF031430) from rat lung and rat cerebellum cDNA lambda ZAPII libraries (Stratagene). The PCR products were subcloned into pBS vector (Stratagene).
For eukaryotic expression, vti1b, syntaxin 7 and syntaxin 8 were expressed with an N-terminal HA tag under the control of an SV40 promoter using the pSI vector system (Promega, Madison, WI). Endobrevin was expressed with an N-terminal myc tag using the same vector system. For PCR amplification the following oligonucleotides were used: gccgctcgagccaccatggagtacccctacgacgtgcccgactacgccatggccgcctccgccg and ctagtctagatcaatggtgtcgaaagaatttg for vti1b; gccgctcgagccaccatggagtacccctacgacgtgcccgactacgccatgtcttacactccgggg and ctagtctagatcagcctttcagacccca for syntaxin 7; gccgctcgagccaccatggagtacccctacgacgtgcccgactacgccatggccccagacccctg and ctagtctagatcagttggttggccacac for syntaxin 8; and gccgctcgagccaccatggagcagaagctgatcagcgaggaggacctgatggaggccagtgggag and ctagtctagattaagtggggatggtgcc for endobrevin.
The soluble domain of syntaxin 8 (residues 1–213) was amplified by PCR from a rat lung cDNA l ZAPII library (Stratagene) using the oligonucleotides cggggatccatggccccagacccctg and ggaattctaggaagctgactttctgtcc. To obtain a bacterial expression vector for a GST–syntaxin 8 fusion protein (pBK45), the PCR product was cloned into pGex2T (Pharmacia).
GST fusion proteins of the cytosolic parts of cellubrevin (residues 1–82), syntaxin 7 (residues 1–236) and syntaxin 13 (residues 1–250) were generated using the following oligonucleotides for PCR amplification: gcgggatccatgtctacaggggtgcc and gcgctcgagtcattacatcttgcagttcttcc for cellubrevin; cgcggatccatgtcttacactccggggat and ggcggctcgagctattattttctggatttgcgctg for syntaxin 7; and gcgcccgggaatgtcctacggtccctta and ggcggctcgagctattacttcttgcgagattttttc for syntaxin 13. PCR products were cloned into pGex2T (Pharmacia).
The cytosolic fragments of syntaxin 7 (residues 1–236) and syntaxin 13 (residues 1–250) were subcloned into the pET28a vector (Novagen, Madison, WI), which includes a thrombin cleavage site for the removal of the upstream His6 tag. For PCR amplification, the following oligonucleotides were used: ggcggcatatgtcttacactccggg and ggcggctcgagctattattttctggatttgcgctg for syntaxin 7; and ggcggcatatgtcctacggtccctta and ggcggctcgagctattacttcttgcgagattttttc for syntaxin 13. The cytosolic fragment of syntaxin 8 (residues 1–213) was subcloned into a pET30b vector (Novagen), which includes an enterokinase cleavage site for the removal of the upstream His6 tag, means of excising a NcoI–EcoRI fragment from pBK45 and insertion into the target vector cleaved by the same enzymes.
The SNARE motif containing fragments of vti1b (residues 130–206), syntaxin 7 (residues 159–236) and syntaxin 8 (residues 136–213) was subcloned into pET28a vector (Novagen). For PCR amplification, the following oligonucleotides were used: gcgcggcatatggcattactcctacaaggc and gccgctcgagttacttgttggttatcacttttc for vti1b; gaggcacatatggaaatcacagaggatgac and ggcggctcgagctattattttctggatttgcgctg for syntaxin 7; and gaggcacatatgggtttcgatgagatccgg and ggaattctaggaagctgactttctgtcc for syntaxin 8.
All recombinant proteins were expressed as His6-tagged or GST-tagged fusion proteins and purified by Ni2+–agarose or GSH–Sepharose, respectively. With exception of the cytoplasmic expression constructs of syntaxin 8 and vti1b, the tags of all proteins were removed using thrombin. All proteins were further purified using Mono-Q or Mono-S columns on an FPLC system (Pharmacia). All proteins were 95% pure as judged by SDS–PAGE and Coomassie Blue staining. The cytoplasmic domain of syntaxin 8 is protease sensitive. We were not able to purify an untagged version of the protein.
Purification of cytoplasmic and core complexes
For assembly of cytoplasmic complexes, equimolar amounts of syntaxin 7 (residues 1–236), syntaxin 8 (residues 1–213 plus an N-terminal sequence containing the His6 tag), vti1b (residues 1–206 plus an N-terminal His6 tag) and endobrevin (residues 1–74) were incubated overnight at 4°C. For the assembly of core complexes, equimolar amounts of the SNARE motifs of endobrevin (residues 1–74), syntaxin 8 (residues 136–213), syntaxin 7 (residues 159–236) and vti1b (residues 130–206) were incubated overnight at 4°C. Both complexes were purified by ion exchange chromatography using a Mono-Q column on an FPLC system (Pharmacia).
Limited proteolysis of SNARE complexes
One hundred micrograms of cytoplasmic complex were incubated for 30 min with 1 µg of trypsin. The reaction was stopped by adding phenylmethylsulfonyl fluoride (PMSF) to a final concentration of 1 mM, and the sample analyzed by size-exclusion chromatography using a Superdex 75 PC 3.2/30 column (Pharmacia). Samples were analyzed by SDS–PAGE with and without boiling.
Antibodies
Antisera were raised in rabbits against recombinant SNAP-29, soluble syntaxin 7 (residues 1–236) and soluble syntaxin 13 (residues 1–250) that were expressed as GST fusion proteins, thrombin cleaved, and separated from GST by ion exchange chromatography. For the generation of antisera specific for syntaxin 8, a fusion protein of GST with the soluble part of syntaxin 8 (residues 1–213, pBK45) was injected into rabbits.
The following antibodies were described previously: cellubrevin (rabbit antiserum R54) (Annaert et al., 1997); endobrevin (rabbit antiserum) (Fasshauer et al., 1999); synaptobrevin 2 (monoclonal antibody, Cl69.1) (Edelmann et al., 1995); vti1a and vti1b (Antonin et al., 2000b). The following antibodies were kind gifts: VAMP-4 (Steegmaier et al., 1999) and VAMP-7 (Advani et al., 1999). Antibodies for syntaxin 6 were obtained from Transduction Laboratories (San Diego, CA)
Assembly of mixed core complexes
Purified SNARE motifs of the neuronal core complex, including synaptobrevin 2 (residues 1–96), SNAP-25 (residues 1–83), SNAP-25 (residues 120–206) and syntaxin 1 (residues 180–262), and of the endosomal core complex, including endobrevin (residues 1–74), syntaxin 8 (residues 136–213), syntaxin 7 (residues 159–236) and vti1b (residues 130–206), were incubated at a final concentration of 6 µM in standard buffer (20 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM EDTA) in the indicated mixtures, overnight at 4°C. The next day the samples were analyzed for the presence of SDS-resistant core complexes by 15% SDS–PAGE.
Disassembly of complexes
Five micromolar cytoplasmic and core complexes, respectively, were disassembled by the addition of 4 µM NSF, 36 µM α-SNAP, 3 mM MgCl2, 3 mM ATP, and incubation for 30 min at 30°C in standard buffer. The reaction was stopped by adding SDS sample buffer, and the samples were separated by 15% SDS–PAGE. As controls, the reaction was either carried out in the absence of NSF and α-SNAP, or the ATPase activity of NSF was abolished by replacing MgCl2 with 10 mM EDTA. For immunodetection of SDS-resistant complexes, an antiserum specific for endobrevin was used.
Electrophoretic procedures
Routinely, SDS–PAGE was carried out as described by Laemmli (1970). When testing for SDS resistance, samples were solubilized in SDS sample buffer [10 mM Tris pH 6.8, 2% SDS, 10% sucrose, 25 mM dithiothreitol (DTT)] and incubated at room temperature (not boiled) or 95°C (boiled) for 5 min before analysis by 15% SDS–PAGE.
Inhibition of in vitro assembly of SNARE proteins by Fab fragments
Recombinant SNARE motifs of the endosomal core complex (see above) were incubated in phosphate-buffered saline (PBS) for 1 h on ice, with a 10- or 30-fold molar excess of Fab fragments specific for the respective protein. Then the missing three SNARE motifs were added to each sample, followed by incubation overnight at 4°C in standard buffer. Final concentrations were 0.06 µM for the SNARE motifs, and 0.6 or 1.8 µM for the Fab fragments, respectively. To stop the reaction, SDS sample buffer without DTT was added. As an assay for assembly, the formation of an SDS-resistant core was monitored by SDS–PAGE and immunoblotting using the antibody specific for endobrevin. An anti-rabbit Fc antibody was used as a secondary antibody to exclude a crossreactivity with the light chains of the antibodies used for inhibition. For control, endobrevin was pre-incubated either without antibodies or with anti-cellubrevin Fabs.
Immunoprecipitations
A crude membrane fraction was prepared from rat liver. Liver was homogenized by 10 strokes with a glass teflon homogenizer at 1000 r.p.m. in 280 mM sucrose, 50 mM Tris–HCl pH 7.4, 1 mM EDTA, 0.1 mM PMSF, followed by centrifugation at 1000 gav for 10 min. The supernatant was centrifuged for 1 h at 200 000 gav, and the resulting pellet was resuspended in 50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1 mM PMSF. The sample was solubilized in extraction buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1 mM PMSF, 1% Triton X-100) at a final protein concentration of 0.3 mg/ml. Insoluble material was removed by centrifugation at 200 000 gav for 60 min. For immunoprecipitation, excess amounts of antibodies with the following specificities were added to separate aliquots: endobrevin, vti1b, syntaxin 7, syntaxin 8, syntaxin 6, synaptobrevin 2, SNAP-29 and syntaxin 13. After incubation for 2 h at 4°C, protein A–Sepharose (Pharmacia) was added in amounts sufficient to bind all IgG quantitatively, followed by incubation for 1 h. Protein in the unbound material was precipitated according toWessel and Flügge (1984). Bound material was obtained by the addition of SDS sample buffer following eight wash cycles of the protein A–Sepharose with extraction buffer. Both fractions were analyzed by SDS–PAGE and immunoblotting with the indicated antibodies. Anti-mouse Fc antibodies and anti-rabbit Fc antibodies were used for detection to exclude binding to the antibodies used for immunoprecipitation.
NRK cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum, 100 U/ml penicillin and 100 µg/ml streptomycin, in humidified incubators with 5% CO2 at 37°C. Cells were transfected by electroporation following the procedure described by Lang et al. (1997). After 48 h, the cells were collected and homogenized in a stainless ball homogenizer with 20 passages (clearance 0.0009 inches) in 280 mM sucrose, 50 mM Tris–HCl pH 7.4, 1 mM EDTA, 0.1 mM PMSF. A crude membrane fraction was prepared as described above for liver. The samples were solubilized and centrifuged as described above. Where indicated, membranes derived from different sets of transfected cells were mixed before solubilization. Immunoprecipitations were carried out using antibodies specific for the myc- and HA-epitopes, respectively, and analyzed as described above.
Microinjection
HeLa cells were grown in DMEM supplemented with 10% FCS, 100 U/ml penicillin and 100 µg/ml streptomycin, in humidified incubators with 5% CO2 at 37°C. For microinjection, cells were grown on poly-l-lysine-coated coverslips. Fab fragments, dialyzed against 20 mM Tris–HCl pH 7.4 and containing 25 µg/ml DAPI, were microinjected using a Transjector 5246 (Eppendorf, Germany). Cells were then incubated in 0.1% bovine serum albumin (BSA)/DMEM for 1 h at 37°C. For labeling of the endocytic pathway, 1 µg/ml Texas-Red EGF (Molecular Probes, Eugene, OR) in 0.1% BSA/DMEM was then added, followed by incubation for 1 h at 4°C to allow for receptor binding. Cells were washed with PBS and incubated in 0.1% BSA/DMEM for the indicated time at 37°C. After these treatments, cells were washed, fixed with 4% paraformaldehyde and observed by immunofluorescence microscopy.
Endobrevin, syntaxin 7, syntaxin 8, vti1b and cellubrevin antisera were affinity purified using the respective recombinant protein coupled to CNBr–Sepharose 4B (Pharmacia). Preparation of Fab fragments, cell free assays for homotypic fusion of PC12 cell-derived early and late endosomes, and the inhibition of endosome fusion with Fab fragments were carried out as described previously (Antonin et al., 2000a). Multi-angle laser light scattering measurements and CD spectroscopy were performed as described (Fasshauer et al., 1999).
Acknowledgments
Acknowledgements
We are greatly indebted to Dr R.H.Scheller (Stanford) for the gift of antibodies and cDNAs; Martin Margittai (Göttingen) for providing recombinant NSF; Dr T.C.Südhof (Dallas), J.E.Rothman and S.Whiteheart (New York) for gifts of molecular reagents; Uwe Pleßmann for peptide sequencing; and Dagmar Diezmann, Maria Druminski, Beate Köhler and Ursel Ries for excellent technical assistance. This work was supported by grants from the Deutsche Forschungsgemeinschaft to G.F.v.M. and R.J. (SFB 523, TP B6 and B7) and from the Volkswagenstiftung to G.F.v.M.
References
- Advani R.J., Bae,H.R., Bock,J.B., Chao,D.S., Doung,Y.C., Prekeris,R., Yoo,J.S. and Scheller,R.H. (1998) Seven novel mammalian SNARE proteins localize to distinct membrane compartments. J. Biol. Chem., 273, 10317–10324. [DOI] [PubMed] [Google Scholar]
- Advani R.J., Yang,B., Prekeris,R., Lee,K.C., Klumperman,J. and Scheller,R.H. (1999) VAMP-7 mediates vesicular transport from endosomes to lysosomes. J. Cell Biol., 146, 765–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annaert W.G., Becker,B., Kistner,U., Reth,M. and Jahn,R. (1997) Export of cellubrevin from the endoplasmic reticulum is controlled by BAP31. J. Cell Biol., 139, 1397–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W., Holroyd,C., Tikkanen,R., Höning,S. and Jahn,R. (2000a) The R-SNARE enobrevin/VAMP-8 mediates homotypic fusion of early and late endosomes. Mol. Biol. Cell, 11, 3289–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W., Riedel,D. and von Mollard,G.F. (2000b) The SNARE Vti1a-β is localized to small synaptic vesicles and participates in a novel SNARE complex. J. Neurosci., 20, 5724–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock J.B. and Scheller,R.H. (1997) Protein transport. A fusion of new ideas. Nature, 387, 133–135. [DOI] [PubMed] [Google Scholar]
- Edelmann L., Hanson,P.I., Chapman,E.R. and Jahn,R. (1995) Synaptobrevin binding to synaptophysin: a potential mechanism for controlling the exocytotic fusion machine. EMBO J., 14, 224–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasshauer D., Otto,H., Eliason,W.K., Jahn,R. and Brunger,A.T. (1997) Structural changes are associated with soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor complex formation. J. Biol. Chem., 272, 28036–28041. [DOI] [PubMed] [Google Scholar]
- Fasshauer D., Eliason,W.K., Brunger,A.T. and Jahn,R. (1998a) Identification of a minimal core of the synaptic SNARE complex sufficient for reversible assembly and disassembly. Biochemistry, 37, 10354–10362. [DOI] [PubMed] [Google Scholar]
- Fasshauer D., Sutton,R.B., Brunger,A.T. and Jahn,R. (1998b) Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl Acad. Sci. USA, 95, 15781–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasshauer D., Antonin,W., Margittai,M., Pabst,S. and Jahn,R. (1999) Mixed and non-cognate SNARE complexes. Characterization of assembly and biophysical properties. J. Biol. Chem., 274, 15440–15446. [DOI] [PubMed] [Google Scholar]
- Fernandez I., Ubach,J., Dulubova,I., Zhang,X., Sudhof,T.C. and Rizo,J. (1998) Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell, 94, 841–849. [DOI] [PubMed] [Google Scholar]
- Ferro-Novick S. and Jahn,R. (1994) Vesicle fusion from yeast to man. Nature, 370, 191–193. [DOI] [PubMed] [Google Scholar]
- Fischer von Mollard G. and Stevens,T.H. (1999) The Saccharomyces cerevisiae v-SNARE Vti1p is required for multiple membrane transport pathways to the vacuole. Mol. Biol. Cell, 10, 1719–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer von Mollard G., Nothwehr,S.F. and Stevens,T.H. (1997) The yeast v-SNARE Vti1p mediates two vesicle transport pathways through interactions with the t-SNAREs Sed5p and Pep12p. J. Cell Biol., 137, 1511–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli T., Chilcote,T., Mundigl,O., Binz,T., Niemann,H. and De Camilli,P. (1994) Tetanus toxin-mediated cleavage of cellubrevin impairs exocytosis of transferrin receptor-containing vesicles in CHO cells. J. Cell Biol., 125, 1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli T., Zahraoui,A., Vaidyanathan,V.V., Raposo,G., Tian,J.M., Karin,M., Niemann,H. and Louvard,D. (1998) A novel tetanus neurotoxin-insensitive vesicle-associated membrane protein in SNARE complexes of the apical plasma membrane of epithelial cells. Mol. Biol. Cell, 9, 1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg J. and Howell,K.E. (1989) Membrane traffic in endocytosis: insights from cell-free assays. Annu. Rev. Cell Biol., 5, 453–481. [DOI] [PubMed] [Google Scholar]
- Hanson M.A. and Stevens,R.C. (2000) Cocrystal structure of synaptobrevin-II bound to botulinum neurotoxin type B at 2.0 Å resolution. Nature Struct. Biol., 7, 687–692. [DOI] [PubMed] [Google Scholar]
- Hanson P.I., Heuser,J.E. and Jahn,R. (1997) Neurotransmitter release—four years of SNARE complexes. Curr. Opin. Neurobiol., 7, 310–315. [DOI] [PubMed] [Google Scholar]
- Holroyd C., Kistner,U., Annaert,W. and Jahn,R. (1999) Fusion of endosomes involved in synaptic vesicle recycling. Mol. Biol. Cell, 10, 3035–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis J.C., Nichols,B.J., Dhruvakumar,S. and Pelham,H.R. (1998) Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J., 17, 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R. and Südhof,T.C. (1999) Membrane fusion and exocytosis. Annu. Rev. Biochem., 68, 863–911. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lang T., Wacker,I., Steyer,J., Kaether,C., Wunderlich,I., Soldati,T., Gerdes,H.H. and Almers,W. (1997) Ca2+-triggered peptide secretion in single cells imaged with green fluorescent protein and evanescent-wave microscopy. Neuron, 18, 857–863. [DOI] [PubMed] [Google Scholar]
- Martinez-Arca S., Alberts,P., Zahraoui,A., Louvard,D. and Galli,T. (2000) Role of tetanus neurotoxin insensitive vesicle-associated membrane protein (TI-VAMP) in vesicular transport mediating neurite outgrowth. J. Cell Biol., 149, 889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride H.M., Rybin,V., Murphy,C., Giner,A., Teasdale,R. and Zerial,M. (1999) Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell, 98, 377–386. [DOI] [PubMed] [Google Scholar]
- Mellman I. (1996) Endocytosis and molecular sorting. Annu. Rev. Cell Dev. Biol., 12, 575–625. [DOI] [PubMed] [Google Scholar]
- Misura K.M., Scheller,R.H. and Weis,W.I. (2000) Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature, 404, 355–362. [DOI] [PubMed] [Google Scholar]
- Mullock B.M. et al. (2000) Syntaxin 7 is localized to late endosome compartments, associates with Vamp 8 and is required for late endosome–lysosome fusion. Mol. Biol. Cell, 11, 3137–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N., Yamamoto,A., Wada,Y. and Futai,M. (2000) Syntaxin 7 mediates endocytic trafficking to late endosomes. J. Biol. Chem., 275, 6523–6529. [DOI] [PubMed] [Google Scholar]
- Otto H., Hanson,P.I. and Jahn,R. (1997) Assembly and disassembly of a ternary complex of synaptobrevin, syntaxin and SNAP-25 in the membrane of synaptic vesicles. Proc. Natl Acad. Sci. USA, 94, 6197–6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst S., Hazzard,J.W., Antonin,W., Südhof,T.C., Jahn,R., Rizo,J. and Fasshauer,D. (2000) Selective interaction of complexin with the neuronal SNARE complex. Determination of the binding regions. J. Biol. Chem., 275, 19808–19818. [DOI] [PubMed] [Google Scholar]
- Poirier M.A., Hao,J.C., Malkus,P.N., Chan,C., Moore,M.F., King,D.S. and Bennett,M.K. (1998) Protease resistance of syntaxin– SNAP-25–VAMP complexes. Implications for assembly and structure. J. Biol. Chem., 273, 11370–11377. [DOI] [PubMed] [Google Scholar]
- Prekeris R., Klumperman,J., Chen,Y.A. and Scheller,R.H. (1998) Syntaxin 13 mediates cycling of plasma membrane proteins via tubulovesicular recycling endosomes. J. Cell Biol., 143, 957–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prekeris R., Yang,B., Oorschot,V., Klumperman,J. and Scheller,R.H. (1999) Differential roles of syntaxin 7 and syntaxin 8 in endosomal trafficking. Mol. Biol. Cell, 10, 3891–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman J.E. (1994) Mechanisms of intracellular protein transport. Nature, 372, 55–63. [DOI] [PubMed] [Google Scholar]
- Simonsen A., Gaullier,J.M., D’Arrigo,A. and Stenmark,H. (1999) The Rab5 effector EEA1 interacts directly with syntaxin-6. J. Biol. Chem., 274, 28857–28860. [DOI] [PubMed] [Google Scholar]
- Söllner T., Bennett,M.K., Whiteheart,S.W., Scheller,R.H. and Rothman,J.E. (1993) A protein assembly–disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation and fusion. Cell, 75, 409–418. [DOI] [PubMed] [Google Scholar]
- Spang A. and Schekman,R. (1998) Reconstitution of retrograde transport from the Golgi to the ER in vitro. J. Cell Biol., 143, 589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steegmaier M., Klumperman,J., Foletti,D.L., Yoo,J.S. and Scheller,R.H. (1999) Vesicle-associated membrane protein 4 is implicated in trans-Golgi network vesicle trafficking. Mol. Biol. Cell, 10, 1957–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam V.N., Loh,E., Horstmann,H., Habermann,A., Xu,Y., Coe,J., Griffiths,G. and Hong,W. (2000) Preferential association of syntaxin 8 with the early endosome. J. Cell Sci., 113, 997–1008. [DOI] [PubMed] [Google Scholar]
- Sutton R.B., Fasshauer,D., Jahn,R. and Brunger,A.T. (1998) Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature, 395, 347–353. [DOI] [PubMed] [Google Scholar]
- Tang B.L., Tan,A.E., Lim,L.K., Lee,S.S., Low,D.Y. and Hong,W. (1998) Syntaxin 12, a member of the syntaxin family localized to the endosome. J. Biol. Chem., 273, 6944–6950. [DOI] [PubMed] [Google Scholar]
- Ungermann C., von Mollard,G.F., Jensen,O.N., Margolis,N., Stevens,T.H. and Wickner,W. (1999) Three v-SNAREs and two t-SNAREs, present in a pentameric cis-SNARE complex on isolated vacuoles, are essential for homotypic fusion. J. Cell Biol., 145, 1435–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D.M., Pevsner,J., Scullion,M.A., Vaughn,M. and Kaplan,J. (2000) Syntaxin 7 and VAMP-7 are soluble N-ethylmaleimide-sensitive factor attachment protein receptors required for late endosome–lysosome and homotypic lysosome fusion in alveolar macrophages. Mol. Biol. Cell, 11, 2327–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimbs T., Mostov,K., Low,S.H. and Hofmann,K. (1998) A model for structural similarity between different SNARE complexes based on sequence relationships. Trends Cell Biol., 8, 260–262. [DOI] [PubMed] [Google Scholar]
- Wessel D. and Flügge,U.I. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem., 138, 141–143. [DOI] [PubMed] [Google Scholar]
- Xu Y., Wong,S.H., Tang,B.L., Subramaniam,V.N., Zhang,T. and Hong,W. (1998) A 29-kilodalton Golgi soluble N-ethylmaleimide-sensitive factor attachment protein receptor (Vti1-rp2) implicated in protein trafficking in the secretory pathway. J. Biol. Chem., 273, 21783–21789. [DOI] [PubMed] [Google Scholar]
- Yang B., Gonzalez,L.,Jr, Prekeris,R., Steegmaier,M., Advani,R.J. and Scheller,R.H. (1999) SNARE interactions are not selective. Implications for membrane fusion specificity. J. Biol. Chem., 274, 5649–5653. [DOI] [PubMed] [Google Scholar]
- Yoshimori T., Yamamoto,A., Moriyama,Y., Futai,M. and Tashiro,Y. (1991) Bafilomycin A1, a specific inhibitor of vacuolar-type H+-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J. Biol. Chem., 266, 17707–17712. [PubMed] [Google Scholar]
- Yoshimori T. et al. (2000) The mouse SKD1, a homologue of yeast Vps4p, is required for normal endosomal trafficking and morphology in mammalian cells. Mol. Biol. Cell, 11, 747–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., von Mollard,G.F., Kovaleva,V., Stevens,T.H. and Raikhel,N.V. (1999) The plant vesicle-associated SNARE AtVTI1a likely mediates vesicle transport from the trans-Golgi network to the prevacuolar compartment. Mol. Biol. Cell, 10, 2251–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]