An important objective of de novo protein design is the preparation of metalloproteins, as many natural systems contain metals which play crucial roles for the function and/or structural integrity of the biopolymer.[1, 2] Metalloproteins catalyze some of the most important processes in nature, ranging from energy generation and transduction to complex chemical transformations. At the same time, metals in excess can be deleterious to cells and some ions are purely toxic, having no known beneficial effects (e.g., HgII or PbII). One hopes to use a first principles approach for realizing both known metallocenters and also to prepare novel sites which may lead to exciting new catalytic transformations. However, designing novel metalloproteins is a challenging and complex task, especially if one desires to prepare asymmetric metal environments.
Numerous metalloprotein systems have been designed over the past fifteen years, typically using unassociated peptides that assemble into three- stranded coiled coils or helix-loop-helix motifs that form anti-parallel four- stranded bundles. In terms of metal ion binding these systems have been functionalized with heme,[3, 4] and non-heme mononuclear,[5] and binuclear centers.[6, 7] It is often difficult to prepare non-symmetrical metal sites through these strategies due to the symmetry of the systems relying on homo-oligomerization. Thus, the preparation of a single polypeptide chain capable of controlling a metal coordination environment is a key objective.
Previously, we have designed soft, thiol-rich metal binding sites into the interior of parallel, three-stranded α-helical coiled coils involving cysteine and/or penicillamine as the ligating amino acid residues.[8, 9] These systems have served as hallmarks for understanding the metallobiochemistry of different heavy metals such as CdII, HgII, AsIII, and PbII.[8-11] We have shown how to control the geometry and coordination number of metals such as CdII and HgII at the protein interior as well as fine-tuning the physical properties of the metals, which has led to site-selective molecular recognition of CdII.[12-14] While these homotrimeric assemblies have been very useful, the production of heterotrimeric systems that are capable of fine tuning metal environments controllably or introduction of a site specific H-bond has been elusive.[15] Therefore, we have chosen an alternative strategy to satisfy this objective using a single polypeptide chain instead of multiple self associating peptides.
Existing designed heteromeric helical bundles and coiled-coils show energetic preferences of several kcal/mol for the desired heteromeric versus homomeric assemblies.[16, 17] However, the energy gap between a hetero and homomeric assembly often depends critically on ionic strength, pH, and other environmental parameters. Moreover, the objective of many studies in de novo protein design is to impart an energetically sub-optimal coordination geometry on the metal ion, and the degree to which this strategy will be successful depends on the size of the energy gap between the desired heteromeric assembly and other homomeric or misfolded states. Also, even when heterooligomeric bundles have been used to successfully identify specific environmental effects that influence substrate binding or metal ion cofactor reactivity,[18] the non-covalently assembled complexes have often been difficult to characterize structurally, possibly due to small populations of alternately assembled species. In this case, combining the active site residues into a construct with linked helices greatly facilitated structural analysis and catalytic characterization.[19]
An attractive starting scaffold to meet our objectives is the de novo designed three-helix bundle α3D. The NMR structure of this protein is known and it has been proven that the helices are oriented in a counter-clockwise topology.[20] Although the α3D protein originated from a coiled-coil, its helices were shortened to the point where it might be better considered as a globular protein whose repetitive structure makes each of the heptads very similar to one another (in the absence of end-effects). The stability of α3D is similar to that of natural proteins and thus α3D should be tolerant to mutations, allowing this protein to serve as an excellent framework to engineer specific metal binding sites. Additionally, with this protein scaffold we can study the effect of the ligating residue located on the second helix which is antiparallel to the first and third helices of the bundle.
Before attempting to prepare asymmetric metal coordination environments or site specific H-bonds, we felt it was important to first redesign α3D so as to introduce symmetric metal binding sites involving cysteine residues. This approach would allow us to exploit the extensive body of work defining heavy metal complexation using the TRI and Coil Ser peptides,[8, 15] to assess whether a specific metal structure could be achieved in the modified α3D construct. It would also allow us to probe the physical properties of metals such as CdII, HgII and PbII in a more natural antiparallel helical system. A significant amount of literature exists where metal binding sites have been designed in existing four-helix bundles and in a mixture of α/β protein structural frameworks.[21, 22] However, there are no such examples where novel metal binding site(s) have been engineered within an antiparallel single chain three-helix bundle by rational design. The three-helix bundle occurs ubiquitously in nature as a versatile and robust scaffold ranging from the helical IgG-binding domains,[23] to DNA-binding proteins, structural proteins, and enzymes..[24] Despite its widespread occurrence in nature only a few attempts have been made to prepare single chain three-helix bundles. [25, 26]
Based on visual inspection of the α3D structure, four potential sites along the bundle were identified where three cysteines, one from each helix, could be introduced. Out of these four mutants, α3DIV (Figure 1), located at the C-terminal end of the bundle, seemed to be optimal based on the properties of the starting protein.[20, 27-29] Previous NMR structural and dynamic investigations showed a gradient in the dynamic behavior and malleability of the protein, with the C-terminal end of the bundle being most amenable to amino acid substitutions. The selected location has a well-ordered backbone conformation, but the side chains of the residues to be mutated are less well ordered than residues in other locations of the bundle. The 3-Cys site, which is largely sequestered from solvent, occupies a “box”; the sides are formed by the backbone of the helices and the bottom by the apolar side chains of Phe 31, Ile 14, and Ile 63. The aromatic residue of Phe 31 lies directly over the predicted metal-binding site and lines most of the bottom of the box. The top is formed by the main-chain and side chains of residues in the non-helical loops including Leu 21, as well as the apolar portion of Tyr 70 at the terminus of helix 3. His 72, which was entirely disordered in the NMR solution structure, also lies proximal to the site. Moreover, after introduction of the Cys side chains in one of the two preferred rotamers for Cys, the thiol SG atoms formed a nearly equilateral triangle with the side chains well oriented to form the desired site (inter SG distances = 3.5 − 4.5 Å). Overall, the location is ideal to explore the effects of hydrophobic sequestration in the present work. The sequence of α3DIV is shown in Table 1. After expression of a synthetic gene of α3DIV in E. coli followed by purification by HPLC (see Materials and Methods) the MW of α3DIV was determined to be 7945.1 Da by Electrospray Ionization Mass Spectrometry (ESI), which corresponds to α3DIV with the deletion of the N-terminal Met (calc MW = 7946.9 Da). The folding behavior of α3DIV was studied in solution by CD and NMR spectroscopy. The CD spectrum of 5 μM α3DIV has double minima at 208 and 222 nm at pH 8 with the molar ellipticity [θ] values characteristic of a well-folded α-helical construct (97% folded based on [θ]222) (Figure S1A). Furthermore, α3DIV remained well-folded between pH 3 and 9. The 1H-1H NOESY spectrum of 3 mM α3DIV shows chemical shift dispersions characteristic of a well-folded α-helical structure at pH 6 (Figure S1B). GuHCl-induced unfolding of α3DIV was performed by monitoring the change in ellipticity at 222 nm as a function of GuHCl concentration at pH 8. The resulting titration curve was plotted as concentration of folded protein vs. GuHCl concentration (Figure S2) and was fit to a two-state equilibrium,[30-32] yielding the free energy of unfolding (ΔGu) to be 2.5 kcal mole−1, with the degree of cooperativity (m) being 1.4 kcal mole−1 MGuHCl−1. The midpoint of the transition (Cm) occurred at a 1.8 M GuHCl concentration. These results indicate that replacing three Leu residues of the WT α3D with cysteines resulted in a loss of unfolding free energy of ~ 2.5 kcal mole−1.[20]
Figure 1.

PyMol model of α3DIV generated from the NMR structure of α3D. Cys residues, located at the C-terminal end of the bundle are shown as spheres. The protein backbone is shown in orange. The Cys site can be considered to be located in a hydrophobic “box” formed by the hydrophobic residues F31, I14, I63, L21, and Y70, shown as sticks.
Table 1.
Sequence of α3DIV.a
| Peptide | Sequence |
|---|---|
| α3DIV | MGS WAEF K QR LAAIKTR CQAL GG SEAECAAF E KE IAAFESE LQAY KGKGNPE |
| V E AL R KE AAAIRDE CQAY RHN |
Residues in red represent mutations to the WT α3D
Having established that α3DIV is well folded and stable in solution, we investigated the complexation of HgII, CdII, and PbII to this peptide. Metal ion binding titrations were performed by adding aliquots of stock solutions of metals to peptide solutions at pH values where each metal fully coordinated to α3DIV (pH 8.6 for HgII, and pH 8 for CdII and PbII). The progress of the titrations was monitored by the appearance of characteristic absorption bands due to Ligand to Metal Charge Transfer (LMCT) transitions at characteristic wavelengths,[15, 33, 34] upon formation of metal-thiolate bonds in the complexes HgII(α3DIV)−, CdII(α3DIV)−, and PbII(α3DIV)−. The resulting UV/Vis absorption spectra (Figure S3) and the molar absorption coefficients (Δε), at various wavelengths (Table 2) are consistent with the assignment that all three Cys thiolates of α3DIV are incorporated into the first coordination sphere of the metal ions.[15, 33, 34] CdII and PbII binding constants were determined to be 2.0×107 M−1 and 3.1×107 M−1, respectively, from the analysis of the titration data.[35] Due to the high affinity binding, the association constant of HgII could not be determined. Next, we examined pH dependent complexation of these metals to α3DIV by monitoring changes in the LMCT band as a function of pH. Titration data of HgII were fit to the release of one thiol proton upon forming a HgS3 complex from linear HgS2(SH) complex of α3DIV,[15] resulting in a pKa of 7.1±0.1 (Table 2, Figure S4). For CdII and PbII the titration curves (Figure S4) were fit to simultaneous dissociation of two Cys thiols,[15, 33, 34, 36] yielding pKa2 of 10.6±0.1 and 10.2±0.1 (Table 2), respectively. These pKa values are slightly more acidic than TRI peptides.[15, 33, 36] Nonetheless, these pKa values are consistent with the coordination modes of HgII as trigonal HgS3, CdII as pseudotetrahedral CdS3O (O being an exogenous water molecule), and PbII as trigonal pyramidal PbS3 complexes.
Table 2.
Physical Parameters of CdII, HgII and PbII complexes of α3DIV.
| Complex | UV/Vis λnm (Δε M−1cm−1) |
NMR δ (ppm) | Apparent pKa |
Binding Constant (Kb) (M−1)a |
|
|---|---|---|---|---|---|
| 113Cd | 199Hg | ||||
| Cd(α3DIV)− | 232 (18, 200) | 583 | 10.6±0.1b | 2.0×107 | |
| 595 | |||||
| Hg(α3DIV)− | 247 (12,500) | −244 | 7.1±0.1c | ||
| 265 (8,400) | |||||
| 295 (3,900) | |||||
| Hg(α3DIV)d | 240 (850) | −938e | |||
| Pb(α3DIV)− | 236 (18,000) | 10.2±0.1b | 3.1×107 | ||
| 260 (14,400) | |||||
| 278 (9,100) | |||||
| 346 (3,150) | |||||
Model used to obtain binding constants is MII + (α3DIV)3− ⇌ MII(α3DIV)− (Kb). These values represent the lower limit of Kb.
Model used to obtain pKa2 values for CdII and PbII is MII(α3DIVS(SH)2)+ ⇌ MII(α3DIV)− + 2H+ (Ka2).
Model used to obtain the pKa for HgII is HgII(α3DIVS2(SH)) ⇌ HgII(α3DIV)− + H+ (Ka).
Linear HgS2 complex of α3DIV.
199Hg NMR δ of linear HgS2 complex at pH 5.8.
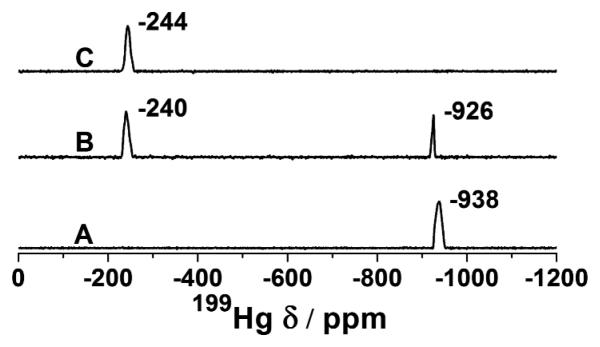
199Hg NMR and 199mHg PAC (Perturbed Angular Correlation) spectroscopies were used to probe the coordination environment around HgII bound to α3DIV at pH 5.8, 8.6, and 7.4. Based on the pKa value, HgII is expected to form a linear HgS2 complex at pH 5.8 and a trigonal HgS3 complex at pH 8.6, with a mixture of linear and trigonal complexes at the intermediate pH. 199Hg NMR chemical shifts (Figure 2) and 199mHg PAC spectral parameters (Figure S5, Table S1) of α3DIV at pH 5.8, 8.6, and 7.4 confirm that α3DIV forms a linear HgS2 complex at pH 5.8 and a trigonal HgS3 complex at pH 8.6, with the formation of both dithiolate- and trithiolate-HgII complexes at pH 7.4 in distorted geometries.[10, 37-43] 113Cd NMR spectrum of α3DIV has two resonances at δ = 595 and 583 ppm at pH 8 (Figure S6) indicating the presence of two CdII species with chemical shift values similar to what has been observed for 4-coordinate pseudotetrahedral CdS3O species.[36] 111mCd PAC spectroscopy was used to confirm the coordination environment and geometry of CdII complexes of α3DIV. 111mCd PAC spectrum of α3DIV has three Nuclear Quadrupole Interactions (NQIs), at ω0 = 0.35, 0.27 and 0.17 rad/ns (Figure S7, Table S2) at pH 8.1. The first two peaks at 0.35 rad/ns and 0.27 rad/ns agree strikingly well with a typical CdS3O signal in exo and endo conformations, respectively, as observed for TRI peptides.[36] The lowest frequency NQI at 0.17 rad/ns can be best assigned to a CdS3N species where N is His 72. Even though the chemical shift of 595 ppm in the 113Cd NMR spectrum (Figure S6) is lower than what has been reported for a CdS3N species,[44] Quantum chemical calculations show that a change in Cd-S bond length of 0.01 Å can cause a change in chemical shift of ~20 ppm (Hemmingsen et.al. unpublished results). Thus, tentatively, the 113Cd NMR resonance at δ = 595 ppm can be best assigned to the CdS3N species.
Figure 2.
199Hg NMR spectra of solutions containing 2.93 mM α3DIV and 0.8 equivalent of 199Hg(NO3)2 at pH 5.8 (A), 7.4 (B), and 8.6 (C).
In conclusion, we have been successful in engineering metal binding sites containing cysteine residues in an existing de novo antiparallel three-helix bundle, yielding α3DIV, a protein which is well folded and stable in solution, and capable of binding heavy metals with high affinity (>107 M−1). Spectroscopic properties of HgII, CdII and PbII complexes of α3DIV are very similar to existing parallel three-stranded coiled coils, showing that we have achieved our objective of preparing a single polypeptide chain capable of binding metal ions with high affinity and predefined coordination geometry. Understanding the biochemistry of binding of heavy metals to a single polypeptide chain can be potentially useful for developing peptide-based water purification systems or specific heavy metal ion sensors. Clearly, having a single peptide chain rather than self-associating helical peptides makes these goals more achievable. Further studies will explore the possibilities of preparing similar constructs containing asymmetric metal binding sites such as those in Type I blue copper proteins, and to explore the effects of the electronic structure of the aromatic residue, Phe 31, or second-shell effects by replacing any of the surrounding residues as well as making catalytic metalloproteins.
Experimental Section
A synthetic DNA of α3DIV was cloned into pET-15b (Celtek Genes) vector and expressed in E. coli BL21(DE3) competent cells (Stratagene) grown in M9 media. After sonication and heat denaturation at 55°C the lyophilized powder was purified on a C18 preparative reverse phase HPLC column over a linear gradient of 100% H2O/0.1% TFA to 10% H2O/90% Acetonitrile/0.1% TFA over 50 minutes. The MW of the pure peptide was determined to be 7945.1 Da by ESI which corresponds to α3DIV without the first Met (calc MW 7946.9 Da). Yield of the pure protein was 17 mg/L. Concentration of the protein was determined by A280nm using the known extinction coefficient ε280 = 8.61 mM−1.[45]
CD spectra were collected on an Aviv model 202 CD spectrometer using rectangular open top quartz cuvettes at 25°C. GuHCl titration experiments were carried out using a Microlab 500 series syringe pump automatic titrator controlled by Aviv software. Titrations were carried out by mixing two separate solutions of 10 μM peptide containing 0.0 and 7.63 M GuHCl in 10 mM phosphate buffer at pH 8. Observed ellipticities in millidegree were converted to molar ellipticities (deg cm2 dmole−1 res−1) as described previously,[33] using 59 amino acids in the helical region of the protein. GuHCl titration data were fit to an equation derived based on a two-state model. [30-32] A 1H-1H NOESY experiment was performed using standard procedures.[46]
CdII, HgII, and PbII binding titrations were performed at room temperature on a Cary 100 Bio UV/Vis spectrometer using anaerobic cuvettes (Starna Inc.) of 1-cm path length by adding aliquots of stock solutions of different metals. Peptide samples of 20 – 30 μM were prepared in 50 mM of appropriate buffers (TRIS for pH 8 and CHES for pH 8.6) and 40 – 60 μM TCEP inside an inert atmosphere box (Vacuum Atmospheres Co., model OMNI-LAB). Stock solutions of 8 mM CdCl2, 7.37 mM HgCl2, and 5.16 mM PbCl2 were also prepared inside the inert atmosphere box. In each case, difference spectra were obtained by subtracting the background spectra of samples containing peptide, buffer, and TCEP. Direct titration data were analyzed by non-linear least squares fits to an equation used previously.[47] The difference molar extinction coefficients (Δε) were determined based on the total metal concentrations after subtracting the background spectra.
pH titrations were performed as described previously,[34, 36] by adding small aliquots of KOH to solutions containing 20 – 30 μM of α3DIV and 1 equivalent of CdCl2, HgCl2′ or PbCl2. In the cases of CdII and PbII, the titration data were analyzed using the model: simultaneous two proton dissociation as described previously.[34, 36] For HgII, the data were analyzed using the model: dissociation of one thiol proton of Cys.[15]
113Cd NMR and 199Hg NMR experiments were performed according to standard procedures.[48] An exponential line broadening of 200 Hz was applied prior to Fourier transformation while processing 199Hg NMR data.
Samples for 111mCd PAC measurement contained 300 μM α3DIV, 1/12 equivalent of CdII, and 55% sucrose (w/w) in 20 mM TRIS buffer at pH 8.1. Sample preparation and data collection were performed at the University of Copenhagen.[36, 48] Samples for 199mHg PAC experiments contained 200 μM α3DIV, 80 μM HgII, and 55% sucrose in 100 mM of an appropriate buffer (phosphate for pH 5.8, 7.4 and CHES for pH 8.6). Sample preparation and data collection were performed at CERN.[41]
Supplementary Material
Footnotes
V.L.P. thanks the National Institute of Health for support of this research (ES012236), P.W.T. and L.H. thank ISOLDE collaboration at CERN for the 199mHg beam time grant (experiment IS488) and The Danish Council for Independent Research | Natural Sciences.
Supporting information for this article is available on the WWW under http://www.angewandte.org
References
- 1.Holm RH, Kennepohl P, Solomon EI. Chemical Reviews. 1996;96:2239. doi: 10.1021/cr9500390. [DOI] [PubMed] [Google Scholar]
- 2.Lu Y, Yeung N, Sieracki N, Marshall NM. Nature. 2009;460:855. doi: 10.1038/nature08304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibney BR, Dutton PL. Adv. Inorg. Chem, Vol 51. 2001;51:409. [Google Scholar]
- 4.Reedy CJ, Gibney BR. Chem. Rev. 2004;104:617. doi: 10.1021/cr0206115. [DOI] [PubMed] [Google Scholar]
- 5.Kennedy ML, Petros AK, Gibney BR. J. Inorg. Biochem. 2004;98:727. doi: 10.1016/j.jinorgbio.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Cochran FV, Wu SP, Wang W, Nanda V, Saven JG, Therien MJ, DeGrado WF. Journal of the American Chemical Society. 2005;127:1346. doi: 10.1021/ja044129a. [DOI] [PubMed] [Google Scholar]
- 7.Ghirlanda G, Osyczka A, Liu W, Antolovich M, Smith KM, Dutton PL, Wand AJ, DeGrado WF. Journal of the American Chemical Society. 2004;126:8141. doi: 10.1021/ja039935g. [DOI] [PubMed] [Google Scholar]
- 8.Peacock AFA, Iranzo O, Pecoraro VL. Dalton Transactions. 2009:2271. doi: 10.1039/b818306f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pecoraro VL, Peacock AFA, Iranzo O, Luczkowski M. Bioinorganic Chemistry. American Chemical Society; 2009. p. 183. [Google Scholar]
- 10.Dieckmann GR, McRorie DK, Tierney DL, Utschig LM, Singer CP, O'Halloran TV, Penner-Hahn JE, DeGrado WF, Pecoraro VL. J. Am. Chem. Soc. 1997;119:6195. [Google Scholar]
- 11.Touw DS, Nordman CE, Stuckey JA, Pecoraro VL. Proc. Natl. Acad. Sci., U.S.A. 2007;104:11969. doi: 10.1073/pnas.0701979104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee K-H, Cabello C, Hemmingsen L, Marsh ENG, Pecoraro VL. Angew. Chem., Int. Ed. 2006;45:2864. doi: 10.1002/anie.200504548. [DOI] [PubMed] [Google Scholar]
- 13.Iranzo O, Cabello C, Pecoraro VL. Angew. Chem., Int. Ed. 2007;46:6688. doi: 10.1002/anie.200701729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peacock AFA, Hemmingsen L, Pecoraro VL. Proceedings of the National Academy of Sciences. 2008;105:16566. doi: 10.1073/pnas.0806792105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iranzo O, Ghosh D, Pecoraro VL. Inorg. Chem. 2006;45:9959. doi: 10.1021/ic061183e. [DOI] [PubMed] [Google Scholar]
- 16.Marsh ENG, DeGrado WF. Proc. Natl. Acad. Sci., U.S.A. 2002;99:5150. doi: 10.1073/pnas.052023199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Summa CM, Rosenblatt MM, Hong J-K, Lear JD, DeGrado WF. J. Mol. Biol. 2002;321:923. doi: 10.1016/s0022-2836(02)00589-2. [DOI] [PubMed] [Google Scholar]
- 18.Kaplan J, DeGrado WF. Proc. Natl. Acad. Sci., U.S.A. 2004;101:11566. doi: 10.1073/pnas.0404387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faiella M, Andreozzi C, de Rosales RTM, Pavone V, Maglio O, Nastri F, DeGrado WF, Lombardi A. Nat. Chem. Biol. 2009;5:882. doi: 10.1038/nchembio.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walsh STR, Cheng H, Bryson JW, Roder H, DeGrado WF. Proc. Natl. Acad. Sci., U.S.A. 1999;96:5486. doi: 10.1073/pnas.96.10.5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Regan L. Advances in Molecular and Cell Biology. 1997;22A:51. [Google Scholar]
- 22.Hellinga HW. Folding & Design. 1998;3:R1. doi: 10.1016/S1359-0278(98)00001-7. [DOI] [PubMed] [Google Scholar]
- 23.Gouda H, Torigoe H, Saito A, Sato M, Arata Y, Shimada I. Biochemistry. 1992;31:9665. doi: 10.1021/bi00155a020. [DOI] [PubMed] [Google Scholar]
- 24.Schneider JP, Lombardi A, DeGrado WF. Folding & Design. 1998;3:R29. doi: 10.1016/S1359-0278(98)00011-X. [DOI] [PubMed] [Google Scholar]
- 25.Bryson JW, Desjarlais JR, Handel TM, DeGrado WF. Protein Sci. 1998;7:1404. doi: 10.1002/pro.5560070617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansson JS, Gibney BR, Skalicky JJ, Wand AJ, Dutton PL. J. Am. Chem. Soc. 1998;120:3881. [Google Scholar]
- 27.Walsh ST, Cheng RP, Wright WW, Alonso DO, Daggett V, Vanderkooi JM, DeGrado WF. Protein Science. 2003;12:520. doi: 10.1110/ps.0223003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walsh STR, Sukharev VI, Betz SF, Vekshin NL, DeGrado WF. Journal of Molecular Biology. 2001;305:361. doi: 10.1006/jmbi.2000.4184. [DOI] [PubMed] [Google Scholar]
- 29.Walsh STR, Lee AL, DeGrado WF, Wand AJ. Biochemistry. 2001;40:9560. doi: 10.1021/bi0105274. [DOI] [PubMed] [Google Scholar]
- 30.Agashe VR, Udgaonkar JB. Biochemistry. 1995;34:3286. doi: 10.1021/bi00010a019. [DOI] [PubMed] [Google Scholar]
- 31.Santoro MM, Bolen DW. Biochemistry. 1988;27:8063. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- 32.Santoro MM, Bolen DW. Biochemistry. 1992;31:4901. doi: 10.1021/bi00135a022. [DOI] [PubMed] [Google Scholar]
- 33.Matzapetakis M, Farrer BT, Weng T-C, Hemmingsen L, Penner-Hahn JE, Pecoraro VL. J. Am. Chem. Soc. 2002;124:8042. doi: 10.1021/ja017520u. [DOI] [PubMed] [Google Scholar]
- 34.Matzapetakis M, Ghosh D, Weng T-C, Penner-Hahn JE, Pecoraro VL. J. Biol. Inorg. Chem. 2006;11:876. doi: 10.1007/s00775-006-0140-7. [DOI] [PubMed] [Google Scholar]
- 35.Chakraborty S, Touw DS, Peacock AFA, Stuckey J, Pecoraro VL. Journal of the American Chemical Society. 2010;132:13240. doi: 10.1021/ja101812c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iranzo O, Lee KH, Jakusch T, Hemmingsen L, Pecoraro VL. Chem. Eur. J., 2009;15:3761. doi: 10.1002/chem.200802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright JG, Natan MJ, MacDonnell FM, Ralston DM, O'Halloran TV. Progress in Inorganic Chemistry:Bioinorganic Chemistry. 1990;38:323. [Google Scholar]
- 38.Huffman DL, Utschig LM, O'Halloran T. In: Metal Ions in Biological Systems. Sigel H, editor. Vol. 34. Marcel Dekker, Inc.; New York: 1997. p. 503. [PubMed] [Google Scholar]
- 39.Butz T, Troger W, Pohlmann T, Nuyken O. Z. Naturforsch. 1992;47a:85. [Google Scholar]
- 40.Dieckmann GR. University of Michigan; Ann Arbor: 1995. Ph.D. Thesis. [Google Scholar]
- 41.Iranzo O, Thulstrup PV, Ryu S-B, Hemmingsen L, Pecoraro VL. Chem. Eur. J. 2007;13:9178. doi: 10.1002/chem.200701208. [DOI] [PubMed] [Google Scholar]
- 42.Lippert C, Troger W, Butz T. In: 30th Zakopane School of Physics. Gorlich EA, editor. Krakow; Poland: 1995. [Google Scholar]
- 43.Troger W. Hyperfine Interactions. 1999;120/121:117. [Google Scholar]
- 44.Coleman JE. In: Metallobiochemistry Part D: Physical and Spectroscopic Methods for Probing Metal Ion Environment in Metalloproteins. R. JF, Vallee BL, editors. Vol. 227. Academic Press; 1993. p. 16. [Google Scholar]
- 45.Aitken A, Learmonth M. l. In: The Protein Protocols Handbook. Walker JM, editor. Humana Press; 1996. p. 3. [Google Scholar]
- 46.Iranzo O, Chakraborty S, Hemmingsen L, Pecoraro VL. Journal of the American Chemical Society. 2010 doi: 10.1021/ja104433n. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chakraborty S, Touw DS, Peacock AFA, Stuckey JA, Pecoraro VL. Journal of the American Chemical Society. 2010;132:13240–13250. doi: 10.1021/ja101812c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luczkowski M, Stachura M, Schirf V, Demeler B, Hemmingsen L, Pecoraro VL. Inorganic Chemistry. 2008;47:10875. doi: 10.1021/ic8009817. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.