Abstract
Recording of four-dimensional (4D) spectra for proteins in the solid state has opened new avenues to obtain virtually complete resonance assignments and three-dimensional (3D) structures of proteins. As in solution state NMR, the sampling of three indirect dimensions leads per se to long minimal measurement time. Furthermore, artifact suppression in solid state NMR relies primarily on radio-frequency pulse phase cycling. For an n-step phase cycle, the minimal measurement times of both 3D and 4D spectra are increased n times. To tackle the associated ‘sampling problem’ and to avoid sampling limited data acquisition, solid state G-Matrix Fourier Transform (SS GFT) projection NMR is introduced to rapidly acquire 3D and 4D spectral information. Specifically, (4,3)D (HA)CANCOCX and (3,2)D (HACA)NCOCX were implemented and recorded for the 6 kDa protein GB1 within about 10% of the time required for acquiring the conventional congeners with the same maximal evolution times and spectral widths in the indirect dimensions. Spectral analysis was complemented by comparative analysis of expected spectral congestion in conventional and GFT NMR experiments, demonstrating that high spectral resolution of the GFT NMR experiments enables one to efficiently obtain nearly complete resonance assignments even for large proteins.
Keywords: Magic-angle spinning, Chemical shift assignments, GB1, Correlation spectroscopy
Introduction
In recent years, solid-state NMR (SSNMR) spectroscopy has advanced as a powerful tool for comprehensive, high-resolution structural studies of proteins (McDermott et al. 2000; Castellani et al. 2002; Bockmann et al. 2003). Significant methodological developments include effective heteronuclear decoupling schemes (Bennett et al. 1995; Fung et al. 2000; Detken et al. 2002), robust, band-selective heteronuclear polarization transfer schemes employing adiabatic (Baldus et al. 1996) and SPECIFIC CP (Baldus et al. 1998) elements, broadband DARR homonuclear polarization transfer methods (Takegoshi et al. 2001; Morcombe et al. 2004) and improvements in MAS probes in terms of both B1 homogeneity (Paulson et al. 2004) and reduction of electric field-induced sample heating (Stringer et al. 2005; Doty et al. 2006).
As a result, it is now possible to determine the three-dimensional (3D) structures of macroscopically disordered solid proteins at atomic-resolution (Franks et al. 2008; Wasmer et al. 2008) and study site-resolved dynamics using SSNMR spectroscopy (Lorieau and McDermott 2006). This has opened new avenues to study important biological systems such as membrane proteins (which constitute up to ~30% of the genomes of living organisms) and amyloid fibrils, which are otherwise very difficult to approach using the traditional structural biology tools. Moreover, SSNMR spectroscopy can provide structural information complementary to that obtained from X-ray crystallography or solution state NMR. Various multidimensional homonuclear and heteronuclear NMR experiments have been developed akin to those used in solution-state NMR spectroscopy (Hohwy et al. 1999; Hong 1999; Rienstra et al. 2000; Detken et al. 2001; Baldus 2002). The improved resolution and additional unique correlations of multidimensional experiments have been employed to assign a number of proteins in the solid state (McDermott et al. 2000; Pauli et al. 2001; Igumenova et al. 2004a, b; Marulanda et al. 2004; Franks et al. 2005; Pintacuda et al. 2007; Sperling et al. 2010). Novel 4D experiments correlating interresidue 13C and 15N chemical shifts to establish unique connectivities and increasing the unique correlation information of a spectrum have been demonstrated (Franks et al. 2007).
Given the unambiguous correlation information and improved resolution of high dimensional correlation experiments, spectra with very high dimensionality are attractive. However, in the “sampling-limited” NMR data acquisition regime, conventional acquisition of multidimensional data leads to use of valuable spectrometer time to sample indirect dimensions but not to achieve sufficient signal-to-noise ratios (Szyperski et al. 2002; Kim and Szyperski 2003; Szyperski and Atreya 2006); typical two-dimensional (2D), three-dimensional (3D), and four-dimensional (4D) NMR spectra need minutes, hours or days, respectively, for completion, whereas five- and six-dimensional (5D, 6D) experiments are not feasible given the time restraints. In view of the staggering increase of minimal measurement time with increasing dimensionality, which constitutes the ‘NMR sampling problem’ (Szyperski and Atreya 2006), an attractive approach is to encode the information of a high-dimensional spectrum in lower-dimensional spectra. This forms the basis of G-matrix Fourier transform (GFT) projection NMR spectroscopy (Kim and Szyperski 2003; Atreya and Szyperski 2004; Atreya et al. 2005; Eletsky et al. 2005; Liu et al. 2005; Shen et al. 2005; Szyperski and Atreya 2006; Atreya et al. 2007; Zhang et al. 2008). GFT NMR is based on joint sampling of chemical shifts in a single dimension, such that linear combinations of chemical shifts are phase-sensitively detected in a pure absorption mode and then edited into sub-spectrum by linear combination. Thereby, GFT NMR enables one to speed up NMR data collection by orders of magnitude. Importantly, the editing with the G-matrix transformation ensures that the number of peaks in each of the sub-spectra is the same as in the original high-dimensional parent spectrum. Moreover, since linear combinations of shifts are registered, the spectral width in the GFT dimension equals the sum of the spectral widths of the spectral widths of the jointly sampled shifts, resulting in increased signal dispersion.
Here, we introduce SS GFT NMR spectroscopy which meets with an urgent demand considering that artifact suppression in solid state NMR relies primarily on radio-frequency pulse phase cycling: for an n-step phase cycle, the minimal measurement times of both 3D and 4D spectra are increased n times. Specifically, we implemented SS GFT NMR experiments for rapid resonance assignment. These experiments were recorded for 56-residue protein GB1, and their utility for resonance assignment is discussed.
Materials and methods
Sample preparation and NMR spectroscopy
Protein GB1 was prepared with 15N,13C-labeling as a microcrystalline solid as described previously (Franks et al. 2005); an 18-mg (2.7 μmol) quantity was packed in 3.2 limited speed rotor (Varian, Inc., Fort Collins, Colorado). NMR experiments were performed on a four-channel, 500 MHz Varian Infinity Pulse Spectrometer, equipped with a Balun™ 1H-13C-15N 3.2-mm MAS probe. The MAS rate was maintained at 11,111 ± 3 Hz, and the variable temperature unit was set to 273 K with a flow rate of 100 standard cubic feet per hour (scfh), resulting in a sample temperature of 280 ± 4 K as determined by methanol calibration (Van Geet 1968).
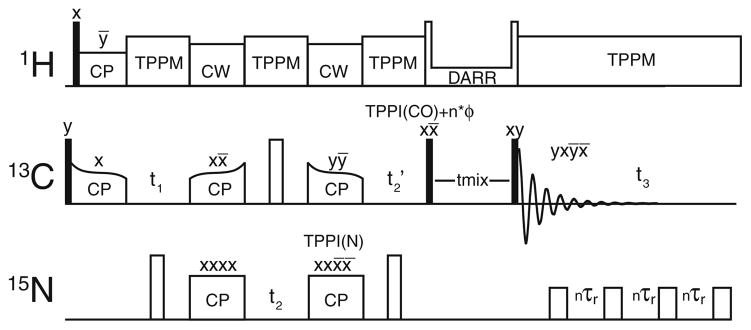
The basis for SS GFT (4,3)D (HA)CANCOCX is the three-channel correlation scheme (Fig. 1) as previously utilized for 4D CNCC spectroscopy (Franks et al. 2007). To indicate which chemical shifts are jointly sampled, the corresponding nuclei are underlined in the name of the experiments (Kim and Szyperski 2003). 13C Boltzmann polarization is combined with the cross polarized signal from 1H, utilizing a short CP time to maximize signal intensity on Cα sites. Following t1 evolution on CA[i], CA[i] to N[i], where i denotes the residue number, SPECIFIC CP (Baldus et al. 1998) transfer is achieved using conditions described below. After frequency labeling with the chemical shifts of N[i] during t2, polarization is transferred again in band-selective manner, principally to CO[i−1]. CO chemical shifts are then jointly sampled with N[i] during t2 but scaled with a factor κ (Kim and Szyperski 2003). Following broadband homonuclear DARR mixing, 13C signals are detected in the direct dimension under rotor-synchronized 15N decoupling. Throughout all chemical shift evolution and detection periods, TPPM 1H-decoupling (Bennett et al. 1995) is applied, whereas high power continuous wave decoupling is utilized during the SPECIFIC CP transfers.
Fig. 1.
Pulse sequence used for (4,3)D HA(CA)NCOCX GFT experiments. Filled and open rectangles represent π/2 and π pulses, respectively. 15N decoupling was applied during acquisition by a π train with each pulse separated by 10 rotor periods. Efficient, band selective 15N–13C (N–CO) and 13C–15N (CA–N) polarization transfer were achieved with adiabatic SPECIFIC CP (Baldus et al. 1998), while DARR (Takegoshi et al. 2003) (RAD, Morcombe et al. 2004) was used for broadband 13C–13C mixing. Time Proportional Phase Incrementation (TPPI) (Marion and Wuthrich 1983) was applied to each pulse following time evolution periods. An additional cumulative phase was added the following t2′ (nφarb) for manipulation of the spectral window (Keeler and Neuhaus 1985). Independent cosine and sine modulated subspectra are recorded by adjusting the starting point of the TPPI phases added to the second 15N CP and the 13C π/2 prior to mixing
The widths for high power π/2 radio-frequency (rf) pulses on 1H, 13C, and 15N were 2.0, 3.0, and 5.0 μs, respectively. TPPM decoupling conditions were , 16°, 7.6 μs. CA-N and N-CO SPECIFIC CP (Baldus et al. 1998) conditions were optimized using tangent ramp cross polarization (CP) was utilized for polarization transfers following the notation of Detken et al. (Detken et al. 2001) with slight modification (Franks et al. 2007):
| (1) |
where the adiabaticity parameter is defined as
| (2) |
Constant amplitude spin lock fields were utilized on 1H and 15N, and tangential amplitude ramps applied to the 13C channel. The first (H–C) CP period was 450 μs with and (Δ/2π = 8 kHz; β/2π = −3.5 kHz), the second (CA-N) CP period was 6 ms with (Δ/2π = 2 kHz; β/2π = 400 Hz) and . The off-resonance N-CO transfer step had a duration of 8 ms with of 36.5 kHz (Δ/2π = 2 kHz; β/2π = −400 Hz) and of ~28 kHz. CW decoupling with was applied during both C–N and N–C CP periods. DARR (Takegoshi et al. 2001, 2003; Morcombe et al. 2004) mixing (18 ms) was applied with the to yield 13C side-chain correlations.
For (4,3)D (HA)CANCOCX, the maximal evolution times were t1(13CA, 32 × 135 μs) = 4.32 ms, t2(15N, 48 × 180 μs) = 8.64 ms, t2′(13CO, 48 × 90 μs; κ = 0.5) = 4.32 ms and t3(Acquisition) = 23.04 ms (1,536 × 15 μs), while the recycle delay was set to 2 s. 4 transients per increment (corresponding to a minimal 4-step phase cycle) were acquired, yielding a total measurement time of 10.8 h.
For (3,2)D (HACA)NCOCX, frequency labeling on CA is omitted and the maximum evolution times were t1(15N, 96 × 180 μs) = 17.28 ms, t1′(13CO, 96 × 90 μs; κ = 0.5) = 8.64 ms, and t2(Acquisition) = 23.04 ms (1,536 × 15 μs). 8 transients were acquired per FID yielding a total measurement time of 1.3 h. Chemical shifts are referenced to the external standard adamantane at 40.48 ppm. Spectra were processed in the program nmrPipe (Delaglio et al. 1995) and analyzed using the program Sparky 3.0 (Goddard and Kneller 2006).
NMR data processing
The (4,3)D experiments were processed as follows. In the direct dimension, three points were backwards linear predicted, Lorentzian–Gaussian transformations with 60 Hz net line-broadening were applied, and the time domain data multiplied with a 72° phase shifted sine-bell function, zero-filled to 2048 points, and Fourier transformed. In t2(15N;13C′), a Lorentzian-Gaussian transformation with 50 Hz net line-broadening was applied, time domain data were multiplied by a 81° phase shifted sine-bell, zero filled to 256 points, and Fourier transformed. In t3(13Cα), time domain data were expanded by 8 points by linear prediction, subject to a Lorentzian–Gaussian transformation with 80 Hz net line-broadening, multiplied with 81° phase shifted sine-bell function, zero-filled to 128 points and Fourier transformed.
The (3,2)D experiments were processed as follows. In the direct dimension, three points were backward linear predicted. The resulting time domain data were (1) subjected to a Lorentzian-Gaussian transformation with 60 Hz net broadening, (2) multiplied by a 72° phase shifted sine-bell function, (3) zero-filled to 4,096 points, (4) Fourier transformed, and (5) subjected to a polynomial baseline correction. The indirect dimensions were processed with Lorentzian–Gaussian transformations with 20 Hz net line-broadening, zero-filled to 1,024 points and Fourier transformed.
The (4,3)D and (3,2)D sub-spectra constituting the GFT NMR experiments were obtained by linearly combination in the frequency domain (Kim and Szyperski 2003).
Simulations
Simulations were performed in order to estimate the hypothetical spectral resolution of GFT NMR spectra for other proteins with known chemical shifts. The procedure followed those employed previously by (Tycko 1996) and (Franks et al. 2007), but with suitable adjustments for the line-widths of peaks in the GFT-dimensions, where the line-widths of jointly sampled shifts add up if non-constant time evolution periods are implemented ((Kim and Szyperski 2003); Fig. 1). As such, several proteins ranging in molecular weight from 6 to 81 kDa, were considered for calculation of the linear combinations of chemical shifts as registered in the GFT NMR experiments. In addition, the definition of spectral degeneracy (and conversely, resolution) is modified as follows. The spin system of residue i is considered ‘degenerate’ if all N, CA, CB and C′ resonance frequencies are within one line width of any other frequency j in dimension m; this is the case when the following inequality.
| (3) |
is satisfied for all values of j and m, where ∧i,m denotes the resonance frequency of interest and lwm represents the line width.
For the GFT dimension, one then has that
| (4) |
where X and Y are the jointly sampled dimensions. The simulations were performed under a variety of assumed linewidths, in order to assess the relative benefits of enhanced digital resolution in indirect dimensions, which is available with SS GFT NMR. The directly detected line-width was assumed to be 0.5 ppm (which is typical for 13C resonances in uniformly 13C labeled solid proteins at 500 MHz).
Results and discussion
Benefits of SS GFT NMR
For both solution and state-state NMR, the benefits of increasing the dimensionality are well known. Arguably, the benefit of increased dimensionality is higher in the solid state than in solution NMR, since resolution in 13C-13C and 15N-13C SS 2D planes is not yet as high as typically observed in 2D 1H-15N planes of proteins in solution. We have previously demonstrated the advantages arising from increasing the number of dimensions from three to four for several solid proteins and with various line widths (Franks et al. 2007). Nonetheless, practical limitations—including finite sampling, probe and instrument stability over long measurement times—prevent one from recording the high dimensional experiments with the desired maximal evolution times in the indirect dimensions. Hence, the use of GFT NMR spectroscopy (Kim and Szyperski 2003) to speed up the acquisition of high-dimensional spectral information in the solid state represents a valuable approach.
To guide implementation of SS GFT NMR experiments, we predicted the number of resolved spin systems in conventional and GFT spectra for protein GB1 (Table 1). For (4,3)D (HA)CANCOCX, which was recorded in 10.8 h, one obtains in agreement with the experimental results, vide infra) that 53 out of 55 spin systems are resolved: only the signals arising from E15 and E42 are not resolved in at least one of the two sub-spectra. If the 4D congener experiment is recorded within the same amount of time, maximal evolution times must be dramatically reduced and the signals of only 41 residues can be expected to be resolved. It would require more than a week to record the 4D experiment with the same maximal evolution times as the (4,3)D GFT NMR experiment; yet no additional peaks would be resolved. An even more pronounced situation is encountered when comparing (3,2)D GFT and conventional 3D experiments: the use of GFT NMR experiments recorded in 1.3 h resolves 54 of 55 possible signals, while the conventional congeners recorded with the same measurement time provides too little resolution to be practically useful. Acquisition of the conventional 3D spectrum with the same maximum evolution time as in the GFT experiment would require more than 30 times as much measurement time. Considering that, in spite of the short measurement times, the GFT spectra exhibit adequate experimental sensitivity (Table 2), we conclude that GFT NMR represents a valuable approach to avoid sampling limited data acquisition (Szyperski et al. 2002) for proteins in the solid state.
Table 1.
Number of spin systems of GB1 expected to be resolved in GFT and conventional NMR spectra recorded with either the same measurement time or maximal evolution times
| Experiment type | Time (h)a | Digital resolution (ppm)b | Number of resolved peaksc |
|---|---|---|---|
| (HA)CANCOCX (4,3)D GFT | 10.8 | (15N) 2.3 (13CA, 13CO) 1.8 |
53 (50, 48, 50, 47) |
| (HA)CANCOCX 4D | 10.8 | (15N) 4.6 (13CA) 4.9 (13CO) 5.5 |
41 |
| (HA)CANCOCX 4D | 172.8 | (15N) 2.3 (13CA, 13CO) 1.8 |
53 |
| (HA)CONCACX (4,3)D GFT | 10.8 | (15N) 2.3 (13CA, 13CO) 1.8 |
53 (50, 48, 50, 39) |
| (HA)CONCACX 4D | 10.8 | (15N) 4.6 (13CA) 4.9 (13CO) 5.5 |
35 |
| (HA)CONCACX 4D | 172.8 | (15N) 2.3 (13CA, 13CO) 1.8 |
53 |
| (HACA)NCOCX (3,2)D GFT | 1.3 | (15N) 1.1 (13CO) 0.9 |
54 (40, 48, 53, 37) |
| (HACA)NCOCX 3D | 1.3 | (15N) 9.1 (13CO) 11.1 |
18 |
| (HACA)NCOCX 3D | 42.6 | (15N) 1.1 (13CO) 0.9 |
54 |
| (HACO)NCACX (3,2)D GFT | 1.3 | (15N) 1.1 (13CA) 0.9 |
52 (52, 37, 41, 28) |
| (HACO)NCACX 3D | 1.3 | (15N) 9.1 (13CA) 11.1 |
18 |
| (HACO)NCACX 3D | 42.6 | (15N) 1.1 (13CA) 0.9 |
55 |
Times were fixed to match data taken for this study
Acquisition linewidth was assumed to be 0.5 ppm, digital resolution values were taken from experiments performed in this study
Number of CA and/or CB resonances for each amino acid residue resolved in any sub-spectrum, parentheses indicate number of sum, difference, central peak, and both sum and difference frequencies resolved in each simulated GFT experiment for GB1
Table 2.
Average signal-to-noise ratios for spectra
| Experiment type | Measurement time (h) | Average signal to noise | Simulated resolved residues | Actual resolved residues |
|---|---|---|---|---|
| (HA)CANCOCX (4,3)D GFT | 10.8 | |||
| Sum | 13.5 | 50 | 55 | |
| Difference | 13.3 | 48 | 55 | |
| (HACA)NCOCX (3,2)D GFT | 1.3 | |||
| Sum | 11.6 | 40 | 41 | |
| Difference | 12.9 | 48 | 39 | |
| Central peaks | 24.0 | 53 | 54 |
Comparable benefits of GFT NMR are expected when considering the hypothetical resolution expected for other proteins (Table 3), including Het-S fibrils (Mw = 5 kDa for the rigid domain), bovine pancreatic trypsin inhibitor (BPTI; Mw = 6 kDa), ubiquitin (Mw = 8.6 kDa), maltose binding protein (MBP; Mw = 42 kDa), and malate synthase G (MSG; Mw = 82 kDa). For the class of small (Mw < 10 kDa) proteins, it is expected that the (3,2)D experiments would resolve ≥80% of the spin systems in at least one sub-spectrum, and approximately half of the spin systems all sub-spectra. With the addition of another indirect dimension in the (4,3)D congeners, one expects a resolution that is either approximately the same or greater than in the conventional 3D and 4D experiment. For example, one can expect that >70% of the spin systems of MBP (42 kDa) are resolved in all (4,3)D sub-spectra, while 90% are expected to be resolved in at least one sub-spectrum. Respectively, 46 and 78% are expected to be resolved for MSG (82 kDa).
Table 3.
Predicted resolution of (3,2)D and (4,3)D SS GFT NMR compared with conventional experiments
| Proteina (size)b | (4,3)D (HA)CANCOCX | (4,3)D (HA)CONCACX | (3,2)D (HACO)NCACX | (3,2)D (HACA)NCOCX | |
|---|---|---|---|---|---|
| Het-Sj 45(43) | Centrald | 42 | 43 | 43 | 42 |
| Sume | 41 | 40 | 33 | 31 | |
| Difff | 42 | 35 | 31 | 30 | |
| Allg | 41 | 34 | 22 | 21 | |
| Anyh | 42 | 43 | 43 | 34 | |
| Conventionali | 42 | 42 | 43 | 42 | |
| GB1 56 (56) | Central | 55 | 55 | 52 | 54 |
| Sum | 55 | 53 | 37 | 41 | |
| Difference | 55 | 55 | 43 | 48 | |
| All | 51 | 53 | 28 | 38 | |
| Any | 55 | 55 | 49 | 54 | |
| Conventional | 55 | 55 | 55 | 55 | |
| BPTI 58 (58) | Central | 53 | 53 | 47 | 48 |
| Sum | 48 | 46 | 28 | 37 | |
| Difference | 53 | 44 | 23 | 30 | |
| All | 46 | 39 | 17 | 24 | |
| Any | 57 | 56 | 47 | 49 | |
| Conventional | 57 | 55 | 53 | 53 | |
| Ubiquitin 76 (76) | Central | 71 | 71 | 69 | 67 |
| Sum | 74 | 67 | 50 | 48 | |
| Difference | 74 | 71 | 48 | 52 | |
| All | 74 | 61 | 38 | 40 | |
| Any | 75 | 75 | 72 | 68 | |
| Conventional | 75 | 75 | 71 | 71 | |
| MBP 370 (356) | Central | 282 | 275 | 144 | 167 |
| Sum | 292 | 222 | 88 | 110 | |
| Difference | 295 | 202 | 76 | 112 | |
| All | 256 | 150 | 43 | 77 | |
| Any | 339 | 306 | 170 | 187 | |
| Conventional | 347 | 320 | 275 | 282 | |
| MSG 723 (711) | Central | 413 | 409 | 162 | 186 |
| Sum | 434 | 296 | 100 | 114 | |
| Difference | 438 | 282 | 81 | 137 | |
| All | 334 | 193 | 48 | 85 | |
| Any | 564 | 506 | 206 | 226 | |
| Conventional | 635 | 604 | 409 | 413 |
Previously unassigned chemical shift values were assumed to be a common (degenerate) value of 999.9 ppm. Het-S assignments are missing 10 13C′ carbons
The chemical shift values for each protein were derived from the following BMRB entries and references: Het-S fibrils (Siemar et al. 2005,2006) GB1 in the solid state (BMRB 15156) (Franks et al. 2005); BPTI in solution (BMRB 5358) (McDermott et al. 2000); ubiquitin in solution (BMRB 5387) (Flynn et al. 2002); maltose binding protein in solution (BMRB 4354) (Gardner et al. 1998) malate synthase G in solution (BMRB 5471) (Tugarinov et al. 2002)
The size indicates the total number of residues present in the protein and, in parentheses, the number resolved with ideal resolution (0.001 ppm). Due to the absence of amide 1H resonances in our simulations, not all residues are resolved in the simulated spectra at this limit
1 ppm linewidths are assumed for each evolution period, with the synchronous dimension having an additive linewidth, and 0.5 ppm in the direct dimension
Central peaks are calculated assuming only X dimension (where X is the first component of the synchronously evolved (X, Y) dimension)
Summation peaks are calculated as ΩX + ΩY
Difference peaks are calculated as ΩX − ΩY
The resonance is counted only if it is resolved in each of the summation, difference, and central peaks spectra
The resonance is counted if it is resolved in one or more of the summation, difference, and central peaks spectrum
Presuming that the full grid of the XD experiment is sampled; this experiment would require an order of magnitude more time than the corresponding set of SS GFT data simulated here
The simulations include amide Nitrogen, α-carbon, β-carbon, and Carbonyl only
In contrast to solution NMR, linewidths in SS NMR do not depend on the molecular weight Mw. Considering that the S/N scales with the number of protein molecules in the sample which itself scales with the inverse of Mw, it is expected that S/N ~1/Mw. Hence, we anticipate that the above prediction of resolved signals constitutes a valuable guide for practical applications of SS GFT NMR.
Implementation of GFT NMR experiments
We consider the minimum dimensionality required to assign a protein with GFT NMR as the lowest number of dimensions with which resolving nearly all (~90%) of the experimentally observed spin systems are resolved. As demonstrated above, conventional 3D or (3,2)D GFT experiments accomplish this goal. For example, even for a small protein such as GB1 with especially well resolved spectra, 2D N–C experiments are not sufficient to obtain unambiguous assignments. Rather, 3D spectra were needed to identify overlapped spin systems arising from the helix, as well as frequently occurring residue types (such as threonine). Therefore, we anticipate that for most proteins with Mw >10 kDa, at least (4,3)D experiments will be required to derive nearly complete assignments.
Hence we adapted the previously reported CANCOCX 4D sequence (Franks et al. 2007) to record (4,3)D (HA)CANCOCX (Fig. 1), where the t2(15N) and t3(13CO) evolution periods are jointly sampled. Independent cosine and sine modulated sub-spectra for GFT NMR data acquisition are formed by shifting the initial TPPI phases marked by φ2 and φ2′ by 0° or 90°, respectively (Fig. 1). φ2 and φ2′ are incremented for each t2 time step to provide the desired modulations. The four-step phase cycle (Fig. 1) was adequate to remove artifacts caused by over the remaining 13C polarization from the initial broadband 1H-13C cross polarization. The carrier was placed at 55 ppm, resulting in an offset of ~120 ppm for the CO region, which is much larger than the desired spectral width, in this case 44 ppm, in order to maintain rotor-synchronization for the isotropic chemical shift evolution. Thus, the CO region is folded three times. Greater flexibility in the choice of the 13C carrier could be afforded by utilizing TPPI (Marion and Wuthrich 1983) with phase increments different from 90° in order to adjust the spectral window (Keeler and Neuhaus 1985). The SPECIFIC CP (Baldus et al. 1998) transfers were >95% selective, i.e. <5% of the polarization before mixing was observed on an unintended nucleus. One can envision using broadband transfers along with GFT encoding given carefully considered joint sampling.
The choice of jointly sampled chemical shifts was made by considering spectral dispersion and linewidth. Correlations of inter-residue CA signals have been shown to be desirable for assignments (Seidel et al. 2004; Heise et al. 2005; Franks et al. 2007), resulting in well resolved spectra. For (4,3)D (HA)CANCOCX, the CA dimension is thus chosen to be an independently sampled dimension. The N dimension also provides critical connectivity information, so that these shifts are measured in the central peak spectrum. Since the CO dimension is used primarily to enhance resolution, the CO dimension was jointly sampled with N to measure Ω(N+CO) and Ω(N−CO) in the two sub-spectra of (4,3)D (HA)CANCOCX. Including 1H chemical shifts is desirable, as this will encode a new information set and enhance sensitivity; however, we made no attempt to do so because both the sample and probe were not designed for such experiments (Chevelkov et al. 2006; Zhou et al. 2007a).
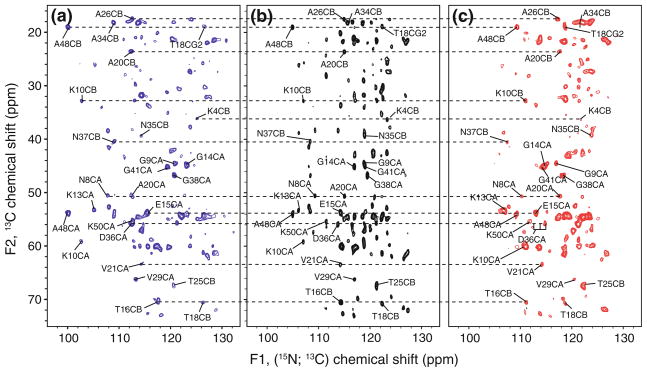
For (3,2)D (HACA)NCOCX, CA evolution is omitted. Despite its short measurement time (26.6 min), however, the central peak (HACA)NCOCX spectrum (Fig. 2b) exhibits average signal-to-noise ratios (SNR) ranging from ~10 to ~40. Peak detection is similar to 2D N(CO)CX, except that sidechains nitrogen signals of Asn (15Nδ2) and Gln (15Nε2) are not observed, since the N-CA SPECIFIC CP mixing times (~6–8 ms) chosen for the current implementation are not sufficiently long to transfer substantial polarization from Cβ to Nδ2 (in Asn) or Cγ to Nε2 (in Gln).
Fig. 2.
(3,2)D (HACA)NCOCX: subspectra comprising signals located at sums (a) and differences (b) of 15N and 13C′ chemcial shifts along F1 are shown on the left and right, respectively, of the central peak spectrum comprising signals at 15N chemical shifts. Positive contours above 6 times the RMS noise floor are drawn for each spectrum with contour separation factor of 1.3. The total measurement time was 87 min (t1 96 × 180 μs; t1′ 96 × 90 μs). As an illustration, selected peaks are labeled and connected. Labels correspond to the destination peak with the source atom consisting of the succeeding residue’s amide 15N and a 13CO component as appropriate. Dashed horizontal lines were added to connect peaks from the same residue
In the two sub-spectra of (3,2)D (HACA)NCOCX, the expected improvement in resolution yields that 78% (43) of the possible (55) residues in the protein GB1 are readily identified in both sub-spectra (Fig. 2a, c). For all but one case, overlapping peaks in one sub-spectrum are resolved in the other. For example, the difference spectrum peaks of G41 and G14 are severely overlapped. However, the summation peaks are well resolved. In the case of A26 CB, the opposite is true: the summation peak is not entirely resolved, but the difference peak is resolved. In cases where ambiguity remained in the sum and difference spectra, central peaks often aided in confirming the appropriate peaks.
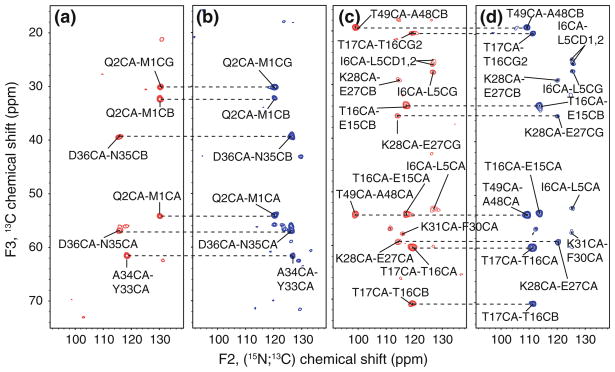
The inclusion of a third, independently sampled dimension resulted in the resolution of all possible spin systems. Among all the N, C′, CA, and CB resonances of GB1, only the Y33 CB resonance (which is near a rotational resonance condition at this MAS rate) was not observed. In addition, signals from many CG and CD signals are observed; only residues with long side chains consisting of more than 4 carbon atoms (e.g., aromatic residues and lysines) could not be completely assigned. It has been shown, however, that the use of a longer DARR mixing time would enable one to observe the missing signals (Franks et al. 2007). Two representative planes of the (4,3)D (HA)CANCOCX experiment are shown in Fig. 3, where three residues are easily identified due to the distinct peak pattern (Fig. 3a, b). The corresponding spin systems have nearly identical CA chemical shifts (within 0.1 ppm of 56.03 ppm, A34 CA = 56.1, D36 and Q2 = 55.9 pm). The mean position of the chemical shift doublet detected in the GFT dimension corresponds to the previously reported 15N chemical shifts of D36 (121.1 ppm), A34 (122.7 ppm) and Q2 (125.2 ppm). In a second, more congested plane (Fig. 3c, d), the signals of 5 residues are readily identified. The resolution introduced by the simultaneous sampling of 13CO is demonstrated for the T49CA-A48CB and T17CA-T16CG2 peaks. The two peaks are relatively close to one another (~2 ppm) in the difference (blue, Fig. 3d) sub-spectrum, but are separated by ~20 ppm in the sum (red, Fig. 3c) sub-spectrum.
Fig. 3.
Planes taken along F1(CA) at CA chemical of 56.0 ppm (a, b) and 60.2 ppm (c, d) from (4,3)D (HA)CANCOCX recorded for protein GB1. The CA and atoms with correlated chemical shifts are indicated. The subspectra with red contour levels comprise signals at the sum of 15N and 13C′ chemcial shifts, while the subspectra with blue contour lines comprise corresponding differences of shifts. Contours are cut at 6 time the RMS noise floor, with a spacing factor of 1.3. The spectra shown here were acquired in 7.2 h, inclusion of a central peaks spectrum extends the measurement time to 10.8 h (t1 32 × 135 μs; t2 48 × 180 μs; t2′ 48 × 90 μs)
The chemical shifts derived from the GFT spectrum (Supporting Information, Table 1) agree well with the previously reported solid state NMR chemical shifts for GB1 (Franks et al. 2005). The GFT derived 15N shifts, agree to 0.2 ± 0.3 ppm with the previously reported values, with two outlying differences of 0.9 and 0.8 ppm, and a median difference of 0.1 ppm. The GFT derived chemical shifts show a 0.7 ± 2.0 ppm with liquid state NMR, while the ssNMR derived shifts are 0.6 ± 2.0 ppm from the liquid state NMR values. There was a small linear phase encoded in the 13C′ component of the joint 15N-13C′ dimension caused by an incorrect initial delay. Still, the absolute maximum difference between ssNMR and GFT derived chemical shifts is only 1.3 ppm, with an average diference of 0.0 ± 0.6 ppm, and a median difference of 0.0 ppm. As for the 15N chemical shifts, there is no significant change to the agreement with the solution NMR chemical shifts; −0.3 ± 1.1 ppm for GFT, and −0.2 ± 0.8 ppm for ssNMR. The offset caused by the phase can be compensated by simple linear regression as a function of the effective carrier offset using directly detected carbonyl peaks from the GFT experiment as calibration points. We have chosen to report the uncorrected chemical shifts obtained from the usual GFT analysis.
Conclusions
We have demonstrated the value of recording GFT NMR spectra for assigning proteins in the solid state, thereby reducing measurement times for to obtain 3D or 4D spectral information by about an order of magnitude. In a series of simulated spectra, we have demonstrated that GFT NMR is well suited for establishing highly resolved correlations in large proteins. Experimentally, we have demonstrated with GB1 that the encoded information is complete and accurate, and highly resolved spectra can be acquired in a small fraction of the time required for the conventionally sampled experiment. GFT experiments are most beneficial when data acquisition is otherwise pursued in the ‘sampling limited’ regime, as true when using micromole quantities of GB1 with current NMR instrumentation.
We anticipate that a much larger range of protein samples will come into this regime as the sensitivity of SSNMR experiments for biological solids continues to improve. Substantial sensitivity benefits have been demonstrated now by proton detection (Morcombe et al. 2005; Chevelkov et al. 2006; Zhou et al. 2007a, b), dynamic nuclear polarization (Bajaj et al. 2003), and cryogenic MAS (Doty et al. 2006). In particular, for highest magnetic field strengths (corresponding to 900 MHz 1H resonance frequency or higher) spectral widths and thus sampling demand scale up accordingly. Recording of (4,2)D SS GFT NMR experiments may then become attractive, and for systems showing very high chemical shift degeneracy (such as membrane proteins) recording of G2FT SS NMR spectra (Atreya et al. 2005) may well become beneficial. Moreover, we expect that future implementations of SS GFT NMR experiments will profit from acquiring data in ‘clean absorption mode’ (Wu et al. 2009) to eliminate residual dispersive signal components. Taken together, we anticipate that a broad range of studies will benefit from SS GFT NMR in the future.
Supplementary Material
Acknowledgments
The authors thank the National Institute of Health for funding through NIGMS NIGMS/Roadmap Initiative (R01GM075937 and R01GM073770 to C.R.) and the National Science Foundation (MCB 0817857 to T.S.), and Lindsay J. Sperling and Andrew J. Nieuwkoop for assistance in preparing the manuscript.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10858-010-9451-7) contains supplementary material, which is available to authorized users.
Contributor Information
W. Trent Franks, Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA.
Hanudatta S. Atreya, Department of Chemistry, State University of New York at Buffalo, Buffalo, NY 14260, USA
Thomas Szyperski, Department of Chemistry, State University of New York at Buffalo, Buffalo, NY 14260, USA.
Chad M. Rienstra, Email: rienstra@scs.uiuc.edu, Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA. Department of Biochemistry, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA. Center for Biophysics and Computational Biology, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA
References
- Atreya HS, Szyperski T. G-matrix Fourier transform NMR spectroscopy for complete protein resonance assignment. Proc Natl Acad Sci USA. 2004;101:9642–9647. doi: 10.1073/pnas.0403529101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atreya HS, Eletsky A, Szyperski T. Resonance assignment of proteins with high shift degeneracy based on 5D spectral information encoded in G(2)FT NMR experiments. J Am Chem Soc. 2005;127:4554–4555. doi: 10.1021/ja042562e. [DOI] [PubMed] [Google Scholar]
- Atreya HS, Garcia E, Shen Y, Szyperski T. J-GFT NMR for precise measurement of mutually correlated nuclear spin-spin couplings. J Am Chem Soc. 2007;129:680–692. doi: 10.1021/ja066586s. [DOI] [PubMed] [Google Scholar]
- Bajaj VS, Farrar CT, Mastovsky I, Vieregg J, Bryant J, Elena B, Kreischer KE, Temkin RJ, Griffin RG. Dynamic nuclear polarization at 9T using a novel 250 GHz gyrotron microwave source. J Magn Reson. 2003;160:85–90. doi: 10.1016/j.jmr.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Baldus M. Correlation experiments for assignment and structure elucidation of immobilized polypeptides under magic angle spinning. Prog Nucl Magn Reson Spectrosc. 2002;41:1–47. [Google Scholar]
- Baldus M, Geurts DG, Hediger S, Meier BH. Efficient N-15-C-13 polarization transfer by adiabatic-passage Hartmann-Hahn cross polarization. J Magn Reson A. 1996;118:140–144. [Google Scholar]
- Baldus M, Petkova AT, Herzfeld JH, Griffin RG. Cross polarization in the tilted frame: assignment and spectral simplification in heteronuclear spin systems. Mol Phys. 1998;95:1197–1207. [Google Scholar]
- Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear decoupling in rotating solids. J Chem Phys. 1995;103:6951–6958. [Google Scholar]
- Bockmann A, Lange A, Galinier A, Luca S, Giraud N, Juy M, Heise H, Montserret R, Penin F, Baldus M. Solid state NMR sequential resonance assignments and conformational analysis of the 2 × 10.4 kDa dimeric form of the Bacillus subtilis protein Crh. J Biomol NMR. 2003;27:323–339. doi: 10.1023/a:1025820611009. [DOI] [PubMed] [Google Scholar]
- Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H. Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature. 2002;420:98–102. doi: 10.1038/nature01070. [DOI] [PubMed] [Google Scholar]
- Chevelkov V, Rehbein K, Diehl A, Reif B. Ultra-High Resolution in Proton Solid-State NMR Spectroscopy at High Levels of Deuteration. Angew Chem Int Ed. 2006;45:3878–3881. doi: 10.1002/anie.200600328. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe: a multidimensional spectral processing system based on Unix Pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Detken A, Hardy EH, Ernst M, Kainosho M, Kawakami T, Aimoto S, Meier BH. Methods for sequential resonance assignment in solid, uniformly C-13, N-15 labelled peptides: Quantification and application to antamanide. J Biomol NMR. 2001;20:203–221. doi: 10.1023/a:1011212100630. [DOI] [PubMed] [Google Scholar]
- Detken A, Hardy EH, Ernst M, Meier BH. Simple and efficient decoupling in magic-angle spinning solid- state NMR: the XiX scheme. Chem Phys Lett. 2002;356:298–304. [Google Scholar]
- Doty FD, Kulkarni J, Turner C, Entzminger G, Bielecki A. Using a cross-coil to reduce RF heating by an order of magnitude in triple-resonance multinuclear MAS at high fields. J Magn Reson. 2006;182:239–253. doi: 10.1016/j.jmr.2006.06.031. [DOI] [PubMed] [Google Scholar]
- Eletsky A, Atreya HS, Liu GH, Szyperski T. Probing structure and functional dynamics of (large) proteins with aromatic rings: L-GFT-TROSY (4, 3)D HCCHNMR spectroscopy. J Am Chem Soc. 2005;127:14578–14579. doi: 10.1021/ja054895x. [DOI] [PubMed] [Google Scholar]
- Flynn PF, Milton MJ, Babu CR, Wand AJ. A simple and effective NMR cell for studies of encapsulated proteins dissolved in low viscosity solvents. J Biomol NMR. 2002;23:311–316. doi: 10.1023/a:1020229307552. [DOI] [PubMed] [Google Scholar]
- Franks WT, Zhou DH, Wylie BJ, Money BG, Graesser DT, Frericks HL, Sahota G, Rienstra CM. Magic-angle spinning solid-state NMR spectroscopy of the beta-1 immunoglobulin binding domain of protein G (GB1): 15N and 13C chemical shift assignments and conformational analysis. J Am Chem Soc. 2005;127:12291–12305. doi: 10.1021/ja044497e. [DOI] [PubMed] [Google Scholar]
- Franks WT, Kloepper KD, Wylie BJ, Rienstra CM. Four-dimensional heteronuclear correlation experiments for chemical shift assignment of solid proteins. J Biomol NMR. 2007;39:107–131. doi: 10.1007/s10858-007-9179-1. [DOI] [PubMed] [Google Scholar]
- Franks WT, Wylie BJ, Schmidt HLF, Nieuwkoop AJ, Mayrhofer RM, Shah GJ, Graesser DT, Rienstra CM. Dipole tensor-based atomic-resolution structure determination of a nanocrystalline protein by solid-state NMR. Proc Natl Acad Sci USA. 2008;105:4621–4626. doi: 10.1073/pnas.0712393105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung BM, Khitrin AK, Ermolaev K. An improved broadband decoupling sequence for liquid crystals and solids. J Magn Reson. 2000;142:97–101. doi: 10.1006/jmre.1999.1896. [DOI] [PubMed] [Google Scholar]
- Gardner KH, Zhang XC, Gehring K, Kay LE. Solution NMR studies of a 42 kDa Escherichia coli maltose binding protein beta-cyclodextrin complex: Chemical shift assignments and analysis. J Am Chem Soc. 1998;120:11738–11748. [Google Scholar]
- Goddard TD, Kneller DG. Sparky. Vol. 3. University of California; San Francisco: 2006. [Google Scholar]
- Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M. Molecular-level secondary structure, polymorphism, and dynamics of full-length alpha-synuclein fibrils studied by solid-state NMR. Proc Natl Acad Sci USA. 2005;102:15871–15876. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohwy M, Rienstra CM, Jaroniec CP, Griffin RG. Fivefold symmetric homonuclear dipolar recoupling in rotating solids: application to double quantum spectroscopy. J Chem Phys. 1999;110:7983–7992. [Google Scholar]
- Hong M. Resonance assignment of C-13/N-15 labeled solid proteins by two- and three-dimensional magic-angle-spinning NMR. J Biomol NMR. 1999;15:1–14. doi: 10.1023/a:1008334204412. [DOI] [PubMed] [Google Scholar]
- Igumenova TI, McDermott AE, Zilm KW, Martin RW, Paulson EK, Wand AJ. Assignments of carbon NMR resonances for microcrystalline ubiquitin. J Am Chem Soc. 2004a;126:6720–6727. doi: 10.1021/ja030547o. [DOI] [PubMed] [Google Scholar]
- Igumenova TI, Wand AJ, McDermott AE. Assignment of the backbone resonances for microcrystalline ubiquitin. J Am Chem Soc. 2004b;126:5323–5331. doi: 10.1021/ja030546w. [DOI] [PubMed] [Google Scholar]
- Keeler J, Neuhaus D. Comparison and evaluation of methods for two-dimensional Nmr-spectra with absorption-mode line-shapes. J Magn Reson. 1985;63:454–472. [Google Scholar]
- Kim S, Szyperski T. GFT NMR, a new approach to rapidly obtain precise high-dimensional NMR spectral information. J Am Chem Soc. 2003;125:1385–1393. doi: 10.1021/ja028197d. [DOI] [PubMed] [Google Scholar]
- Liu GH, Shen Y, Atreya HS, Parish D, Shao Y, Sukumaran DK, Xiao R, Yee A, Lemak A, Bhattacharya A, Acton TA, Arrowsmith CH, Montelione GT, Szyperski T. NMR data collection and analysis protocol for high-throughput protein structure determination. Proc Natl Acad Sci USA. 2005;102:10487–10492. doi: 10.1073/pnas.0504338102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieau JL, McDermott AE. Conformational flexibility of a microcrystalline globular protein: Order parameters by solid-state NMR spectroscopy. J Am Chem Soc. 2006;128:11505–11512. doi: 10.1021/ja062443u. [DOI] [PubMed] [Google Scholar]
- Marion D, Wuthrich K. Application of phase sensitive two-dimensional correlated spectroscopy (Cosy) for measurements of H-1-H-1 spin-spin coupling-constants in proteins. Biochem Biophys Res Commun. 1983;113:967–974. doi: 10.1016/0006-291x(83)91093-8. [DOI] [PubMed] [Google Scholar]
- Marulanda D, Tasayco ML, McDermott A, Cataldi M, Arriaran V, Polenova T. Magic angle spinning solid-state NMR spectroscopy for structural studies of protein interfaces. Resonance assignments of differentially enriched Escherichia coli thioredoxin reassembled by fragment complementation. J Am Chem Soc. 2004;126:16608–16620. doi: 10.1021/ja0464589. [DOI] [PubMed] [Google Scholar]
- McDermott A, Polenova T, Bockmann A, Zilm KW, Paulsen EK, Martin RW, Montelione GT. Partial NMR assignments for uniformly (C-13, N-15)-enriched BPTI in the solid state. J Biomol NMR. 2000;16:209–219. doi: 10.1023/a:1008391625633. [DOI] [PubMed] [Google Scholar]
- Morcombe CR, Gaponenko V, Byrd RA, Zilm KW. Diluting abundant spins by isotope edited radio frequency field assisted diffusion. J Am Chem Soc. 2004;126:7196–7197. doi: 10.1021/ja047919t. [DOI] [PubMed] [Google Scholar]
- Morcombe CR, Paulson EK, Gaponenko V, Byrd RA, Zilm KW. H-1-N-15 correlation spectroscopy of nanocrystalline proteins. J Biomol NMR. 2005;31:217–230. doi: 10.1007/s10858-005-0527-8. [DOI] [PubMed] [Google Scholar]
- Pauli J, Baldus M, van Rossum B, de Groot H, Oschkinat H. Backbone and side-chain 13C and 15N resonance assignments of the alpha-spectrin SH3 domain by magic angle spinning solid state NMR at 17.6 Tesla. Chem Biol Chem. 2001;2:101–110. doi: 10.1002/1439-7633(20010401)2:4<272::AID-CBIC272>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Paulson EK, Martin RW, Zilm KW. Cross polarization, radio frequency field homogeneity, and circuit balancing in high field solid state NMR probes. J Magn Reson. 2004;171:314–323. doi: 10.1016/j.jmr.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Pintacuda G, Giraud N, Pierattelli R, Bockmann A, Bertini I, Emsley L. Solid-state NMR spectroscopy of a paramagnetic protein: assignment and study of human dimeric oxidized CuII-ZnII superoxide dismutase (SOD) Angew Chem Int Ed. 2007;46:1079–1082. doi: 10.1002/anie.200603093. [DOI] [PubMed] [Google Scholar]
- Rienstra CM, Hohwy M, Hong M, Griffin RG. 2D and 3D 15N-13C-13C NMR chemical shift correlation spectroscopy of solids: assignment of MAS spectra of peptides. J Am Chem Soc. 2000;122:10979–10990. [Google Scholar]
- Seidel K, Lange A, Becker S, Hughes CE, Heise H, Baldus M. Protein solid-state NMR resonance assignments from (C-13, C-13) correlation spectroscopy. Phys Chem Chem Phys. 2004;6:5090–5093. [Google Scholar]
- Shen Y, Atreya HS, Liu GH, Szyperski T. G-matrix Fourier transform NOESY-based protocol for high-quality protein structure determination. J Am Chem Soc. 2005;127:9085–9099. doi: 10.1021/ja0501870. [DOI] [PubMed] [Google Scholar]
- Siemar AB, Ritter C, Ernst M, Riek R, Meier BH. High-resolution solid-state NMR spectroscopy of the prion protein HET-s in its amyloid conformation. Angew Chem Int Ed. 2005;44:2441–2444. doi: 10.1002/anie.200462952. [DOI] [PubMed] [Google Scholar]
- Siemer AB, Arnold AA, Ritter C, Westfeld T, Ernst M, Riek R, Meier BH. Observation of highly flexible residues in amyloid fibrils of the HET-s prion. J Am Chem Soc. 2006;128:13224–13228. doi: 10.1021/ja063639x. [DOI] [PubMed] [Google Scholar]
- Sperling LJ, Berthold DA, Sasser TL, Jeisy-Scott V, Rienstra CM. J Mol Biol. 2010;399:268–282. doi: 10.1016/j.jmb.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer JA, Bronnimann CE, Mullen CG, Zhou DHH, Stellfox SA, Li Y, Williams EH, Rienstra CM. Reduction of RF-induced sample heating with a scroll coil resonator structure for solid-state NMR probes. J Magn Reson. 2005;173:40–48. doi: 10.1016/j.jmr.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Szyperski T, Atreya HS. Principles and applications of GFT projection NMR spectroscopy. Magn Reson Chem. 2006;44:S51–S60. doi: 10.1002/mrc.1817. [DOI] [PubMed] [Google Scholar]
- Szyperski T, Yeh DC, Sukumaran DK, Moseley HNB, Montelione GT. Reduced-dimensionality NMR spectroscopy for high-throughput protein resonance assignment. Proc Natl Acad Sci USA. 2002;99:8009–8014. doi: 10.1073/pnas.122224599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takegoshi K, Nakamura S, Terao T. C-13-H-1 dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett. 2001;344:631–637. doi: 10.1063/1.2364503. [DOI] [PubMed] [Google Scholar]
- Takegoshi K, Nakamura S, Terao T. C-13-H-1 dipolar-driven C-13-C-13 recoupling without C-13 rf irradiation in nuclear magnetic resonance of rotating solids. J Chem Phys. 2003;118:2325–2341. [Google Scholar]
- Tugarinov V, Muhandiram R, Ayed A, Kay LE. Four-dimensional NMR spectroscopy of a 723-residue protein: chemical shift assignments and secondary structure of malate synthase G. J Am Chem Soc. 2002;124:10025–10035. doi: 10.1021/ja0205636. [DOI] [PubMed] [Google Scholar]
- Tycko R. Prospects for resonance assignments in multidimensional solid-state NMR spectra of uniformly labeled proteins. J Biomol NMR. 1996;8:239–251. doi: 10.1007/BF00410323. [DOI] [PubMed] [Google Scholar]
- Van Geet AL. Calibration of the methanol and glycol nuclear magnetic resonance thermometers with a static thermistor probe. Anal Chem. 1968;42:2227–2229. [Google Scholar]
- Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218–289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- Wu YB, Ghosh A, Szyperski T. Clean absorption-mode NMR data acquisition. Angew Chem Int Ed. 2009;48:1479–1483. doi: 10.1002/anie.200804927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Atreya HS, Kamen DE, Girvin ME, Szyperski T. GFT projection NMR based resonance assignment of membrane proteins: application to subunit c of E-coli F1F0 ATP synthase in LPPG micelles. J Biomol NMR. 2008;40:157–163. doi: 10.1007/s10858-008-9224-8. [DOI] [PubMed] [Google Scholar]
- Zhou DH, Shah G, Cormos M, Mullen C, Sandoz D, Rienstra CM. Proton-detected solid-state NMR spectroscopy of fully protonated proteins at 40 kHz magic-angle spinning. J Am Chem Soc. 2007a;129:11791–11801. doi: 10.1021/ja073462m. [DOI] [PubMed] [Google Scholar]
- Zhou DH, Shea JJ, Nieuwkoop AJ, Franks WT, Wylie BJ, Mullen C, Sandoz D, Rienstra CM. Solid-state protein-structure determination with proton-detected triple-resonance 3D magic-angle spinning NMR spectroscopy. Angew Chem Int Ed. 2007b;46:8380–8383. doi: 10.1002/anie.200702905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.