Abstract
Brain-derived neurotrophic factor (BDNF), neurotrophin-4 (NT4) and their TrkB receptor regulate taste system development. To determine where and when gustatory neurons come in contact with these important factors, temporospatial expression patterns of Bdnf, Ntf4/5 and TrkB in the peripheral taste system were examined using RT-PCR. In the lingual epithelium, Ntf4/5 mRNA expression was higher than that of Bdnf at embryonic day 12.5 (E12.5), and the expression of both factors decreased afterwards. However, Ntf4/5 expression decreased at an earlier age than Bdnf. Bdnf and Ntf4/5 are expressed in equal amounts at E12.5 in geniculate ganglion, but Bdnf expression increased from E14.5 to birth, whereas Ntf4/5 expression decreased. These findings indicate that NT4 functions at early embryonic stages and is derived from different sources than Bdnf. TrkB expression in the geniculate ganglion is robust throughout development and is not a limiting factor for neurotrophin function in this system.
Keywords: Taste, Development, BDNF, NT4, TrkB, Tongue, Soft palate, Geniculate ganglion, Fungiform papillae, Real-time PCR
INTRODUCTION
In the peripheral taste system, taste buds within the anterior two-thirds of the tongue and palate are innervated by geniculate ganglion neurons through the chorda tympani nerves and greater superficial petrosal (GSP) nerves. The development of these geniculate neurons is regulated by neurotrophins (Conover et al., 1995; Liu et al., 1995; Liebl et al., 1997; Al-Hadlaq et al., 2003; Patel and Krimm, 2010). Neurotrophins are a family of proteins that regulate cell survival, differentiation, and neurite growth in the central and the peripheral nervous system (Bibel and Barde, 2000; Huang and Reichardt, 2001). The neurotrophins, brain-derived neurotrophic factor (BDNF) and neurotrophin-4 (NT4), together with their common high-affinity tyrosine kinase TrkB receptor, control taste system development (Fritzsch et al., 1997; Zhang et al., 1997; Krimm et al., 2001; Agerman et al., 2003).
Geniculate neuron number is regulated by both BDNF and NT4. Specifically, overexpression of either BDNF or NT4 increases the number of geniculate ganglion neurons (Ringstedt et al., 1999; Krimm et al., 2001). In mutant mice lacking either BDNF or NT4, half of the neurons are lost from the geniculate ganglion, whereas in mice that lack both BDNF and NT4 almost all neurons are lost (Conover et al., 1995; Liu et al., 1995; Liebl et al., 1997; Patel and Krimm, 2010). However, the neuron loss in Bdnf and Ntf4/5 knockout mice occurs at different times during development (Patel and Krimm, 2005; Krimm, 2007; Patel and Krimm, 2010), indicating that the taste system may be exposed to these factors at different times or they may be expressed in different locations. In mice lacking the primary receptor for these neurotrophins; i.e., TrkB, only about 5% of geniculate ganglion neurons remain (Fritzsch et al., 1997). Thus, BDNF/TrkB and NT4/TrkB signaling pathways regulate neuronal number in geniculate ganglion, but with differences in the timing of this regulation.
In addition to differentially regulating the timing of neuron loss, BDNF and NT4 differentially regulate neurite outgrowth (Rochlin et al., 2000), target innervation, and taste bud development. For example, BDNF expressed specifically in taste epithelia is required for gustatory target innervation while NT4 is not (Ma et al., 2009). Mice lacking BDNF lose more than half of their fungiform taste buds by birth (Nosrat et al., 1997; Mistretta et al., 1999; Patel et al., 2008), whereas mice lacking NT4 lose very few fungiform papillae or taste buds by birth (Liebl et al., 1999; Patel et al., 2008).
The observed differences in the timing of geniculate neuron loss, target innervation and taste bud loss all indicate that BDNF and NT4 could each function uniquely in regulating gustatory development. The differing roles of BDNF and NT4 might be due to differences in the timing, level or location of their gene expression. Bdnf is known to be expressed in developing fungiform placodes or papillae and taste bud containing regions in the palate (Nosrat and Olson, 1995; Nosrat et al., 1996). However, changes in its expression level have not been quantified across different ages. Within the geniculate ganglion, there is disagreement in the literature as to whether Bdnf is expressed at all during development (Ernfors et al., 1992; Schecterson and Bothwell, 1992). Almost no information exists concerning the pattern and levels of Ntf4/5 expression. Ntf4/5mRNA is not detectable in taste papillae of embryonic and postnatal rats when using in situ hybridization (Nosrat et al., 1996; Nosrat et al., 2001). Furthermore, Ntf4/5 expression in the developing geniculate ganglion has not been examined. Thus, detailed information concerning changes in the timing and levels of neurotrophin expression in the developing taste system is lacking.
The interpretation of mutant mouse studies is complicated by the possibility that neurotrophic factors may regulate each other. For example, removal of functional BDNF could result in changes in Ntf4/5 expression in the gustatory system. This would complicate interpretation of Bdnf −/− data because it would be unclear whether the effects were due to the absence of BDNF or due to the change in NT4 expression. In vitro studies have shown that administration of BDNF, NT4 or NT-3 reciprocally increases each other’s mRNA levels (Xiong et al., 2002; Patz and Wahle, 2004). BDNF and NT4 function via the same receptors and they may be influenced by similar feedback mechanisms. Therefore, it is important to determine whether BDNF and NT4 are capable of regulating each other.
Here, we examined the embryonic timing of Bdnf, Ntf4/5 and TrkB mRNA expression in the developing taste system of wild type and transgenic mice. Ntf4/5 expression is highest at E12.5 in the geniculate ganglion and tongue, where it may be involved in early developmental events, such as regulation of early geniculate neuron survival and axonal branching as neurons initially innervate the tongue. By the time gustatory axons reach their targets; i.e., fungiform placodes, Bdnf expression is higher than Ntf4/5 in the lingual epithelium indicating that BDNF may have an important target-derived role. Bdnf expression continues to increase in the ganglion until birth, where it may have a late autocrine or paracrine role.
RESULTS
Expression of Bdnf increases while Ntf4/5 decreases in the geniculate ganglion as a function of embryonic age
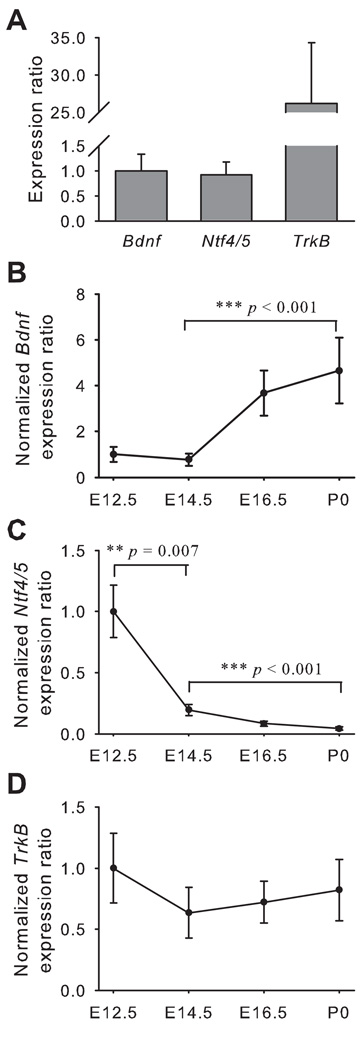
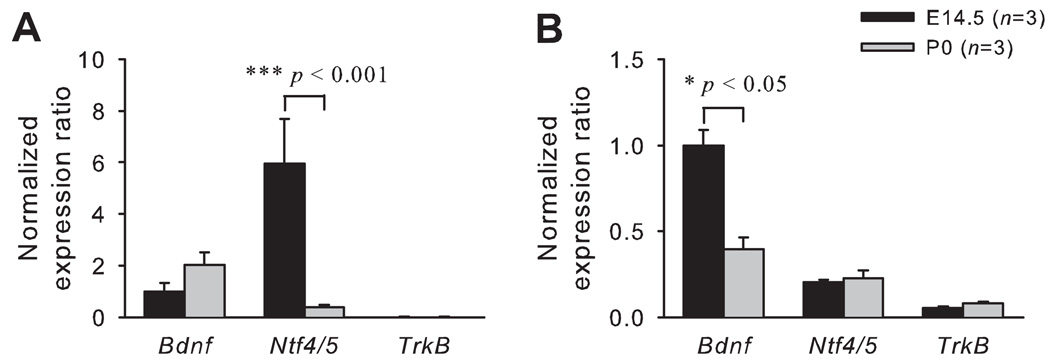
Laser capture microdissection (LCM) was used to remove the geniculate ganglion from sections of the head (Figs. 1A and 1B). Real-time PCR was performed to determine the time course of Bdnf, Ntf4/5 and TrkB expression in geniculate ganglion during embryonic development. E12.5 was the youngest age studied since at E12.5 the axons from geniculate ganglion extend to the tongue, but do not innervate their targets (Mbiene, 2004). At E12.5, the expression levels of Bdnf and Ntf4/5 mRNA in geniculate ganglion were equivalent. TrkB mRNA expression was more than 25-fold higher than Bdnf mRNA expression (Fig. 2A). During embryonic development, Bdnf expression increased from E14.5 to birth, with the expression level at birth being 6-fold higher than that at E14.5 (p < 0.001; Fig. 2B). Ntf4/5 expression decreased from E12.5 to E14.5, and the level was 81% lower at E14.5 (p < 0.01). By birth, Ntf4/5 mRNA was almost undetectable (Fig. 2C). TrkB expression was maintained at a high level through embryonic development from E12.5 to birth (Fig. 2D).
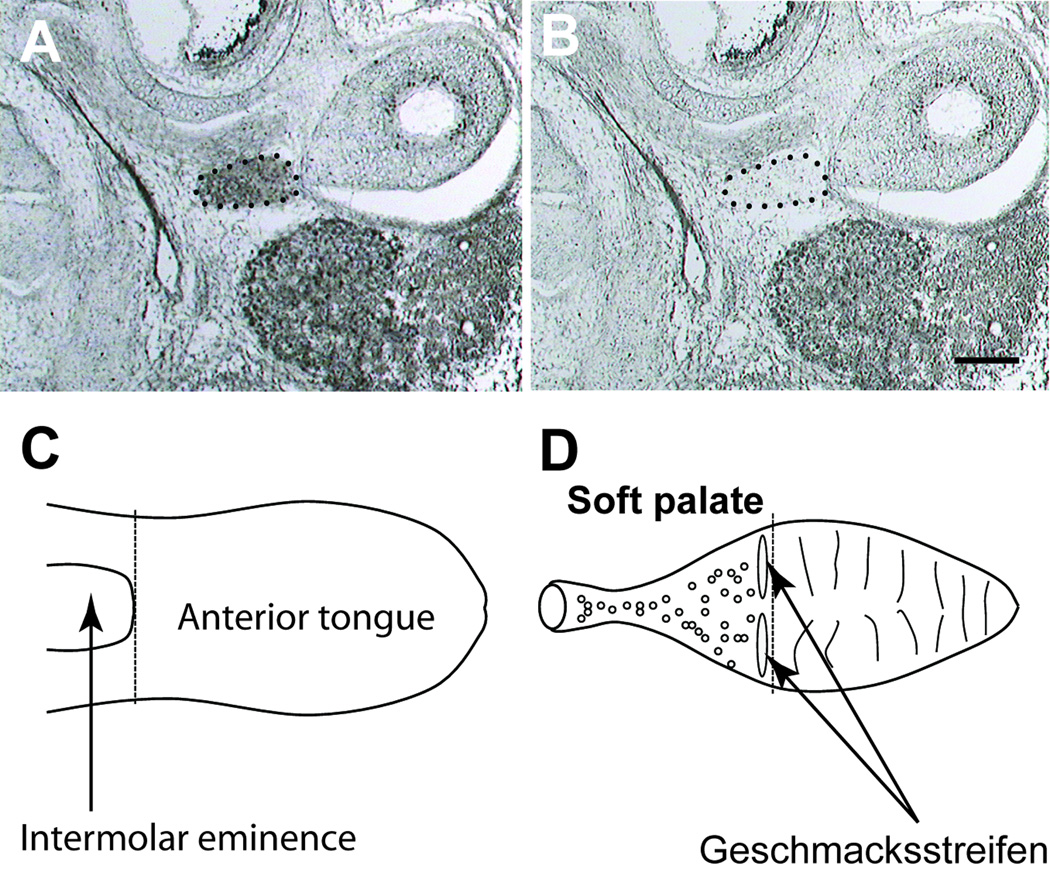
Figure 1. Neurotrophin expression was examined in the geniculate ganglion, the anterior tongue and soft palate.
After dehydration, the geniculate ganglion (outlined by dotted line) was visualized in transverse tissue sections (A), and could be easily captured using laser capture microdissection (LCM). LCM successfully removed the ganglion but not the surrounding tissue (B). The anterior tongue, defined as the portion of the tongue immediately rostral to the intermolar eminence (C), was dissected and the epithelium and mesenchyme were separated for analysis. The soft palate was dissected by making a cut between the hard and soft palate, and then the tissue caudal to this cut was removed (D). The epithelium was separated from the connective tissue and developing glands underneath the epithelium for analysis. Scale bar in A equals 200 µm and applies to A and B.
Figure 2. Expression of Bdnf increases while Ntf4/5 decreases in the geniculate ganglion during embryonic development.
The expression levels at E12.5, the youngest age studied, were calibrated against bdnf expression. At this age, Bdnf and Ntf4/5 have similar expression levels in geniculate ganglion, while TrkB expression was much greater than Bdnf and Ntf4/5 (A). To compare expression levels across different ages, they were normalized to each sample’s respective reference gene expression and then reported as a fold of E12.5. The expression levels for Bdnf in the geniculate ganglion increased (B), Ntf4/5 decreased (C) and TrkB remained unchanged (D) as embryonic development progresses (E12.5 to P0). Error bars show standard error.
Bdnf and Ntf4/5 expression decreases in the anterior tongue during embryonic development
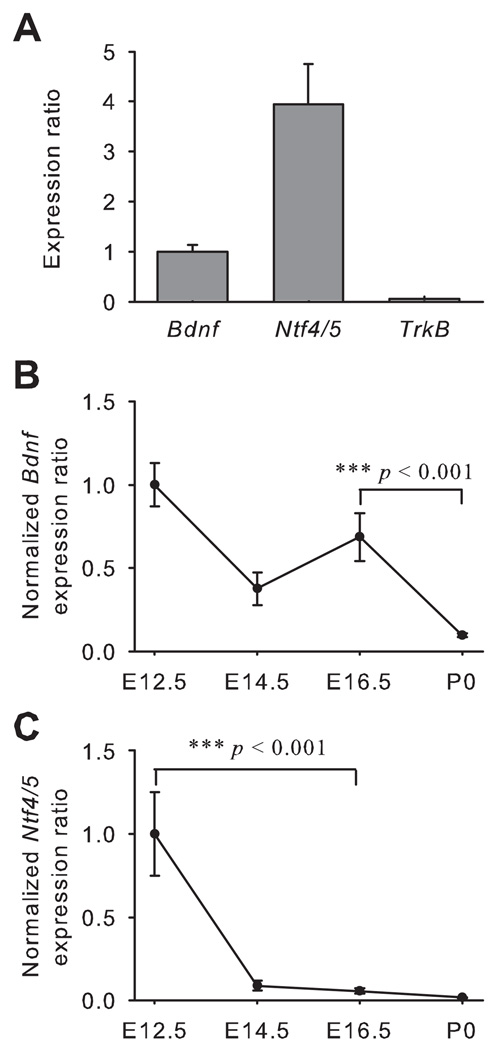
Real-time PCR was performed on lingual epithelium from the front of the tongue (Fig. 1C). We chose to examine the entire lingual epithelium rather than limiting our analysis to taste epithelium because neurotrophins are released factors and expression of these factors in non-gustatory epithelia likely influences gustatory neurons at early ages (before target innervation). In particular, NT4 expression is unlikely to be limited to gustatory epithelium and we wanted to compare its expression with that of BDNF. In the anterior tongue epithelium, Ntf4/5 expression was about 4-fold higher than Bdnf expression at E12.5, but TrkB expression was much lower than that of Bdnf and Ntf4/5 at this age (Fig. 3A). The expression of Bdnf was not different among E12.5, E14.5 and E16.5, but it was reduced from E16.5 to birth (p < 0.001; Fig. 3B). Ntf4/5 expression decreased at an earlier age than for Bdnf, between E12.5 and E16.5 (p < 0.001; Fig. 3C), as the axons from geniculate ganglion are reaching the lingual epithelium. However, the relative TrkB expression remained constant across the embryonic ages from E12.5 to birth.
Figure 3. In the tongue epithelium, expression of both Bdnf and Ntf4/5 decreases during embryonic development.
The expression ratio for Ntf4/5 was about 4-fold higher than for Bdnf at E12.5 (A), while TrkB expression was almost undetectable. As development progressed (E12.5 to P0) expression levels for both Bdnf (B) and Ntf4/5 (C) decreased from E12.5 to birth. Error bars show standard error.
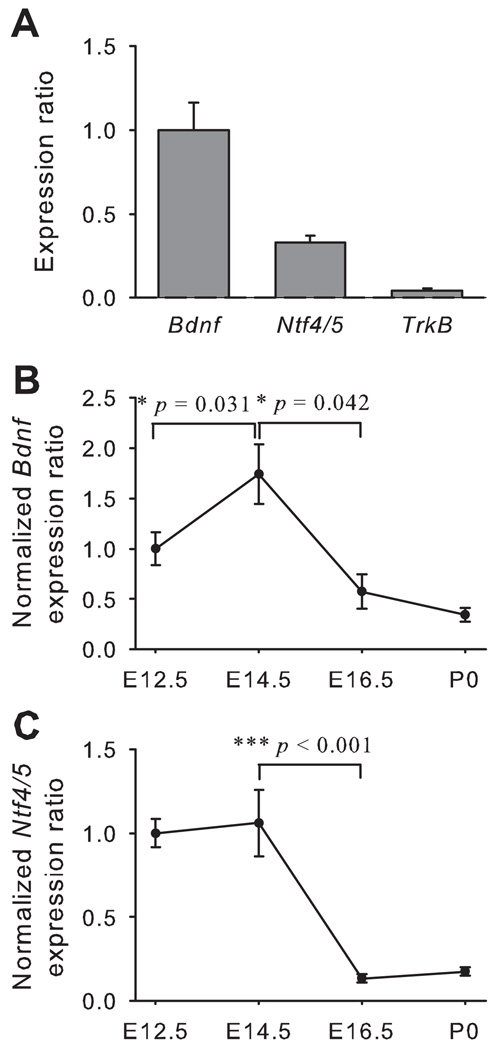
In the mesenchyme and developing muscle from the anterior tongue, there was more Bdnf than Ntf4/5 at E12.5, while TrkB expression was much lower compared with that of Bdnf and Ntf4/5 (Fig. 4A). The expression level of Bdnf mRNA reached its peak in the tongue mesenchyme at E14.5 (E12.5 vs E14.5, p = 0.031), and then the expression decreased by E16.5 (p = 0.042; Fig. 4B). The expression of Ntf4/5 remained constant between E12.5 and E14.5, but it was greatly reduced after E14.5 (p < 0.001; Fig. 4C).
Figure 4. In the tongue mesenchyme, the Bdnf and Ntf4/5 expression decreases with embryonic age.
The expression ratio for Bdnf was greater than for Ntf4/5 at E12.5 while TrkB expression was almost undetectable (A). The expression of Bdnf (B) and Ntf4/5 (C) in the tongue mesenchyme decreased across embryonic age from E12.5 to birth. Error bars show standard error.
Expression of Ntf4/5 decreases during the development of the soft palate
Because the soft palate is not yet formed at E12.5, we began our expression analysis at E14.5. The epithelium included the geschmacksstreifen and the posterior palatine field (Fig. 1D). In the soft palate epithelium, Ntf4/5 expression at E14.5 was 6-fold higher than Bdnf expression, but TrkB expression was almost absent. This expression pattern was similar to that of the lingual epithelium at E12.5. Although the expression of Bdnf was not different between E14.5 and birth, Ntf4/5 expression at birth was reduced by 94% compared with that at E14.5 (Fig. 5A).
Figure 5. The expression of Ntf4/5 in soft palate (SP) epithelium, and the expression of Bdnf in the lamina propria/submucosa decreases from E14.5 to birth.
In the palatal epithelium, the expression levels of Ntf4/5 were greater than those of Bdnf and TrkB at E14.5, but the levels were greatly reduced at birth (A). In the submucosa there was more Bdnf than Ntf4/5 at E14.5, but Bdnf expression decreased by birth (B).
In the connective tissue and developing glands underneath the soft palate epithelium, the expression pattern of Bdnf, Ntf4/5 and TrkB at E14.5 was similar to that in the anterior tongue mesenchyme at E12.5. Bdnf expression was 4.9-fold higher than Ntf4/5 expression, and the expression level of TrkB was low. Bdnf mRNA expression at birth was reduced compared with that at E14.5 (p < 0.05), while the expression of Ntf4/5 mRNA remained constant between E14.5 and birth (Fig. 5B).
Developmental expression of endogenous BDNF is revealed by β-gal expression
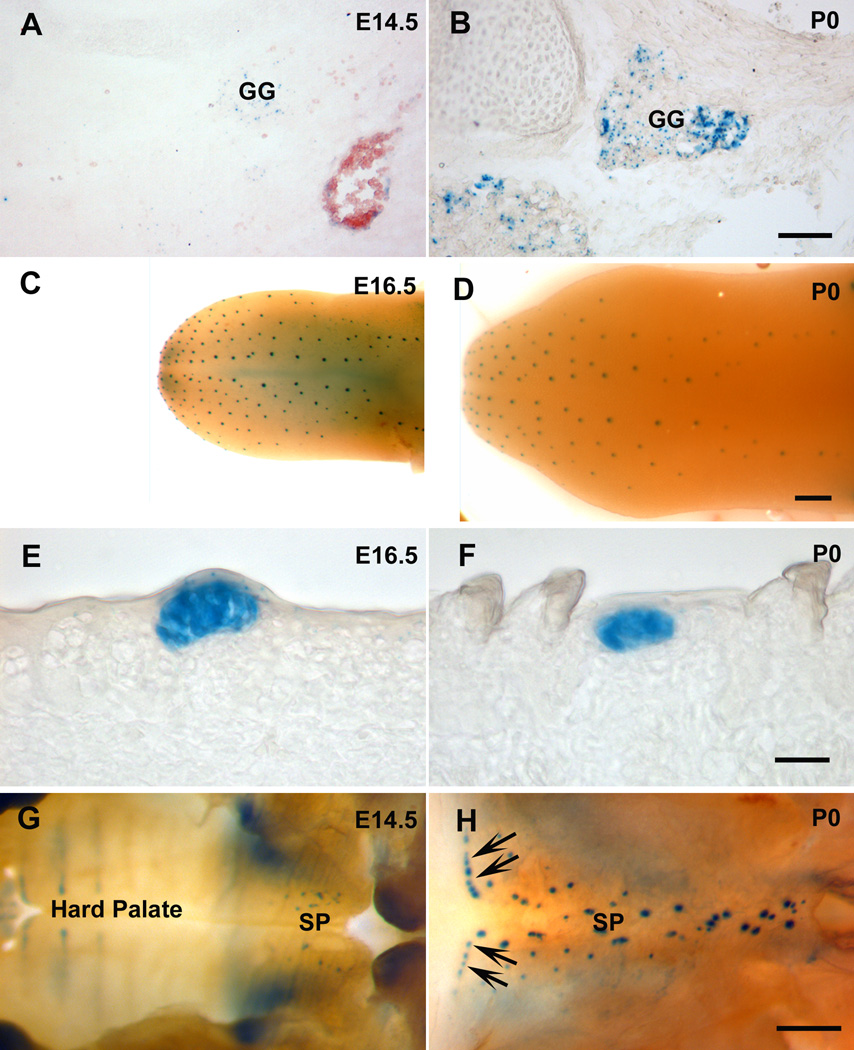
To confirm the time course of BDNF expression in the peripheral taste system, β-gal staining of Bdnf lacZ/+ mice was performed on geniculate ganglion, the anterior tongue and soft palate. Consistent with our RT-PCR findings in the geniculate ganglion, there was weak β-gal staining in geniculate ganglion at E14.5, but the β-gal labeling at birth was much more intense (Figs. 6A and 6B). In the lingual epithelium, β-gal labeling was limited to specific regions and the labeled spots had a distribution similar to that of fungiform papillae (Figs. 6C and 6D). The decreased Bdnf expression levels from E16.5 to birth, revealed by real-time PCR, could be due to fewer cells expressing BDNF or due to less BDNF expression per cell (Figs. 6E and 6F), and/or an increase in the size of the non Bdnf expressing epithelium between placodes. The decrease becomes more apparent in postnatal taste buds (our unpublished data). Unlike the tongue epithelium, in the soft palate epithelium β-gal positive spots increase in number as the soft palate grows (Figs. 6G and 6H). This might explain why there was not a reduction in Bdnf expression in the palatal epithelium between E14.5 and birth. Taken together, the β-gal staining results were consistent with the results of RT-PCR.
Figure 6. The changes of β-gal staining in BdnfLacZ/+ mice are consistent with the changes revealed by RT-PCR.
β-gal was detected in geniculate ganglion (GG) at both E14.5 (A) and birth (B), but much stronger staining was observed at birth. Whole mount β-gal staining of the tongue demonstrated that the distribution pattern of β-gal-positive spots was similar between E16.5 (C) and birth (D), but the intensity of staining for β-gal in E16.5 tongue was stronger than that at birth (E and F). β-gal-positive signals were found in soft palate (SP) at both E14.5 (G) and birth (H), but more at birth, especially in the region corresponding to the Geschmacksstreifen (arrows). Scale bar is 100 µm for A and B; 1000 µm for C and D; 20 µm for E and F; 400 µm for G and H.
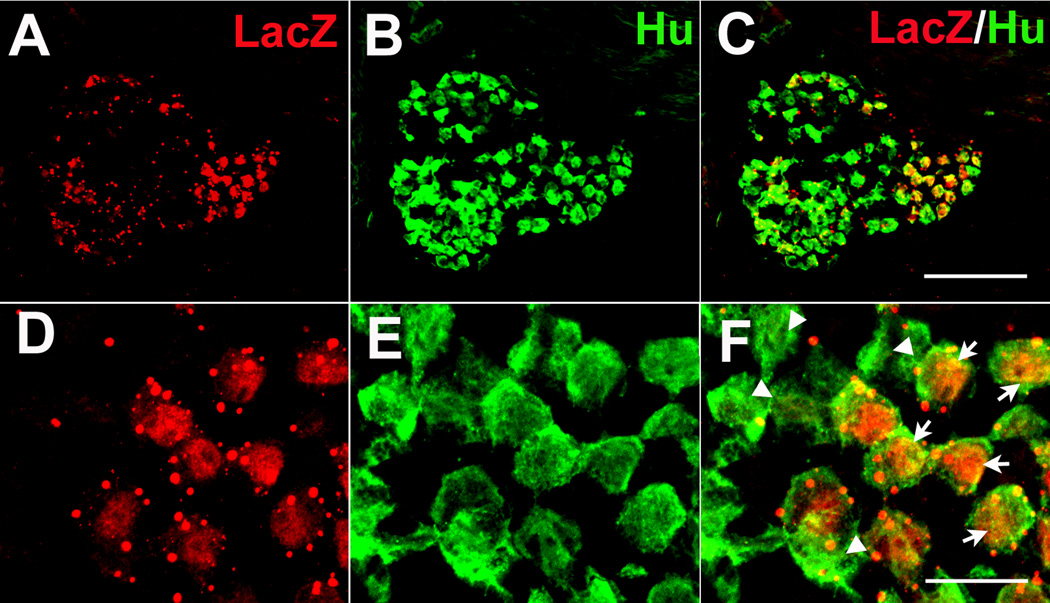
In order to determine if neurons in the geniculate ganglion express BDNF, the ganglion was labeled with antibodies directed against β-gal proteins and a neuronal specific marker; i.e., Hu. β-gal immunoreactivity was detected in a subset of geniculate ganglion cells. In addition, some β-gal was observed outside of clearly labeled neurons (Fig. 7). Therefore, many geniculate neurons clearly express BDNF, but non-neuronal cells in the geniculate ganglion may also be capable of expressing BDNF.
Figure 7. BDNF is expressed in the geniculate ganglion neurons at birth.
Double label immunohistochemistry at birth illustrates that BDNF (A: anti-β-gal) is expressed in some neurons (B: anti-Hu) but not all (C). This can be seen more clearly at higher magnification (D, E, F). The arrows indicate cells that co-express β-gal and Hu. Arrowheads indicate β-gal negative neurons. Scale bar in C, 100 µm, applies for A, B and C; Scale bar in F, 20 µm applies for D, E and F.
BDNF and NT4 do not regulate each other
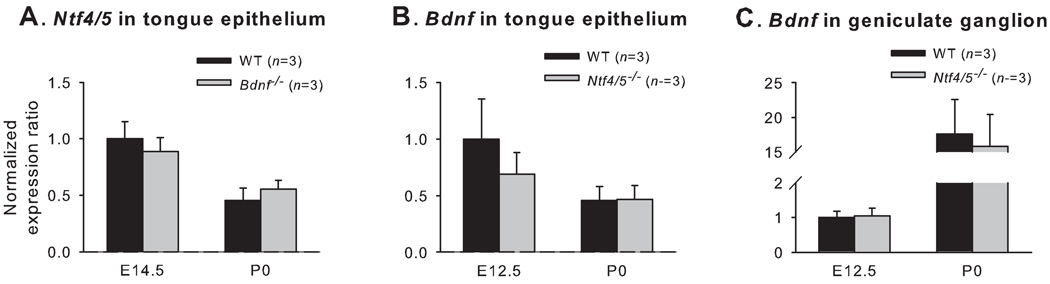
To determine whether BDNF and NT4 are capable of regulating each other, Bdnf and Ntf4/5 expression was examined in the peripheral taste system, using real-time PCR in Ntf4/5−/− and Bdnf−/− mice. We examined E14.5 in Bdnf −/− mice because BDNF influences cell survival and target innervation at this age (Ma et al., 2009; Patel and Krimm, 2010) and could also regulate Ntf4/5 expression. We also examined Ntf4/5 at birth in Bdnf −/− mice after these major roles of BDNF were completed. In Bdnf −/− mice, no Bdnf mRNA was detected in the lingual epithelium and the geniculate ganglion. Removal of functional BDNF had no effect on the expression of Ntf4/5 in the tongue epithelium from transgenic mice at either E14.5 or birth as compared with that from wild type mice (p > 0.05; Fig. 8A). The expression of Ntf4/5 in geniculate ganglion was not compared statistically since it was expressed in very low amounts in both Bdnf −/− and wild type mice. NT4 regulates geniculate neuron number early in development by E12.5, and therefore Ntf4/5−/− mice were examined at this age and at birth. In Ntf4/5−/− mice, there was no Ntf4/5 mRNA expression, and the expression of Bdnf in the tongue epithelium (Fig. 8B) and geniculate ganglion (Fig. 8C) was not different from that in wild type mice. Taken together, BDNF and NT4 do not appear to regulate each other during gustatory development.
Figure 8. BDNF and NT4 do not regulate each other.
The expression of Ntf4/5 in the lingual epithelium was not different between Bdnf −/− and wild type mice at either E14.5 or birth (A). Ntf4/5 expression in geniculate ganglion was too low to be compared. The expression of Bdnf in the lingual epithelium of Ntf4/5−/− mice, was not different from that of wild type mice at E12.5 and birth (B). The expression of Bdnf in the geniculate was not different between Ntf4/5−/− and wild type mice at E12.5 and birth (C).
DISCUSSION
We examined the embryonic time course of Bdnf, Ntf4/5 and TrkB expression in the peripheral taste system. We found that Ntf4/5 expression in the peripheral taste system is robust initially but decreases rapidly during development. Although previous in situ hybridization experiments demonstrated that Ntf4/5 mRNA is undetectable in the rat tongue (Nosrat et al., 2001), this might be because Ntf4/5 mRNA is evenly distributed throughout the tongue epithelium and it is unlikely to be detected using in situ hybridization. The higher expression of Bdnf mRNA in the earlier embryonic tongue epithelium is generally consistent with previous in situ hybridization experiments in rats, in which Bdnf mRNA is detected in the embryonic fungiform papillae and the expression starts to diminish after E17.5 (Nosrat and Olson, 1995; Nosrat et al., 1996). In the geniculate ganglion, Bdnf mRNA expression actually increases during embryonic development. The common receptor for BDNF and NT4; i.e., TrkB, is highly expressed in the geniculate ganglion, which is consistent with expression of TrkB during the embryonic development of the rat geniculate ganglion (Yamout et al., 2005). However, in the tongue and soft palate TrkB mRNA expression levels are very low and do not change during development. These data provide clues for the functional roles of the TrkB and its ligands in peripheral taste development.
One well established function of neurotrophins is their ability to regulate neuron survival. The changes in neurotrophin expression that we observed correspond closely with peak normal developmental cell death in geniculate ganglion. Geniculate ganglion neuron survival is dependent on BDNF, NT4 or both. Specifically, overexpression of either BDNF or NT4 leads to an increased number of geniculate ganglion neurons (Ringstedt et al., 1999; Krimm et al., 2001), whereas removal of BDNF or NT4 leads to a reduction of neuron number (Conover et al., 1995; Liu et al., 1995; Liebl et al., 1997; Patel and Krimm, 2010). During development, the total number of geniculate ganglion neurons remains fairly constant, indicating that new neurons are differentiating at the same rate as others are dying. In rats, a high occurrence of neuronal degeneration has been observed in geniculate ganglion during the period between E12.5 and E16.5, and with peak neuron death at E16.5 (Carr et al., 2005) (corresponding to E14.5 in the mouse). Between E12.5 and E14.5, Ntf4/5 expression decreases in all of the locations where it is expressed. This decrease in Ntf4/5 expression may contribute to the increased neuron death at E14.5 because at this age NT4 expression is reduced, but BDNF expression in the ganglion has not yet increased and gustatory fibers invading the lingual epithelia may not have fully innervated their targets (Mbiene, 2004; Ma et al., 2009), which could limit BDNF from the periphery. After E14.5, the expression level of Bdnf in the geniculate ganglion starts to increase and the chorda tympani fibers have established connections with fungiform papillae, where Bdnf expression remains high until E16.5.
In addition to predicting the changes in normal cell death, differences in neurotrophin expression predict that neurons become BDNF and NT4 dependent at different developmental stages. In the geniculate ganglion, Ntf4/5 decreases after E12.5, and in the anterior tongue epithelium, Ntf4/5 expression is greater than BDNF expression at E12.5, but then is greatly reduced. The reduction in Ntf4/5 expression after E12.5 suggests that NT4 in the geniculate ganglion and the tongue is mainly involved in early development events that occur at E12.5 or earlier. Consistent with these findings, NT4 mutant mice lose geniculate ganglion neurons from E11.5, which is earlier than that in BDNF mutants (Patel and Krimm, 2005). At this age, gustatory axons have reached the tongue, but have not innervated specific taste regions at the tongue surface. The ganglion, tongue mesenchyme, and lingual epithelia may be important sources of NT4 to the developing neurons, but the target (i.e. taste placode) is not. NT4 expressed in the ganglion may rescue neuronal precursors from cell death or increase their proliferation. Alternatively, NT4 in the mesenchyme and from the epithelia may influence the survival of differentiated neurons before they reach their target. This early role of NT4 is not unique to the taste system. For example, in Ntf4/5−/− nodose/petrosal (N/P) ganglion neuronal loss mainly occurs between E12.5 and E14.5, whereas in BDNF−/− N/P ganglion the loss mainly occurs between E14.5 and birth (ElShamy and Ernfors, 1997).
In addition to regulating neuron survival, BDNF and NT4 are expressed at the correct time and place to regulate axon growth and branching. In the mouse, the chorda tympani nerves enter the tongue and extend along the base of the tongue, and fiber bundles branch from the chorda tympani nerves at the tongue base. Chorda tympani axon directional growth, toward the surface of the tongue, occurs as soon as gustatory fibers enter the tongue. However, axon fibers do not enter the apical epithelia of the developing fungiform papillae until around E14.5 to E15.5 (Mbiene, 2004; Lopez and Krimm, 2006b). We found that Bdnf and Ntf4/5 expression in the tongue mesenchyme is highest during innervation, and then reduced after E14.5, which may indicate that these neurotrophins are involved in the axon growth to the tongue epithelial surface and branching in the mesenchyme. Indeed, in vitro studies have shown that both BDNF and NT4 are capable of stimulating geniculate axon outgrowth (Rochlin et al., 2000). Axon branching from the chorda tympani nerve is normal in either Bdnf or Ntf4/5 null mutant mice, but almost absent in double knockout mice, lacking both BDNF and NT4. Therefore, BDNF and NT4 function interchangeably to support branching toward the lingual surface. Once chorda tympani fibers reach their target, BDNF and NT4 along the project pathway are no longer needed and therefore expression is reduced.
There is substantial evidence that correct spatial expression of BDNF is crucial for appropriate gustatory target innervation (Nosrat et al., 1997; Ringstedt et al., 1999; Krimm et al., 2001; Lopez and Krimm, 2006a). BDNF expressed in epithelial placodes, which later form fungiform papillae and taste buds, acts as a short-range guidance molecule for growing gustatory nerve fibers during initial target innervation. BDNF-mediated targeting in the anterior tongue requires that BDNF is expressed on or before E13.5 in mice (Ma et al., 2009). Consistent with this role, we observed high BDNF expression by E12.5 in the lingual epithelium and BDNF-lacZ staining revealed that BDNF is expressed in specific locations where fungiform papillae will develop. Therefore, BDNF expressed in the embryonic tongue epithelium participates in initial gustatory innervation of fungiform papillae.
In addition to influencing gustatory neurons, BDNF may directly influence taste bud development. In our experiments, both BDNF and TrkB amplification products were obtained from the anterior tongue epithelial cDNA. This is consistent with previous reports from hamsters, in which BDNF and TrkB are co-expressed within fungiform taste buds (Ganchrow et al., 2003). BDNF expressed in the mouse fungiform taste buds could exert an autocrine and/or paracrine influence on taste bud development or maintenance.
Another source of BDNF for taste bud maintenance is the geniculate ganglion, where BDNF expression increases after target innervation as taste buds are differentiating. In rodents, transection of the chorda tympani nerves results in fewer and smaller taste buds (Farbman, 1969; Cheal and Oakley, 1977; Cain et al., 1996; Sollars and Bernstein, 2000; Sollars et al., 2002; Guagliardo and Hill, 2007), and taste buds reappear after reinnervation (Cheal et al., 1977; Cheal and Oakley, 1977; Cain et al., 1996). It is possible that the transection of gustatory nerves blocks the transport of BDNF from geniculate ganglion neurons to taste buds because BDNF can function as an anterograde survival factor (Altar and DiStefano, 1998).
BDNF and NT4 function via the same set of receptors, and the primary receptor for these ligands is TrkB. In the geniculate ganglion, TrkB expression is much higher than Bdnf and Ntf4/5 expression and the high expression level is maintained throughout embryonic development. Consistently, most or all cells in the rat geniculate ganglion express TrkB mRNA by E13, and the expression level does not change at the later embryonic ages (Ernfors et al., 1992; Yamout et al., 2005). In the mammalian nervous system, there are three TrkB isoforms. Only the full-length isoform contains a tyrosine kinase domain, which mediates the downstream signaling transduction of TrkB activated by BDNF or NT4 (Klein et al., 1990; Middlemas et al., 1991). In our experiments, the primers for TrkB were designed to detect the full-length rather than the truncated isoforms. Therefore, the geniculate ganglion contains high level of the TrkB isoform that mediates BDNF and NT4 signaling. This high expression level of TrkB in the geniculate ganglion suggests that its ligands, BDNF and NT4, but not TrkB, are the limiting factors for neurotrophic functions in the developing geniculate ganglion.
In conclusion, the expression pattern of neurotrophins in the peripheral taste system suggests that NT4 has an early role in development, while BDNF has a later embryonic role. Ntf4/5 expression is greater than Bdnf at E12.5 or earlier, indicating that NT4 is the primary neurotrophin influencing gustatory neuron development at early embryonic stages, before target innervation. Therefore NT4 does not function as a target derived neurotrophic factor. Instead, high levels of NT4 in the geniculate ganglion and along the projection pathway may play an important role in regulating gustatory neuron number in undifferentiated neuronal precursors or in differentiated neurons before they reach their final target. It is likely that BDNF expressed in the taste target is important for regulating ganglion development and target innervation at E14.5 and later. Increasing levels of BDNF in the geniculate ganglion as it matures may regulate neuron differentiation, continued survival, taste bud or CNS development.
EXPERIMENTAL PROCEDURES
Animals
C57Bl/6J (wild type) mice and mice heterozygous for targeted mutations of Bdnf and Ntf4/5 (Stock #002266 and Stock #002497, Bar Harbor, Maine) were acquired from Jackson Laboratories (Ernfors et al., 1994; Liu et al., 1995). For BdnflacZ/+ mice, the BDNF coding sequence at one allele is replaced by the E. coli galactosidase (lacZ) gene, which serves as a reporter gene (Jones et al., 1994). Embryonic mice were obtained from time-bred females that were placed with males just before the 8-h dark period and examined for plugs the following morning. The day a plug was found was defined as E0.5. Ages were verified using morphological features for each embryo stage (Kaufman, 1995). Animals were cared for and used in accordance with guidelines of the U.S. Public Health Service Policy on Humane Care and Use of Laboratory Animals and NIH Guide for the Care and Use of Laboratory Animals.
Tissue Isolation for real-time PCR
Tongues were isolated from embryonic and newborn mice at the following ages: E12.5, E14.5, E16.5 and birth (n=3 at each age). Laser capture microdissection (LCM) was used to isolate the geniculate ganglion from serial sections at E12.5 (n=3), E14.5 (n=4), E16.5 (n=3) and birth (n=4). First, tissues containing geniculate ganglia were cut into 10 µm thick sections on a cryostat and directly mounted onto glass slides. Sections were processed to allow for visual identification of the geniculate ganglion. Specifically, the sections were fixed in a solution of 75% ethyl alcohol (ETOH) for 30 seconds and then rinsed in nuclease-free water. The slides were then taken through an alcohol dehydration series of 75%, 95% and 100% ETOH for 30 seconds each, followed by immersion in xylene for 5 minutes. After the slides were removed from xylene and the remaining xylene had evaporated, they were placed onto a laser capture microscope (Arcturus) and the geniculate ganglia were identified and captured onto CapSure Macro LCM Caps (Molecular Devices). Figure 1A shows the images of a tissue section before and after geniculate ganglion cells were captured. For each animal, all sections containing geniculate ganglia were collected and the entire geniculate ganglion was captured for RNA isolation. The captured samples were then collected into tubes containing total RNA isolation reagent (Qiagen) for later RNA isolation.
Lingual epithelium and mesenchyme were isolated by enzymatic dissociation from the anterior portion of the tongue (Fig. 1B), where fungiform papillae are located. Briefly, tongues were dissected, rinsed with cold PBS and then incubated in sterile dispase I-solution (BD Biosciences) for 15 min – 40 min depending on age. After incubation, the tongues were placed into PBS-BSA solution. The epithelial sheets were peeled off from the underlying mesenchyme, and both epithelium and mesenchyme were transferred into separate tubes containing RNAlater (Ambion). The epithelium and mesenchyme from soft palate (Fig. 1C) were isolated in the same way. All samples were stored at −80°C until RNA extraction.
RNA extraction and real-time PCR
Total RNA from each geniculate ganglion, and the epithelia and mesenchyme of the tongue and the soft palate, was extracted using RNeasy micro kit or RNeasy mini kit (Qiagen). DNase I treatment was applied to eliminate traces of DNA during the procedure. After the extraction, RNA was analyzed with RNA 6000 Pico/Nano Chip kits in Bioanalyzer 2100 (Agilent Technologies) and RNA Integrity Number (RIN) and 28S/18S ratio were used to estimate the RNA quality. Only RNA samples with RNA integrity number (RIN) more than 8.0 were used in this study. Reverse transcription was performed using 200 U Superscript III Reverse Transcriptase (Invitrogen) and 50 ng random hexamers (Invitrogen) in 25 µl reaction volume containing 1 × First strand buffer (Invitrogen), 0.5 mM dNTPs, and 40 U of RNase inhibitor. All samples produced sufficient amounts of RNA for real-time PCR without pooling. To control differences in the amount of RNA isolated from different ages, the same amount of RNA from each sample was used for geniculate ganglion (3 ng) and for lingual epithelium (50 ng). After incubation for 50 min at 50°C, the reaction was stopped by heating (5 min at 85°C). To check if there was genomic DNA contamination, RNA was also treated in parallel in the absence of reverse transcriptase.
Real-time PCR was performed by ABI PRISM/7900HT Sequence detection systems (Applied Biosystems), using either QuantiFas SYBR Green PCR kit (Qiagen) or TaqMan Universal PCR kit (Applied Biosystems), and oligonucleotide primer/probe sets, which were designed from the sequences in the GenBank database using Beacon Designer software (PREMIER Biosoft International). When it is possible, the primers were chosen to span an intron to avoid the detection of any contamination of genomic DNA. TaqMan probes were labeled at the 5´-end with a fluorescent reporter dye (FAM) and at the 3´-end with a quencher dye,(TAMRA). The sequences of primers and probes are shown in Table 1.
Table 1.
Sequences of primer pairs and probes used for real-time RT-PCR
| Gene | Fragment | |
|---|---|---|
| GenBank Accession # | Sequence 5'-3' | size (bp) |
| BDNF (X55573) | 110 | |
| Forward primer | TGCAGGGGCATAGACAAAAGG | |
| Reverse primer | CTTATGAATCGCCAGCCAATTCTC | |
| Taqman Probe | ACTGGAACTCGCAATGCCGAACTACCCA | |
| NT4 (NM_198190) | 95 | |
| Forward primer | AGCGTTGCCTAGGAATACAGC | |
| Reverse primer | GGTCATGTTGGATGGGAGGTATC | |
| Taqman Probe | TGAGCAGTGAACCCGACCACCCAGG | |
| TrkB (X17647) | 86 | |
| Forward primer | AAGGACTTTCATCGGGAAGCTG | |
| Reverse primer | TCGCCCTCCACACAGACAC | |
| Taqman Probe | CCAACCTCCAGCACGAGCACATTGTCAA | |
| 18S rRNA (X00686) | 76 | |
| Forward primer | CAGGATTGACAGATTGATAGCTCTTTC | |
| Reverse primer | ATCGCTCCACCAACTAAGAACG | |
| Taqman Probe | CCATGCACCACCACCCACGGAATCG | |
| GAPDH (NM_008084) | 130 | |
| Forward primer | AATGTGTCCGTCGTGGATCTG | |
| Reverse primer | CAACCTGGTCCTCAGTGTAGC | |
| Taqman Probe | CGTGCCGCCTGGAGAAACCTGCC | |
| β-Actin (NM_007393) | 144 | |
| Forward primer | CTGGGACGACATGGAGAAGATC | |
| Reverse primer | GTCTCAAACATGATCTGGGTCATC | |
| Taqman Probe | ACCTTCTACAATGAGCTGCGTGTGGCC |
The real-time PCR reactions were conducted using 20 µl total volume with 1 × Master Mix, 200 nM primers (SYBR Green PCR kit) or 720/200 nM primer/probe sets (TaqMan PCR kit) and the same amount of target cDNA across different ages. To determine the PCR efficiencies, mouse brain cortex cDNA were serially (10-fold) diluted to perform PCR in parallel. For normalization of cDNA loading, all samples were run in parallel with the housekeeping gene, 18S ribosomal RNA, mouse glyceraldehyde 3 phosphate dehydrogenase (GAPDH) and β-actin. Each assay was carried out in triplicate. Amplification of cDNA was performed for 40 cycles of 95°C for 15 s and 60°C for 1 min.
X-Gal staining
To detect LacZ, embryos (E14.5 and E16.5, n=2) and newborn mice (n=2) were perfused and post-fixed with 0.5% glutaraldehyde, and rinsed three times in PBS/MgCl2. The dissected tissues were then placed in freshly prepared β-galactosidase (β-gal) staining solution including 6 mM potassium ferricyanide, 6 mM potassium ferrocyanide, 1 mg/ml X-Gal, 2 mM MgCl2, 0.02% Igepal and 0.01% sodium deoxycholate in PBS (InvivoGen) for 1 h – 5 h at 37°C. Images of whole-mount and tissue section (25 µm) staining were taken using a Retiga 1300 digital camera (Qimaging) mounted to a microscope (Leica).
Immunohistochemistry
Embryos and newborn mice were perfused and post-fixed with 0.2% paraformaldehyde. The target tissues were embedded in O.C.T. compound. Tissue blocks were sectioned at 16 µm thickness. Immunofluorescence staining was performed using rabbit anti-β-gal (1:500, Cappel) and mouse anti-Hu (1:500, Molecular Probes) primary antibodies. Cryostat sections were blocked in 5% normal goat serum and 0.25% Triton X-100 in PBS, followed by incubation overnight at RT in primary antibodies. Before incubation with anti-β-gal antibody, tissue sections were post-fixed with 4% paraformaldehyde for 1 h. After washing, the sections were incubated at RT with secondary goat anti-rabbit and goat anti-mouse antibodies, conjugated to AlexaFluor 555 or AlexaFluor 488 (1:500; Molecular Probes). Primary and secondary antibodies were diluted in PBS containing 5% normal goat serum and 0.25% Triton X-100. Coverslips were mounted with DPX mounting medium (Fluka).
Statistical analysis of data
Results are expressed as mean ± SEM. For real-time PCR, the comparative 2−ΔΔCT method was used to determine target gene expression levels (Livak and Schmittgen, 2001). The normalized expression of the target gene was calculated as normalized expression = (Etarget)ΔCTtarget (control – sample) /(Eref)ΔCTref (control – sample), where Etarget is reaction efficiency of the gene of interest; Eref, reaction efficiency of the reference gene; and ΔCT, the cycle difference between the control and the sample. The significant difference between groups was evaluated by REST (Pfaffl et al., 2002). A p value of less than 0.05 was considered statistically significant.
ACKNOWLEDGEMENTS
This work was supported by a grant from the National Institutes of Health to RFK (DC007176).
Grant: NIH DC007176 (to R. F. K.)
REFERENCES
- Agerman K, Hjerling-Leffler J, Blanchard MP, Scarfone E, Canlon B, Nosrat C, Ernfors P. BDNF gene replacement reveals multiple mechanisms for establishing neurotrophin specificity during sensory nervous system development. Development. 2003;130:1479–1491. doi: 10.1242/dev.00378. [DOI] [PubMed] [Google Scholar]
- Al-Hadlaq SM, Bradley RM, MacCallum DK, Mistretta CM. Embryonic geniculate ganglion neurons in culture have neurotrophin-specific electrophysiological properties. Neuroscience. 2003;118:145–159. doi: 10.1016/s0306-4522(02)00814-x. [DOI] [PubMed] [Google Scholar]
- Altar CA, DiStefano PS. Neurotrophin trafficking by anterograde transport. Trends Neurosci. 1998;21:433–437. doi: 10.1016/s0166-2236(98)01273-9. [DOI] [PubMed] [Google Scholar]
- Bibel M, Barde YA. Neurotrophins: key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 2000;14:2919–2937. doi: 10.1101/gad.841400. [DOI] [PubMed] [Google Scholar]
- Cain P, Frank ME, Barry MA. Recovery of chorda tympani nerve function following injury. Exp Neurol. 1996;141:337–346. doi: 10.1006/exnr.1996.0169. [DOI] [PubMed] [Google Scholar]
- Carr VM, Sollars SI, Farbman AI. Neuronal cell death and population dynamics in the developing rat geniculate ganglion. Neuroscience. 2005;134:1301–1308. doi: 10.1016/j.neuroscience.2005.05.034. [DOI] [PubMed] [Google Scholar]
- Cheal M, Dickey WP, Jones LB, Oakley B. Taste fiber responses during reinnervation of fungiform papillae. J Comp Neurol. 1977;172:627–646. doi: 10.1002/cne.901720406. [DOI] [PubMed] [Google Scholar]
- Cheal M, Oakley B. Regeneration of fungiform taste buds: temporal and spatial characteristics. J Comp Neurol. 1977;172:609–626. doi: 10.1002/cne.901720405. [DOI] [PubMed] [Google Scholar]
- Conover JC, Erickson JT, Katz DM, Bianchi LM, Poueymirou WT, McClain J, Pan L, Helgren M, Ip NY, Boland P, et al. Neuronal deficits, not involving motor neurons, in mice lacking BDNF and/or NT4. Nature. 1995;375:235–238. doi: 10.1038/375235a0. [DOI] [PubMed] [Google Scholar]
- ElShamy WM, Ernfors P. Brain-derived neurotrophic factor, neurotrophin-3, and neurotrophin-4 complement and cooperate with each other sequentially during visceral neuron development. J Neurosci. 1997;17:8667–8675. doi: 10.1523/JNEUROSCI.17-22-08667.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernfors P, Lee KF, Jaenisch R. Mice lacking brain-derived neurotrophic factor develop with sensory deficits. Nature. 1994;368:147–150. doi: 10.1038/368147a0. [DOI] [PubMed] [Google Scholar]
- Ernfors P, Merlio JP, Persson H. Cells Expressing mRNA for Neurotrophins and their Receptors During Embryonic Rat Development. Eur J Neurosci. 1992;4:1140–1158. doi: 10.1111/j.1460-9568.1992.tb00141.x. [DOI] [PubMed] [Google Scholar]
- Farbman AI. Fine structure of degenerating tast buds after denervation. J Embryol Exp Morphol. 1969;22:55–68. [PubMed] [Google Scholar]
- Fritzsch B, Sarai PA, Barbacid M, Silos-Santiago I. Mice with a targeted disruption of the neurotrophin receptor trkB lose their gustatory ganglion cells early but do develop taste buds. Int J Dev Neurosci. 1997;15:563–576. doi: 10.1016/s0736-5748(96)00111-6. [DOI] [PubMed] [Google Scholar]
- Ganchrow D, Ganchrow JR, Verdin-Alcazar M, Whitehead MC. Brain-derived neurotrophic factor-, neurotrophin-3-, and tyrosine kinase receptor-like immunoreactivity in lingual taste bud fields of mature hamster. J Comp Neurol. 2003;455:11–24. doi: 10.1002/cne.2162. [DOI] [PubMed] [Google Scholar]
- Guagliardo NA, Hill DL. Fungiform taste bud degeneration in C57BL/6J mice following chorda-lingual nerve transection. J Comp Neurol. 2007;504:206–216. doi: 10.1002/cne.21436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF. Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KR, Farinas I, Backus C, Reichardt LF. Targeted disruption of the BDNF gene perturbs brain and sensory neuron development but not motor neuron development. Cell. 1994;76:989–999. doi: 10.1016/0092-8674(94)90377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman M. The Atlas of Mouse Development. London: Academic Press; 1995. [Google Scholar]
- Klein R, Conway D, Parada LF, Barbacid M. The trkB tyrosine protein kinase gene codes for a second neurogenic receptor that lacks the catalytic kinase domain. Cell. 1990;61:647–656. doi: 10.1016/0092-8674(90)90476-u. [DOI] [PubMed] [Google Scholar]
- Krimm RF. Factors that regulate embryonic gustatory development. BMC Neurosci. 2007;8 Suppl 3:S4. doi: 10.1186/1471-2202-8-S3-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimm RF, Miller KK, Kitzman PH, Davis BM, Albers KM. Epithelial overexpression of BDNF or NT4 disrupts targeting of taste neurons that innervate the anterior tongue. Dev Biol. 2001;232:508–521. doi: 10.1006/dbio.2001.0190. [DOI] [PubMed] [Google Scholar]
- Liebl DJ, Mbiene JP, Parada LF. NT4/5 mutant mice have deficiency in gustatory papillae and taste bud formation. Dev Biol. 1999;213:378–389. doi: 10.1006/dbio.1999.9385. [DOI] [PubMed] [Google Scholar]
- Liebl DJ, Tessarollo L, Palko ME, Parada LF. Absence of sensory neurons before target innervation in brain-derived neurotrophic factor-, neurotrophin 3-, and TrkC-deficient embryonic mice. J Neurosci. 1997;17:9113–9121. doi: 10.1523/JNEUROSCI.17-23-09113.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ernfors P, Wu H, Jaenisch R. Sensory but not motor neuron deficits in mice lacking NT4 and BDNF. Nature. 1995;375:238–241. doi: 10.1038/375238a0. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lopez GF, Krimm RF. Epithelial overexpression of BDNF and NT4 produces distinct gustatory axon morphologies that disrupt initial targeting. Dev Biol. 2006a;292:457–468. doi: 10.1016/j.ydbio.2006.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez GF, Krimm RF. Refinement of innervation accuracy following initial targeting of peripheral gustatory fibers. J Neurobiol. 2006b;66:1033–1043. doi: 10.1002/neu.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Lopez GF, Krimm RF. Epithelial-derived brain-derived neurotrophic factor is required for gustatory neuron targeting during a critical developmental period. J Neurosci. 2009;29:3354–3364. doi: 10.1523/JNEUROSCI.3970-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbiene JP. Taste placodes are primary targets of geniculate but not trigeminal sensory axons in mouse developing tongue. J Neurocytol. 2004;33:617–629. doi: 10.1007/s11068-005-3331-1. [DOI] [PubMed] [Google Scholar]
- Middlemas DS, Lindberg RA, Hunter T. trkB, a neural receptor protein-tyrosine kinase: evidence for a full-length and two truncated receptors. Mol Cell Biol. 1991;11:143–153. doi: 10.1128/mcb.11.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistretta CM, Goosens KA, Farinas I, Reichardt LF. Alterations in size, number, and morphology of gustatory papillae and taste buds in BDNF null mutant mice demonstrate neural dependence of developing taste organs. J Comp Neurol. 1999;409:13–24. [PMC free article] [PubMed] [Google Scholar]
- Nosrat CA, Blomlof J, ElShamy WM, Ernfors P, Olson L. Lingual deficits in BDNF and NT3 mutant mice leading to gustatory and somatosensory disturbances, respectively. Development. 1997;124:1333–1342. doi: 10.1242/dev.124.7.1333. [DOI] [PubMed] [Google Scholar]
- Nosrat CA, Ebendal T, Olson L. Differential expression of brain-derived neurotrophic factor and neurotrophin 3 mRNA in lingual papillae and taste buds indicates roles in gustatory and somatosensory innervation. J Comp Neurol. 1996;376:587–602. doi: 10.1002/(SICI)1096-9861(19961223)376:4<587::AID-CNE7>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Nosrat CA, MacCallum DK, Mistretta CM. Distinctive spatiotemporal expression patterns for neurotrophins develop in gustatory papillae and lingual tissues in embryonic tongue organ cultures. Cell Tissue Res. 2001;303:35–45. doi: 10.1007/s004410000271. [DOI] [PubMed] [Google Scholar]
- Nosrat CA, Olson L. Brain-derived neurotrophic factor mRNA is expressed in the developing taste bud-bearing tongue papillae of rat. J Comp Neurol. 1995;360:698–704. doi: 10.1002/cne.903600413. [DOI] [PubMed] [Google Scholar]
- Patel AV, Huang T, Krimm RF. BDNF and NT4 both are essential for the survival of developing gustatory neurons but differentially regulate the development of taste buds in the tongue vs the soft palate. International Symposium on Olfaction and Taste; Abstract viewer and Itinerary Planner; San Francisco. 2008. [Google Scholar]
- Patel AV, Krimm RF. Society for Neuroscience. Washington DC: Abstract viewer and Itinerary Planner; 2005. BDNF and NT4 regulate geniculate ganglion neuron number at different developmental time points. [Google Scholar]
- Patel AV, Krimm RF. BDNF is required for the survival of differentiated geniculate ganglion neurons. Dev Biol. 2010;340:419–429. doi: 10.1016/j.ydbio.2010.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patz S, Wahle P. Neurotrophins induce short-term and long-term changes of cortical neurotrophin expression. Eur J Neurosci. 2004;20:701–708. doi: 10.1111/j.1460-9568.2004.03519.x. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringstedt T, Ibanez CF, Nosrat CA. Role of brain-derived neurotrophic factor in target invasion in the gustatory system. J Neurosci. 1999;19:3507–3518. doi: 10.1523/JNEUROSCI.19-09-03507.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochlin MW, O'Connor R, Giger RJ, Verhaagen J, Farbman AI. Comparison of neurotrophin and repellent sensitivities of early embryonic geniculate and trigeminal axons. J Comp Neurol. 2000;422:579–593. [PubMed] [Google Scholar]
- Schecterson LC, Bothwell M. Novel roles for neurotrophins are suggested by BDNF and NT-3 mRNA expression in developing neurons. Neuron. 1992;9:449–463. doi: 10.1016/0896-6273(92)90183-e. [DOI] [PubMed] [Google Scholar]
- Sollars SI, Bernstein IL. Neonatal chorda tympani transection permanently disrupts fungiform taste bud and papilla structure in the rat. Physiol Behav. 2000;69:439–444. doi: 10.1016/s0031-9384(99)00259-0. [DOI] [PubMed] [Google Scholar]
- Sollars SI, Smith PC, Hill DL. Time course of morphological alterations of fungiform papillae and taste buds following chorda tympani transection in neonatal rats. J Neurobiol. 2002;51:223–236. doi: 10.1002/neu.10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H, Futamura T, Jourdi H, Zhou H, Takei N, Diverse-Pierluissi M, Plevy S, Nawa H. Neurotrophins induce BDNF expression through the glutamate receptor pathway in neocortical neurons. Neuropharmacology. 2002;42:903–912. doi: 10.1016/s0028-3908(02)00043-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamout A, Spec A, Cosmano J, Kashyap M, Rochlin MW. Neurotrophic factor receptor expression and in vitro nerve growth of geniculate ganglion neurons that supply divergent nerves. Dev Neurosci. 2005;27:288–298. doi: 10.1159/000086708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Brandemihl A, Lau D, Lawton A, Oakley B. BDNF is required for the normal development of taste neurons in vivo. Neuroreport. 1997;8:1013–1017. doi: 10.1097/00001756-199703030-00039. [DOI] [PubMed] [Google Scholar]