Abstract
Objectives
Microvillus inclusion disease (MVID) is a rare autosomal recessive enteropathy characterized by intractable diarrhea and malabsorption. Recently, various MYO5B gene mutations have been identified in MVID patients. Interestingly, several MVID patients showed only a MYO5B mutation in one allele (heterozygous) or no mutations in the MYO5B gene, illustrating the need to further functionally characterize the cell biological effects of the MYO5B mutations.
Methods
The genomic DNA of nine patients diagnosed with microvillus inclusion disease was screened for MYO5B mutations, and qPCR and immunohistochemistry on the material of two patients was performed to investigate resultant cellular consequences.
Results
We demonstrate for the first time that MYO5B mutations can be correlated with altered myosin Vb mRNA expression and with an aberrant subcellular distribution of the myosin Vb protein. Moreover, we demonstrate that the typical and myosin Vb–controlled accumulation of rab11a-and FIP5-positive recycling endosomes in the apical cytoplasm of the cells is abolished in MVID enterocytes, which is indicative for altered myosin Vb function. Also, we report 8 novel MYO5B mutations in 9 MVID patients of various etnic backgrounds, including compound heterozygous mutations.
Conclusions
Our functional analysis indicate that MYO5B mutations can be correlated with an aberrant subcellular distribution of the myosin Vb protein and apical recycling endosomes which, together with the additional compound heterozygous mutations, significantly strengthen the link between MYO5B and MVID.
Keywords: microvillus inclusion disease, myosin Vb, apical recycling endosome, brush border
Introduction
A structurally, compositionally, and functionally distinct plasma membrane at the apex of the intestinal epithelial cell monolayer provides a selective and protective barrier that regulates the uptake of nutrients from the lumen. The inability of intestinal cells to maintain an apical brush border and the consequence it has on human functioning becomes particularly apparent in patients diagnosed with microvillus inclusion disease (MVID; OMIM 251850)
Microvillus inclusion disease is a rare autosomal recessive disease presenting with severe intractable diarrhea and malabsorption in neonates [1–5]. At the cellular level, variable brush border atrophy with accumulation of lysosomal granules and microvillus inclusions is observed in the apical cytoplasm of MVID enterocytes [1,2,6,7]. Periodic acid-Shiff-stained and other apical brush border components (e.g. CD10) are typically absent from the cell surface and accumulate in compartments in the apical cytoplasm [8]. In contrast to the apical proteins, basolateral proteins display a normal polarized distribution at the surface of MVID enterocytes [8,9] which appear normally arranged in monolayers with distinguishable cell-cell adhesion junctions.
MVID is often described in children born of consanguineous parents and this allowed Müller et al. [10] to map the MVID locus to 18q21 using homozygosity mapping in an extended Turkish kindred. Mutation analysis of a positional candidate gene from the region of homozygosity, MYO5B, revealed a homozygous in-frame insertion in the MVID patients from the Turkish kindred. To date, 25 different nonsense, missense, splice site, or in-frame insertion mutations in the MYO5B gene (OMIM# 606540) have been identified in 28 MVID patients from consanguineous and unrelated marriages [10,11,12]. The MYO5B gene encodes myosin Vb, which is an actin filament-based motor protein that interacts with and regulates among others the subcellular spatial distribution of recycling endosomes that express small GTPase proteins such as Rab11a on their cytoplasmic surface.
In several MVID patients MYO5B mutations were found in only one allele (heterozygous) or no MYO5B mutation was found [10]. Moreover, although knockdown of myosin Vb in human epithelial colorectal adenocarcinoma (Caco-2) cells recapitulates most of the cellular phenotypes of MVID [12], it is not known whether myosin Vb mRNA and protein expression and myosin Vb function is affected in MVID patients. Such information would support MYO5B gene screening as a diagnostic tool for this difficult to recognize rare disease, and allow reliable genetic counseling and prenatal screening. Supporting evidence that MYO5B mutations have consequences for the expression and/or function of the myosin Vb protein in MVID enterocytes as well as mutational analyses of additional MVID patients are therefore imperative. In this study we have used small intestine biopsies to demonstrate that MVID-associated MYO5B mutations affect the expression and function of the myosin Vb protein in MVID enterocytes. In addition, we have performed mutation analyses of 9 additional MVID patients of various ethnic background and report 8 new MYO5B mutations; three homozygous and five heterozygous mutations which include stop codons/nonsense mutations, missense mutations, splice site mutations, large deletions, and compound heterozygous mutations.
Materials and methods
Description of patients and clinical history
Nine patients in whom histological examination of small intestine mucosa confirmed the diagnosis of MVID were included in this study. Patients 1–6 were collected from a larger patient cohort that received a bowel transplant via the Liver/Small Bowel Transplant Program of the University Of Nebraska Medical Center (USA). Patient 1 is a 1 year old Hispanic male with early-onset MVID from reported non-consanguineous parents. His brother died of MVID at 21 months of age. Patient 2 is a 3 year old Hispanic female with early-onset MVID of consanguineous parents (first cousins). She has two sisters with MVID one of which is patient 3. Patient 3 is a 5 year old Hispanic female with early-onset MVID from consanguineous parents (first cousins). She has two sisters with MVID one of which is patient 2. Patient 4 is a 1 year old Navajo Indian male with early-onset MVID of related parents (died of sepsis). Patient 5 is a 12 year old Navajo Indian female with early-onset MVID from related parents (died of sepsis with multi-organ system failure). She has two healthy sisters. Patient 6 is a 0 year old Caucasian female with early onset MVID from reported non-consanguineous parents (died of sepsis from aspergillis and continuing acute rejection). She has one healthy sibling and two siblings died with unknown cause. Patient 7 is a 1 year old Polish-Caucasian female with early-onset MVID from reported non-consanguineous parents. Patient 8 was a 5 year old Moroccan boy with early-onset MVID from consanguineous parents (first-degree cousins). Patient 9 is a 5 year old Dutch-Caucasian boy from unrelated parents who was diagnosed with late-onset MVID. Unaffected parents and siblings of patient 7, 8 and 9 were also recruited and, after informed consent, saliva samples were collected and genomic DNA was extracted. In addition, available duodenal tissue from patient 8 and 9 and age-matched normal control patients was obtained and processed for immunohistochemistry. Also two 2 year old Dutch girls (twin) of non-consanguineous parents who presented severe secretory diarrhea and nutrient malabsorption directly after birth, but did not display the diagnostic light and electron microscopical hallmarks of MVID, were included in the study (patients 10 and 11). Written consent was obtained for all patients. This study has been reviewed and approved by the University Medical Center Groningen review board.
DNA and RNA isolation
DNA was isolated from peripheral blood samples using standard laboratory procedures. DNA and RNA from saliva was also collected and isolated (OrageneDNA and OrageneRNA, DNA Genotek Inc, Ottawa, Canada). RNA from in liquid nitrogen snap-frozen biopsy samples was isolated after homogenization using 1 mm glass beads using Trizol (Invitrogen, Carlsbed, CA). Concentration and purity were determined with NanoDrop ND-1000 (Isogen Life Science, De Meern, the Netherlands).
RT-PCR
Real-time PCR reaction was performed for the quantification of MYO5B. RNA was isolated from duodenum biopsies of twelve controls and patients 8 and 9. cDNA was generated with a High Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA) using 1μg total RNA. Primers were designed with Primer Express v.3 (Applied Biosystems); RT_MYO5Bfor: TTGGAAGTGTGGCGATTCAG; RT_MYO5Brev: GCAGTCGGCAGAAGTTGCTT. For GUSB expression, we used a TaqMan Pre-Developed Assay (Applied Biosystems). Reactions consisted of 1xSYBR Green (or Universal) PCR Mastermix, 1mM of each primer and 1μl cDNA. Cycling conditions were 50°C for 2 min, 95°C for 10 min and 40 cycles of 95°C for 15 s and 60°C for 1 min. Results were analyzed using SDS v.2.3 (Applied Biosystems).
Sequencing
The MYO5B coding region and splice sites were PCR amplified and directly sequenced in probands. Their relatives and approximately 50 control individuals (~100 chromosomes) were screened for the detected mutations. Primers for PCR amplification (supplementary tables 1 and 2 show primers used for amplification of genomic DNA and cDNA, respectively) were designed using Primer321 on the genomic sequence of MYO5B (NC_000018.8) and its mRNA (NM_001080467). The PCR reaction was performed with 50ng genomic DNA in 20ul reaction volume which included 1xPCR buffer-A (GE Healthcare, Piscataway/NJ), 2.5 mM dNTPs, 1mM primers (Eurogentec, San Diego, CA), 0.5U Taq polymerase (GE Healthcare). For exons 1, 2, 12, 17, and 18, PCR reaction was performed with 150ng of genomic DNA in 25ul reaction volume with 1xPCR buffer (Buffer-B) made of 0.1M Tris-HCL (pH8.8), 0.1M MgCl2, 0.01M mercaptoethanol, 0.05M ethylenediaminetetraacetic acid/0.1M (NH4)2SO4. The PCR conditions differed with respect to the annealing temperatures and buffers used. Initial denaturation at 95°C for 5 min (4 min Buffer-B); 40 cycles (33 cycles Buffer-B) of denaturation at 95°C for 30 s (1 min Buffer-B), annealing for 30 s (1 min Buffer-B), and extension at 72°C for 30 s (2min Buffer-B); final extension at 72°C for 5 min (7 min Buffer-B). PCR products were purified (37°C for 15 min, 80°C for 15 min) with ExoSap-IT (USB, Cleveland, Ohio) and Sephadex columns. Sequencing reactions were performed using BigDye terminator mix (Applied Biosystems). Sequences were read on a 3730 DNA analyzer and 3130 Genetic analyzer (Applied Biosystems) and we aligned sequencing data with control and reference sequences using ContigExpress software (Invitrogen, Carlsbed, CA).
Deletion detection
In order to detect large deletions in MYO5B in patient 7, real-time quantitative PCR was used to determined copy numbers of the exons. Reactions consisted of 1xSYBR Green PCR Mastermix (Applied Biosystems), 1mM of each primer (supplementary table 2) and 25ng of genomic DNA. Cycling conditions were 50°C for 2 min, 95°C for 10 min and 40 cycles of denaturation at 95°C for 15s, and annealing for 1 min.
Immunohistochemistry
Duodenal biopsies of MVID patients 8 and 9 and age-matched controls were fixed in paraffin, and cut in 3 μm thick sections. Slides were dried overnight in 60°C and deparaffinized in xylol-100%-96%-70%ethanol and demiwater. Epitopes were retrieved by protease digestion or in citric acid pH6.0 (autoclaved; 5 min, 120°C). Endogenous peroxidase was deactivated with 3.5% H2O2. Following blocking of non-specific binding sites in 4% normal-goat-serum, slides were incubated with primary antibodies, washed, and incubated with appropriate horseradish peroxidase-conjugated secondary antibodies. Diaminobenzidine was used as a substrate for peroxidase. Hematoxyline was used to stain the nuclei. Slides were dehydrated with ethanol, dried and mounted. Antibodies used: polyclonal antibodies raised against a synthetic peptide derived from the C-terminal hypervariable region of the human Rab11a sequence (Zymed Laboratories Inc); polyclonal antibodies against Rip11/FIP5 [13]; polyclonal antibodies raised against a synthetic peptide corresponding to C- or N-terminal residues (amino acids 1093-1112 or 23–41, respectively) of human myosin Vb (Antagene Inc; 60B923) that recognizes a single band of the appropriate molecular mass of ~214 kDa on Western blot; Horseradish peroxidase-conjugated donkey anti-rabbit, sheep anti-mouse antibodies (GE Healthcare). Immunohistochemistry images show villus cells.
Results
Eight new MYO5B mutations associated with nine microvillus inclusion disease patients
MYO5B is composed out of 40 coding exons which were separately amplified and subjected to sequence analysis. All eleven patients were included in the mutation analysis by direct sequencing of the entire gene in both forward and reverse directions. Six patients revealed homozygous mutations. Patient 6 revealed one heterozygous change, while patient 7 and 9 carry compound heterozygous mutations (Table 1). Patients 10 and 11, who presented nutrient malabsorption and intractable secretory diarrhea after birth but were not diagnosed with MVID, did not reveal MYO5B mutations.
Table 1.
Summary of MYO5B mutations associated with MVID as reported in this study.
| Subject | Ancestry | Parental Consang. | Sex/onset | MYO5B mutation | Homo/heterozyguos | MYOVb domain | Predicted effect on RNA and/or protein |
|---|---|---|---|---|---|---|---|
| 1 | Hispanic | n.r. | Male/early | c.946 G>A (p.Gly316Arg, exon8) | Homozygous | head | Nonconservative substitution, evolutionary conserved |
| 2, 3 | Hispanic | yes | Female/early | c.2330_del G (p.Gly777AsnfsX6, exon 19) | Homozygous | Neck (IQ1) | Nonconservative substitution, evolutionary conserved, PMT, loss of dimerization and cargo-binding domains |
| 4, 5 | Navajo Indian | yes | Male/early; Female/early | c.1979C>T (p.Pro660Leu, exon 16) | Homozygous | Head | Nonconservative substitution, evolutionary conserved |
| 6 | Caucasian | No | Female/Early | c.2246 C>T (p.Arg749X, exon 19) | Compound Heterozygous | Neck (IQ1) | Nonconservative substitution, evolutionary conserved, PMT, loss of dimerization and cargo-binding domains |
| 7 | Polish | No | Female/early | c.28-?_1545+?del (deletion exon 2–12) | Compound Heterozygous | Head | Shortened protein, in frame deletion residues 10-515 |
| 7 | Polish | No | Female/early | c.1367A>G (p.Asn456Ser; exon 11) | Compound Heterozygous | Head | Nonconservative substitution, evolutionary conserved |
| 8 | Moroccan | Yes | Male/early | c.4366C>T (p.Gln1456X, exon 33) | homozygous | Tail | NMD; PMT, loss of distal Rab11a BD |
| 9 | Dutch | No | Male/late | c.4460-1G>C (splicing, intron 33) | Compound Heterozygous | Tail | Partial intron 33 insertion, NMD; PMT, loss of distal Rab11a BD |
| 9 | Dutch | No | Male/late | c.1540T>C (p.Cys514Arg, exon 12) | Compound Heterozygous | Head | Nonconservative substitution, evolutionary conserved |
NMD, nonsense-mediated RNA decay; PMT, prematurely terminated protein; Rab11a BD (binding domain); n.r., not reported.
Patient 1 carries a homozygous non-conservative missense mutation in exon 8 (c.946G>A, p.Gly316Arg), which replaces a small aliphatic glycine (conserved in myosin Va and Vc; supplementary figure 1) with a large and charged arginine in the protein’s conserved head domain region. In patients 2 and 3 we found a shared homozygous deletion in exon 19 (c.2330_del G; supplementary figure 2). This mutation disturbs the reading frame and leads to a premature stop codon (p.Gly777AsnfsX6; supplementary figure 3) in the first calmodulin-binding IQ1 motif of myosin Vb. Any resultant protein will therefore not be able to dimerize and function as a processive motor protein, and lacks the entire cargo-binding tail domain. Patients 4 and 5 are homozygous for a non-conservative missense mutation in exon 16 (c.1979C>T, p.Pro660Leu). This mutation was recently described [11] in 7 Navajo MVID patients. In patient 6 we found onecvx heterozygous mutation in exon 19 which results in a premature stop codon (c.2246C>T, p.Arg749X) in the head domain of myosin Vb (p.Arg749 is conserved in myosin Va and Vc; supplementary figure 4). Resultant protein will not be able to dimerize and function as a processive motor protein, and lack the entire cargo-binding tail domain.
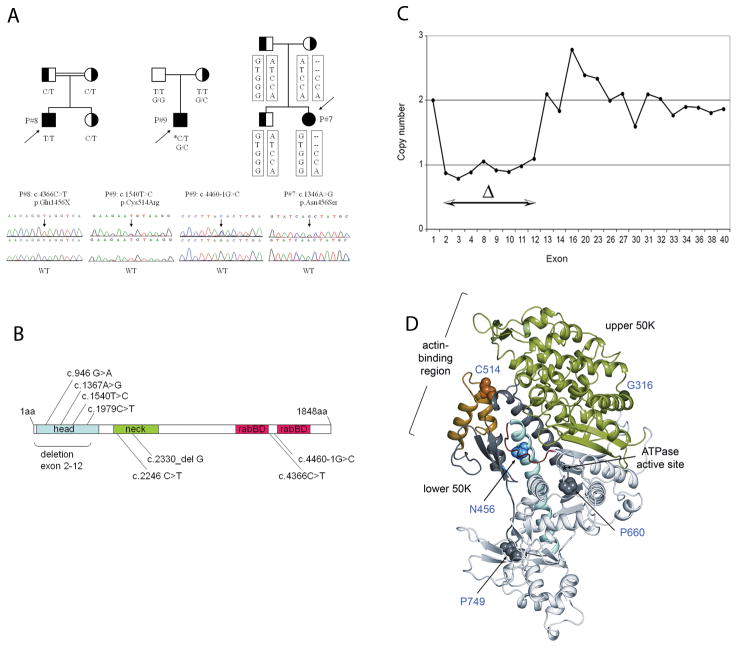
For patient 7, 8 and 9, we also obtained DNA samples from unaffected siblings and/or parents. Patient 7 reveals a compound heterozygous mutation, which includes a paternal allele with a non-conservative asparagine-to-serine (c.1367A>G, p.Asn456Ser) substitution in exon 11 of the head domain (figure 1A, B, D) (conserved in myosin Va and Vc; supplementary figure 5), together with a missense variant p.Met1688Val (c.5062A>G) in exon 37 (p.Met1688 is substituted in MYO5A and MYO5C; supplementary figure 6). p.Met1688Val represents an infrequent polymorphism, as it was found in Polish and Dutch controls with allele frequencies of 5.8% (6/104) and 1.7% (2/116), respectively. When searching for a maternally transmitted mutation in patient 7, a Mendelian inconsistency in the inheritance of the exon11 variant c.1367A>G was observed: the mother appeared to be homozygous A/A while the patients was homozygous G/G (figure 1A). This could possibly point towards a maternal transmission of a deletion. Using real-time PCR to determine the copy number of the MYO5b gene, we found that the maternal allele in patient 7 contained a deletion involving exons 2–12 of MYO5B (figure 1C), rendering any protein formed incapable of binding actin and function as a motor protein. Sequencing of MYO5B in patient 8 revealed a homozygous stop codon in exon 33 (c.4366C>T, p.Gln1456X) (Figure 1A) which removes the terminal Rab11a-binding sites (1799–1814) (figure 1B), which only functions in unison with the more proximal Rab11a-binding site (1400–1415). Sequencing of MYO5B in patient 9 showed that this patient is a compound heterozygote carrying a de novo non-conservative substitution mutation in exon 12 (c.1540T>C, p.Cys514Arg), and a maternally derived mutation in intron 33 (c.4460-1G>C) that destroys the canonical splice acceptor (SA) site (Figure 1A, B). Intron 33 harbors three clusters of potent candidate cryptic SA-sites (supplementary Figure 7). PCR on the patient’s intestinal cDNA with primers for intron 33 and exon 35 demonstrated retention of >100 bp of intron 33 immediately upstream of exon 34. This ‘extended exon 34’ contains nine stop codons, at least one in each of the three reading frames (supplementary Figure 8). The p.Cys514 residue forms part of the helix-turn-helix motif in the motor domain that is associated with actin-binding [14–17] (figure 1E). All mutations identified in this study are listed in Table 1. The position of all mutated residues in the crystal structure of the myosin Vb head domain are depicted in figure 1E.
Figure 1.
Identification and mutation analysis of MYO5B in MVID patients and parents/siblings. A) Pedigrees in three families with MVID patients 7, 8 and 9 (designated P7, P8, and P9). The c.1540T>C substitution in patient 9 is a de novo mutation on the paternal chromosome (indicated with asterisk). Haplotype analysis in patient 7 and her family was indicative for a deletion in MYO5B on the maternal chromosome. The MYO5B variants tested were (top to bottom): c.1367A>G (exon 11), c.3276+11 (rs2276176, intron 24), c.4222-73 (rs490648, intron 31), c.4315+5 (rs488890 intron 32), c.5062A>G (exon 37), c.5313+72(rs621101, intron38) B) MYO5B mutations identified in MVID patients. C) Deletion mapping results of patient 7. The maternally derived deletion, spanning exons 2–12, was determined by quantitative PCR using DNA from the proband’s brother (2 copies for each exon) to normalize the signal. D) Ribbon diagram of the nucleotide-free structure of the motor domain of chicken myosin V (rigor-like/strong actin-binding state, PDB code: 1OE9), indicating the locations of mutated residues (figure prepared with PyMOL (DeLano Scientific LLC)).
Resequencing mutation-containing exons revealed that none of the identified mutations were detected in 50 ethnically matched controls, or have been reported as known variants (in HapMap, dbSNP, and the 1000 genome database), unless stated otherwise.
MYO5B mutations affect the expression and function of the myosin Vb protein in MVID enterocytes
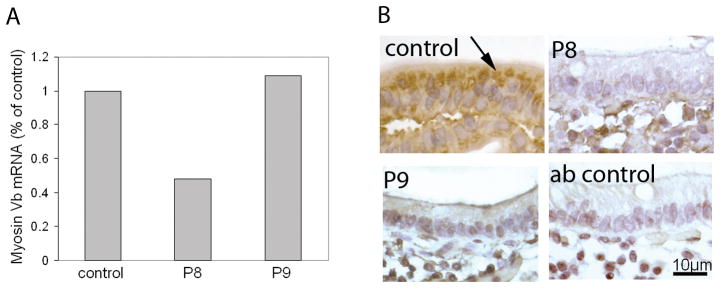
We analyzed the expression levels of myosin Vb mRNA from biopsies of patients 8 and 9 by real-time PCR and compared these to control patients. In patient 8, myosin Vb mRNA expression was reduced by 50% when compare to 14 non MVID control patients. (Figure 2A), which is in agreement with the identified nonsense mutation p.Gln1456X which is predicted to result in nonsense-mediated RNA decay [18]. In patient 9 myosin Vb mRNA levels were comparable to controls (Figure 2A).
Figure 2.
Real-time PCR detection of myosin Vb mRNA and immunohistochemical labeling of myosin Vb. A) RNA was extracted from small intestinal biopsies of MVID patient 8 and 9 (designated P8 and P9) as described in Materials and methods, and relative myosin Vb mRNA levels was determined with real-time PCR. B) small intestinal biopsies of MVID patient 8 and 9 and age-matched controls were labeled with antibodies against human myosin Vb. Negative (non-immune first antibody) control staining are shown. The accumulation of myosin Vb in the apical cytoplasm in control enterocytes (arrows) is lost in MVID enterocytes.
We also analyzed the cellular expression pattern of myosin Vb protein in duodenal biopsies of patients 8 and 9 and age-matched controls. The myosin Vb protein is present in the villus enterocytes and mainly concentrated at their apical aspect below the brush border of control enterocytes (figure 2B, arrow). In contrast, no or little specific myosin Vb signal was detected in the enterocytes of patient 8 and 9, respectively (figure 2B). The MYO5B mutations did not involve residues that were used in the synthetic peptides to generate these antibodies, and the antibodies should recognize the mutant protein if present. The lack of clear myosin Vb signal in MVID enterocytes may reflect the absence of the protein (in accordance with the reduced myosin Vb mRNA levels in patients 8; see above), and/or may reflect a dispersion of remaining myosin Vb protein throughout the cells rendering myosin Vb below the detection limit.
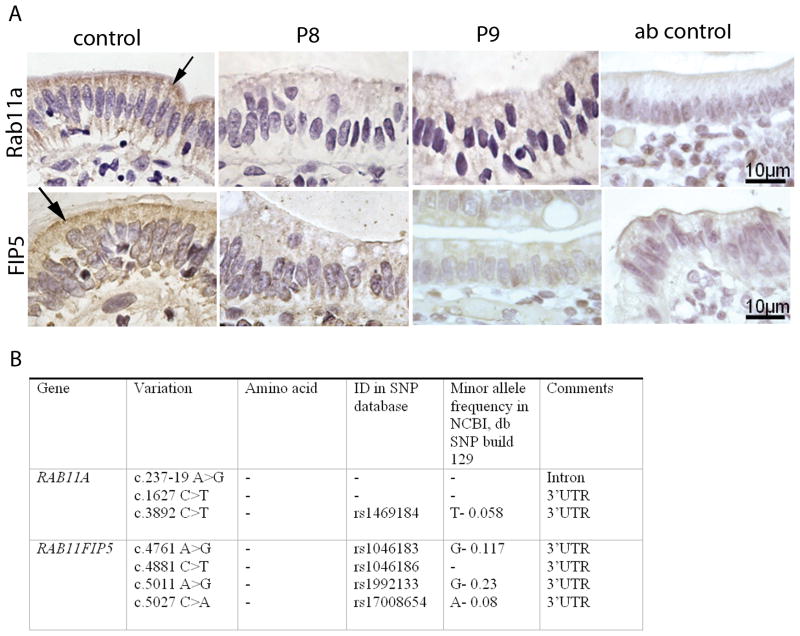
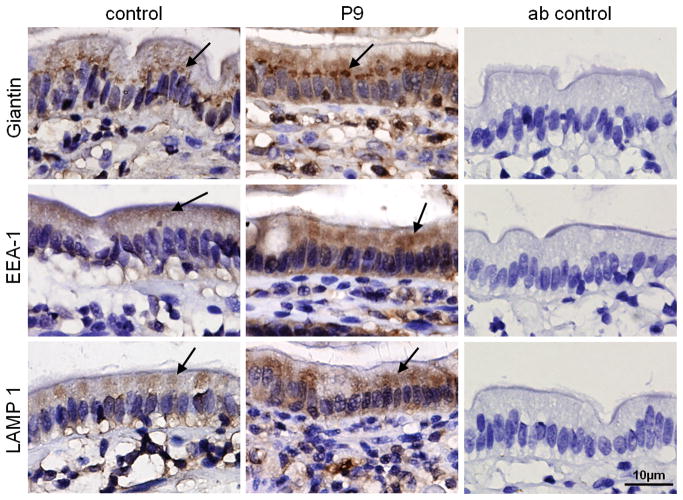
Myosin Vb regulates the subcellular positioning of recycling endosomes by binding to small GTPase Rab proteins such as Rab11a at the cytosolic surface of endosomes and attaching these endosomes to and moving them along actin filaments [19–24]. Alterations in the typical spatial organization of recycling endosomes in MVID enterocytes can therefore be used as a read-out for altered myosin Vb function. To address this, the expression and distribution of recycling endosome-associated proteins Rab11a [19–24] and the Rab11a effector protein FIP5 (Rip11) [13] was investigated. The recycling endosome-associated proteins Rab11a and FIP5 (also known as Rip11) accumulate just below the enterocyte brush border close to the apical membrane in control duodenal tissue, similar to myosin Vb (Figure 3A, arrows). In contrast, in MVID enterocytes, Rab11a and FIP5 did not accumulate in the apical region and, instead, no specific staining pattern (compare to negative antibody (ab) control), could be observed (Figure 3A). Sequence analysis revealed single nucleotide polymorphisms but no functional mutations in the coding regions of the genes RAB11A and RAB11FIP5 in patients 8, and 9 (summarized in figure 3B). Early endosomal antigen 1 (EEA1), a marker of early sorting endosomes and typically excluded from myosin Vb-positive recycling endosomes, and the late endosome- and lyosome-associated protein LAMP-1 displayed comparable staining patterns in controls and MVID patient 9 (Figure 4, arrows). The distribution of the Golgi complex was also apparently unaffected in MVID enterocytes, although it appeared somewhat more concentrated in the supranuclear region (Figure 4).
Figure 3.
Immunohistochemical labeling and variant analysis of Rab11a and FIP5(/Rip11). A) The distribution of Rab11a and FIP5(/Rip11) in control and MVID enterocytes of patients 8 and 9 (designated P8 and P9) is shown. Negative (non-immune first antibody) staining is shown (Ab control). Note that the accumulation of Rab11a in the apical cytoplasm of control enterocytes (arrows) is lost in MVID enterocytes. B) Sequence variants in the genes RAB11A and RAB11FIP5.
Figure 4.
Immunohistochemical labeling of early endosome-, late endosome/lyosome, and Golgi-associated proteins. The subcellular distributions of giantin, EEA1 and LAMP-1 in control and MVID enterocytes (patient 9 (P9)) are shown. Negative (non-immune first antibody) control staining are shown (ab control).
Discussion
We have analyzed the sequence of MYO5B in 9 MVID patients and identified 8 new mutations (~25% of all reported mutations) including a large deletion, a single nucleotide deletion, two missense mutations, and one nonsense mutation. We also report two additional compound heterozygous MYO5B mutations. In two patients we found a homozygous missense mutation that has been described previously [11]. Our study adds 8 mutations to the 25 earlier reported by Müller and colleagues (24 mutations/21 patients) [10,12] and Erickson and colleagues (one mutation shared by 7 Navajo patients) [11], yielding a total of 33 distinct MYO5B mutations in 37 MVID patients that have been identified to date. With our data, providing 25% of all currently reported MYO5B mutations and patients, we make a first analysis of the current MYO5B mutation spectrum. Of the 33 thus far published MYO5B mutations, 24 are localized in the N-terminal head domain that includes actin-binding and ATP catalytic sites, 2 in calmodulin-binding IQ motifs that form the light chain-binding lever arm domain, 1 in a potential coiled-coil regions that mediates the association of the heavy chain into dimers, and 6 are localized in the cargo-binding globular tail domain. All of the MYO5B mutations are distinct from those reported in MYO5A and other nonconventional myosins. Furthermore, the reported heterozygous mutations are exclusively found in patients of Caucasian origin (Polish, Irish, French, and USA), and include at least one nonsense mutation or large deletion. Interestingly, all but one missense mutations cluster in the myosin Vb head domain, whereas the nonsense, splice-site and deletions/insertions are found randomly in the motor, lever arm, and tail domain. While some of the mutations are predicted to result in nonsense-mediated RNA decay (e.g. the homozygous p.Gln1456X mutation in patient 8 and the c.4460-1G>C mutation in patient 9), which is supported by the observed reduction in myosin Vb mRNA levels in patient 8, other MYO5B mutations involve residues that are important for the function of the myosin Vb protein. Indeed, the N456 residue mutated in patient 7, for instance, is part of a set of conserved motifs shared in all myosins that participate in coupling changes in the ATPase active site (P-loop and switch I) to conformational changes in the actin-binding and force-generating domains, and proposed to have a pivotal role in motor function as mediator of allosteric communication [14–17].
We demonstrate that a nonsense MYO5B mutation correlates with reduced myosin Vb mRNA expression, and that MYO5B mutations correlate with an aberrant cellular expression pattern of the myosin Vb protein. A main function of myosin Vb is to regulate the subcellular distribution and positioning of recycling endosomes. It does so by interacting with small GTPase Rab proteins such as Rab11a at the cytosolic surface of recycling endosomes and coupling these endosomes to and positioning them along actin filaments. We demonstrate that the typical and myosin Vb–controlled accumulation of rab11a- and FIP5-positive recycling endosomes in the apical cytoplasm of the cells is abolished in MVID enterocytes. These data are indicative for an altered myosin Vb function in MVID enterocytes. It should be noted that our conclusions are based on two cases of this rare disease and that future experiments are necessary to further consolidate these. Further in-depth analysis of MYO5B mutations and their molecular and cell biological consequences are warranted to expand our understanding of how they are related to MVID pathogenesis.
It is encouraging that all MVID patients (except for one [10]) that have been screened thus far carry mutations in their MYO5B gene, and the discovery of additional compound heterozygous mutations by Ruemmele and colleagues [12] and us (this study) significantly strengthens the correlation between MYO5B and MVID. This correlation is further supported by a recent study [12] in which knockdown of myosin Vb in human epithelial colorectal adenocarcinoma (Caco-2) cells recapitulates most of the cellular phenotypes of MVID, and by our observation that MYO5B mutations were not found in two patients that presented with secretory diarrhea and malabsorption after birth but were not diagnosed with MVID. A firm association of MYO5B mutations with MVID is a major advance in the diagnosis of this rare but fatal disease, in which variable phenotypes are seen among patients. It will also facilitate reliable genetic counseling and prenatal screening. Because total parenteral nutrition and bowel transplants are, at best, non-permanent solutions for treating this devastating disease, the continuing identification of MYO5B mutations will pave the way for the development of alternative therapeutic strategies.
Supplementary Material
Acknowledgments
This research was sponsored by the Dutch Digestive Foundation (MLDS), the Jan Kornelis de Cock Foundation, and De Drie Lichten Foundation. MG was supported by the Ubbo Emmius Foundation. CW was supported by a grant from the Netherlands Organization for Scientific Research (NWO-VICI). SvIJ and ER were supported by the Royal Dutch Academy of Arts and Sciences.
We thank the patients, their parents and siblings, and the transplantation teams of the UMC Groningen and the Children’s Memorial Health Institute in Warsaw. We thank Carolien Gijsbers (Juliana Children’s Hospital, The Hague, The Netherlands) and Marc Benninga (Academic Medical Center Amsterdam, The Netherlands) for the shared care of the two Dutch patients with MVID, and the Dutch Digestive Diseases Foundation (MLDS) for supporting the national collaboration for patients with intestinal failure. We thank Julius Baller and Mathieu Platteel for expert technical assistance, and Hilda Keuning for analysis of histological findings. Control DNA samples were provided by Yvonne Vos (Dutch and Moroccan) and Anna Rybak (Polish). Hanna Romanowska collected DNA samples from the Polish patient and her family. We are indebted to Henkjan Verkade for critically reading the manuscript. This research was sponsored by the Dutch Digestive Foundation (MLDS), the Jan Kornelis de Cock Foundation, and De Drie Lichten Foundation. MG was supported by the Ubbo Emmius Foundation. CW was supported by a grant from the Netherlands Organization for Scientific Research (NWO-VICI). SvIJ and ER were supported by the Royal Dutch Academy of Arts and Sciences.
References
- 1.Phillips AD, Jenkins P, Raafat F, Walker-Smith JA. Congenital microvillous atrophy: specific diagnostic features. Arch Dis Child. 1985;60:135–40. doi: 10.1136/adc.60.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phillips AD, Schmitz J. Familial microvillous atrophy: a clinicopathological survey of 23 cases. J Pediatr Gastroenterol Nutr. 1992;14:380–96. [PubMed] [Google Scholar]
- 3.Cutz E, Rhoads JM, Drumm B, et al. Microvillus inclusion disease: an inherited defect of brush-border assembly and differentiation. N Engl J Med. 1989;320:646–51. doi: 10.1056/NEJM198903093201006. [DOI] [PubMed] [Google Scholar]
- 4.Sherman PM, Mitchell DJ, Cutz E. Neonatal enteropathies: defining the causes of protracted diarrhea of infancy. J Pediatr Gastroenterol Nutr. 2004;38:16–26. doi: 10.1097/00005176-200401000-00007. [DOI] [PubMed] [Google Scholar]
- 5.Goulet O, Ruemmele F, Lacaille F, et al. Irreversible intestinal failure. J Pediatr Gastroenterol Nutr. 2004;38:250–69. doi: 10.1097/00005176-200403000-00006. [DOI] [PubMed] [Google Scholar]
- 6.Ruemmele FM, Schmitz J, Goulet O. Microvillous inclusion disease (microvillous atrophy) Orphanet J Rare Dis. 2006;1:22. doi: 10.1186/1750-1172-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iancu TC, Mahajnah M, Manov I, et al. Microvillous inclusion disease: ultrastructural variability. Ultrastruct Pathol. 2007;31:173–88. doi: 10.1080/01913120701350712. [DOI] [PubMed] [Google Scholar]
- 8.Ameen NA, Salas PJ. Microvillus inclusion disease: a genetic defect affecting apical membrane protein traffic in intestinal epithelium. Traffic. 2000;1:76–83. doi: 10.1034/j.1600-0854.2000.010111.x. [DOI] [PubMed] [Google Scholar]
- 9.Michail S, Collins JF, Xu H, et al. Abnormal expression of brush-border membrane transporters in the duodenal mucosa of two patients with microvillus inclusion disease. J Pediatr Gastroenterol Nutr. 1998;27:536–42. doi: 10.1097/00005176-199811000-00008. [DOI] [PubMed] [Google Scholar]
- 10.Müller T, Hess MW, Schiefermeier N, et al. MYO5B mutations cause microvillus inclusion disease and disrupt epithelial cell polarity. Nat Genet. 2008;40:1163–5. doi: 10.1038/ng.225. [DOI] [PubMed] [Google Scholar]
- 11.Erickson RP, Larson-Thomé K, Valenzuela RK, et al. Navajo microvillous inclusion disease is due to a mutation in MYO5B. Am J Med Genet Part A. 2008;146A:3117–19. doi: 10.1002/ajmg.a.32605. [DOI] [PubMed] [Google Scholar]
- 12.Ruemmele FM, Müller T, Schiefermeier N, et al. Loss-of-function of MYO5B is the main cause of microvillus inclusion disease: 15 novel mutations and a CaCo-2 RNAi cell model. Hum Mutat. 2010;31:544–551. doi: 10.1002/humu.21224. [DOI] [PubMed] [Google Scholar]
- 13.Prekeris R, Klumperman J, Scheller RH. Rab11/Rip11 protein complex regulates apical membrane trafficking via recycling endosomes. Mol Cell. 2000;6:1437–48. doi: 10.1016/s1097-2765(00)00140-4. [DOI] [PubMed] [Google Scholar]
- 14.Holmes KC, Schroder RR, Sweeney HL, et al. The structure of the rigor complex and its implications for the power stroke. Philos Trans R Soc Lond B Biol Sci. 2004;359:1819–28. doi: 10.1098/rstb.2004.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coureux PD, Sweeney HL, Houdusse A. Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J. 2004;23:4527–37. doi: 10.1038/sj.emboj.7600458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang S, Liao JC, Dunn AR, et al. Predicting allosteric communication in myosin via a pathway of conserved residues. J Mol Biol. 2007;373:1361–73. doi: 10.1016/j.jmb.2007.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cecchini M, Houdusse A, Karplus M. Allosteric communication in myosin V: from small conformational changes to large directed movements. PLoS Comput Biol. 2008;4:e1000129. doi: 10.1371/journal.pcbi.1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isken O, Maquat LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat Rev Genet. 2000;9:699–712. doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lapierre LA, Kumar R, Hales CM, et al. Myosin vb is associated with plasma membrane recycling systems. Mol Biol Cell. 2001;12:1843–57. doi: 10.1091/mbc.12.6.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swiatecka-Urban A, Talebian L, Kanno E, et al. Myosin Vb is required for trafficking of the cystic fibrosis transmembrane conductance regulator in Rab11a-specific apical recycling endosomes in polarized human airway epithelial cells. J Biol Chem. 2007;282:23725–36. doi: 10.1074/jbc.M608531200. [DOI] [PubMed] [Google Scholar]
- 21.Nedvetsky PI, Stefan E, Frische S, et al. A Role of myosin Vb and Rab11-FIP2 in the aquaporin-2 shuttle. Traffic. 2007;8:110–23. doi: 10.1111/j.1600-0854.2006.00508.x. [DOI] [PubMed] [Google Scholar]
- 22.Roland JT, Kenworthy AK, Peranen J, et al. Myosin Vb interacts with Rab8a on a tubular network containing EHD1 and EHD3. Mol Biol Cell. 2007;18:2828–37. doi: 10.1091/mbc.E07-02-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoekstra D, Tyteca D, van IJzendoorn SC. The subapical compartment: a traffic center in membrane polarity development. J Cell Sci. 2004;117:2183–92. doi: 10.1242/jcs.01217. [DOI] [PubMed] [Google Scholar]
- 24.van IJzendoorn SC. Recycling endosomes. J Cell Sci. 2006;19:1679–81. doi: 10.1242/jcs.02948. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.