Abstract
The Saccharomyces cerevisiae NHP6A and NHP6B proteins are chromatin architectural factors, functionally and structurally related to the mammalian high mobility group (HMG)-1 and -2 proteins, a family of non-sequence-specific DNA binding proteins. nhp6a nhp6b mutants have various morphological defects and are defective in the induced expression of several RNA polymerase II-transcribed genes. We found that NHP6A/B proteins are also required for full induction of the yeast CHA1 gene. Importantly, CHA1 basal level expression is increased 10-fold in an nhp6a nhp6b double deletion mutant. Micrococcal nuclease and DNase I analysis of the CHA1 gene in this strain showed an open promoter structure, characteristic of the activated state of this promoter, even under non-inducing conditions. To address the possible function of the NHP6A/B proteins in chromatin-mediated gene regulation, we performed whole-genome transcriptional profiling of a Δnhp6a Δnhp6b yeast strain. Our results suggest that NHP6A/B proteins play an important regulatory role, repressing as well as potentiating expression of genes involved in several cellular processes, and that NHP6A/B control is exerted at the level of the individual gene.
Keywords: CHA1/chromatin/HMG-like proteins/Saccharomyces cerevisiae/transcriptional regulation
Introduction
The bulk of eukaryotic DNA is found assembled into chromatin. Although one important function of chromatin is to compact DNA, it has become apparent that chromatin structure also plays an important role in transcriptional regulation. Nucleosomes, the structural subunits of chromatin, provide a means to control the accessibility of regulatory factors to cognate sites and potentiate interactions between distant regulatory elements (Grunstein, 1992; Felsenfeld et al., 1996; Wolffe et al., 1997). Sequence-specific regulatory factors might function, at least in part, by counteracting or enhancing chromatin-mediated repression. To date, studies on chromatin structure have focused primarily on histone– DNA interactions. Non-histone chromatin proteins provide an extra layer of possible interactions, adding to the functional and structural complexity of the chromatin fiber (Bustin and Reeves, 1996; Bustin, 1999; Bianchi and Beltrame, 2000).
The Saccharomyces cerevisiae CHA1 gene encodes the catabolic l-serine (l-threonine) dehydratase, which is responsible for biodegradation of serine and threonine. We have shown that expression of the CHA1 gene is transcriptionally induced by serine and threonine, and that two promoter elements, UAS1CHA and UAS2CHA, are required and sufficient to confer serine/threonine inducibility to yeast genes (Bornæs et al., 1993). Furthermore, the two cis-acting sequences are bound by Cha4p, a gene-specific transcriptional activator (Holmberg and Schjerling, 1996). In previous studies we used accessibility to modifying nucleases, such as micrococcal nuclease (MNase) and DNase I, to determine the in vivo chromatin structure of the CHA1 chromosomal locus, both in the non-induced state and upon induction. Thus, we reported that upon activation, a precisely positioned nucleosome (nuc-1) occluding the TATA box and the transcription start site at CHA1 is removed, and that this structural transition is independent of the SWI/SNF or ADA complex (Moreira and Holmberg, 1998).
Recent studies have shown that transcriptional regulation by some eukaryotic enhancers requires the assembly of a specific nucleoprotein complex likely to involve architectural factors (Paull et al., 1993; Tjian and Maniatis, 1994; Yie et al., 1999). To further our understanding of the molecular mechanisms behind the structural transitions at the CHA1 promoter and their relationship to the process of gene activation, we investigated the role of architectural factors in this process.
Two non-sequence-specific DNA-binding proteins, non-histone proteins 6A and 6B (NHP6A/B), have been identified in S.cerevisiae in a search for proteins that could functionally replace HU in Hin-mediated DNA inversion (Paull and Johnson, 1995). Two highly related genes, NHP6A and NHP6B, encode the NHP6A/B proteins and deletion of both genes is required for any observable phenotype (Costigan et al., 1994). NHP6A/B are members of the high mobility group (HMG)-1 and -2 family proteins, and are required for activated gene expression, both in vitro and in vivo, of a subset of RNA polymerase II-transcribed genes (Paull et al., 1996). HMG-1 and -2 are very abundant chromatin-binding proteins, shown to bind substrates with a wide minor groove, such as four-way junctions, severely undertwisted DNA, cis-platinated DNA, and DNA at the entry and exit points of nucleosomes (Wright and Dixon, 1988; Bianchi et al., 1992; Pil and Lippard, 1992; Bustin, 1999; Bianchi and Beltrame, 2000). Although the exact cellular function of HMG-1/-2 proteins remains unknown, stimulatory (Stoute and Marzluff, 1982; Tremethick and Molloy, 1988; Singh and Dixon, 1990; Shykind et al., 1995) and inhibitory (Ge and Roeder, 1994; Stelzer et al., 1994) effects on transcription, as well as on in vitro nucleosome assembly and chromatin organization, DNA repair and recombination, have been described (Bonne-Andrea et al., 1984; Nightingale et al., 1996; van Gent et al., 1997; Melvin and Edwards, 1999).
In this study we report on the effect of deleting the yeast genes encoding HMG-1/-2-like proteins (NHP6A/B) on CHA1 chromatin structure and transcriptional activity, as well as on global gene expression.
Results
Basal and activated CHA1 gene expression in a Δnhp6a Δnhp6b double mutant
The yeast NHP6A/B proteins are members of the HMG-1/2 family of proteins, a group of proteins reportedly involved in both the repression and mediation of in vitro nucleosome assembly (reviewed in Bustin et al., 1990). The conflicting reports from various in vitro experimental approaches have long obscured the biological function(s) played by HMG-1/-2 proteins. It is widely accepted, nonetheless, that these proteins play a structural role in the regulation of chromatin (Bustin, 1999).
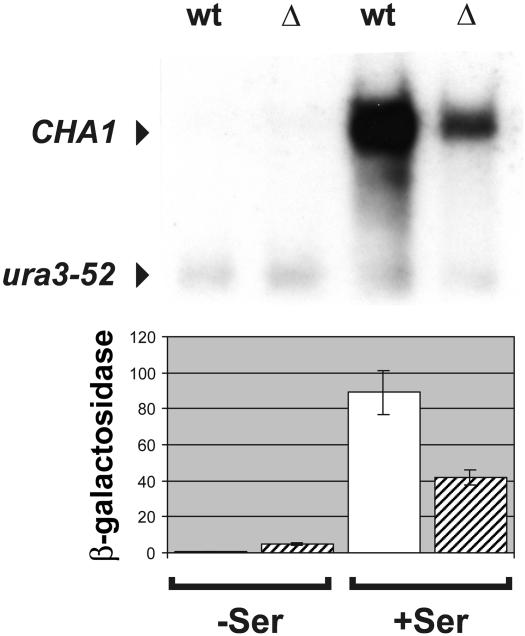
Expression of the yeast CHA1 gene is induced ∼100-fold by the presence of serine in the growth medium (Ramos and Wiame, 1982; Petersen et al., 1988). Northern blot analysis of total RNA isolated from wild type (wt) and a Δnhp6a Δnhp6b (Δ) strain showed that CHA1 transcript levels in cells grown under inducing conditions were reduced by ∼70% in the mutant strain compared with the isogenic wild type (Figure 1, upper panel, Δ +Ser and wt +Ser, respectively). We also analyzed transcript levels from the CHA1 gene in cells grown under non-inducing conditions, and detected no difference between the wild-type and mutant cells (Figure 1, upper panel, wt –Ser and Δ –Ser, respectively). Thus, activated expression of CHA1 depends on the presence of NHP6A and/or NHP6B proteins.
Fig. 1. Transcriptional activity of the CHA1 gene in cells lacking the NHP6A and NHP6B proteins. Upper panel: northern blot analysis of a Δnhp6A Δnhp6B mutant strain (RJY6012; Δ) and an isogenic wild-type strain (SEY6210; wt). Ten micrograms of total RNA isolated from cells grown in the absence (–Ser) or presence (+Ser) of serine were electrophoresed in a 1.5% formaldehyde–agarose gel, blotted and hybridized with 32P-labeled probes for the CHA1 and URA3 genes. Lower panel: β-galactosidase assay of a reporter plasmid, pTK120, containing a CHA1::lacZ translational fusion. Cells grown in the absence (–Ser) or presence (+Ser) of serine were harvested and β-galactosidase activity determined.
Any moderate effect of the NHP6A/B proteins on CHA1 basal level transcription would not be noticeable in our northern blot analysis since CHA1 basal transcription is very low. To address this point we examined the expression pattern of the CHA1 gene in the Δnhp6a Δnhp6b strain using a reporter construct. We transformed the mutant and its isogenic wild type with a centromeric reporter plasmid, pTK120 (a CHA1::lacZ translational fusion described in Bornæs et al., 1993). Levels of β-galactosidase activity were measured with or without the addition of serine to the growth medium (Figure 1, lower panel). Consistent with the results for CHA1 mRNA levels, activation of the CHA1–lacZ construct was reduced by ∼50% in nhp6b nhp6b cells compared with its wild-type counterpart (Figure 1, lower panel, compare standard deviation of Δ +Ser with wt +Ser). Interestingly, the basal level expression of the CHA1–lacZ fusion construct was increased 10-fold in the mutant strain (Figure 1, lower panel, compare standard deviations of Δ with wt). These results show that NHP6A/B proteins play a dual role in the regulation of the CHA1 gene. They potentiate CHA1 transcriptional activation in the presence of serine and repress CHA1 basal expression under non-inducing growth conditions.
Chromatin structure of the CHA1 gene in a Δnhp6a Δnhp6b strain
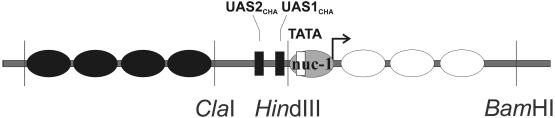
Induction of CHA1 expression in the presence of serine in the growth medium is mediated by the activator Cha4p through two cis-acting elements, UAS1CHA and UAS2CHA, present in the promoter region (Holmberg and Schjerling, 1996). A striking chromatin transition takes place at the CHA1 gene upon induction (Moreira and Holmberg, 1998), with the remodeling of a nucleosome (nuc-1) positioned over the putative TATA box (positions –132 and –82) and transcription start site (position –20), and disarrangement of the nucleosomes arrayed over the coding sequence (schematized in Figure 2). The observed chromatin remodeling in this promoter is independent of the SWI/SNF and ADA/GCN5 complexes (Moreira and Holmberg, 1998), two multimeric complexes implicated in counteracting chromatin-mediated repression (reviewed in Winston and Carlson, 1992; Pazin and Kadonaga, 1997a,b; Struhl, 1998).
Fig. 2. Chromatin organization of the CHA1 gene. Nucleosomes are depicted as filled ellipses, or as open ellipses for nucleosomes rearranged upon induction, and the nucleosome covering the TATA box, displaced upon induction, is marked nuc-1. Relevant restriction sites and cis-acting sequences are shown. UAS1CHA and UAS2CHA are represented by filled rectangles, and the TATA element by an open rectangle.
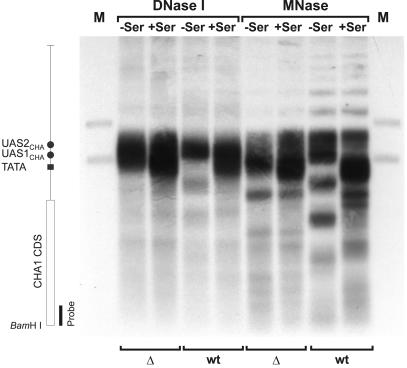
To investigate whether the decrease in activated expression of CHA1 in the nhp6a nhp6b mutant reflects a defective activation-dependent chromatin remodeling of the CHA1 promoter, MNase and DNase I digests of RJY6012 (Δnhp6a Δnhp6b) and SEY6210 (wild type) cells were carried out (Figure 3, Δ and wt, respectively). We obtained the previously observed band pattern in the wild type in the absence (Figure 3, wt –Ser) or presence (Figure 3, wt +Ser) of serine, namely a strong hypersensitive site in the 5′ flank and an ordered nucleosomal array covering both the coding region and the TATA box in uninduced cells. Upon induction, the nucleosome covering the TATA region is remodeled and the pattern of the coding region becomes diffuse. The mutant strain, however, showed a drastic effect of the NHP6 proteins on CHA1 chromatin. MNase and DNase I digests of mutant nuclei (Figure 3, Δ) yielded patterns different from those of the isogenic wild type (Figure 3, compare Δ with wt). Mutant cells grown under non-inducing as well as inducing conditions show an expanded hypersensitive region with absence of nuc-1 (Figure 3, Δ) and display a disarrayed nucleosomal assembly over the coding sequence. The observed chromatin structure at the CHA1 promoter in cells grown under non-inducing conditions correlates well with the 10-fold increase in basal expression in the nhp6a nhp6b double mutant. The fact that nuc-1 is remodeled in nhp6a nhp6b cells grown under non-inducing conditions could explain the measured increase in basal level expression. We have shown previously that in the CHA1 promoter the process of nucleosome disruption precedes that of transcriptional initiation (Moreira and Holmberg, 1998). Our observation that the CHA1 promoter is remodeled under non-inducing conditions in a strain lacking NHP6A and NHP6B suggests that these proteins participate in control of the stability of the TATA-occluding nucleosome in the CHA1 promoter.
Fig. 3. Chromatin analysis of the CHA1 gene in the Δnhp6a Δnhp6b double mutant (RJY6012; Δ) and the isogenic wild-type strain (SEY6210; wt). MNase- and DNase I-based mapping of nucleosome organization was carried out as described previously (Moreira and Holmberg, 1998). Cells were grown in the absence (–Ser) or presence (+Ser) of serine, nuclei purified and digested for 10 min with 20 U/ml DNase I or 100 U/ml MNase. DNA was isolated, digested with BamHI, separated on a 1% agarose gel, blotted and hybridized with a 32P-labeled CHA1-specific PCR amplificate. Lanes M contain restriction enzyme double digests of genomic DNA with BamHI and ClaI or BamHI and HindIII, to generate position marker fragments. The vertical map indicates the relative positions of the various cis-acting sequences and the CHA1 coding sequence (CDS).
Chromatin structure of the PHO5 gene in a Δnhp6a Δnhp6b strain
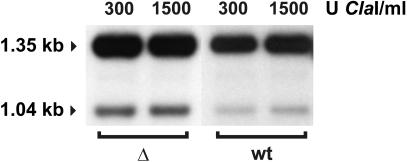
The yeast PHO5 gene is a well studied case of a promoter possessing a nucleosome placed over the TATA box, which is involved in gene regulation (reviewed in Svaren and Hörz, 1997). Gene-specific activator binding and nucleosome disruption are two separate events in the CHA1 promoter, with the former preceding the latter (Moreira and Holmberg, 1998). In the remodeling of the PHO5 promoter by the activator Pho4p, however, these two functions seem to be linked (Svaren and Hörz, 1997). Paull et al. (1996) demonstrated that induced expression of the PHO5 gene is marginally decreased (80% of wild type) and that basal level expression is very slightly increased (∼2.5-fold) in an nhp6a nhp6b strain. To discover whether the increase in basal transcription of the PHO5 gene correlated with remodeling of the nucleosomal structure of its promoter, we examined the structure of the PHO5 gene in nhp6a nhp6b cells grown under non-inducing conditions by MNase analysis. In spite of the slight increase in basal transcription in the nhp6a nhp6b strain (Paull et al., 1996; data not shown) we could not observe any difference between the PHO5 promoter structure in this strain and its wild-type counterpart (data not shown). Since the effect that the deletion of NHP6A/B has on PHO5 basal transcription is quite small (∼2.5-fold), it is possible that only a fraction of cells have increased expression and chromatin remodeling of the PHO5 gene. Our analysis could then conceivably reflect a mixture of two structures, with a minor remodeled form being obscured by a predominant assembled form. To investigate this possibility, we used accessibility of the PHO5 promoter to the restriction endonuclease ClaI, an assay shown previously to reflect the extent of disruption of nucleosome –2 at the PHO5 promoter (Almer et al., 1986). In both the nhp6a nhp6b strain (Δ) and its wild-type counterpart (wt), accessibility to ClaI was <10% (Figure 4), showing that the PHO5 promoter is not remodeled in the mutant strain. Thus, the alteration in chromatin structure observed at the CHA1 promoter in nhp6a nhp6b cells is not due to a defect in a general chromatin component.
Fig. 4. Chromatin accessibility analysis of the PHO5 promoter in cells lacking NHP6A and NHP6B. Restriction enzyme-based mapping of nucleosome organization was carried out essentially as described in the legend for Figure 3, except that isolated nuclei were digested for 30 min with ClaI. DNA was isolated, digested with ApaI, separated on a 1% agarose gel, blotted, and hybridized with a 32P-labeled PHO5-specific PCR amplificate. A 1.35 kb ApaI fragment is generated in the absence of cleavage by ClaI, and a 1.04 kb fragment is generated if the ClaI site is accessible.
Genome-wide expression patterns in cells lacking the NHP6A/B proteins
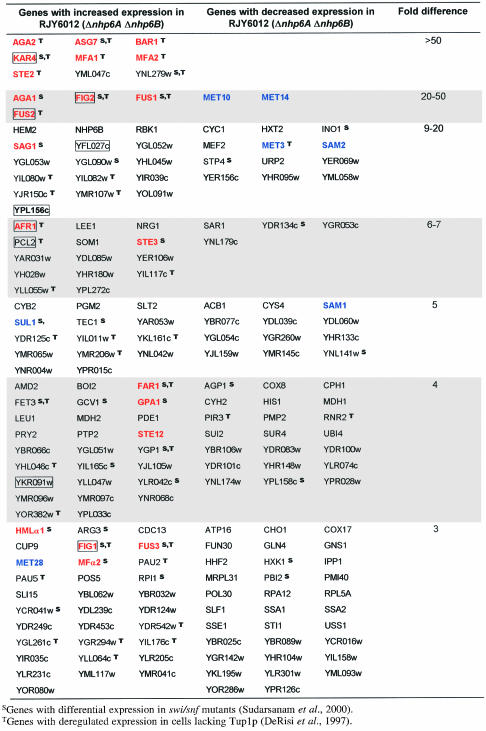
The completion of the sequencing project for the S.cerevisiae genome made it possible to quantitate mRNA levels for all identified open reading frames (ORFs) (DeRisi et al., 1997; Wodicka et al., 1997). We used commercially available high-density arrays containing a total of 6144 yeast ORFs spotted on two nylon membrane microarrays (filters I and II) to quantitate global mRNA levels in yeast cells devoid of NHP6A/B proteins. If one considers only those genes whose expression changed >3-fold in two independent experiments, 197 genes corresponding to 3.2% of the yeast genome are affected in the nhp6a nhp6b double mutant compared with the isogenic wild-type strain (Table I). Of these, 114 (1.9%) are up-regulated and 83 (1.4%) are down-regulated in the nhp6a nhp6b double mutant. Analysis of the affected ORFs revealed some interesting patterns of expression. The ORFs that show the greatest differences in expression (>50-fold) are mostly a-specific genes, genes involved in mating-type determination that are normally expressed at very low levels or not at all in α cells (e.g. ASG7 or MFA1). One field from a microarray membrane is shown in Figure 5, for both RJY6012 (nhp6a nhp6b; Δ) and SEY6210 (wild type; wt), with differentially expressed mating genes highlighted for comparison. In total, 21 genes involved in the pheromone response pathway show differential expression in the nhp6a nhp6b mutant (Table I, shown in red, bold typeface). Moreover, nine pheromone-regulated genes required for yeast mating differentiation (Erdman et al., 1998) are up-regulated in the mutant strain (Table I, boxed genes). Interestingly, both sets of genes show only transcriptional derepression in nhp6a nhp6b. A third group of genes identified in our analysis was that of genes involved in methionine biosynthesis (Table I, shown in blue, bold typeface), with both up- and down-regulated genes. We could not discern any further functional grouping in the remaining genes, which could reveal an effect of NHP6A/B proteins on any additional cellular process. Thus, NHP6A/B proteins affect the transcription of several genes, positively as well as negatively, involved in a variety of cellular processes, but particularly those implicated in the mating response and methionine biosynthesis, and pheromone-regulated genes.
Table I. Genes that display differential expression in a Δnhp6A Δnhp6B mutant strain.
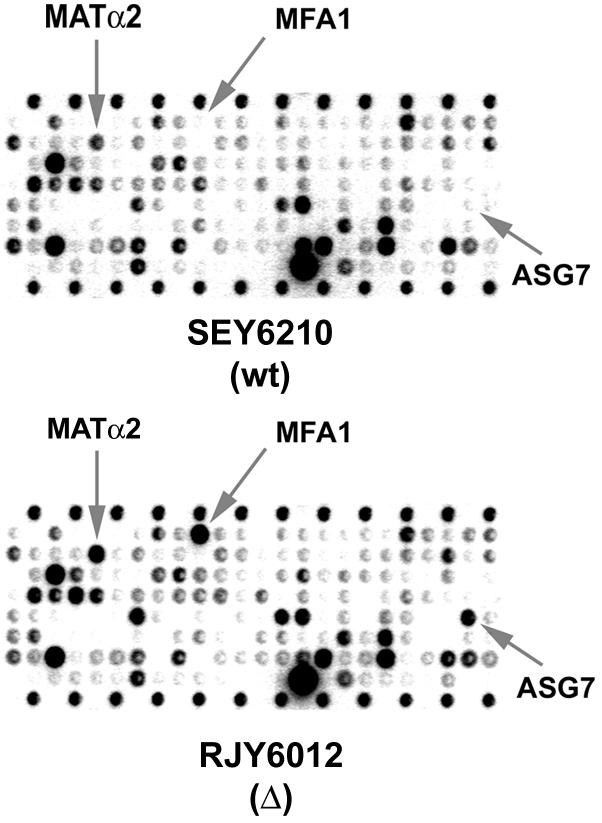
Fig. 5. Transcriptional analysis by microarray hybridization. Membranes containing 6144 yeast ORFs were hybridized with labeled RNA from strains RJY6012 (MATα Δnhp6a Δnhp6b) or SEY6210 (MATα wt). One field from such a membrane is shown, with genes involved in the mating response and found to be deregulated in the mutant strain indicated by gray arrows.
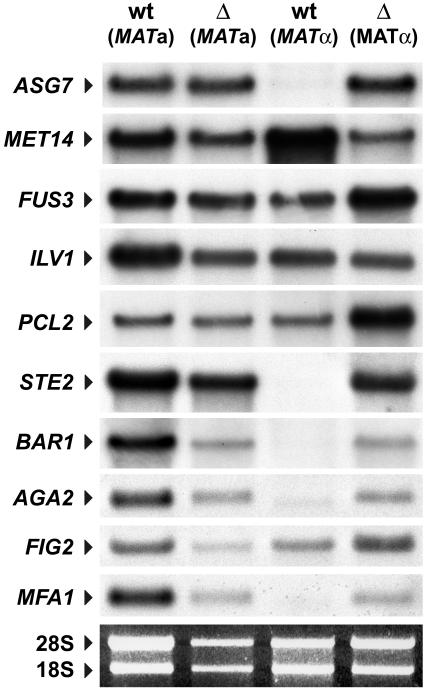
Northern blot analysis of selected loci in nhp6a nhp6b double mutants
To assess the validity of the results obtained through our genome-wide analysis, we examined the expression of some of the deregulated loci using northern blot analysis. One locus, ILV1, which had shown no difference in expression in our microarray analysis, was used as hybridization control. We isolated total RNA from strains RJY6012 (MATα Δnhp6a Δnhp6b; Δ MATα) and SEY6210 (isogenic wild-type; wt MATα) and investigated the transcript levels of various genes by northern blotting. Additionally, since we had observed a considerable effect on a-specific genes, we included a MATa Δnhp6a Δnhp6b mutant strain (DY2382; Δ MATa) and its isogenic wild-type strain (DY150; wt MATa) in this analysis. Northern blot analysis of these strains (Figure 6) reiterates the effects we had observed with the high-density array analysis (Table I). Thus, a-specific genes such as ASG7, STE2, BAR1, AGA2 and MFA1 display very low or undetectable levels of transcription in SEY6210 (MATα wt), but robust expression in RJY6012 (MATα Δnhp6a Δnhp6b), resulting in very large ratios of induction, similar to those observed in our microarray assay. Interestingly, with the exception of ASG7, which shows a slight increase (3-fold), these genes show no difference (STE2, AGA2 and FIG2) or even a slight decrease (2-fold for BAR1 and MFA1) in expression in the a mating-type strain (Figure 6; cf. DY150 and DY2382). Other loci, such as FUS3 and PCL2, show ratios of induction in MATα nhp6a nhp6b of 4- and 6-fold, respectively, and somewhat lower levels of derepression (2- and 3-fold, respectively) in MATa nhp6a nhp6b. As expected from the microarray analysis, MET14 expression is decreased in MATα nhp6a nhp6b (12-fold). Also, in the case of MET14, transcript levels are unaltered in the MATa nhp6a nhp6b mutant. We concluded that the effects we had observed in our whole genome analysis correctly reflect the patterns of gene expression in yeast cells lacking NHP6 proteins. Furthermore, for some genes the deregulation is mating-type dependent.
Fig. 6. Expression of various loci in cells lacking NHP6A and NHP6B. Northern blot analysis of a MATα Δnhp6a Δnhp6b mutant strain [Δ (MATα); RJY6012] and its isogenic wild-type counterpart [wt (MATα); SEY6210] plus a MATa Δnhp6a Δnhp6b mutant strain [Δ (MATa); DY150] and its isogenic wild-type counterpart [wt (MATa); DY2382]. Ten micrograms of total RNA isolated from cells grown in minimal medium were electrophoresed in a 1.5% formaldehyde–agarose gel, blotted and hybridized with 32P-labeled probes for the different genes. An ethidium bromide staining of the gel is shown as loading control.
Pheromone production in Δnhp6a Δnhp6b double mutants
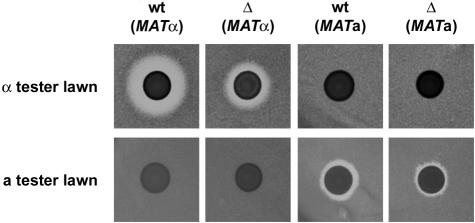
Our genome-wide analysis of the Δnhp6a Δnhp6b mutant strain revealed substantial differences in the expression of genes involved in the pheromone response pathway. To determine whether the observed transcriptional effects had a physiological effect, we used a previously described plate assay for pheromone production (Sprague, 1991). Briefly, the strains to be tested are spotted onto a lawn of cells from an a-factor tester strain (MATα sst2) and an α-factor tester strain (MATa bar1). These tester strains are hypersensitive to pheromone and will not grow in the immediate vicinity of a pheromone-producing strain of the opposite mating type, thus resulting in a clear zone or halo around the spotted cells.
We spotted strains RJY6012 (MATα Δnhp6a Δnhp6b; Δ MATα), SEY6210 (MATα wild-type; wt MATα), DY2382 (MATa Δnhp6a Δnhp6b; Δ MATa) and DY150 (MATa wild type; wt MATa) onto a lawn of either MT503 (MATα sst2; a-factor tester strain) or XP635-10C (MATa bar1; α-factor tester strain) cells (Figure 7). Strains RJY6012 and SEY6210 show a halo in the α-factor tester strain but not in the a-factor tester strain, corresponding to production of α- but not a-pheromone by these strains. This is not surprising in the case of SEY6210 since this is an α strain; however, in the case of RJY6012 we had observed transcription of a-specific genes including MFA1 and MFA2 (Table I), the genes encoding a-pheromone, and would thus expect this strain to produce a-pheromone as well. Seemingly, expression of MFA1 and MFA2 is not sufficient to allow RJY6012 to secrete biologically active a-pheromone. Additionally, the halo produced by RJY6012 is substantially smaller (∼2- to 3-fold) than that of its isogenic counterpart. Even though this plate-based assay is of a qualitative rather than quantitative nature, the greatly reduced size of the halo implies a considerable reduction in α-pheromone secretion, which is probably due to the increased expression of BAR1 (the barrier protease that degrades α factor) in nhp6a nhp6b. Conversely, DY150 and DY2382 show a halo in the a-factor tester strain but not in the α-tester strain, corresponding to production of a- but not α-pheromone by these strains. Again, we observed a reduction (∼50%) in the size of the halo produced by the MATa nhp6a nhp6b mutant strain (DY2382) compared with the isogenic wild type (DY150), consistent with the down-regulation of MFA1 in this strain. These results lead us to conclude that even though expression of a-specific genes, including those coding for a-pheromone, is greatly up-regulated in MATα nhp6a nhp6b cells, the strain maintains its apparent mating type and secretes only α-pheromone, albeit to a lesser extent. Similarly, the MATa nhp6a nhp6b mutant maintains its apparent mating type and secretes only a-pheromone.
Fig. 7. Halo assay for production of mating pheromone. Ten microliters of a cell suspension (∼1 × 1010 cells/ml) from strains RJY6012 (MATα Δnhp6a Δnhp6b), SEY6210 (MATα wt), DY2382 (MATa Δnhp6a Δnhp6b) and DY150 (MATa wt) were spotted onto a plate, grown overnight at 30°C and replica plated onto a lawn of 1–5 × 107 cells of either MT503 (a-factor tester strain) or XP635-10C (α-factor tester strain) cells. Plates were then incubated at room temperature for 2–4 days.
Discussion
In this article we present data showing that NHP6A and/or NHP6B proteins are required not only for full activation but also for repression of basal expression of the CHA1 gene, and for nucleosome positioning under non-inducing conditions, suggesting a role for these proteins in chromatin organization in vivo. Furthermore, a genome-wide analysis of cells lacking NHP6A/B identified various genes with deregulated levels of expression, with some discrete subsets of genes being affected. The pattern of gene expression in nhp6a nhp6b cells, with 114 up-regulated genes and 83 down-regulated genes, shows that these proteins are generally required for both activation and repression of target genes. These data suggest an important role of NHP6A/B in chromatin-mediated gene regulation, both positive and negative, in several yeast cellular processes.
Regulation of CHA1 gene expression and chromatin structure by NHP6A/B
The stimulatory function of NHP6A and/or NHP6B on CHA1-activated transcription, determined by northern blot analysis and β-galactosidase measurements (Figure 1, cf. wt +Ser and Δ +Ser), is in accord with the previously reported observation that expression of a number of RNA polymerase II-transcribed genes is impaired in an nhp6a nhp6b mutant. This effect was proposed to be due to the lack of formation at the TATA box of a TBP–TFIIA– NHP6A/B complex with increased affinity for TFIIB (Paull et al., 1996), a process probably similar to that observed with HMG-1/2 and the in vitro activated transcription of the adenovirus major late promoter (Shykind et al., 1995). Activated transcription of the CHA1 gene is dependent on the presence of a functional TATA element (at position –82), and activation-impaired TBP mutants, defective in interactions with TFIIA, TFIIB and the TATA box, abrogate activated CHA1 expression (Moreira and Holmberg, 1998). Thus, the drastic decrease in induced CHA1 expression observed in the nhp6a nhp6b mutant is readily explained, assuming a similar mechanism of activation for this gene to that already suggested, with NHP6A/B proteins participating in a TBP–TFIIA– NHP6A/B ternary complex with increased affinity for TFIIB. But what of the difference in CHA1 basal transcription and chromatin structure in Δnhp6a Δnhp6b cells?
The increase in CHA1 basal expression in Δnhp6a Δnhp6b cells under non-inducing growth conditions (Figure 1, cf. wt –Ser and Δ –Ser) suggests the NHP6A/B proteins somehow to be involved in the repression of this gene. HMG-1/-2 proteins have previously been shown to act as repressors of class II transcription, either by forming an HMG1–TBP–DNA complex capable of inhibiting pol II transcription in vitro (Ge and Roeder, 1994) or by HMG-2-mediated repression after assembly of a TBP–TFIIA–promoter complex (Stelzer et al., 1994). However, these two mechanisms are unlikely to explain the observed loss of repression at CHA1. The CHA1 gene has a precisely positioned nucleosome (nuc-1; see Figure 2) that occludes the TATA box under non-inducing growth conditions (Figure 3, wt –Ser), thus reducing basal transcription of this gene to undetectable levels. Therefore, formation of a repressive complex including NHP6A/B, and TBP–TFIIA–DNA or TBP–DNA cannot be viewed as a possible mechanism of action at the CHA1 gene, since formation of such a complex is incompatible with the observed presence of nuc-1 at the CHA1 promoter in a non-induced, wild-type strain (Figure 3, wt –Ser). A more likely function for NHP6A/B at the CHA1 promoter is that these proteins are required either for assembly of a repressive nucleoprotein complex (e.g. nucleosome) or function as auxiliary co-repressors, facilitating the recruitment/binding of a repressive factor(s) working through chromatin (e.g. histone deacetylase activity).
Alternatively, the expression of some general chromatin factor might be impaired in the nhp6a nhp6b double deletant, which would also result in an altered chromatin structure at the CHA1 promoter and elevated levels of basal expression due to increased accessibility of the TATA box. If this were the case, then we would expect to see a similar situation at other loci, also normally associated with nucleosomes under non-inducing conditions. We chose to examine the PHO5 gene, where a nucleosome positioned over the TATA element is involved in gene regulation (reviewed in Svaren and Hörz, 1997). We compared the structure of the PHO5 gene in the nhp6a nhp6b double deletion mutant with an isogenic wild type grown under non-inducing conditions. A restriction enzyme accessibility assay (Figure 4) showed no difference in Δnhp6a Δnhp6b PHO5 promoter accessi-bility relative to its wild-type counterpart. In conclusion, although a slight derepression of basal transcription occurs in the PHO5 gene in Δnhp6a Δnhp6b (Paull et al., 1996; data not shown), in contrast to CHA1 no structural change could be detected in the PHO5 promoter region. These results argue that the effect of NHP6A/B on the CHA1 promoter is specific rather than generalized, as one would expect should the levels of some general chromatin component, such as histones, be impaired in the nhp6a nhp6b deletion mutant.
Another possible explanation for the observed up-regulation of CHA1 transcript levels is that expression of some factor directly involved in modulating CHA1 chromatin structure is down-regulated in Δnhp6a Δnhp6b. We have shown previously that CHA1 expression is up-regulated and chromatin structure altered in a sir4 strain (Moreira and Holmberg, 1998). Furthermore, we have also shown that RSC, an essential ATP-driven chromatin-remodeling complex, is required for chromatin-mediated transcriptional repression of the yeast CHA1 (Moreira and Holmberg, 1999). Should expression of any of these genes be defective in Δnhp6a Δnhp6b, the outcome would be the exact phenotype we observed, namely showing increased basal expression and altered chromatin structure at the CHA1 gene.
A genomic view of NHP6A/B cell function
To determine whether the effects we observed at the CHA1 gene in nhp6a nhp6b were due to impaired expression of some repressive factor, and to gain some insight into the cellular function(s) of NHP6A/B proteins, we performed transcriptional whole-genome analysis in an nhp6a nhp6b double deletion mutant. We could identify 197 genes, corresponding to 3.2% of the yeast genome, differentially expressed in nhp6a nhp6b (Table I). Of these, 114 (1.9%) are up-regulated and 83 (1.4%) are down-regulated, suggesting that NHP6A/B are involved in transcriptional repression at other loci than CHA1. Some interesting patterns of expression emerged in this analysis, with various genes involved in the mating response (Table I, shown in red, bold typeface), pheromone-regulated genes required for yeast mating differentiation (Table I, boxed genes), and genes involved in methionine biosynthesis (Table I, shown in blue, bold typeface) deregulated in nhp6a nhp6b. However, we did not observe any difference in the expression of SIR4, RSC components, or of any other factor that could otherwise explain the deregulation at CHA1, leading us to conclude that NHP6A and B have a function in chromatin-mediated gene regulation.
HMG-1/-2 proteins have been shown to be involved in nucleosome assembly in vitro and chromatin organization (Bonne-Andrea et al., 1984; Nightingale et al., 1996). Additionally, a recent report has shown that expression of the S.cerevisiae HO gene, where chromatin structure plays an important regulatory role, requires NHP6A/B. Moreover, a gcn5 nhp6a nhp6b triple mutant has severe growth defects, suggesting that the SAGA histone acetyltransferase complex and NHP6A/B proteins function in parallel pathways, and indicating a possible role for NHP6A/B proteins in chromatin-mediated gene regulation (Yu et al., 2000).
The patterns of gene expression we observed in Δnhp6a Δnhp6b cells raise an interesting question as to the global in vivo function(s) of these proteins. The large number of genes affected in a common cellular process is striking (Table I, genes that are red bold typeface, boxed, or blue bold typeface). These effects were confirmed by northern blot analysis of the expression of several genes (Figure 6), and therefore can be construed as real and not an artifact of the microarray analysis. Furthermore, the biological assay for pheromone production (Figure 7) showed a quantifiable physiological difference between Δnhp6a Δnhp6b and isogenic wild-type cells, clearly reflecting the differences we detected at the transcriptional level. Thus, even considering the possibility that only one gene in a particular pathway is an actual target for NHP6A/B proteins, with the remaining genes being indirect effects, the profile of gene expression in nhp6a nhp6b is provocative, providing a wealth of information and suggesting a possible mechanism of action for these proteins.
Analysis of the affected genes in nhp6a nhp6b (Table I) reveals no clustering in the positions of these genes, but rather that they are spread throughout the genome. These data show that the control of gene expression by NHP6A/B proteins is gene specific rather than affecting larger chromosomal domains. Furthermore, to determine whether some common sequence could be associated with dependence on NHP6A/B, we used two sequence analysis programs, MEME (http://atlas.med.harvard.edu; Bailey and Gribskov, 1998) and AlignACE (http://meme.sdsc.edu; Hughes et al., 2000), to analyze the promoter regions of the affected genes (up to 800 bp 5′ of the ATG codon), but were unable to identify any common motifs that could serve as cognate sites for NHP6A/B. This negative result suggests that NHP6A/B may be targeted to specific promoters by gene-specific transcription factors.
We also compared the set of genes identified in the genome-wide expression analysis we performed in an nhp6a nhp6b double mutant with data sets for mutants involved in chromatin-mediated gene regulation available in the literature. Interestingly, the published datasets for swi/snf (Sudarsanam et al., 2000) and tup1 mutants (DeRisi et al., 1997) show a high degree of overlap with our dataset. In fact, 40 out of the 197 genes (20%) affected in the nhp6a nhp6b mutant are also affected in a strain lacking tup1, with 32 of the 197 (16%) affected in swi–snf mutants (Table I, genes labeled with superscript S and T for swi–snf and tup1, respectively). If one considers only those genes that are up-regulated in the nhp6a nhp6b mutant, then the relative proportion of common affected genes increases to 35/114 (30%) and 25/114 (21%) for the tup1 and swi–snf mutants, respectively.
Overexpression of NHP6A/B suppresses defects in the Slt2/Mpk1 MAP kinase (MAPK) pathway, and genetic and phenotypic analysis of nhp6a nhp6b mutants suggests that NHP6A/B function downstream of Slt2 (Costigan et al., 1994). Interestingly, we find that SLT2 expression is up-regulated 5-fold in nhp6a nhp6b (Table I). Moreover, the SBF transcription activator complex (composed of Swi4p and Swi6p) is itself a target of the Slt2/Mpk1 MAP kinase (MAPK) pathway (Madden et al., 1997), and a high copy suppressor screen for proteins required for full activity of the SBF complex identified NHP6A (Sidorova and Breeden, 1999). The SBF complex binds to cognate elements in target genes such as HO and CLN1, the expression of which requires NHP6A/B (Sidorova and Breeden, 1999; Yu et al., 2000). No direct interaction between NHP6A and the SBF complex could be demonstrated (Sidorova and Breeden, 1999). However, the experiment was not carried out in the context of chromatin, and it is conceivable that the effect of NHP6A on Swi6-regulated promoters is through chromatin modulation.
We propose that NHP6A/B proteins function in vivo as co-regulatory factors, directly involved in recruiting or in stabilizing interactions of trans-acting factors with cognate sequences. NHP6A/B proteins could be responsible for modulating chromatin structure at target promoters, allowing additional regulatory factors to be recruited. This function might be particularly important in cases where factors have no DNA-sequence specificity, such as for Ssn6–Tup1, or weak sequence specificity, such as for the SWI/SNF complex. This model would also explain the effect we observed at the CHA1 gene. We have previously reported that a SWI/SNF homologous complex termed RSC is required for repression of CHA1 basal expression (Moreira and Holmberg, 1999). Should NHP6A/B be involved in recruitment of the RSC complex to the CHA1 promoter, one would expect that in the absence of NHP6A/B proteins, targeting would be impaired and deregulation of the CHA1 gene would occur, as indeed we have observed.
Materials and methods
Strains
Strains RJY6012 (MATα his3-Δ200 leu2-3,112 ura3-52 trp1-Δ201 lys2-801 suc2-Δ9 gal3 nhp6A::ura– nhp6B::LEU2) and SEY6210 (MATα his3-Δ200 leu2-3,112 ura3-52 trp1-Δ201 lys2-801 suc2-Δ9 gal3) were provided by Reid C.Johnson. Strains MT503 (MATα sst2-1 leu2-3,112 his3 can1) and XP635–10C (MATa bar1 gal2 leu2-3,112) were provided by Vivian Mckay. Strains DY150 (MATa ade2 can1 his3 leu2 ura3 trp1) and DY2382 (MATa ade2 can1 his3 leu2 ura3 trp1 nhp6A::URA3 nhp6B::HIS3) were provided by David J.Stillman.
Cells were grown in minimal medium (0.67% bacto yeast nitrogen base without amino acids, 2% glucose, buffered with 10 g succinic acid and 6 g NaOH per liter) supplemented with the required amino acids at appropriate concentrations, in the absence (–Ser) or presence (+Ser) of serine as inducer (1 g/l).
Restriction endonucleases and DNA-modifying enzymes were purchased from Boehringer Mannheim (Mannheim, Germany), Taq polymerase was from Pharmacia (Amersham Pharmacia Biotech) and radiolabeled nucleotides were from ICN Pharmaceuticals, Inc. (Costa Mesa, CA).
Indirect end-labeling chromatin analysis
Isolation of nuclei and nuclease digestion were carried out as described (Moreira and Holmberg, 1998). Restriction endonuclease cleavage was performed under similar conditions as for MNase and DNase I, except that nuclei were treated for 1 h at 37°C. Subsequently, samples were treated with proteinase K and genomic DNA isolated. After secondary digestion with the appropriate restriction enzyme, the treated DNA samples were electrophoresed in 1% agarose gels in 1× TBE, transferred onto Positive™ nylon membranes (Oncor, Gaithersburg, MD) and hybridized following standard protocols (Sambrook et al., 1989).
Northern blot analysis
Total RNA was isolated from untreated nuclei using the Qiagen RNeasy Total RNA Kit (Qiagen, Germany) according to the manufacturer’s instructions. Ten micrograms of RNA per sample were loaded onto a 1.4% agarose–formaldehyde gel and electrophoresed in 1× MOPS, transferred onto Positive™ nylon membranes and hybridized following standard protocols (Sambrook et al., 1989). Transcript levels were quantified using a Cyclone Storage Phosphor System (Packard Instrument Company, Meriden, CT).
Radiolabeling of probes
Labeling was carried out as described (Espelund et al., 1990). A 32P-labeled PCR fragment covering positions +300 to +554 relative to the translational initiation codon was used for chromatin and northern blot analysis of the CHA1 gene. A 32P-labeled PCR fragment covering positions –1300 to –1086 relative to the translational initiation codon was used as probe for chromatin and restriction enzyme analysis of the PHO5 gene. The URA3 messenger was detected with a 32P-labeled PCR amplificate covering positions –23 to +151. All other transcripts were detected with a 32P-labeled PCR amplificate encompassing the entire ORF, from position +1 to the last nucleotide in the stop codon.
β-galactosidase activity
β-galactosidase measurements were performed as described previously (Remacle and Holmberg, 1992), with all values shown in Miller units (Miller, 1972).
Genome-wide expression analysis using high-density microarrays
Wild-type yeast cells (SEY6210) and cells bearing a double Δnhp6A Δnhp6B deletion (RJY6012) were grown in parallel in minimal medium at 30°C. Cultures were harvested at OD600 = 1. Total RNA was isolated as previously described. One microgram of total RNA per sample was labeled and hybridized to Yeast GeneFilters® Microarrays filters I and II (Research Genetics, Huntsville, AL), according to the manufacturer’s instructions. Quantitation was performed on a Cyclone Storage Phosphor System (Packard Instrument Company, Meriden, CT). Overall hybridization results from the two strains were normalized by setting the hybridization intensities of the control spots present in a given filter to be identical in both samples. Two independent hybridizations were performed for each sample. The results from the two hybridizations were averaged and only those genes that showed a similar effect in both hybridizations were considered in our analysis.
Acknowledgments
Acknowledgements
We are grateful to Reid C.Johnson, Vivian Mckay and David J.Stillman for providing yeast strains. This work was supported by a grant from the Carlsberg Foundation to J.M.A.M. and S.H., and grants from the Danish Research Councils, the Novo-Nordisk Foundation, the Lundbeck Foundation and Løvens Kemiske Fabriks Forskningsfond to S.H.
References
- Almer A., Rudolph,H., Hinnen,A. and Hörz,W. (1986) Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J., 5, 2689–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L. and Gribskov,M. (1998) Methods and statistics for combining motif match scores. J. Comp. Biol., 5, 211–221. [DOI] [PubMed] [Google Scholar]
- Bianchi M.E. and Beltrame,M. (2000) Upwardly mobile proteins. EMBO rep., 1, 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M.E., Falciola,L., Ferrari,S. and Lilley,D.M.J. (1992) The DNA binding site of HMG1 protein is composed of two similar segments (HMG boxes), both of which have counterparts in other eukaryotic regulatory proteins. EMBO J., 11, 1055–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonne-Andrea C., Harper,F., Sobczak,J. and De Recondo,A.-M. (1984) Rat liver HMG1: a physiological nucleosome assembly factor. EMBO J., 3, 1193–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornæs C., Ignatovic,M.W., Schjerling,P., Kielland-Brandt,M.C. and Holmberg,S. (1993) A regulatory element in the CHA1 promoter which confers inducibility by serine and threonine on Saccharomyces cerevisiae genes. Mol. Cell. Biol., 13, 7604–7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M. (1999) Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol., 19, 5237–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M. and Reeves,R. (1996) High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog. Nucleic Acid Res. Mol. Biol., 54, 35–100. [DOI] [PubMed] [Google Scholar]
- Bustin M., Lehn,D.A. and Landsman,D. (1990) Structural features of the HMG chromosomal proteins and their genes. Biochim. Biophys. Acta, 1049, 231–243. [DOI] [PubMed] [Google Scholar]
- Costigan C., Kolodrubetz,D. and Snyder,M. (1994) NHP6A and NHP6B, which encode HMG1-like proteins, are candidates for downstream components of the yeast SLT2 mitogen-activated protein kinase pathway. Mol. Cell. Biol., 14, 2391–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi J.L., Iyer,V.R. and Brown,P.O. (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science, 278, 680–686. [DOI] [PubMed] [Google Scholar]
- Erdman S., Lin,L., Malczynski,M. and Snyder,M. (1998) Pheromone-regulated genes required for yeast mating differentiation. J. Cell Biol., 140, 461–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espelund M., Stacy,R.A. and Jakobsen,K.S. (1990) A simple method for generating single-stranded DNA probes labeled to high activities. Nucleic Acids Res., 18, 6157–6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G., Boyes,J., Chung,J., Clark,D. and Studitsky,V. (1996) Chromatin structure and gene expression. Proc. Natl Acad. Sci. USA, 93, 9384–9388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H. and Roeder,R.G. (1994) The high mobility group protein HMG1 can reversibly inhibit class II gene transcription by interaction with the TATA-binding protein. J. Biol. Chem., 269, 17136–17140. [PubMed] [Google Scholar]
- Grunstein M. (1992) Histones as regulators of genes. Sci. Am., 267, 40–47. [DOI] [PubMed] [Google Scholar]
- Holmberg S. and Schjerling,P. (1996) Cha4p of Saccharomyces cerevisiae activates transcription via serine/threonine response elements. Genetics, 144, 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J.D., Estep,P.W., Tavazoie,S. and Church,G.M. (2000) Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol., 296, 1205–1214. [DOI] [PubMed] [Google Scholar]
- Madden K., Sheu,Y.-J., Baetz,K., Andrews,B. and Snyder,M. (1997) SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science, 275, 1781–1784. [DOI] [PubMed] [Google Scholar]
- Melvin V.S. and Edwards,D.P. (1999) Coregulatory proteins in steroid hormone receptor action: the role of chromatin high mobility group proteins HMG-1 and -2. Steroids, 64, 576–586. [DOI] [PubMed] [Google Scholar]
- Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 201–205. [Google Scholar]
- Moreira J.M.A. and Holmberg,S. (1998) Nucleosome structure of the yeast CHA1 promoter: analysis of activation-dependent chromatin remodeling of an RNA-polymerase-II-transcribed gene in TBP and RNA pol II mutants defective in vivo in response to acidic activators. EMBO J., 17, 6028–6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira J.M.A. and Holmberg,S. (1999) Transcriptional repression of the yeast CHA1 gene requires the chromatin-remodeling complex RSC. EMBO J., 18, 2836–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nightingale K., Dimitrov,S., Reeves,R. and Wolffe,A.P. (1996) Evidence for a shared structural role for HMG1 and linker histones B4 and H1 in organizing chromatin. EMBO J., 15, 548–561. [PMC free article] [PubMed] [Google Scholar]
- Paull T.T. and Johnson,R.C. (1995) DNA looping by Saccharomyces cerevisiae high mobility group proteins NHP6A/B. Consequences for nucleoprotein complex assembly and chromatin condensation. J. Biol. Chem., 270, 8744–8754. [DOI] [PubMed] [Google Scholar]
- Paull T.T., Haykinson,M.J. and Johnson,R.C. (1993) The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev., 7, 1521–1534. [DOI] [PubMed] [Google Scholar]
- Paull T.T., Carey,M. and Johnson,R.C. (1996) Yeast HMG proteins NHP6A/B potentiate promoter-specific transcriptional activation in vivo and assembly of preinitiation complexes in vitro. Genes Dev., 10, 2769–2781. [DOI] [PubMed] [Google Scholar]
- Pazin M.J. and Kadonaga,J.T. (1997a) SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein–DNA interactions? Cell, 88, 737–740. [DOI] [PubMed] [Google Scholar]
- Pazin M.J. and Kadonaga,J.T. (1997b) What’s up and down with histone deacetylation and transcription? Cell, 89, 325–328. [DOI] [PubMed] [Google Scholar]
- Petersen J.G.L., Kielland-Brandt,M.C., Nilsson-Tillgren,T. and Holmberg,S. (1988) Molecular genetics of serine and threonine catabolism in Saccharomyces cerevisiae. Genetics, 119, 527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pil P.M. and Lippard,S.J. (1992) Specific binding of chromosomal protein HMG1 to DNA damaged by the anticancer drug cisplatin. Science, 256, 234–237. [DOI] [PubMed] [Google Scholar]
- Ramos F. and Wiame,J.-M. (1982) Occurrence of a catabolic l-serine (l-threonine) deaminase in Saccharomyces cerevisiae. Eur. J. Biochem., 123, 571–576. [DOI] [PubMed] [Google Scholar]
- Remacle J.E. and Holmberg,S. (1992) A REB1-binding site is required for GCN4-independent ILV1 basal level transcription and can be functionally replaced by an ABF1-binding site. Mol. Cell. Biol., 12, 5516–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Shykind B.M., Kim,J. and Sharp,P.A. (1995) Activation of the TFIID–TFIIA complex with HMG-2. Genes Dev., 9, 1354–1365. [DOI] [PubMed] [Google Scholar]
- Sidorova J. and Breeden,L. (1999) The MSN1 and NHP6A genes suppress SWI6 defects in Saccharomyces cerevisiae. Genetics, 151, 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J. and Dixon,G.H. (1990) High mobility group proteins 1 and 2 function as general class II transcription factors. Biochemistry, 29, 6295–6302. [DOI] [PubMed] [Google Scholar]
- Sprague G.F. Jr (1991) Assay of yeast mating reaction. Methods Enzymol., 194, 77–93. [DOI] [PubMed] [Google Scholar]
- Stelzer G., Goppelt,A., Lottspeich,F. and Meisterernst,M. (1994) Repression of basal transcription by HMG2 is counteracted by TFIIH-associated factors in an ATP-dependent process. Mol. Cell. Biol., 14, 4712–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoute J.A. and Marzluff,W.F. (1982) HMG-proteins 1 and 2 are required for transcription of chromatin by endogenous RNA polymerase. Biochem. Biophys. Res. Commun., 107, 1279–1284. [DOI] [PubMed] [Google Scholar]
- Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev., 12, 599–606. [DOI] [PubMed] [Google Scholar]
- Sudarsanam P., Iyer,V.R., Brown,P.O. and Winston,F. (2000) Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 97, 3364–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svaren J. and Hörz,W. (1997) Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem. Sci., 22, 93–97. [DOI] [PubMed] [Google Scholar]
- Tjian R. and Maniatis,T. (1994) Transcriptional activation: a complex puzzle with few easy pieces. Cell, 77, 5–8. [DOI] [PubMed] [Google Scholar]
- Tremethick D.J. and Molloy,P.L. (1988) Effects of high mobility group proteins 1 and 2 on initiation and elongation of specific transcription by RNA polymerase II in vitro. Nucleic Acids Res., 16, 11107–11123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent D.C., Hiom,K., Paull,T.T. and Gellert,M. (1997) Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J., 16, 2665–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F. and Carlson,M. (1992) Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet., 8, 387–391. [DOI] [PubMed] [Google Scholar]
- Wodicka L., Dong,H., Mittmann,M., Ho,M.-H. and Lockhart,D.J. (1997) Genome-wide expression monitoring in Saccharomyces cerevisiae. Nature Biotechnol., 15, 1359–1367. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P., Wong,J. and Pruss,D. (1997) Activators and repressors: making use of chromatin to regulate transcription. Genes Cells, 2, 291–302. [DOI] [PubMed] [Google Scholar]
- Wright J.M. and Dixon,G.H. (1988) Induction by torsional stress of an altered DNA conformation 5′ upstream of the gene for a high mobility group protein from trout and specific binding to flanking sequences by the gene product HMG-T. Biochemistry, 27, 576–581. [DOI] [PubMed] [Google Scholar]
- Yie J., Senger,K. and Thanos,D. (1999) Mechanism by which the IFN-β enhanceosome activates transcription. Proc. Natl Acad. Sci. USA, 96, 13108–13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Eriksson,P. and Stillman,D. (2000) Architectural factors and the SAGA complex function in parallel pathways to activate transcription. Mol. Cell. Biol., 20, 2350–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]