Abstract
Multisubunit RNA polymerase is an essential enzyme for regulated gene expression. Here we report two Escherichia coli RNA polymerase structures: an 11.0 Å structure of the core RNA polymerase and a 9.5 Å structure of the σ70 holoenzyme. Both structures were obtained by cryo-electron microscopy and angular reconstitution. Core RNA polymerase exists in an open conformation. Extensive conformational changes occur between the core and the holoenzyme forms of the RNA polymerase, which are largely associated with movements in β′. All common RNA polymerase subunits (α2, β, β′) could be localized in both structures, thus suggesting the position of σ70 in the holoenzyme.
Keywords: core RNA polymerase/cryo-electron microscopy/σ70 holoenzyme
Introduction
Control over gene expression is exerted mainly at the level of transcription. DNA-dependent RNA polymerase (RNAp) is the vital enzyme in transcription and therefore the main target of transcriptional regulation. The multisubunit RNA polymerases of eukaryotes (Pols I, II and III), bacteria, archaea and chloroplasts exhibit unambiguous similarities in sequence, structure and function (Archambault and Friesen, 1993; Cramer et al., 2000). Therefore an understanding of the bacterial RNAp system provides an insight into the general mechanisms of all multisubunit RNAps. The recent X-ray structures of the Thermus aquaticus (Taq) core RNAp enzyme and the yeast Pol II enzyme provide a framework against which functional studies can be interpreted (Zhang et al., 1999; Cramer et al., 2000).
An essential step in bacterial transcription is the binding of one of a number of dissociable accessory proteins, termed σ factors, to a core RNAp [in Escherichia coli (Ec): α2ββ′ω, mass 379 kDa], to form a holoenzyme (e.g. α2ββ′ωσ70, mass 449 kDa). Only as a holoenzyme can RNAp specifically initiate transcription (Burgess et al., 1969). The RNAp α subunits (36 kDa) form dimers (α2) that act as the platform onto which the β and β′ subunits bind, and which play a role in transcription activation (Zhang and Darst, 1998). The α subunit consists of two functional domains: a C-terminal domain (αCTD), and an N-terminal domain (αNTD) responsible for the dimerization of the α subunits (Igarashi and Ishihama, 1991). The αCTD and αNTD are joined by a flexible linker of ∼14 amino acids (Jeon et al., 1997). The β and β′ subunits (150 and 155 kDa, respectively) together form the catalytic centre of the enzyme.
We have used cryo-electron microscopy (cryo-EM) (Dubochet et al., 1988) of single particles in combination with angular reconstitution (van Heel, 1987) to derive two related three-dimensional (3D) structures at ∼10 Å resolution: the Ec core RNAp and σ70 holoenzyme. Comparison of the two structures allows a detailed description of the conformational changes that occur upon binding of σ70. Since many functional studies have been performed on Ec RNAp, our structures reflect a well characterized multisubunit RNAp.
Results
The core RNAp reconstruction
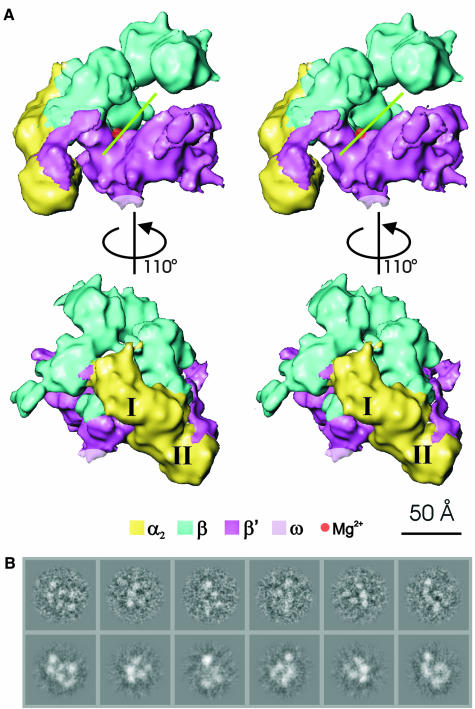
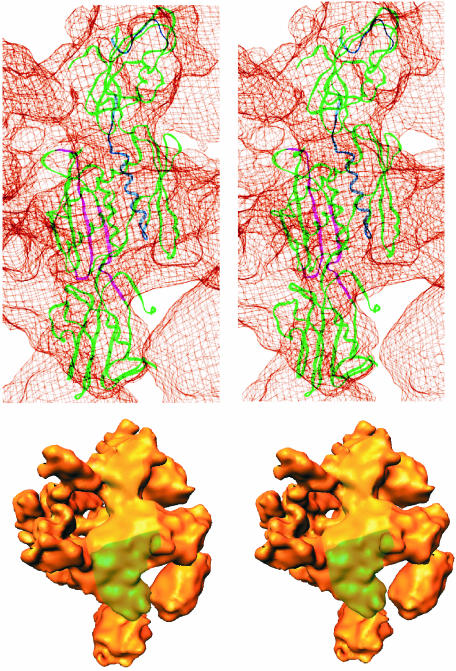
The structure of the Ec core RNAp (Figure 1A) was determined to 11.0 Å resolution (Figure 2). Representative characteristic views and reprojections of the Ec core RNAp are shown in Figure 1B. The features of the Ec RNAp core structure (Figures 1 and 3) are similar to those seen in the X-ray crystallographic structure of Taq core RNAp [Protein Data Bank (PDB) accession code 1DDQ] (Zhang et al., 1999). The ‘crab claw’ shape, used to describe eubacterial core RNAps (Polyakov et al., 1995; Zhang et al., 1999), is discernible. An internal channel, with a diameter of ∼32 Å (green line in Figure 1), runs between the jaws of the claw. This diameter, although slightly larger than reported previously (Darst et al., 1998; Zhang et al., 1999), is consistent with the proposed DNA binding function associated with the channel. The overall dimensions of the Ec RNAp are 120 Å along the direction of the channel, 150 Å from the back of the complex to the tips of the jaws, and 115 Å crossing between the jaws of the claw and perpendicular to the other two axes (see Figure 1).
Fig. 1. Stereoviews of the E.coli core RNAp structure. (A) View approximately down the axis of the RNAp channel (top), illustrating the ‘crab claw’ shape. Each jaw of the claw consists primarily of a β or a β′ subunit. The green line indicates the orientation of the channel, which is thought to be a site of DNA interaction. The deduced position of the chelated Mg2+ ion at the active centre is indicated by the red sphere. (Bottom) Top view rotated about the vertical axis by 110° to reveal the distinctive shape of the NTD dimer and β flexible flap. The subunit locations were identified by applying subunit boundaries derived from those found in Taq core RNAp (see text). The subunits are colour coded as indicated. The two α subunits, αI and αII, are indicated. (B) Representative characteristic views used for core RNAp reconstruction are shown in the top row. Below are the reprojections of the Ec core RNAp along the Euler angle assigned to the characteristic view.
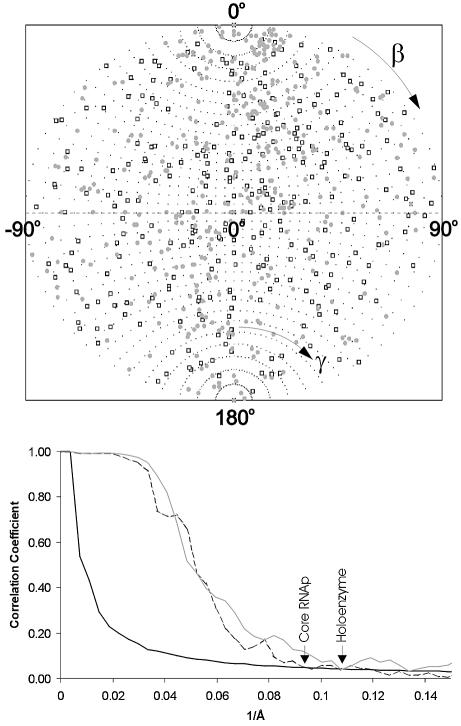
Fig. 2. Angular distributions and resolutions. (Top) Angular distribution of the class averages used for the final 3D reconstructions of Ec core RNAp (open square) and σ70 holoenzyme (grey circle). The ‘asymmetric triangles’ of these asymmetric structures covers the full unit sphere: the β angles range from 0° to 180°; the γ angles range from –90° to + 90°. (Bottom) Resolution of the 3D reconstructions determined by Fourier shell correlation (FSC). The first cross-over with the 3σ threshold curve (solid line) indicates the resolutions achieved in the experiments. The FSC function measures the normalized cross-correlation between two 3D reconstructions as a function of spatial frequency (see Materials and methods). The FSC from the Ec core RNAp reconstruction (dashed line) crosses the 3σ threshold at a spatial frequency of 0.093 Å–1 (10.75 Å), whereas the FSC from the σ70 holoenzyme reconstruction (grey line) crosses the 3σ threshold at a spatial frequency of 0.107 Å–1 (9.35 Å).
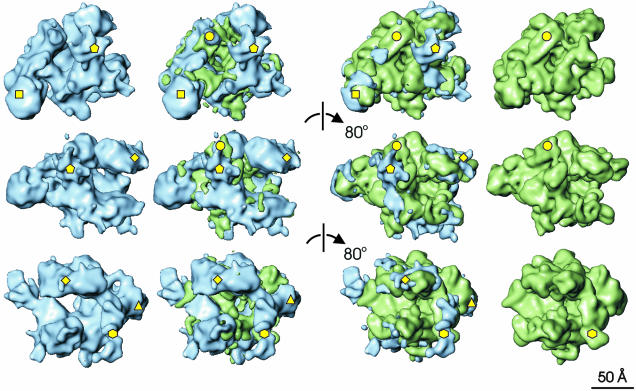
Fig. 3. Comparison of the E.coli and T.aquaticus core RNAps. Each row of this composite illustration shows surface rendered views of Ec core RNAp in blue (far left) and Taq core RNAp (far right). The difference map between Taq and Ec is shown in green and superimposed on Ec core RNAp (middle left). The difference map between Ec and Taq is shown in blue and superimposed on Taq core RNAp (middle right). Each successive row is related to the previous one by an 80° clockwise rotation about a vertical axis. The most significant differences between the two structures, discussed in detail in the main text, are marked by yellow shapes. Pentagon, Ec insertion in the C-terminus of β′; circle, β′ residues 945–977 (Taq); triangle, Ec β DRII insertion; diamond, β conserved regions A, B and C; hexagon, β flexible flap; square, αII-associated density.
Core RNAp shows an overall morphology similar to previous lower resolution Ec structures (Darst et al., 1998). The earlier Ec structures, however, had been interpreted as a closed conformation of the core RNAp (Polyakov et al., 1995; Darst et al., 1998). The X-ray crystallographic Taq core RNAp structure, on the other hand, was described as an open conformation (see Mooney and Landick, 1999). Here we present the Ec core RNAp in a conformation closely resembling the open conformation of Taq core RNAp (Zhang et al., 1999). We suggest that the open state of the enzyme is the predominant state of the core RNAp.
The similarities between mesophilic and thermophilic core RNAps
The mesophilic Ec core RNAp (Figures 1 and 3) strongly resembles the thermophilic Taq core RNAp (Zhang et al., 1999). An 11 Å density map of the Taq core RNAp was generated based on the published atomic co-ordinates (PDB accession code 1DDQ) (Zhang et al., 1999). The Ec and Taq core RNAp densities were then aligned using cross-correlation functions (Orlova et al., 2000). The high cross-correlation coefficient (0.81) between the aligned structures is consistent with the visual resemblance between the Taq and Ec core RNAp. The similarity between these two core RNAp structures, in combination with the sequence similarity between corresponding subunits in these two species (α: 40% identity, 55% similarity; β: 41% identity, 57% similarity; β′: 37% identity, 49% similarity), suggests that the Taq subunit boundaries are also applicable to Ec core RNAp. Taq subunit structures were thus used to localize the Ec core RNAp subunits. As a result, the region of density connecting the two jaws of the core RNAp has been identified as the α2 dimer (the NTD portion of each α subunit only, labelled I and II in Figure 1). One arm of the claw is largely comprised of the β subunit and the other of the β′ subunit.
Having found the α2, β and β′ subunits in the cryo-EM map, these individual subunits were extracted from the 3D reconstruction into separate maps. The αNTD dimer comprises ∼21% of the total mass (compared with an expected 19%), the β subunit corresponds to ∼42% of the total mass (expected 40%) and the β′ subunit to ∼37% of the mass (expected 41%). The separate subunit maps were also compared with their subunit equivalents in the Taq structure and found to have correlation coefficients in the range 0.84–0.89.
The small size of the ω subunit and its tentative assignment within the Taq core RNAp (Zhang et al., 1999) makes it difficult to localize the corresponding ω subunit accurately in the Ec map. Based on the protruding density in which the Taq ω subunit was placed, the location of the Ec ω subunit is marked in grey in Figure 1, where a similar protruding density is found. The most likely position of the active-centre Mg2+ is indicated by a red sphere in Figure 1.
The differences between mesophilic and thermophilic core RNAps
Difference maps between the two aligned core RNAps are shown in Figure 3. Comparisons show that the trend is for more mass within the channel of Taq and more mass to be found on the outer surface of Ec. This observation accounts for the widening of the internal channel in Ec. Many of the differences are small and difficult to interpret. Also, flexible structures will not have been resolved in the Ec map. However, it is possible to try and account for some of the larger differences.
There is a substantial difference marked by the pentagon on the Ec structure. Two possibilities may account for this density. The first, and favoured, is that it is due to the extra ∼200 amino acids in the C-terminus of the β′ subunit in Ec, found between the conserved region G and H (see Zhang et al., 1999 for β and β′ subunit alignments and nomenclature). The alternative is that the density comprising the two long parallel α-helices in Taq (Figure 3, marked by a circle) not evident in the Ec core RNAp map is found at a distinctly different location in the Ec structure. Such reorganization seems unlikely for two reasons: (i) a linkage to the N-terminal portion of β′ would be long, and is not evident in the Ec structure; and (ii) structural reorganization of β′ around the highly conserved catalytic centre would be required for such a conformational change to occur.
A redistribution of the β subunit mass is observed in the Ec core RNAp (Figure 3, marked by a diamond). This region corresponds to the conserved regions ‘A’, ‘B’ and ‘C’ of the β subunit (Severinov et al., 1996). The increased mass observed in this area of the Ec map may be caused by an insertion of ∼200 amino acids between conserved regions βB and βC in Ec, termed dispensable region I (DRI) (Severinov et al., 1996). A second dispensable region, DRII, exists within the Ec β subunit, corresponding to an insertion of ∼100 amino acids. The DRII insertion in Ec is likely to correspond to the position marked by the triangle (Figure 3). This is in agreement with the work of Opalka et al. (2000).
The β flexible flap (Figure 3, marked by a hexagon) consists of conserved region βG (Zhang et al., 1999). It appears to have moved away from the axis of the channel by 15° compared with its position within the Taq core. This region of the β subunit in Taq core RNAp is involved in the crystal-packing contacts (Zhang et al., 1999) and may thus have moved with respect to its position in Ec. Zhang et al. (1999) proposed that this flap-like structure is linked flexibly to the rest of the β subunit, and our results support this idea.
A significant increase in density associated with αII is observed in the Ec core RNAp structure (Figure 3, indicated by a square). An interaction between domain II of αII and a sequence of β′—between conserved regions β′D and β′E —was seen in the Taq core RNAp (Zhang et al., 1999). The observed increase in mass may be attributed to a more extensive interaction of the Ec αNTD with the β′ subunit. An alternative interpretation is that this increase in mass is due to the αCTD. However, the failure to see the αCTD in the crystal structure, indicative of its flexible linkage with the rest of the α dimer, is not consistent with this view. Overall, most differences between Ec and Taq core RNAp structures are small or can otherwise be rationalized.
The holoenzyme RNAp reconstruction
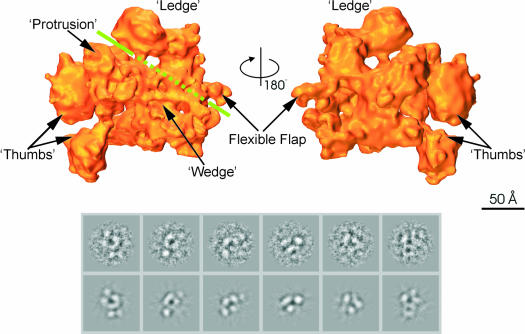
Unlike core RNAp, our current structural understanding of the holoenzyme is limited, with the previous best model being resolved to 27 Å (Darst et al., 1989). We show that substantial conformational changes occur in the RNAp structure upon binding of σ70. The structure of the Ec σ70 holoenzyme, resolved to 9.5 Å (Figure 2), is shown in Figure 4 (top). Representative characteristic views and reprojections of the Ec core RNAp are shown in Figure 4 (bottom). Overall, the structure of the holoenzyme is quite different from that of the core enzyme. The complicated interpretation of the holoenzyme based on the core enzyme is presented below. A number of structural details of the holoenzyme were named, including the ‘thumbs’, ‘ledge’ and ‘flexible flap’ (Figure 4, top). The thumbs and ledge of the holoenzyme are held apart by the wedge-shaped density, which, in the third dimension, is placed above the rest of the surrounding density. Running between the wedge and ledge densities is a groove, which could correspond to the ‘channel’ in the core enzyme (green line). Between the ‘upper’ thumb and ledge, a ‘protrusion’ is visible within the groove.
Fig. 4. Structure of E.coli σ70 holoenzyme. (Top) Surface views of the σ70 holoenzyme; the front view and back view are related through a 180° rotation around the vertical axis. Features discussed in the main text are labelled. The green line indicates the position of the groove. Where the line is dashed, the groove is obscured by the presence of the ‘wedge’-shaped density in front. (Bottom) Representative characteristic views used for core RNAp reconstruction. Below are the reprojections of the Ec holoenzyme along the Euler angle assigned to the characteristic view.
The overall dimensions of the holoenzyme are ∼160 Å from the flexible flap to the thumbs, 120 Å between the ledge and the far tip of the wedge, and 135 Å along the ‘back’ (from the bottom tip of αII to the ledge). These dimensions are somewhat larger than the size of the Ec core structure. To locate the subunits within the σ70 holoenzyme, we compared the holoenzyme with the available subunit structures.
Location of the αNTD dimer within the holoenzyme
Within the σ70–holoenzyme density map, the αNTD dimer is located across the ‘back’ of the holoenzyme (Figure 5). The αNTD dimer is found at a position comparable with the position of the αNTD dimer in the Ec core RNAp. Within the holoenzyme, the αNTD was readily located by placement of the Ec αNTD dimer from the Ec core structure or from the αNTD dimer atomic structure (PDB accession code 1BDF) (Zhang and Darst, 1998). The fitting of the distinctive shape of the αNTD dimer into the holoenzyme density map was unambiguous (Figure 5) and could be achieved with both the α2 dimers tried (the Ec αNTD dimer atomic structure and the Taq αNTD dimer). The better fit (cross-correlation coefficient of 0.85) was achieved with the Ec αNTD dimer.
Fig. 5. Identification of the αNTD dimer in E.coli σ70 holoenzyme. (Top) Stereoview of the αNTD dimer within the Ec σ70 holoenzyme. The αNTD dimer backbone is represented by green ribbons, except for the αI residues protected from hydroxyl radical cleavage by the binding of β (shown in blue) and the αII residues protected by the binding of β′ (shown in magenta) (Heyduk et al., 1996). The holoenzyme density is shown in a ‘chicken wire’ representation. The αNTD fits well within the cryo-EM density, with the exception of the region of the αNTD dimer that interacts with β′. Some domain movement may be induced by the binding of β′. (Bottom) This stereoview of the back of σ70 holoenzyme illustrates the placement of the αNTD dimer. The holoenzyme is orientated such that the αNTD dimer is in a vertical orientation, comparable to its orientation in the chicken-wire model. The position of each αNTD subunit is indicated in colour: αI in yellow and αII in green.
For the holoenzyme, the Ec αNTD dimer (1BDF) gave the better fit, suggesting that upon holoenzyme formation, domain II of each αNTD subunit bends away from the main channel and adopts a conformation more like that of the free Ec αNTD dimer. Some flattening was observed in domain I of each αNTD in the holoenzyme when compared with the Ec core RNAp. Recall that, compared with the structure of the isolated αNTD dimer in our Ec core RNAp structure (see Figure 1), the relative orientation of the subunits had changed slightly. Domain II of each αNTD subunit is bent in towards the main (DNA-binding) channel. This modest rearrangement of αNTD was also observed in the Taq structure (Zhang et al., 1999).
The placement of the αNTD dimer in the holoenzyme (Figure 5) is supported by two further lines of evidence. (i) The fitting of the αNTDs into the holoenzyme density map identifies the interface between the α subunits and the β and β′ subunits (Figure 5). The orientation of the αNTD shown is consistent with the known interactions between β and β′ (Figure 5) (Heyduk et al., 1996; Zhang and Darst, 1998). (ii) This orientation of the α subunit places the αCTD on the outer face of RNAp, where αCTD interacts with upstream promoter sequences and transcription activators (Niu et al., 1996). However, at the current levels of resolution (and probably in part due to its flexible linkage), it has not been possible to clearly resolve the αCTD within the Ec core (see above) and holoenzyme density maps.
Location and movements of the β and β′ subunits
Based on sequence alignments, biochemical and structural evidence, it is clear that β and β′ have a modular domain composition (Severinov et al., 1996; Zhang et al., 1999). Indeed, this modular organization has been further demonstrated by physically breaking up the β and β′ subunits into smaller domain modules and then reconstituting active RNA polymerases (Severinov et al., 1996). Thus, the functions of the domains do not require an intact subunit. Many allosteric changes in proteins involve domain movements and since σ70 is a tight core-binding ligand, its binding could easily drive such domain movements. We applied movements to whole subunits and to discrete domains to interpret the holoenzyme map.
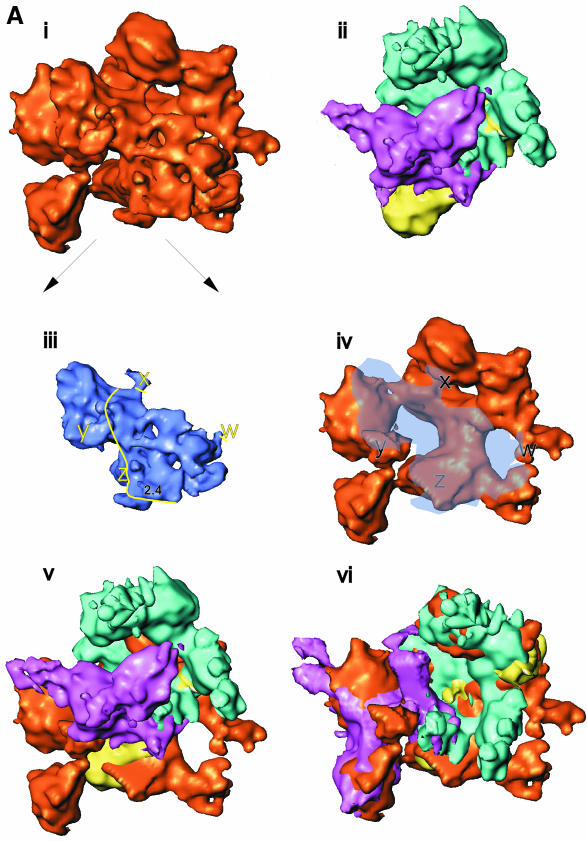
The core and holoenzyme were aligned based on the location of the αNTD dimer [see Figure 6A(i), (ii) and (iv)]. The placement of the αNTD dimer allowed the localization of α2 residues protected by the binding of β and β′ in hydroxyl radical footprinting experiments (Heyduk et al., 1996). The β- and β′-protected sites on α2 are indicated on the αNTD chain traces by blue and magenta, respectively (Figure 5). These contact sites provided additional anchor points for docking the β and β′ subunits. Translations and rotations were applied to the core RNAp β and β′ subunits (see below) in order to match the density distributions within the holoenzyme (Figure 6A).
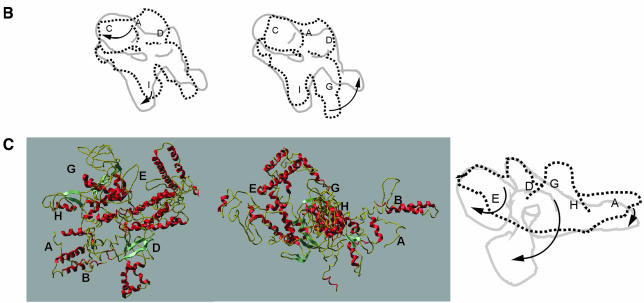
Fig. 6. Subunit locations and subunit movements upon σ70 binding. (A) The putative intermediates used to locate β and β′ subunits within the holoenzyme. See the main text for a detailed description. (i) Holoenzyme. (ii) Core RNAp (subunits coloured as in Figure 1). Both structures are orientated with the αNTD dimer along the ‘back-face’. (iii) Holoenzyme ‘wedge’ and ‘protrusion’ density made transparent for display purposes. After locating the positions of β and β′ subunits within the holoenzyme, it was possible to assign the wedge density to include σ70. The density we assign to σ70 is delineated by the yellow line (right). The positions marked w, x, y and z indicate likely core–σ70 subunit interaction regions. Also marked is the probable location of the –10 DNA interacting region of σ70, region 2.4. (iv) Holoenzyme with wedge and protrusion made translucent, to reveal masked structures. (v) The lack of overall superimposition of β and β′ between the aligned core RNAp and holoenzyme. By adjusting the positions (both rotationally and translationally) of β and β′ (also see B and C), superimposition was greatly enhanced (see vi). (vi) Placement of β and β′ (originating from the Ec core RNAp, but having been conformationally adjusted, see B and C) into the holoenzyme density. A single movement of β′ could not account for the holoenzyme position of β′ (see also C). One of the density matching rotations is shown in (vi). (v) and (vi) are displayed with transparent ‘wedge’ and ‘protrusion’ density, necessary to prevent this region of the holoenzyme obscuring the placement of β and β′ subunits within the holoenzyme. (B) Schematics of the conformation changes of the β subunit. The arrows indicate the deduced relative direction of movements in β. The locations of some of the conserved regions in β are indicated. The translationally aligned (but not rotational aligned) core (----) and holoenzyme (—) positions of each subunit are shown for β (left). After rotational alignment (right) of the β subunit, it became obvious that the β flexible flap was bent back by some 90° in the holoenzyme when compared with the core subunits, upon the binding of the σ70 subunit. (C) (left) A ribbon representation of β′ subunit (from Taq), demonstrating its modular domain organization. The positions of some conserved regions for β′ are indicated. This view was then rotated ∼120° about the vertical axis, then 15° in the plane of the page (middle), such that it is in the same location as the schematic of β′ (right). The aligned core (----) and holoenzyme (—) positions of each subunit are shown in for β′ (right). The position of the core β′ conserved regions are indicated (core locations). Arrows indicate the likely domain movements.
The characteristic bi-lobed shape of the β subunit in the core RNAp appeared to be moderately well conserved in the holoenzyme (Figure 4, ‘ledge’). Comparison of the aligned core RNAp and holoenzyme reconstructions [Figure 6A(i), (ii) and (iv)] indicates that the β subunit (blue in Figure 6A) has undergone a repositioning relative to the α subunit (yellow in Figure 6A). A simple rotation of the ‘core’ β subunit density (blue), by some ∼30°, away from the axis of the DNA binding channel, results in an alignment to its position within the holoenzyme map [Figure 6A(vi)]. A pronounced, extended density (∼33% of the total volume) within our holoenzyme map thus fits the overall shape of the β subunit in the core RNAp. As a consequence of this β subunit movement, a more open conformation is observed in the holoenzyme than in the core RNAp. To illustrate such movement better, we removed the density comprising the wedge and the protrusion from the holoenzyme [blue density in Figure 6A(iii) and translucent blue in the holoenzyme, Figure 6A(iv)].
The β flexible flap, consisting of the residues between conserved region βF and βH which includes conserved domain G (Zhang et al., 1999), is clearly discernable in both Ec structures (see Figures 3 and 3). The relative position of the tip of the β flexible flap appears to have moved in the holoenzyme with respect to its position in the core structure [Figure 6A(v), (vi) and B]. Having applied the gross 30° rotation to the (core) β subunit such that the main β subunit densities were aligned (Figure 6A and B), the tip of the flap was found to have undergone a rotation of ∼90° away from the rest of the β subunit. In the holoenzyme, the point of bending of the β flexible flap appears to contact the σ subunit directly (see below).
Similar translational and rotational shifts were applied to the core location of the β′ subunit in order to account for the unassigned densities (β′, purple, Figure 6A). A single gross movement of the core β′ subunit could not account for the β′ subunit within the holoenzyme. Separate movements allowed matching of substantial β′ density to the unassigned holoenzyme density (see below). One of the movements, allowing assignment of the lower thumb to β′, is shown in Figure 6A(vi). As shown in Figure 6C (left), several well developed structural domains in the Taq β′ structure are evident, which assisted the assignment of β′ in the Ec holoenzyme.
In the Taq and Ec core RNAp structures, the β′ subunit is U-shaped with the N- and C-termini in close proximity to each other (Zhang et al., 1999). In the Ec core RNAp structure, the C-terminal portion of the β′ subunit (residues 900–1407 approximately) binds the DNA-interacting channel. In the holoenzyme, the β′ C-terminal portion (domains G and H) has been rotated away by ∼100° from the floor of the channel towards the tip of domain II of αII (Figure 6A and C). Consistent with this rotation is the fact that the lower thumb structure in the holoenzyme and the Ec core RNAp β′ C-terminal domains (in particular the sequence comprising β′ conserved domain H) have very similar shapes and volumes. Moreover, our holoenzyme β′ conformation is indirectly supported by hydroxyl radical footprinting experiments (Fe-BABE) (Owens et al., 1998) in which no interactions were found to occur between σ70 and the C-terminus of the β′ subunit. Based on such experiments, the β′ subunit had been proposed to ‘fall away’ from the RNAp structure upon holoenzyme formation. Although these domain rotations are very large, domain motions of a similar magnitude have been observed in different systems (Gerstein and Krebs, 1998).
The N-terminal domains (∼500 residues, including β′ regions A, B, C and D; Figure 6A and C) of the β′ subunit have undergone little movement in the holoenzyme compared with their position in the core structure (Figure 6C). β′D contains the conserved motif NADFDGD, necessary for chelating the active site Mg2+. This site lies behind the wedge-shaped density in the holoenzyme. The upper thumb contains mainly β′E. The density connecting the two thumbs, the protrusion in the holoenzyme, is likely to correspond to conserved regions β′F and/or β′G. This assignment is required not only to accommodate the remaining β′ density, but also to connect the β′ density in the lower thumb to the rest of the β′ density.
Assignment of σ70
Extensive interactions are known to exist between the σ70, core RNAp subunits and promoter DNA, essential for promoter-specific initiation (Craig et al., 1998; Sharp et al., 1999). In particular, specific contacts are made by the σ70 subunit to the promoter DNA at positions –35 and –10, relative to the +1 transcription start. The densities in the RNAp holoenzyme, which have not been assigned to either α, β or β′, i.e. the wedge density [Figure 6A(iii)], can now largely be attributed to the σ70 subunit.
Deletion analysis has indicated that the region 2.1 of σ70 is necessary and sufficient for core interaction (Lesley and Burgess, 1989). Recent evidence implies that multiple interactions occur between σ and the core (Gross et al., 1998; Sharp et al., 1999). A number of studies (Burgess et al., 1998 and references therein) suggest that major sites for σ70–core binding are found in the N-terminus of β′ and the C-terminus of β. In core RNAp, these two regions are proximal to each other (Zhang et al., 1999). In particular, the main core–σ70 binding determinant was found to lie within the β′ residues 260–309, overlapping with conserved β′ region B. This region forms a coiled coil, which may interact with the coiled coil formed by regions 1.2 and 2.1 of σ70. When the wedge density is excised from the map, we observe four points of contact with the core (marked w–z in Figure 6A). The largest area of contact (point z in Figure 6A) covers β′ conserved regions B and C, and includes a σ70 binding determinant, β′ residues 260–309.
We attempted to locate a large portion of σ70 (Malhotra et al., 1996) within the wedge-shaped density by docking the known crystal structure of a fragment of Ec σ70 (PDB accession code 1SIG, residues 114–448) (Malhotra et al., 1996). A mathematically precise fit was not possible. This suggested that 1SIG does not quite reflect the conformation of σ70 in the holoenzyme. This may not be surprising, since σ70 is known to undergo significant conformational changes upon holoenzyme formation (Callaci et al., 1999). A number of points were highlighted in the fitting process. (i) The size of the σ70 protein, as inferred from 1SIG, suggests that σ70 can be accommodated within the wedge-shaped density. (ii) The size of 1SIG, together with its flat shape, indicates that the larger section of the wedge-shaped density corresponds to 1SIG density. (iii) A notable local ridge in the wedge-shaped density is evident, which may correspond to σ70 conserved region 2.4 (a prominent helix, see 1SIG), which recognizes promoter –10 DNA [marked 2.4, Figure 6A(iii)].
Rotation of σ70 unconserved sequences (residues 137–353) with respect to the rest of the 1SIG allows placement of 1SIG within the wedge piece of density, but not uniquely. Notably, placements of 1SIG positioned the core-interacting helices of region 2.1 at the wedge– holoenzyme area [z, Figure 6A(iii)]. This is an area of β′ believed to bind σ70 (Arthur et al., 2000). Additionally, a part of the β flexible flap touches an area close to the wedge density corresponding to the region we assign to σ70 [w, Figure 6A(iii) and (iv)]. The region associated with the β flexible flap is implicated in core–σ70 interactions (Fisher and Blumenthal, 1980; Borukhov et al., 1991).
Consistent with our assignment of the wedge density to σ70 is the proximity of both β and β′ to the wedge density. This assignment is in good agreement with the multiple core–σ70 interactions (Gross et al., 1998 and references therein) and with the protein–protein footprints (Owens et al., 1998). Furthermore, other densities (i.e. thumbs) that are not assigned to α2 or β correspond more closely in size and morphology to the domains that form β′ than to 1SIG (see Discussion). We do assign the likely location of the σ70–10 DNA recognition helix, but are cautious in assigning further wedge density to σ70 conserved regions 3 and 4. Defining a precise boundary for σ70 is difficult. We have tentatively outlined the σ70 subunit within the holoenzyme density map, based on four points (yellow line, Figure 6C): (i) the mass of each subunit; (ii) position and boundaries of other subunits; (iii) protein footprinting and cross-linking data; (iv) a σ70 subunit boundary refined by comparison to another Ec holoenzyme (our unpublished results).
Discussion
Subunit interactions in the holoenzyme
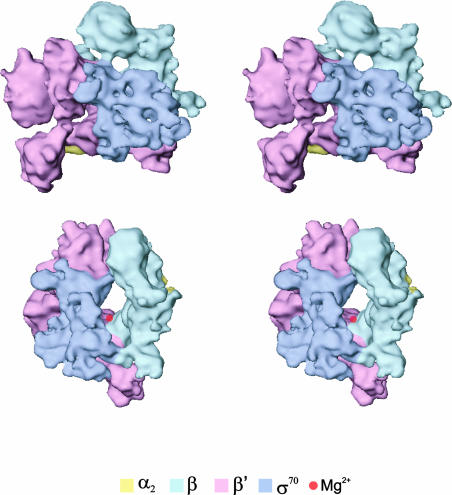
It has been possible to locate α2ββ′ and suggest the location for σ70 within the holoenzyme (see Figure 7). The mass of each assigned subunit is within 5% of the expected. Overall, β′ (and to a lesser extent β) appears to undergo substantial positional and conformational changes upon binding of the σ70 subunit (Figure 6). The β subunit in the holoenzyme has an extended interface with α2 via β regions F, G, H and I, as was the case in the core RNAp. However, the β′ interactions with α have changed. The contacts made by β′ regions C and D and α2 are conserved in the core and holoenzyme forms. Proposed domain movements are likely to disrupt the interactions made by β′ regions G and H to α2. In the holoenzyme, domains II of αII are covered by density. This was also the case in the core RNAp structure, but here these domains are covered by a different region of β′ due to conformational changes in β′.
Fig. 7. Stereoviews of the σ70–holoenzyme. (Top) A front view of the holoenzyme, showing the subunit assignment (see text). The holoenzyme was then rotated about the vertical axis by ∼100° (clockwise), followed by rotation of 25° about the horizontal (bottom). The subunit assignments are coloured as indicated. The deduced position of the chelated Mg2+ ion at the active centre is indicated by the red sphere.
Our proposed domain movements in β′ do not include β′D, which is the domain intimately associated with the active site Mg2+ through the NADFDGD motif. The position of βI, the β′ recruitment motif (Wang et al., 1997), is also unchanged. Therefore, these sites are preserved between the core and holoenzyme. In the core RNAp, the middle of β (Taq residues 998–1008) contacts the β′ regions CGH, and the C-terminal part of β (Taq residues 1009–1099) is surrounded by β′ regions BCDH. To achieve the proposed holoenzyme movements in β′, some contacts between β and β′ will be lost, in particular those contributed by β′G and β′H. Overall, the σ-driven conformational changes in the core subunits appear to result in intra- and inter-subunit structural changes involving a limited set of β′ regions (Wu et al., 1976).
An extensive σ70–β′ interaction is present at point z (see Figure 6A). There also appear to be further interactions between the β′ density (marked y in Figure 6A), i.e. the protrusion, and σ70. Although σ70 appears to interact less substantially with the β subunit than the β′ subunit, we have identified point w as a contact site between the β flexible flap and σ70. Another possible σ70–β interaction is located at point x. This point of interaction was suggested by Owens et al. (1998) between residues 383 and 554 of σ70 and between β regions C and D. No interactions between σ70 and the α subunits have been found in our cryo-EM map.
Transitions from the core to holoenzyme
The new Ec RNA polymerase holo- and core enzyme structures allow description of the conformational changes occurring in the core RNAp subunits upon binding of σ70. The functional significance of the conformational changes in β and β′ is not known, but these could relate to diminishing non-specific DNA binding and to the adoption of conformations that favour the ordered progress of the holoenzyme along the reaction pathway to initiation. Finally, reconstructions of other Ec holoenzymes reveal similar complex conformational changes when compared with the core RNAp (our unpublished observations).
Large changes in σ70 structure contributing to the density we have assigned to β′ seem improbable. This would result in σ70 being assigned to the thumbs or the protrusion, and would suggest that σ70 adopts a completely different fold to that of the crystallized σ70 fragment, and would contradict the vast body of independent biochemical evidence. For example, locating σ70 in the thumbs would mean that σ70 was remotely located from the catalytic centre.
Thermodynamic analysis of the conversion of the closed complex to the open complex suggests that this involves the burying of a large amount of non-polar surface, and, by inference, a major conformational change in RNA polymerase (reviewed in deHaseth et al., 1998). We therefore expect that the holoenzyme structure will change significantly during open-complex formation. It may be significantly different when DNA is bound in the closed complex. To understand conformational changes during initiation, structures of promoter-bound complexes are required. A comparison of different functional states of the holoenzyme associated with the different classes of σ and their respective DNA complexes will be informative. Further comparisons of the DNA-bound complexes with those described for the T7 single-subunit RNAp (Cheetham and Steitz, 2000) should reveal common features and strategies used in transcription.
Materials and methods
Protein purification
Ec σ70 holoenzyme (α2ββ′σω) and core RNA polymerase (α2ββ′ω) were purified from exponentially growing Ec MRE600 cells cultured at 37°C with vigorous aeration in Luria broth. The enzymes were prepared at 4°C by polymin-P precipitation, ammonium sulfate precipitation, and chromatography on Q Sepharose and heparin–Sepharose (Pharmacia) columns. A BioRex-70 (Bio-Rad) column was used to fractionate core and holoenzyme (Burgess and Jendrisak, 1975). The integrity of the purified polymerases was determined by native gel assays (Gallegos and Buck, 1999). The presence of ω in core and holoenzyme was established by N-terminal sequencing of subunits from isolated complexes and showed the Met-1 to be cleaved. Purified σ70 holoenzyme was active in a standard single-round transcription assay at the Ec glnHP1 promoter. Core RNA polymerase was active in poly(dA–dT)-dependent RNA formation. The purified enzymes were dialysed into storage buffer (10 mM Tris–HCl pH 8.0, 250 mM NaCl, 50% glycerol, 0.1 mM EDTA, 1 mM dithiothreitol) until cryo-EM.
Cryo-EM
The enzyme solutions were diluted to 0.4 mg/ml such that the final NaCl concentration was reduced to 100 mM and the glycerol content to 5%. After dilution, each solution was applied to a holey carbon grid and dialysed in situ (Cyrklaff et al., 1994) to further reduce the glycerol content to ∼2%. The samples were vitrified on the holey carbon grids (Dubochet et al., 1988) and transferred to a Philips CM200 FEG electron cryomicroscope using a Gatan side-entry cryo holder and cryo-transfer system. Micrographs were taken under low-dose conditions (∼10 e–/Å2), at an underfocus of 22 000–30 000 Å and a magnification of 50 000.
Image processing
Good quality micrographs, selected by optical diffraction, were digitized on an Image Science GmbH (Berlin, Germany) patchwork densitometer using a step size of 2.1 Å on the specimen scale. Single-molecule images (∼10 000) were selected interactively from six micrographs, and subsequently extracted into frames of 128 × 128 pixels. Contrast transfer function (CTF) correction was performed for each micrograph separately. Precise defocus measurements were achieved by determining the positions of the zero crossings between the first approximately nine Thon rings. The influence of the CTF was corrected by phase flipping up to spatial frequencies of ∼1/7 Å.
Reference-free alignment by classification was used to obtain a set of first references for multi-reference alignment (MRA) (Dube et al., 1993) of the initial data set. Multivariate statistical analysis (MSA) and classification (van Heel, 1989) were used to generate characteristic views (class averages; Figure 1B). The relative orientations of the best class averages (i.e. those made up from very similar raw images) were determined using angular reconstitution (van Heel, 1987). A 3D map was generated by exact-filter back projections (van Heel and Harauz 1986a; Radermacher, 1988). This 3D map was reprojected (see Figure 1B), and the reprojections were used as references for subsequent iterations of MRA, MSA and classification (Schatz et al., 1995). Euler angle assignments were made with respect to an anchor set of projections generated from the first reconstruction. The 3D analysis was refined iteratively until a stable reconstruction was obtained.
For the σ70–holoenzyme reconstruction, a preliminary analysis was performed on an initial set of 1000 images. This procedure was repeated with an independent set of 1000 images to ensure that the previous reconstruction was a true representation of the structure, free of artefacts. The two independent initial reconstructions proved to be very similar (correlation coefficient >0.95 up to 1/15 Å spatial frequency), and the two data sets were then combined to generate a 3D model that was then reprojected to generate the references for the iterative MRA of the full 10 000 image data set. The final reconstruction was generated using the best 400 class averages with a good homogeneous distribution of Euler angles over the unit sphere (Figure 2). For the core RNAp reconstruction, we used the σ70–holoenzyme 3D reconstruction as a starting point to avoid the lengthy reference-free alignment procedures. The reconstruction iterative procedure rapidly converged towards the specific core RNAp properties. The final data set contained ∼8000 particles, selected from four micrographs. The final reconstruction was generated using the best 300 class averages, again with a good homogeneous distribution of Euler angles (Figure 2). The resolutions achieved in the final reconstructions were assessed by Fourier shell correlation (FSC) (Harauz and van Heel, 1986), using the 3σ threshold criterion (Figure 2).
The Taq core RNAp structure (PDB accession code 1DDQ) was converted into densities in IMAGIC-5 with a sampling frequency of 2.1 Å per pixel. This structure was then low-pass filtered to 11 Å resolution. Separate subunit densities for the Taq core RNAp and the αNTD dimer were generated in an identical fashion to the Taq core RNAp structure. The fittings were determined by the highest cross-correlation coefficients between the EM map and the low-pass-filtered atomic structures (Orlova et al., 2000). Fittings were further refined using the ‘O’ software package (Jones and Kjeldgraad, 1997). All densitometry and image processing were performed within the IMAGIC-5 software system (van Heel et al., 1996) under the Linux and Compaq Tru64 UNIX operating systems. All illustrations were generated using the ‘O’ software package (Jones and Kjeldgraad, 1997), ICMlite and IRIS Explorer.
The surface presentations were displayed at a threshold level corresponding to ∼100% of the expected volume. This volume was calculated assuming a mass of the complex of ∼380 kDa for core RNAp and ∼450 kDa for the holoenzyme, and a specific density of 0.833 kDa/Å3.
Acknowledgments
Acknowledgements
We thank Ralf Schmidt, Michael Schatz and Ardan Patwardhan for support with the IMAGIC software system, J.Schumacher for ω subunit analysis and Steve Matthews for critical reading of the manuscript. Our work was financed by the European Union and a BBSRC project grant. Work was conducted in the Imperial College Centre for Structural Biology (www.ic-csb.ic.ac.uk). The CM-200/FEG microscope was funded by BBRSC/HEFCE Joint Research Equipment Initiative grant.
References
- Archambault J. and Friesen,J.D. (1993) Genetics of eukaryotic RNA polymerases I, II and III. Microbiol. Rev., 57, 703–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur T.M., Anthony,L.C. and Burgess,R.R. (2000) Mutational analysis of ‘β260–309’ a σ70 binding site located on E.coli core RNA polymerase. J. Biol. Chem., 275, 23113–23119. [DOI] [PubMed] [Google Scholar]
- Borukhov S. et al. (1991) Mapping of trypsin cleavage and antibody-binding sites and delineation of a dispensable domain in the β subunit of E.coli RNA polymerase. J. Biol. Chem., 266, 23921–23926. [PubMed] [Google Scholar]
- Burgess R. and Jendrisak,J. (1975) A procedure for the rapid, large scale purification of E.coli DNA-dependent RNA polymerase involving polymin P precipitation and DNA–cellulose chromatography. Biochemistry, 14, 4634–4638. [DOI] [PubMed] [Google Scholar]
- Burgess R.R., Travers,A.A., Dunn,J.J. and Bautz,E.K.F. (1969) Factor stimulating transcription by RNA polymerase. Nature, 221, 43–44. [DOI] [PubMed] [Google Scholar]
- Burgess R.R., Arthur,T.M. and Pietz,B.C. (1998) Interaction of E.coli σ70 with core RNA polymerase. Cold Spring Harb. Symp. Quant. Biol., 63, 277–287. [DOI] [PubMed] [Google Scholar]
- Callaci S., Heyduk,E. and Heyduk,T. (1999) Core RNA polymerase from E.coli induces a major change in the domain arrangement of the σ70 subunit. Mol. Cell, 3, 229–238. [DOI] [PubMed] [Google Scholar]
- Cheetham G.M. and Steitz,T.A. (2000) Insights into transcription: structure and function of single-subunit DNA-dependent RNA polymerases. Curr. Opin. Struct. Biol., 10, 117–123. [DOI] [PubMed] [Google Scholar]
- Craig M.L., Tsodikov,O.V., McQuade,K.L., Schlax,P.E.,Jr, Capp,M.W., Saecker,R.M. and Record,M.T.,Jr (1998) DNA footprints of the two kinetically significant intermediates in formation of an RNA polymerase–promoter open complex. J. Mol. Biol., 283, 741–756. [DOI] [PubMed] [Google Scholar]
- Cramer P. et al. (2000) Architecture of RNA polymerase II and implications for the transcription mechanism. Science, 288, 640–649. [DOI] [PubMed] [Google Scholar]
- Cyrklaff M., Roos,N., Gross,H. and Dubochet,J. (1994) Particle–surface interaction in thin vitrified films for cryo-electron microscopy. J. Microsc., 175, 135–142. [Google Scholar]
- Darst S.A., Kubalek,E.W. and Kornberg,R.D. (1989) Three-dimensional structure of E.coli RNA polymerase holoenzyme determined by electron crystallography. Nature, 340, 730–732. [DOI] [PubMed] [Google Scholar]
- Darst S.A., Polyakov,A., Richter,C. and Zhang,G. (1998) Structural studies of E.coli RNA polymerase. Cold Spring Harb. Symp. Quant. Biol., 63, 269–276. [DOI] [PubMed] [Google Scholar]
- deHaseth P.L., Zupancic,M.L. and Record,M.T.,Jr (1998) RNA polymerase–promoter interactions: the comings and goings of RNA polymerase. J. Bacteriol., 180, 3019–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube P., Tavares,P., Lurz,R. and van Heel,M. (1993) The portal protein of bacteriophage SPP1: a DNA pump with 13-fold symmetry. EMBO J., 12, 1303–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubochet J., Adrian,M., Chang,J.J., Homo,J.C., Lepault,J., McDowall,A.W. and Schultz,P. (1988) Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys., 21, 129–228. [DOI] [PubMed] [Google Scholar]
- Fisher R. and Blumenthal,T. (1980) Analysis of RNA polymerase by trypsin cleavage. Evidence for a specific association between subunits σ and β involved in the closed to open complex transition. J. Biol. Chem., 255, 11056–11062. [PubMed] [Google Scholar]
- Gallegos M.-T. and Buck,M. (1999) Sequences in σN determining holoenzyme formation and properties. J. Mol. Biol., 288, 539–553. [DOI] [PubMed] [Google Scholar]
- Gerstein M. and Krebs,W. (1998) A database of macromolecular motions. Nucleic Acids Res., 26, 4280–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C.A., Chan,C., Dombroski,A., Gruber,T., Sharp,M., Tupy,J. and Young,B. (1998) The functional and regulatory roles of σ factors in transcription. Cold Spring Harb. Symp. Quant. Biol., 63, 141–155. [DOI] [PubMed] [Google Scholar]
- Harauz G. and van Heel,M. (1986) Resolution criteria for three dimensional reconstruction. Optik, 73, 146–156. [Google Scholar]
- Heyduk T., Heyduk,E., Severinov,K., Tang,H. and Ebright,R.H. (1996) Determinants of RNA polymerase α subunit for interaction with β, β′ and σ subunits: hydroxyl-radical protein footprinting. Proc. Natl Acad. Sci. USA, 93, 10162–10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K. and Ishihama,A. (1991) Bipartite functional map of the E.coli RNA polymerase α subunit: involvement of the C-terminal region in transcription activation by cAMP–CRP. Cell, 65, 1015–1022. [DOI] [PubMed] [Google Scholar]
- Jeon Y.H., Yamazaki,T., Otomo,T., Ishihama,A. and Kyogoku,Y. (1997) Flexible linker in the RNA polymerase α subunit facilitates the independent motion of the C-terminal activator contact domain. J. Mol. Biol., 267, 953–962. [DOI] [PubMed] [Google Scholar]
- Jones T. and Kjeldgaard,M. (1997) Electron-density map interpretation. Methods Enzymol., 277, 173–207. [DOI] [PubMed] [Google Scholar]
- Lesley S.A. and Burgess,R.R. (1989) Characterization of the E.coli transcription factor σ70: localization of a region involved in the interaction with core RNA polymerase. Biochemistry, 28, 7728–7734. [DOI] [PubMed] [Google Scholar]
- Malhotra A., Severinova,E. and Darst,S.A. (1996) Crystal structure of a σ70 subunit fragment from E.coli RNA polymerase. Cell, 87, 127–136. [DOI] [PubMed] [Google Scholar]
- Mooney R.A. and Landick,R. (1999) RNA polymerase unveiled. Cell, 98, 687–690. [DOI] [PubMed] [Google Scholar]
- Niu W., Kim,Y., Tau,G., Heyduk,T. and Ebright,R.H. (1996) Transcription activation at class II CAP-dependent promoters: two interactions between CAP and RNA polymerase. Cell, 87, 1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalka N., Mooney,R.A., Richter,C., Severinov,K., Landick,R. and Darst,S.A. (2000) Direct localization of a β-subunit domain on the three-dimensional structure of E.coli RNA polymerase. Proc. Natl Acad. Sci. USA, 97, 617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova E.V., Rahman,M.A., Gowen,B., Volynski,K.E., Ashton,A.C., Manser,C., van Heel,M. and Ushkaryov,Y.A. (2000) Structure of α-latrotoxin oligomers reveals that divalent cation-dependent tetramers form membrane pores. Nature Struct. Biol., 7, 48–53. [DOI] [PubMed] [Google Scholar]
- Owens J.T., Miyake,R., Murakami,K., Chmura,A.J., Fujita,N., Ishihama,A. and Meares,C.F. (1998) Mapping the σ70 subunit contact sites on E.coli RNA polymerase with a σ70-conjugated chemical protease. Proc. Natl Acad. Sci. USA, 95, 6021–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyakov A., Severinova,E. and Darst,S.A. (1995) Three-dimensional structure of E.coli core RNA polymerase: promoter binding and elongation conformations of the enzyme. Cell, 83, 365–373. [DOI] [PubMed] [Google Scholar]
- Radermacher M. (1988) Three-dimensional reconstruction of single particles from random and nonrandom tilt series. J. Electron Microsc. Tech., 9, 359–394. [DOI] [PubMed] [Google Scholar]
- Schatz M., Orlova,E.V., Dube,P., Jager,J. and van Heel,M. (1995) Structure of Lumbricus terrestris hemoglobin at 30 Å resolution determined using angular reconstitution. J. Struct. Biol., 114, 28–40. [DOI] [PubMed] [Google Scholar]
- Severinov K., Mustaev,A., Kukarin,A., Muzzin,O., Bass,I., Darst,S.A. and Goldfarb,A. (1996) Structural modules of the large subunits of RNA polymerase. J. Biol. Chem., 271, 27969–27974. [DOI] [PubMed] [Google Scholar]
- Sharp M.M., Chan,C.L., Lu,C.Z., Marr,M.T., Nechaev,S., Merritt,E.W., Severinov,K., Roberts,J.W. and Gross,C.A. (1999) The interface of σ with core RNA polymerase is extensive, conserved and functionally specialized. Genes Dev., 13, 3015–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M. (1987) Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy, 21, 111–124. [DOI] [PubMed] [Google Scholar]
- van Heel M. (1989) Classification of very large electron microscopical image data sets. Optik, 82, 114–126. [Google Scholar]
- van Heel M and Harauz,G. (1986) Exact filters for general geometry three-dimensional reconstruction. Optik, 73, 119–122. [Google Scholar]
- van Heel M., Harauz,G., Orlova,E.V., Schmidt,R. and Schatz,M. (1996) A new generation of the IMAGIC image processing system. J. Struct. Biol., 116, 17–24. [DOI] [PubMed] [Google Scholar]
- Wang Y., Severinov,K., Loizos,N., Fenyo,D., Heyduk,E., Heyduk,T., Chait,B.T. and Darst,S.A. (1997) Determinants for E.coli RNA polymerase assembly within the β subunit. J. Mol. Biol., 270, 648–662. [DOI] [PubMed] [Google Scholar]
- Wu F.Y., Yarbrough,L.R. and Wu,C.W. (1976) Conformational transition of E.coli RNA polymerase induced by the interaction of the σ subunit with core enzyme. Biochemistry, 15, 3254–3258. [DOI] [PubMed] [Google Scholar]
- Zhang G. and Darst,S.A. (1998) Structure of the E.coli RNA polymerase α subunit amino-terminal domain. Science, 281, 262–266. [DOI] [PubMed] [Google Scholar]
- Zhang G., Campbell,E.A., Minakhin,L., Richter,C., Severinov,K. and Darst,S.A. (1999) Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell, 98, 811–824. [DOI] [PubMed] [Google Scholar]