Abstract
BACKGROUND
p14ARF plays a critical role in crosstalk between p53 and Rb pathways and in cellular anticancer mechanisms. We, therefore, investigated the association between the single nucleotide polymorphisms (SNPs) of p14ARF and risk of second primary Malignancy (SPM) after index squamous cell carcinoma of the head and neck (SCCHN).
METHODS
We used Log-rank test and Cox proportional hazards models to assess the association of the two p14ARF SNPs (rs3731217 and rs3088440) with the SPM-free survival and SPM risk among 1,287 incident SCCHN patients.
RESULTS
We found that patients with either p14ARF variant genotypes of the two polymorphisms had a significantly reduced SPM-free survival, compared with patients with no variant genotypes (Log-rank test, P = 0.006). Compared with p14ARF TT and GG genotypes, the variant genotypes of p14ARF TG/GG and GA/AA were associated with significantly moderately increased risk of developing SPM [adjusted hazard ratio (aHR), 1.48, 95% confidence interval (CI), 1.00–2.19 for p14ARF-rs3731217 and aHR, 1.61, 95% CI, 1.07–2.43 for p14ARF-rs3088440), respectively]. Moreover, after combining the variant genotypes of the two SNPs, patients with variant genotypes had a significantly greater risk for SPM compared to patients with no variant genotypes (aHR, 3.07, 95% CI, 1.54–6.12), and the risk was particularly pronounced in several subgroups.
CONCLUSIONS
Our results suggest a modestly increased risk of SPM after index SCCHN with each p14ARF polymorphism and an even greater risk of SPM with combined variant genotypes of the two SNPs. Therefore, p14ARF polymorphisms may be a susceptible marker to SPM risk for SCCHN patients.
Keywords: p14ARF, Squamous cell carcinoma of head and neck, Second primary malignancy, Genetic susceptibility, Polymorphism
INTRODUCTION
Squamous cell carcinoma of the head and neck (SCCHN), which arise from sites of the oral cavity, oropharynx, hypopharynx and larynx, is the sixth most common cancer worldwide with moderately low survival, high recurrence, and high second primary malignancy (SPM) rates.1 There were approximately 48,010 new cases of and 11,260 deaths from SCCHN in 2009 in the United States.2 Nationally, approximately 70% and 91% of SCCHN patients are men and whites, respectively,3 compared to those in our institution (approximately 76% in men and 85% in whites). The occurrence of SPMs in SCCHN patients still remains one of major factors that contribute to the poor overall survival of SCCHN patients.4–6 While cigarette smoking and alcohol use have been found to be associated with the risk of SPM7, 8, most SCCHN patients never develop SPM, implying that there is an inter-individual variation in genetic susceptibility to SPM among SCCHN patients.4 Several previous studies have reported that single nucleotide polymorphisms (SNPs) of genes involved in carcinogen metabolism, DNA repair, cell cycle control, and apoptosis were associated with the risk of SPM after primary SCCHN.9–15
p14ARF, a tumor suppressor gene, is located in the INK4a locus on chromosome 9p21, which encodes two distinct tumor suppressor proteins of p16INK4a and p14ARF and is one of the most common regions mutated in approximately 50% of all human cancers, second only to p53.16, 17 p14ARF interacts directly with MDM2, thereby suppressing the ubiquitin ligase activity of MDM2, and subsequently inhibiting MDM2–mediated degradation of p53. Such interaction leads to stabilization and accumulation of p53. Thus, genetic alteration of p14ARF may affect cell cycle regulation and apoptosis by disrupting the p53 pathway.18, 19 Therefore, p14ARF plays an important role in the ARF–MDM2–p53 pathway. In addition, independent of p53, p14ARF has multiple other tumor suppressor functions which involve interaction with several proteins in other cellular activities such as cell proliferation.20 Furthermore, p14ARF is involved in the ATM/ATR-CHK signaling pathway in response to DNA damage21, indicating that p14ARF may affect cell cycle and DNA repair. It has been demonstrated that p14ARF also interacts with transcription factors, such as E2F-1, E2F-2, in the Rb pathway to prevent Rb proteasomal degradation and trigger its antiproliferative function.22–24 Thus, p14ARF acts in maintaining genomic stability by mediating cellular activities in both p53 and Rb pathways.
Alterations or mutations of p14ARF are frequent events in the development of SCCHN.25–29 However, the roles of p14ARF polymorphisms in the etiology of SPM after index SCCHN have not been investigated. We hypothesized that p14ARF polymorphisms contribute to genetic susceptibility to SPMs after index SCCHN, and these polymorphisms may be genetic markers to identify the subgroups of SCCHN patients at high SPM risk, who might benefit from targeted follow-up and SPM screening as well as consideration of tobacco/alcohol cessation and/or chemoprevention protocols. To test this hypothesis, we first identified two tagging SNPs [those with minor allele frequency (MAF) > 5% and the linkage disequilibrium (LD) measure r2 threshold at 0.8] in the p14ARF gene (rs3731245 and rs3088440) and included additional one reported SNP30, 31 for a pilot study of 400 subjects. Although the allele frequency of all three SNPs in the overall population is more than 5% MAF, we found that p14ARF-rs3731245 had less than 5% MAF in our study patients. Therefore, we finally genotyped the two common (i.e., MAF > 5%) p14ARF-rs3731217 and rs3088440 SNPs in a cohort of 1,287 incident SCCHN patients, and evaluated the association between each or in combination of the two polymorphisms and risk of SPM.
MATERIALS AND METHODS
Study subjects
In this study, 1,667 patients with incident SCCHN were consecutively recruited from May 1995 to January 2007 at the University of Texas M. D. Anderson Cancer Center as described previously elsewhere.12–15 This cohort of patients had newly diagnosed, histopathologically confirmed, and untreated SCCHN, who completed an Institutional Review Board-approved informed consent, without the restriction of age, sex, ethnicity, or clinical stage. The exclusion criteria included any prior cancer history (except for nonmelanoma skin cancer), distant metastases at presentation, primary sinonasal tumors, salivary gland tumors, cervical metastases of unknown origin, and tumors outside the upper aerodigestive tract. Approximately 95% of contacted patients consented to enrollment in the study. Some blood samples for p14ARF genotyping were not available for the patients recruited early in the study, and these patients were not included in the analysis, as were patients without follow-up and patients who underwent only palliative treatment. Therefore, the final analysis included 1,287 patients.
At our institution, SCCHN patients are typically followed and monitored through their treatment and post-treatment courses with regularly scheduled clinical and radiographic examinations. Based on modified criteria of Warren and Gates32, SPMs were considered if the second lesions were different histopathologic type, or if they occurred more than 5 years following treatment for the index tumor, and/or clearly separated by normal epithelium based on clinical and radiographic assessment. Pulmonary lesions were considered as a SPM if they had a non-squamous histology; or if they were isolated squamous lesions greater than 5 years from initial SCCHN and felt to be SPM by the thoracic oncologist and thoracic surgeon. If there was discrepancy or differing of opinions regarding the origin of the tumor (i.e., recurrence vs. SPM), the second lesion was classified as a local recurrence rather than a SPM.
Clinical data, including overall stage at presentation of index tumor, site of index tumor, and treatment, were obtained at initial presentation and through follow-up examinations. Index cancer stage was then dichotomized into the early stage (I and II) and late stage (III and IV). The treatment was grouped into four categories: surgery only, surgery with radiotherapy and/or chemotherapy, radiotherapy, and radiotherapy plus chemotherapy. The epidemiological data, including alcohol and smoking, were obtained from all patients during the visit. Patients who had drunk at least one alcoholic beverage/per day for at least one year during their lifetime were defined as ever drinkers and those who never had such a pattern of drinking were defined as never drinkers. Those patients who had smoked at least 100 cigarettes in their lifetime were defined as ever smokers; otherwise, they were considered never smokers.
p14ARFgenotyping
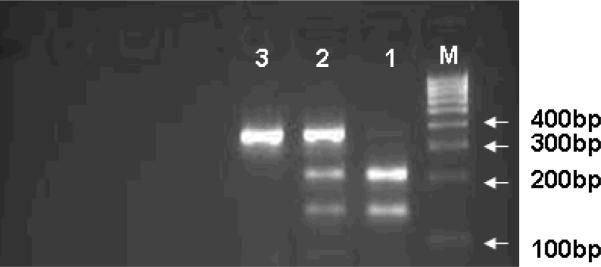
Genomic DNA was isolated from a leukocyte cell pellet of the blood samples by using the QIAGEN DNA Blood Mini Kit (QIAGEN Inc., Valencia, CA) according to the manufacturer's instructions. We genotyped for p14ARF polymorphisms by polymerase chain reaction-restriction fragment length polymorphism. The primers we used for initial three SNPs were as follows: 1) p14ARF-rs3731217: (forward) 5'-AAAAGG GGGACAACCATTCTC-3' and (reverse) 5'-CCCCTCTCAAATATGCTGTCC-3'; 2) p14ARF-rs3088440: (forward) 5'-TGCTCA CTCCAGAAAACTCCA-3' and (reverse) 5'-ATGTGCCACACATCTTTGACC-3'; and 3) p14ARF-rs3731245: (forward) 5'-CAAAAATGGGTCCACAAGGTT-3' and (reverse) 5'-CCC AACATAACCCCAAGT GTT-3'. For p14ARF-rs3731217, the 280-bp PCR products were digested with MvaI (New England BioLabs, Inc.,Beverly, Mass) at 37°C overnight, the T allele was uncut and the G allele was cut into 126 and 154 bp bands; for p14ARF-rs3088440 and rs3731245, the 356 and 323-bp PCR products were digested with HaeIII (New England BioLabs, Inc.,Beverly, Mass) at 37°C overnight, the A allele was uncut and the G allele being cut into 215 and 141-bp bands (rs3088440) (Fig. 1) and 125 and 198-bp bands (rs3731245), respectively. There was 100% concordance when at least 10% of the random samples were retested.
Fig. 1.
PCR-based RFLP genotyping for the p14ARF-rs3088440 polymorphism. Lane M, 100-bp DNA ladder; Lane 1, GG genotype with 141- and 251-bp bands; lane 2, GA genotype with 141-, 215-, and 356-bp bands; and lane 3, AA genotype with 356-bp band.
Statistical Analysis
In this study, the Statistical Analysis System software (version 9.1.3; SAS Institute) was used to perform all statistical analyses. Statistical significance was set at P < 0.05, and all tests were two-sided. SPM occurrence was considered as the primary endpoint of the study. The Student's t test was used to compare the mean age and follow-up time of the patients who developed a SPM and those who did not. The differences in distributions of demographic, epidemiological, and clinical variables, as well as genotypes between the two groups were evaluated using the chi-squared test. Time-to-event was calculated from the date of diagnosis of the index SCCHN to the date of SPM occurrence. Patients, who were not known to have an event at the date of last contact or who died, were censored. Kaplan-Meier curve was used to estimate SPM-free survival, and the log-rank statistic was used to evaluate significant difference (α = 0.05) in SPM-free survival between the two groups with and without variant genotypes of both polymorphisms. The associations between individual epidemiological risk factors, clinical characteristics, and genotypes and time to the occurrence of SPMs, were assessed using both univariate and multivariable Cox proportional hazards regression models. We used a stepwise search strategy to build the multivariable models. A final multivariable proportional hazards model was built using the variables that had prognostic potential suggested by the univariate analysis, and always retained the variables, including age, sex, and ethnicity, due to epidemiological and clinical considerations as described previously.12–15 We assessed associations using hazard ratio (HR) and their 95% confidence interval (CI) for a SPM development in the final Cox regression models with adjustment for age, sex, ethnicity, and smoking and alcohol status.
RESULTS
Patient Characteristics
The demographics, risk exposure and clinical variables for the 1,287 patients are summarized in Table 1. The 1,287 patients were followed with a median follow-up time of 29.7 months (range 0–142.4 months), of whom 1,167 patients did not develop SPM, while 120 (9.3%) patients developed SPM. The mean age at index cancer diagnosis for all patients was 57.5 years (range, 18–94 years, median, 57 years), and the mean age of patients at index SCCHN who developed SPM was significantly older compared with that of patients who did not develop SPM (60.8 years vs. 57.1 years, respectively; P < 0.001). Although this patient cohort predominantly consisted of male (75.9%), sex was not associated with SPM development (P = 0.515). We did not observe significant differences between patients with and without SPM, regarding smoking (P = 0.121), alcohol drinking (P = 0.345), index cancer site (P = 0.316), index cancer stage (P = 0.693), and treatment (P = 0.889). However, compared with the patients without SPM, patients who developed SPM were more likely to be non-Hispanic whites (P = 0.050).
Table 1.
Distribution of selected characteristics of the patient cohort (n =1,287)
| Total | SPM-Free | SPM | |||||
|---|---|---|---|---|---|---|---|
| Variable | No. | % | No. | % | No. | % | P-valuesc |
| Total patients | 1,287 | 100 | 1,167 | 90.7 | 120 | 9.3 | |
| Age | |||||||
| ≤ median (57 years) | 664 | 51.6 | 625 | 53.6 | 39 | 32.5 | < .001 |
| > median (57 years) | 623 | 48.4 | 542 | 46.4 | 81 | 67.5 | |
| Sex | |||||||
| Male | 977 | 75.9 | 883 | 75.7 | 94 | 78.3 | 0.515 |
| Female | 310 | 24.1 | 284 | 24.3 | 26 | 21.7 | |
| Ethnicity | |||||||
| Non-Hispanic White | 1,089 | 84.6 | 995 | 85.3 | 94 | 78.3 | 0.050 |
| Other | 198 | 15.4 | 172 | 14.7 | 26 | 21.7 | |
| Smoking | |||||||
| Never | 345 | 26.8 | 320 | 27.4 | 25 | 20.8 | 0.121 |
| Ever | 942 | 73.2 | 847 | 72.6 | 95 | 79.2 | |
| Alcohol | |||||||
| Never | 336 | 26.1 | 309 | 26.5 | 27 | 22.5 | 0.345 |
| Ever | 951 | 73.9 | 858 | 73.5 | 93 | 77.5 | |
| Index Cancer Site | |||||||
| Oral cavity | 416 | 32.3 | 378 | 32.3 | 38 | 31.7 | 0.316 |
| Oropharynx | 575 | 44.7 | 527 | 45.2 | 48 | 40.0 | |
| Larynx/Hypopharynx | 296 | 23.0 | 262 | 22.5 | 34 | 28.3 | |
| Index Cancer Stage | |||||||
| I or II | 324 | 25.2 | 292 | 25.0 | 32 | 26.7 | 0.693 |
| III or IV | 963 | 74.8 | 875 | 75.0 | 88 | 73.3 | |
| Treatment | |||||||
| Surgery only | 229 | 17.8 | 208 | 17.8 | 21 | 17.5 | 0.889 |
| Surgery + Adjuvant Txa | 320 | 24.9 | 287 | 24.6 | 33 | 27.5 | |
| XRTb | 329 | 25.5 | 301 | 25.8 | 28 | 23.3 | |
| XRT + Chemotherapy | 409 | 31.8 | 371 | 31.8 | 38 | 31.7 | |
Adjuvant Treatment: adjuvant radiotherapy and/or chemotherapy
XRT: radiotherapy
P values were calculated from chi-square test
Of the 120 patients with SPM, 81 patients developed SPMs at tobacco-associated sites including 44 SCCHN and 37 other tobacco-associated cancers (34 lung cancer, 2 esophagus cancer, and 1 bladder cancer); 35 developed SPMs at other sites (10 prostate cancer, 8 papillary thyroid carcinoma, 4 colon adenocarcinoma, 3 lymphoma, 3 hepatic adenocarcinoma, 2 breast cancer, and 1each for the remainder including sarcoma, renal cell carcinoma, endometrial carcinoma, leukemia, and maxillary sinus adenocarcinoma); and 4 developed SPMs at 2 sites (2 patients with both SCCHN and prostate cancer and 2 patients with both SCCHN and papillary thyroid carcinoma). Of the 44 patients with second SCCHN, 24 were synchronous SCCHN primaries. Of these 24 patients with synchronous SCCHN, two patients had bilateral oral cavity cancers, three had bilateral oropharyngeal cancers, one had bilateral hypopharyngeal cancers, and the remainder had simultaneous cancers of more than one head and neck subsite.
Association of p14ARF polymorphisms with risk of SPM after index SCCHN
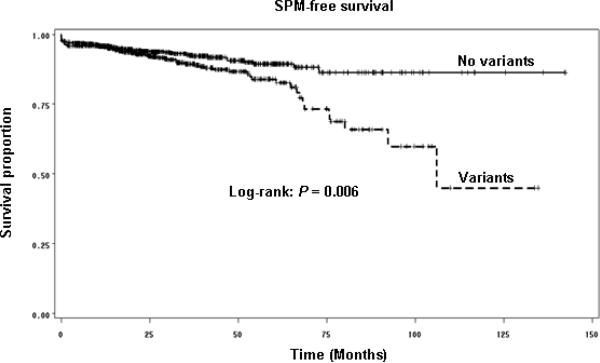
Table 2 shows distributions of p14ARF-rs3088440 and rs3731217 genotypes between patients who did and did not develop SPM and the associations with risk of SPM development. The distribution of p14ARF-rs3731217 genotypes was not significantly different between patients who developed SPM and those who did not, while the significant difference was observed for the p14ARF-rs3088440 polymorphism (P = 0.002). For each polymorphism, compared with patients with the corresponding homozygous wild-type genotypes, patients who possessed the variant genotypes had a significantly approximately 1.5-fold increased risk to develop SPM after multivariable adjustment for age, sex, ethnicity, smoking and drinking (Table 2). To evaluate the combined effect of both p14ARF polymorphisms on risk of SPM, patients who were wild-type homozygous for both genotypes were grouped as a “no variant” reference group, and the reminders with other combined variant genotypes including variant homozygous and heterozygous genotypes were the “variant” group (Table 2). We found that the distribution of the combined genotypes was significantly different between patients who developed SPM and those who did not (P < 0.001 for trichotomized and P = 0.007 for dichotomized). Moreover, patients possessing either variant allele (p14ARF G or p14ARF A) had almost a 1.6 -fold increased risk for SPM, compared with patients with the combined p14ARF TT and GG wild-type genotypes (Table 2). There was a trend for increased SPM risk with an increasing number of variant genotypes, and this trend in risk was statistically significant in a dose-response manner (P = 0.002 for the trend, Table 2). Specifically, the patients with 2 variant genotypes had an approximately 3-fold increased risk for developing SPM, compared with patients without any variant genotypes (Table 2). Furthermore, the patients with p14ARF variant genotypes of both polymorphisms experienced a significantly reduced SPM-free survival compared with patients with no p14ARF variant genotypes (log-rank, P = 0.006, Fig. 2).
Table 2.
SPM risk associated with p14ARF polymorphisms after index SCCHN
| Genotypes | Total (No. = 1,287) | SPM-free (No. = 1,167) | SPM (No. = 120) | Pa | HR (95% CI)b | |||
|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | |||
| p14ARF-rs3731217 | ||||||||
| TT (Ref.c) | 966 | 75.1 | 883 | 75.7 | 83 | 69.1 | 0.207 | 1.00 |
| TG | 293 | 22.8 | 258 | 22.1 | 35 | 29.2 | 1.54 (1.03–2.31) | |
| GG | 28 | 2.1 | 26 | 2.2 | 2 | 1.7 | 0.82 (0.20–3.35) | |
| TG+GG | 321 | 24.9 | 284 | 24.3 | 37 | 30.9 | 0.117 | 1.48 (1.00–2.19) |
| p14ARF-rs3088440 | ||||||||
| GG (Ref.c) | 1038 | 80.7 | 955 | 81.8 | 83 | 69.2 | 0.002* | 1.00 |
| GA | 219 | 17.0 | 185 | 15.9 | 34 | 28.3 | 1.69 (1.11–2.56) | |
| AA | 30 | 2.3 | 27 | 2.3 | 3 | 2.5 | 1.05 (0.33–3.37) | |
| GA+AA | 249 | 19.3 | 212 | 18.2 | 37 | 30.8 | 0.001 | 1.61 (1.07–2.43) |
| 0d (Ref.c) | 749 | 58.2 | 693 | 59.4 | 56 | 46.7 | < .001* | 1.00 |
| 1 | 506 | 39.3 | 452 | 38.7 | 54 | 45.0 | 1.44 (0.99–2.10) | |
| 2 | 32 | 2.5 | 22 | 1.9 | 10 | 8.3 | 3.07 (1.54–6.12) | |
| Trend | P = 0.002 | |||||||
| 0 (Ref.c) | 749 | 58.2 | 693 | 59.4 | 56 | 46.7 | 0.007 | 1.00 |
| ≥ 1 | 538 | 41.8 | 474 | 40.6 | 64 | 53.3 | 1.57 (1.09–2.25) | |
χ2 test for differences in the distribution of P14 genotypes between the patients who developed SPM and the patients who did not.
Adjusted for age, sex, ethnicity, tobacco smoking and alcohol drinking in a Cox model.
Ref. = reference group.
Significant at the 5% level after adjusting for multiple comparisons with Bonferroni correction.
0:p14ARF-rs3731217 TT and p14ARF-rs3088440 GG genotypes; 1: p14ARF-rs3731217 TT and p14ARF-rs3088440 GA+AA or p14ARF-rs3731217 TG+GG and p14ARF-rs3088440 GG genotypes; and 2: p14ARF-rs3731217 TG+GG and p14ARF-rs3088440 GA+AA genotypes.
Fig. 2.
Kaplan-Meier SPM-free survival curve stratified by combined p14ARF wild-tpe genotypes and variantgenotypes
Stratification analysis of the combined p14ARF variant genotypes with risk of SPM
Table 3 shows the association between the combined p14ARFvariant genotypes and risk of SPM in each subgroup of age, sex, ethnicity, smoking/drinking status, index cancer site, index cancer stage and index cancer treatment after adjustment with all other potential confounders. When we used those without any combined variant genotypes (p14ARF-rs3731217 TT and rs3088440 GG) as the reference group, there was an significantly approximately 50 to 70% increased SPM risk for those with any p14ARF variant genotypes among males, non-Hispanic whites, drinkers, those with late stages patients, those with non-oropharyngeal cancer, those who received treatment with DNA-damaging agent, and those with tobacco associated SPM (Table 3). Additionally, there was a more than 2-fold significantly elevated SPM risk for those with any p14ARF variant genotypes among younger patients (≤57 years) (Table 3).
Table 3.
Stratification analysis of association between the combined p14ARF polymorphisms and SPM Risk
| No variants | With variants | ||||
|---|---|---|---|---|---|
| SPM-free No. (%) | SPM No. (%) | SPM-free No. (%) | SPM No. (%) | HR (95%CI)a | |
| Age at Presentation of Index Cancer | |||||
| ≤57 years | 375 (54.1) | 15 (26.8) | 250 (52.7) | 24 (37.5) | 2.18 (1.14–4.17) |
| >57 years | 318 (45.9) | 41 (73.2) | 224 (47.3) | 40 (62.5) | 1.34 (0.86–2.09) |
| Sex | |||||
| Male | 513 (74.0) | 41 (73.2) | 370 (78.1) | 53 (82.8) | 1.65 (1.10–2.48) |
| Female | 180 (26.0) | 15 (26.8) | 104 (21.9) | 11 (17.2) | 1.38 (0.63–3.06) |
| Ethnicity | |||||
| Non-HW | 608 (87.7) | 46 (82.1) | 387 (81.7) | 48 (75.0) | 1.56 (1.04–2.35) |
| Others | 85 (12.3) | 10 (17.9) | 87 (18.3) | 16 (25.0) | 1.53 (0.69–3.41) |
| Smoking Status at Presentation | |||||
| Ever | 498 (71.9) | 46 (82.1) | 349 (73.6) | 49 (76.6) | 1.41 (0.94–2.11) |
| Never | 195 (28.1) | 10 (17.9) | 125 (26.4) | 15 (23.4) | 2.19 (0.97–4.93) |
| Drinking Status at Presentation | |||||
| Ever | 505 (72.9) | 43 (76.8) | 353 (74.5) | 50 (78.1) | 1.53 (1.02–2.31) |
| Never | 188 (27.1) | 13 (23.2) | 121 (25.5) | 14 (21.9) | 1.77 (0.81–3.87) |
| Index Cancer Site | |||||
| Oropharnx | 315 (45.5) | 24 (42.9) | 212 (44.7) | 24 (37.5) | 1.29 (0.73–2.28) |
| Non-oropharynx | 378 (54.5) | 32 (57.1) | 262 (55.3) | 40 (62.5) | 1.69 (1.05–2.72) |
| Index Cancer Stage | |||||
| Early (1 or 2) | 170 (24.5) | 14 (25.0) | 122 (25.7) | 18 (28.1) | 1.58 (0.77–3.25) |
| Late (3 or 4) | 532 (75.5) | 42 (75.0) | 352 (74.3) | 46 (71.9) | 1.57 (1.03–2.39) |
| Index Cancer Treatment | |||||
| Surgery only | 130 (18.8) | 9 (16.1) | 78 (16.5) | 12 (18.8) | 1.81 (0.75–4.37) |
| DNA-damaging | 563 (81.2) | 47 (83.9) | 396 (83.5) | 52 (81.2) | 1.55 (1.04–2.31) |
| SPM Site | |||||
| Tobacco-associated sitesb | 693 (50.0) | 39 (45.9) | 474 (50.0) | 46 (54.1) | 1.68 (1.09–2.58) |
| Other sites | 693 (50.0) | 17 (48.6) | 474 (50.0) | 18 (51.4) | 1.51 (0.77–2.94) |
Adjusted for age, sex, ethnicity, tobacco smoking and alcohol drinking in a Cox model.
Tobacco-associated sites = oral cavity, oropharynx, larynx, hypopharynx, esophagus, lung, or bladder.
DISCUSSION
Given the critical roles of p14ARF as a tumor suppressor gene in many cellular activities, inactivation or expression change of this gene may deregulate these activities, and consequently could influence cancer risk. Although many previous studies have focused on the role of alterations in INK4/AFR or CDKN2A locus in development of SCCHN25–29, there have been no previous studies examining the association of genetic variants in p14ARF with risk of SCCHN, particularly SPM after an index SCCHN. In this study, we examined such association. We found that both p14ARF polymorphisms were associated with a significantly moderately increased risk of SPM in patients with an index SCCHN, and the risk was significantly higher in patients with either p14ARF variant genotypes of the two polymorphisms than those with wild-type homozygous p14ARF genotypes. Patients simultaneously having both variant p14ARF genotypes had an approximately 3-fold increased risk for developing SPM compared with patients without any variant genotypes. Although there is unknown functional relevance of these two p14ARF polymorphisms, they are within the functional regions of the gene's promoter (p14ARF-rs3731217) and 3'UTR (p14ARF-rs3088440), and these two polymorphisms could potentially affect p14 expression levels leading to inter-individual differences in susceptibility to SPM after an index SCCHN. To date, despite no studies have examined the association between these two polymorphisms and risk of cancer, our data suggest that these two SNPs may have functional significance and may contribute to genetic susceptibility to SPM after an index SCCHN in this patient cohort.
Additionally, we observed a greater SPM risk associated with the p14ARF variant genotypes in younger (≤57 years) SCCHN patients, with no significant association among the patients older than 57 years, a finding consistent with the concept of genetic susceptibility characteristic of an early age of onset. Furthermore, the SPM risk was significantly associated with the combined p14ARF variant genotypes in men but not in women. In this study, male SCCHN patients were more likely to be ever-smokers than female patients (P < 0.001), and it is possible that male SCCHN patients carrying the combined p14ARF variant genotypes are more sensitive to tobacco carcinogens that may have been responsible for both index cancer and SPM. This speculation is supported by the finding that the risk of SPM associated with the p14ARF variant genotypes was greater for tobacco-associated rather than non-tobacco-associated SPMs.
While continued exposure to tobacco or alcohol appears associated with elevated SPM risk compared with those who avoided such exposure7, 33, only one study has demonstrated that index treatment modality (radiotherapy) may influence SPM risk.34 In this study, we found that these p14ARF variant genotypes appeared to be risk factors for SPM in SCCHN patients, which may be independent of radiation and/or chemotherapy they had for their index cancer treatment as well as smoking and alcohol status. p14ARF is redistributed in the nucleus in response to DNA damage35, and thus p14ARF polymorphisms could influence SPM risk after such treatment exposure. However, the notion that SPM risk associated with p14ARF polymorphisms is dependent on the index treatment type may simply be an artifact of the small sample size of those treated by surgery alone.
Our study also showed that the non-oropharyngeal index SCCHN patients had a greater SPM risk associated with the p14ARF variant genotypes than those with oropharyngeal cancers. This may represent differences in the etiology of index SCCHN at oropharyngeal and non-oropharyngeal sites in relation to both environmental risk factors and genetic susceptibility. Our previous study suggests that squamous cell carcinomas of the oropharynx are more likely driven by human papillomavirus type 16, while squamous cell carcinomas of the oral cavity and larynx are more likely caused by smoking and alcohol use.36 Therefore, the risk of tobacco- or alcohol-induced SPM after index non-oropharyngeal SCC may be modified by p14ARF genotypes, and these genotypes may play an even greater role in SPM of non-oropharyngeal cancers arising in ever smokers and ever drinkers. Supporting this hypothesis was the observation that the p14ARF variant genotypes were more strongly associated with SPM at tobacco-associated sites than SPM at other sites. Although SPM risk associated with p14ARF variant genotypes was greater among the observed subgroups, the significant association could be by chance due to the rather small sample sizes of these subgroups. However, further large studies are needed to validate our findings.
Although our results support an increased risk of SPM after index SCCHN associated with both p14ARF polymorphisms individually and in combination within a large and well-characterized cohort of SCCHN patients treated at a multidisciplinary cancer center, our findings have several inherited limitations. First, there may be a selection bias for study patients due to the hospital-based nature of this study, and inclusion of selected SNPs in this analysis based on allele frequency from these patients may limit the external validity of this study. While the sample size of our study is relatively large, the small number of SPM in subgroups, especially when the patients were stratified, may limit our ability to detect a certain degree of association. The low rate of SPM in this patient cohort likely reflects high prevalence of both never smokers and patients presenting with late stage, as well as our strict criteria in defining SPM. Additionally, this cohort, at present, still has relatively limited follow-up time (30 month) to develop SPM. Furthermore, our patient cohort included approximately 85% non-Hispanic white, and our findings may not be relevant to SPM risk after index SCCHN in other ethnicities. While demographics, exposure, and clinical data for the cohort were collected prospectively, clinical outcomes including SPM were collected retrospectively under a no strictly defined screening or follow-up regimen. Finally, due to the retrospective nature of the original study design, we did not have information on HPV infection and the continued smoking behavior after index SCCHN diagnosis, and these potential confounders could bias the observed association. Therefore, our future studies on the association between genetic polymorphisms and risk of SPM should incorporate HPV tumor status and smoking behavior after index cancer treatment into the study design.
Condensed abstract.
p14ARF polymorphisms may modulate the risk of second primary malignancy in patients with squamous cell carcinoma of the head and neck and these p14ARF polymorphisms could be a risk marker for genetic susceptibility to SPM of patients with primary squamous cell carcinoma of the head and neck.
Acknowledgments
Funded by: Research Training Award, The American Laryngological, Rhinological, and Otological Society (to E.M.S.); U.T. M.D. Anderson Cancer Center Start-up Funds (to E.M.S.) and National Institute of Health Grants R01 ES-11740 and CA-131274 (to Q.W.); N.I.H. P-30 CA-16672 (to The University of Texas M.D. Anderson Cancer Center) and N.I.H. CA135679 (to G.L.) and CA133099 (to G.L.).
Abbreviations
- SCCHN
squamous cell carcinoma of the head and neck
- SPM
second primary malignancies
- HR
hazard ratio
- CI
confidence interval
- HPV
human papillomavirus
REFERENCE
- 1.Ferlay J, Bray F, Pisani P, Parkin D. GLOBOCAN 2002: Cancer Incidence, Mortality and Prevalence Worldwide. IARC Press; Lyon, France: 2004. [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics. 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 3.Surveillance, Epidemiology and Results (SEER) Program . SEER cancer statistics. Surveillance Research Program, National Cancer Institute; SEER*Stat Database-SEER 9 Regs Public-Use, November 2009 Sub (1973–2007) available at http://seer.cancer.gov. July 2010 release, based on the November 2009 submission. [Google Scholar]
- 4.Sturgis EM, Miller RH. Second primary malignancies in the head and neck cancer patient. Ann Otol Rhinol Laryngol. 1995;104:946–954. doi: 10.1177/000348949510401206. [DOI] [PubMed] [Google Scholar]
- 5.Rennemo E, Zätterström U, Boysen M. Impact of second primary tumors on survival in head and neck cancer: an analysis of 2,063 cases. Laryngoscope. 2008;118:1350–1356. doi: 10.1097/MLG.0b013e318172ef9a. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto E, Shibuya H, Yoshimura R, Miura M. Site specific dependency of second primary cancer in early stage head and neck squamous cell carcinoma. Cancer. 2002;94:2007–2014. doi: 10.1002/cncr.10444. [DOI] [PubMed] [Google Scholar]
- 7.Day GL, Blot WJ, Shore RE, et al. Second cancers following oral and pharyngeal cancers: role of tobacco and alcohol. J Natl Cancer Inst. 1994;86:131–137. doi: 10.1093/jnci/86.2.131. [DOI] [PubMed] [Google Scholar]
- 8.Do KA, Johnson MM, Lee JJ, et al. Longitudinal study of smoking patterns in relation to the development of smoking-related secondary primary tumors in patients with upper aerodigestive tract malignancies. Cancer. 2004;101:2837–2842. doi: 10.1002/cncr.20714. [DOI] [PubMed] [Google Scholar]
- 9.Wu X, Spitz MR, Lee JJ, et al. Novel susceptibility loci for second primary tumors/recurrence in head and neck cancer patients: large-scale evaluation of genetic variants. Cancer Prev Res. 2009;2:617–624. doi: 10.1158/1940-6207.CAPR-09-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gal TJ, Huang WY, Chen C, Hayes RB, Schwartz SM. DNA repair gene polymorphisms and risk of second primary neoplasms and mortality in oral cancer patients. Laryngoscope. 2005;115:2221–2231. doi: 10.1097/01.mlg.0000183736.96004.f7. [DOI] [PubMed] [Google Scholar]
- 11.Minard CG, Spitz MR, Wu X, Hong WK, Etzel CJ. Evaluation of glutathione S-transferase polymorphisms and mutagen sensitivity as risk factors for the development of second primary tumors in patients previously diagnosed with early-stage head and neck cancer. Cancer. 2006;106:2636–2644. doi: 10.1002/cncr.21928. [DOI] [PubMed] [Google Scholar]
- 12.Li F, Sturgis EM, Zafereo ME, et al. p73 G4C14-to-A4T14 polymorphism and risk of second primary malignancy after index squamous cell carcinoma of head and neck. Int J Cancer. 2009;125:2660–2665. doi: 10.1002/ijc.24570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zafereo ME, Sturgis EM, Aleem S, Chaung K, Wei Q, Li G. Glutathione S-transferase polymorphisms and risk of second primary malignancy after index squamous cell carcinoma of the head and neck. Cancer Prev Res. 2009;2:432–439. doi: 10.1158/1940-6207.CAPR-08-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zafereo ME, Sturgis EM, Liu Z, Wang LE, Wei Q, Li G. Nucleotide excision repair core gene polymorphisms and risk of second primary malignancy in patients with index squamous cell carcinoma of the head and neck. Carcinogenesis. 2009;30:997–1002. doi: 10.1093/carcin/bgp096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lei D, Sturgis EM, Liu Z, Zafereo ME, Wei Q, Li G. Genetic polymorphisms of p21 and risk of second primary malignancy in patients with index squamous cell carcinoma of the head and neck. Carcinogenesis. 2010;31:222–227. doi: 10.1093/carcin/bgp279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haber DA. Splicing into senescence: the curious case of p16 and p19ARF. Cell. 1997;91:555–558. doi: 10.1016/s0092-8674(00)80441-9. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez S, Serrano M. A new mechanism of inactivation of the INK4/ARF locus. Cell Cycle. 2006;5:1382–1384. doi: 10.4161/cc.5.13.2901. [DOI] [PubMed] [Google Scholar]
- 18.Gjerset RA, Bandyopadhyay K. Regulation of p14ARF through subnuclear compartmentalization. Cell Cycle. 2006;5:686–690. doi: 10.4161/cc.5.7.2623. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, He L, Bruce A, et al. p14ARF inhibits the growth of p53 deficient cells in a cell-specific manner. Biochim Biophys Acta. 2006;1763:787–796. doi: 10.1016/j.bbamcr.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 20.Sherr CJ, Bertwistle D, DEN Besten W, et al. p53-Dependent and -independent functions of the Arf tumor suppressor. Cold Spring Harb Symp Quant Biol. 2005;70:129–137. doi: 10.1101/sqb.2005.70.004. [DOI] [PubMed] [Google Scholar]
- 21.Eymin B, Claverie P, Salon C, et al. p14ARF activates a Tip60-dependent and p53 independent ATM/ATR/CHK pathway in response to genotoxic stress. Mol Cell Biol. 2006;26:4339–4350. doi: 10.1128/MCB.02240-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bates S, Phillips AC, Clark PA, et al. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- 23.Chin L, Pomerantz J, DePinho RA. The INK4a/ARF tumor suppressor: one gene-two products-two pathways. Trends Biochem Sci. 1998;23:291–296. doi: 10.1016/s0968-0004(98)01236-5. [DOI] [PubMed] [Google Scholar]
- 24.Leduc C, Claverie P, Eymin B, et al. p14ARF promotes RB accumulation through inhibition of its Tip60-dependent acetylation. Oncogene. 2006;25:4147–4154. doi: 10.1038/sj.onc.1209446. [DOI] [PubMed] [Google Scholar]
- 25.Ghosh A, Ghosh S, Maiti GP, et al. SH3GL2 and CDKN2A/2B loci are independently altered in early dysplastic lesions of head and neck: correlation with HPV infection and tobacco habit. J Pathol. 2009;217:408–419. doi: 10.1002/path.2464. [DOI] [PubMed] [Google Scholar]
- 26.Weber A, Wittekind C, Tannapfel A. Genetic and epigenetic alterations of 9p21 gene products in benign and malignant tumors of the head and neck. Pathol Res Pract. 2003;199:391–397. doi: 10.1078/0344-0338-00435. [DOI] [PubMed] [Google Scholar]
- 27.Poi MJ, Yen T, Li J, et al. Somatic INK4a-ARF locus mutations: a significant mechanism of gene inactivation in squamous cell carcinomas of the head and neck. Mol Carcinog. 2001;30:26–36. doi: 10.1002/1098-2744(200101)30:1<26::aid-mc1010>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 28.Ohta S, Uemura H, Matsui Y, et al. Alterations of p16 and p14ARF genes and their 9p21 locus in oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2009;107:81–91. doi: 10.1016/j.tripleo.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 29.Bradley G, Irish J, MacMillan C, et al. Abnormalities of the ARF-p53 pathway in oral squamous cell carcinoma. Oncogene. 2001;20:654–658. doi: 10.1038/sj.onc.1204131. [DOI] [PubMed] [Google Scholar]
- 30.Hu WL, Li SJ, Liu DT, et al. Genetic variants on chromosome 9p21 and ischemic stroke in Chinese. Brain Res Bull. 2009;79:431–435. doi: 10.1016/j.brainresbull.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 31.Sun P, Zhang Z, Wan J, Zhao N, Jin X, Xia Z. Association of genetic polymorphisms in GADD45A, MDM2, and p14 ARF with the risk of chronic benzene poisoning in a Chinese occupational population. Toxicol Appl Pharmacol. 2009;240:66–72. doi: 10.1016/j.taap.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 32.Warren S, Gates O. Multiple primary malignant tumors: a survey of the literature and statistical study. Am J Cancer. 1932;51:1358. [Google Scholar]
- 33.Do KA, Johnson MM, Doherty DA, et al. Second primary tumors in patients with upper aerodigestive tract cancers: joint effects of smoking and alcohol (United States) Cancer Causes Control. 2003;14:131–138. doi: 10.1023/a:1023060315781. [DOI] [PubMed] [Google Scholar]
- 34.Hashibe M, Ritz B, Le AD, Li G, Sankaranarayanan R, Zhang ZF. Radiotherapy for oral cancer as a risk factor for second primary cancers. Cancer Lett. 2005;220:185–195. doi: 10.1016/j.canlet.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 35.Lee C, Smith BA, Bandyopadhyay K, et al. DNA damage disrupts the p14ARF-B23 (nucleophosmin) interaction and triggers a transient subnuclear redistributionof p14ARF. Cancer Res. 2005;65:9834–9842. doi: 10.1158/0008-5472.CAN-05-1759. [DOI] [PubMed] [Google Scholar]
- 36.Dahlstrom KR, Adler-Storthz K, Etzel CJ, et al. Human papillomavirus type 16 infection and squamous cell carcinoma of the head and neck in never-smokers: a matched pair analysis. Clin Cancer Res. 2003;9:2620–2626. [PubMed] [Google Scholar]