Abstract
A post-translational modification affecting the translation termination rate was identified in the universally conserved GGQ sequence of release factor 2 (RF2) from Escherichia coli, which is thought to mimic the CCA end of the tRNA molecule. It was shown by mass spectrometry and Edman degradation that glutamine in position 252 is N5-methylated. Overexpression of RF2 yields protein lacking the methylation. RF2 from E.coli K12 is unique in having Thr246 near the GGQ motif, where all other sequenced bacterial class 1 RFs have alanine or serine. Sequencing the prfB gene from E.coli B and MRE600 strains showed that residue 246 is coded as alanine, in contrast to K12 RF2. Thr246 decreases RF2-dependent termination efficiency compared with Ala246, especially for short peptidyl-tRNAs. Methylation of Gln252 increases the termination efficiency of RF2, irrespective of the identity of the amino acid in position 246. We propose that the previously observed lethal effect of overproducing E.coli K12 RF2 arises through accumulating the defects due to lack of Gln252 methylation and Thr246 in place of alanine.
Keywords: post-translational modification/release factor/translation termination
Introduction
During protein synthesis, release of the polypeptide chain from the ribosome occurs when an in-frame stop codon is encountered. Protein release factors (RFs) are required for the termination process (for a review see Buckingham et al., 1997). Class 1 RFs recognize stop codons and promote the hydrolysis of the ester bond between polypeptide and P-site-bound tRNA on the ribosome. It is generally believed that the hydrolytic reaction is catalysed by the peptidyl transferase centre (see Caskey et al., 1971), recently suggested to be a ribosyme (Zhang and Cech, 1997), and that the role of class 1 RFs is to give specificity to this reaction by recognizing stop codons rather than by carrying out the catalytic task themselves. Class 2 RFs stimulate termination by a mechanism that requires hydrolysis of GTP. Escherichia coli contains two class 1 RFs with differing specificity: RF1 recognizes UAA and UAG codons and RF2 recognizes UAA and UGA codons (Scolnick et al., 1968). RF3 in E.coli is a class 2 factor that stimulates dissociation of RF1 and RF2 from the ribosome after cleavage of the peptidyl-tRNA (Freistroffer et al., 1997). In contrast to eubacteria, eukaryotic and archaebacterial cells employ a single class 1 factor at all three stop codons (Frolova et al., 1994).
RF1 and RF2 in E.coli show extensive amino acid sequence similarity to each other (Caskey et al., 1984; Weiss et al., 1984; Craigen et al., 1985) and to class 1 RFs in other eubacteria, but no general similarity to eukaryotic and archaebacterial class 1 RFs. Recently, however, a completely conserved GGQ motif, surrounded by other partially conserved amino acids, was identified in the amino acid sequence of class 1 RFs from all three kingdoms (Frolova et al., 1999). Mutations in either glycine residue impair eRF1 release activity (Frolova et al., 1999). The conserved region is thought to be involved in stop codon-dependent hydrolysis of peptidyl-tRNA on the ribosome and to interact directly with the peptidyl transferase centre on the 50S ribosomal subunit (Frolova et al., 1999; Song et al., 2000). The crystal structure of eRF1 supports this idea, showing that the GGQ motif lies in a loop connecting an α-helix and a β-strand at one extremity of the molecule. Comparison with a tRNA molecule suggests that the region is homologous to the CCA end of a tRNA. It has been proposed that the glutamine residue in the conserved GGQ motif participates in the coordination of the water molecule during hydrolysis of the ester bond of peptidyl-tRNA on the ribosome (Song et al., 2000).
Although in vitro experiments show that both RF1 and RF2 can read UAA codons in E.coli, it has been argued on genetic grounds that UAA codons (at least in some nucleotide contexts) are read far more often in E.coli by RF1 than by RF2 (Nakamura et al., 1990), despite the fact that RF2 is 5-fold more abundant than RF1 (Adamski et al., 1994). Thus, mutations reducing RF2 activity failed to suppress UAA nonsense mutations, whereas mutations affecting RF1 were effective suppressors (Rydén and Isaksson, 1984; Kawakami et al., 1988; Mikuni et al., 1991). This behaviour distinguished E.coli from Salmonella typhimurium, where the situation was largely reversed (Elliott and Wang, 1991). Multicopy expression of RF2 from these two organisms revealed a second difference: overproduction of E.coli RF2 completely inhibited cell growth whereas overproduction of the factor from S.typhimurium did not (Kawakami and Nakamura, 1990; Mikuni et al., 1991; Uno et al., 1996). Experiments by Tate et al. (1993) showed that the specific activity of RF2 from E.coli as measured by the release of fMet from fMet-tRNA in vitro was considerably lower when the factor was overproduced.
These observations led Nakamura and co-workers to search for the amino acid change(s) between the RF2 in E.coli and S.typhimurium responsible for these phenotypic differences (Uno et al., 1996). A single critical residue was identified out of 16 different amino acids, at position 246, four residues towards the N-terminus from the GGQ motif. Residue 246 is threonine in E.coli RF2 and alanine in S.typhimurium RF2. When Thr246 in E.coli RF2 was changed to Ala246 as in S.typhimurium, the toxicity due to overproduction of the factor was abolished and the termination efficiency in vivo was increased (Uno et al., 1996).
We show here the presence of a functionally important post-translational modification to the glutamine residue of the conserved GGQ motif. Gln252 in E.coli RF2 is normally modified to N5-methylglutamine, but the modification is absent when the factor is overproduced. The presence of threonine at position 246 in RF2 is peculiar to E.coli K12 strains and is not found in E.coli B or MRE600 strains, which we show have Ala246, or in any other bacteria for which sequence data are available. In vitro peptide release data confirm the reduced activity of RF2 containing Thr246 compared with Ala246, particularly for very short peptides. Our results suggest that the accumulated negative effects on release activity of Thr246 and unmodified Gln252 are responsible for the growth inhibition following overexpression of E.coli K12 RF2.
Results
Sequencing of the prfB gene in E.coli K12 and other strains
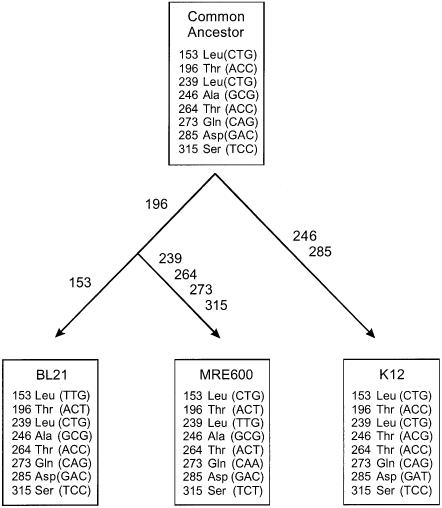
Previous kinetics studies in vitro of translation termination by RF2 using a variety of short peptidyl-tRNAs revealed persistent differences in kinetic parameters according to the E.coli strain employed as a source of RF2 and the mode of preparation of the factor (Dinçbas et al., 1999; Heurgué-Hamard et al., 2000; our unpublished results). Thus, RF2 from E.coli K12 strains was less active than when isolated from MRE600, and overexpression of the factor seemed to reduce its activity, as also observed by others (Tate et al., 1993; Uno et al., 1996). Both biochemical studies and physical characterization by mass spectrometry (see below) suggested the presence of amino acid sequence differences between RF2 isolated from E.coli K12 and other E.coli strains. The prfB gene was therefore sequenced on DNA prepared from two K12 strains (Xac and W3110) and two other E.coli strains (MRE600 and BL21). The sequence from both K12 strains was identical to the prfB K12 sequence in the data banks (EcoGene accession No. EG10762). Several nucleotide differences were found between the E.coli B and MRE600 strains and the K12 sequence, only one of which affected the amino acid sequence. This was an A to G change at codon 246 in both E.coli B and MRE600 strains (ACG in K12), changing Thr246 in K12 RF2 to Ala246. Amino acid 246 in both E.coli B and MRE600 strains is therefore the same as in S.typhimurium (Uno et al., 1996). The complete set of nucleotide changes found is shown in the three lower boxes of Figure 1. The data show that all three strains have accumulated mutations in the prfB gene, silent except for the change affecting residue 246 diverging from a putative common ancestor. Supposing that each mutation occurred once, Figure 1 suggests the codons most likely to be present in a common ancestor of the K12, B and MRE600 strains (upper box), based on a comparison between the three E.coli sequences (lower boxes) and the S.typhimurium prfB sequence (EcoGene accession No. M38590; Kawakami and Nakamura, 1990). This shows that codons 246 and 285 are conserved between S.typhimurium, E.coli B and MRE600 strains, whereas codon 196 is conserved between S.typhimurium and E.coli K12.
Fig. 1. Codon differences between prfB from different E.coli strains. The lower three boxes show the differences revealed by nucleotide sequencing. The upper box shows the codons likely to be present in a common ancestor of the K12, B and MRE600 strains, based on a comparison of the E.coli sequences and the prfB sequence from S.typhimurium (see text).
Efficiency of peptide termination by the different RF2 variants
Chromosomally expressed RF2 was prepared from the E.coli strains W3110 and MRE600 as described (Materials and methods). The proteins will be referred to as RF2(ceThr246) and RF2(ceAla246), respectively, due to the identity of the amino acid at position 246. The wild-type prfB gene of the E.coli W3110 strain and an other wise isogenic mutant (Thr246Ala) were overexpressed (Materials and methods) and purified (Pavlov et al., 1998; Materials and methods). These overexpressed proteins will be referred to as RF2(oeThr246) and RF2(oeAla246), respectively.
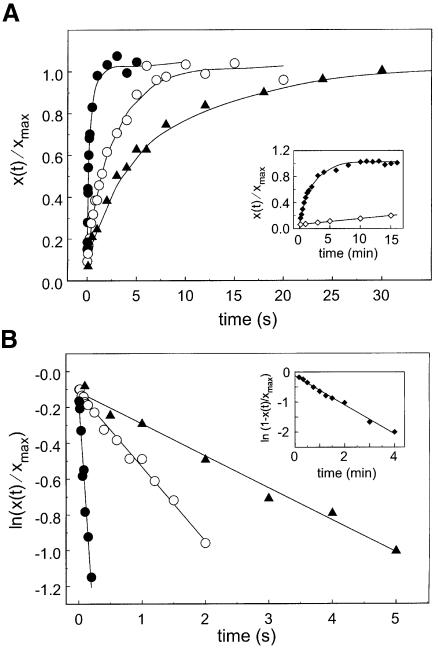
Ribosomes were initiated for translation of three different mRNAs encoding fMet-Ile, fMet-Phe-Ile and fMet-Phe-Thr-Ile, followed by UAA as the termination signal in all cases (Dinçbas et al., 1999; Materials and methods). The mRNAs were translated by the addition of aminoacyl-tRNAs and elongation factors to the initiated ribosomes, but RFs were excluded. Ribosomes stalled at UAA and, ready for termination with RF2, were separated from all other components of the translation system by gel filtration (Freistroffer et al., 1997; Materials and methods). The efficiency of RF2-dependent termination for each of these three release complexes was measured for the four variants of RF2 in two different ways. In one set of experiments, the peptide release reaction was saturated with RF2 to obtain the rate constant (kc) of ester bond hydrolysis (Materials and methods). In a second set of experiments, performed at low concentrations of ribosome and RF2, we obtained the effective association rate constant (kcat/Km) for termination by the different RFs (Freistroffer et al., 1997, 2000; Materials and methods). The results are summarized in Table I and a typical experiment to determine the catalytic rate constant (kc) at release complexes with fMet-Ile-tRNA2Ile in the P-site is shown in Figure 2. Table I shows that RF2(ceAla246), i.e. chromosomally expressed RF2 from MRE600 cells, has the highest efficiency with respect to both kc and kcat/Km. Furthermore, there is little variation in these two parameters when the length of the peptide varies from two to four. RF2(ceThr246), i.e. chromosomally expressed RF2 from K12, is ∼30-fold less efficient in kc and ∼20-fold less efficient in kcat/Km than RF2(ceAla246) for termination with fMet-Ile-tRNA2Ile in the release complex. When the length of the peptide increases, the difference in termination efficiency between the factors decreases to ∼3-fold in kc and 2-fold in kcat/Km for the tetrapeptidyl-tRNA case. This shows that replacing alanine in position 246, as in MRE strains, with threonine, as in K12 strains, is very deleterious for termination on ribosomes carrying peptidyl-tRNAs with very short peptide sequences, e.g. resulting from minigene expression (Hernández et al., 1997; Hernández-Sánchez et al., 1998; Dinçbas et al., 1999).
Table I. Termination rates (kc) and specificity constants (kcat/Km) for four different RF2 proteins for release of the peptide from the ribosome paused at a UAA stop codon and carrying di-, tri- or tetra-peptidyl-tRNA in the P-site.
| RF2 |
kc (s–1) |
kcat/Km (µM–1 s–1) |
|||||
|---|---|---|---|---|---|---|---|
| Peptide released | fMet-Ile | fMet-Phe-Ile | fMet-Phe-Thr-Ile | fMet-Ile | fMet-Phe-Ile | fMet-Phe-Thr-Ile | |
| (ceAla246) | 5.4 (±0.09) | 6.01 (±0.58) | 6.08 (±0.97) | 20.8 (±3.5) | 25.1 (±0.79) | 21.3 (±3.1) | |
| (ceThr246) | 0.167 (±0.02) | 0.79 (±0.09) | 2.01 (±0.29) | 1.19 (±0.05) | 5.25 (±0.49) | 9.67 (±1.57) | |
| (oeAla246) | 0.46 (±0.05) | 0.73 (±0.05) | 1.26 (±0.099) | 1.48 (±0.19) | 2.54 (±0.26) | 4.42 (±0.61) | |
| (oeThr246) | 0.014 (±0.0004) | 0.055 (±0.006) | 0.16 (±0.02) | 0.096 (±0.008) | 0.38 (±0.06) | 4.03 (±0.22) | |
Fig. 2. Release of fMet-Ile peptide from fMet-Ile-tRNA2Ile on the ribosome in the presence of 1.2 µM RF [filled circles, RF2(ceAla246); open circles, RF2(oeAla246); filled triangles, RF2(ceThr246); diamonds, RF2(oeThr246)] and in the absence of RF (open triangles) (see the insert for the last two). (A) The fraction of the peptide released from the ribosomal termination complex plotted as a function of time. (B) Natural logarithm of the fraction of peptidyl-tRNA remaining on the ribosomal termination complex plotted as a function of time.
When RF2 with Ala246 is overexpressed, producing RF2(oeAla246), the termination efficiency with dipeptidyl- tRNAs drops ∼10-fold for kc and even more for kcat/Km. This difference due to overexpression is reduced to ∼3-fold for kc and 5-fold for kcat/Km for tetrapeptidyl-tRNA. The poorest performance of all the protein variants in Table I is displayed by overexpressed RF2 from the K12 strain, i.e. RF2(oeThr246). In the case of ribosomes carrying dipeptidyl-tRNA in the P-site, termination is almost 400 times slower than with RF2(ceAla246) at saturating factor concentrations (kc; see Figure 2) and ∼200 times slower in the kcat/Km range. As the peptide length increases, the performance of RF2(oeThr246) improves considerably, but kc for the factor remains remarkably small even for the tetrapeptidyl-tRNA.
Suspecting that the loss of termination efficiency due to overexpression of RF2 was caused by deficient post-translational modification(s), we turned to mass spectrometry to study more carefully the protein sequences of the K12 and MRE variants of the protein.
Sequence characterization of the four kinetically different RF2 variants
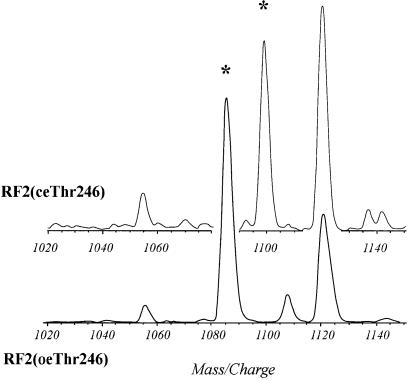
The four RF2 variants described above were characterized by SDS–PAGE, amino acid analysis, N-terminal sequencing and isoelectric focusing (IEF) (Materials and methods). The IEF data show that all protein types have the same isoelectric point (Figure 3); these and other data were in accordance with predictions of the prfB DNA sequences from the W3110 and MRE600 strains. The four protein variants were subjected to tryptic digestion and the digests were analysed by MALDI mass spectrometry (Materials and methods). The spectra for the digests from all four RF2 variants were almost identical, with one clear exception. The m/z values for the 246–256 fragments from the overexpressed RF2(oeAla246) and RF2(oeThr246) variants confirmed the gene sequence predictions for these peptides. However, fragments with the m/z values of the 246–256 peptide were virtually absent in digests obtained from their chromosomally expressed counterparts (Figure 4). These digests instead contained new fragments with m/z values ∼14 units higher than the theoretical values for the 246–256 fragments.

Fig. 3. Isoelectric focusing of six RF2 proteins on a PhastGel IEF4-6.5. All RF2 protein variants focused at the same position on the gel at the end of the run. One sample of E.coli elongation factor G (EF-G) was also applied to the gel as an internal control (uppermost lane).
Fig. 4. MALDI-MS analysis of a tryptic digest of RF2(ceThr246) (upper) overlaid with a section of a MALDI-MS spectrum of a tryptic digest of RF2(oeThr246) (lower). The figure shows the increase in mass of the tryptic peptide 246–256 (marked with an asterisk) from RF2(ceThr246) as compared with the same peptide from RF2(oeThr246).
To characterize these aberrant fragments further, the tryptic peptides from the chromosomally derived RF2s were first separated by reversed phase chromatography. The fractionated aberrant fragments were then identified with MALDI-MS and subjected to amino acid sequence analysis. The sequences agreed with those of the expected 246–256 peptides, except that Gln252 had in each case been changed to a phenylthiohydantoin (PTH) derivative with the retention time of PTH-glutamic acid. Since deamidation of glutamine to glutamic acid leads to an increase in mass of 1 unit, rather than the observed increase in m/z value of ∼14, a substitution of glutamic acid for glutamine at position 252 was not consistent with our data. The two unpredicted fragments from the chromosomally expressed RF2s were therefore analysed further by MS/MS sequence analysis. The m/z in each case was fragmented and the sequence ions showed a sequence corresponding to the theoretical 246–256 peptide, except at position 252, which now had a mass increase of 14 in relation to glutamine (Table II). This strongly suggested that the chromosomally expressed RF2 variants contained methylated Gln252, while the overexpressed variants lacked this post-translational modification. Other experiments show that the corresponding tryptic peptide 229–245 from chromosomally expressed RF1 has the same mass increase of 14 units, compared with the overproduced factor, as the 246–256 RF2 peptide, indicating that Gln235 of the conserved GGQ motif in RF1 is also methylated (results not shown). By analogy with previously described cases, it is likely that Gln252 in RF2 and Gln235 in RF1 are methylated on the amide group (Lhoest and Colson, 1977). To test this prediction, we prepared authentic N5-methylglutamine and applied it to the amino acid sequence analyser. The resulting PTH derivative eluted at the position for PTH-glutamic acid, showing that our sequencing data described above are consistent with an N5-methylated glutamine at position 252 in RF2. The presence of N5-methylglutamine in position 252 was confirmed by repeating the sequence analysis of the tryptic peptide 246–256 from RF2(ceAla246) with the PTH derivatives analysed by reversed phase chromatography on a column optimized for the separation of PTH-glutamic acid from PTH- N5-methylglutamine (see Materials and methods). Under these conditions, authentic PTH-N5-methylglutamine eluted at 12.15 min [with PTH-glycine at 9.92 min and the internal standard dimethylphenylthiourea (DMPTU) at 11.40 min]. The material from cycle 7 of the Edman degradation eluted at 12.18 min (the peak of presumed PTH-N5-methylglutamine), 9.93 min (PTH-glycine from the previous cycle) and 11.38 min (DMPTU), confirming the identity of the N5-methyl-Gln252 in RF2(ceAla246).
Table II. Sequence ions from the MS/MS analysis of the tryptic peptides 246–256 derived from RF2(ceThr246) and RF2(ceAla246).
| Residue No. | RF2(ceThr246) |
RF2(ceAla246) |
|||
|---|---|---|---|---|---|
| m/z | m/z | ||||
| Y1 | 256 | 175.15 | Arg | 175.16 | Arg |
| Y2 | 255 | 289.17 | Asn | 289.20 | Asn |
| Y3 | 254 | 388.25 | Val | 388.27 | Val |
| Y4 | 253 | 525.31 | His | 525.31 | His |
| Y5 | 252 | 667.39 | Gln + 14.02 | 667.37 | Gln + 14.00 |
| Y6 | 251 | 724.41 | Gly | 724.39 | Gly |
| Y7 | 250 | 781.44 | Gly | 781.41 | Gly |
| Y8 | 249 | 852.47 | Ala | 852.44 | Ala |
| Y9 | 248 | 909.50 | Gly | 909.46 | Gly |
| Y10 | 247 | 996.53 | Ser | 996.51 | Ser |
| (M + H)+ | 246 | 1097.59 | Thr | 1067.55 | Ala |
The mass difference between ions Y4 and Y5 is 14 a.m.u. higher than the residual mass of glutamine in both series of sequence ions.
Influence on RF2 activity of a Gln252Glu substitution
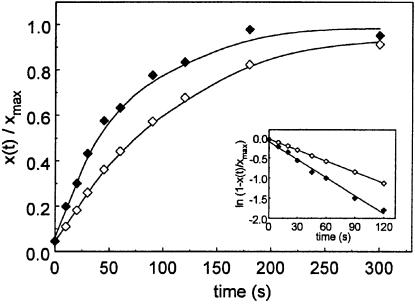
Gln252 in RF2 appears to be universally conserved in all class 1 RFs (Frolova et al., 1999). It has been suggested to play a fundamental role in coordinating the water molecule required for hydrolysis of the ester bond between oligopeptide and tRNA in peptidyl-tRNA that occurs in polypeptide chain termination (Song et al., 2000). To complement the data described above concerning the effect of methylation of Gln252 on the efficiency of termination by RF2, we prepared mutant derivatives of both RF2(Thr246) and RF2(Ala246), in which Gln252 is replaced by glutamic acid. The overproduced proteins were called respectively RF2(oeThr246Glu252) and RF2(oeAla246Glu252) (Materials and methods). The catalytic rate constant (kc) for these two variants was measured (Figure 5). The maximal rate of peptide release from ribosomes carrying dipeptidyl-tRNA was very slow for both mutants [0.010 and 0.016/s for RF2(oeThr246Glu252) and RF2(oeAla246Glu252), respectively], and quite similar to the rate of 0.014/s found for RF2(oeThr246) (see Table I). This implies that the Gln252Glu change reduces the kc of RF2(oeAla246) by ∼30-fold, from 0.46/s to 0.016/s, but has little effect on the already very low efficiency of RF2(oeThr246) (Table I). The effect of the change of glutamine to glutamic acid in the GGQ motif has also been studied in vivo in the case of RF1. The results show that RF1(Glu235) is able to complement a thermosensitive mutation in prfA (Rydén and Isaksson, 1984), showing that this amino acid change does not abolish the activity of the factor, consistent with in vitro observations reported here concerning RF2.
Fig. 5. Hydrolysis of fMet-Ile-tRNA2Ile on the ribosomal complex paused at a UAA stop codon in the presence of 0.8 µM RF. Filled diamonds, RF2(oeAla246Glu252); open diamonds, RF2(oeThr246Glu252). The small insert shows the natural logarithm of the peptidyl-tRNA remaining on the ribosome. The slope gives kc = 0.01/s for RF2(oeThr246Glu252); kc = 0.016/s for RF2(oeAla246Glu252).
Discussion
Functional mapping of class 1 RFs in E.coli has identified two regions thought to mimic vital parts of tRNA. First, there is a GGQ motif also found in class 1 RF from archaebacteria and eukaryotes (Frolova et al., 1999). Secondly, there is a sequence of three amino acids ∼45 residues towards the C-terminus from the GGQ motif that defines the specificity of recognition by the RF similarly to the anticodon of tRNA (Ito et al., 2000). The hypothesis that the GGQ motif mimics the CCA end of tRNA (Frolova et al., 1999) is supported by the recent crystal structure of eukaryotic eRF1 (Song et al., 2000).
We show here that the GGQ motif in E.coli RF2 is modified post-translationally by N5-methylation of the Gln252 residue, and that the modification is functionally important. The amino acid sequence similarity between RF1 and RF2 extends three residues on each side of the GGQ motif, so that these nine residues are conserved between the two proteins. It is therefore not surprising that Gln235 in RF1 also appears to be modified to N5-methylglutamine. This is an unusual modification. Although bacterial proteins (particularly ribosomal proteins) are methylated extensively, we know of only one report of N5-methylglutamine, that of Lhoest and Colson (1977), who demonstrated the presence of this modified amino acid in E.coli ribosomal protein L3. The identity of the gene encoding the L3 methyltransferase is unclear, despite a report describing its mapping (Colson et al., 1979), and there is currently no evidence as to whether the same or a different enzyme may modify RF1 and RF2. Nor do we have any indication of how widespread the modification may be; thus, it is now of considerable interest to study the presence of methylation in class 1 RFs amongst other bacteria and higher organisms.
Frolova et al. (1999) suggested that the GGQ motif may be involved in the hydrolysis of the ester bond in peptidyl-tRNA by interacting directly with the ribosomal peptidyl transferase centre. They offered indirect mutational and functional evidence for this suggestion, which is attractive since it rationalizes the existence of a common RF motif among all organisms that mimics the universally conserved CCA terminus of tRNA. However, the effects of mutating this motif are open to more than one interpretation. One reason for this is that large differences in the catalytic rate constant, kc, of an RF do not necessarily imply that structural perturbations causing these changes must occur in the vicinity of the peptidyl transferase. For instance, it was found by Freistroffer et al. (2000) that kc for both RF1 and RF2 can be reduced as much as 1000-fold when a single base of a stop codon exposed in the ribosomal A-site is mutated to give a sense codon. Although these types of changes were in the mRNA, it can be inferred that variations in protein sequence very remote from the ribosomal peptidyl transferase can also impair not only RF binding but also the catalytic rate constant (kc). Similar findings have been reported for enzymatic binding of aminoacyl-tRNA in complex with EF-Tu and GTP to the ribosomal A-site when there is a codon–anticodon mismatch (Pape et al., 1999). In such cases, the rate of GTP hydrolysis on EF-Tu bound to the ribosome may be reduced dramatically in relation to cases with matching codon–anticodon interactions.
Guided by their crystal structure of human eRF1, Song et al. (2000) propose that Gln252 participates in orientating a water molecule for the hydrolysis of peptidyl-tRNA by the ribosomal peptidyl transferase centre. They suggest that both the carbonyl oxygen and the amide nitrogen of the Gln252 side chain act as proton acceptors. However, a hydrogen bond involving the amide nitrogen as acceptor must be extremely weak, if at all possible. In terms of this model, it is therefore difficult to predict the effects of methylating Gln252 or of substituting glutamine for glutamic acid, or to relate the possible effects to our experimentally obtained data (Table I).
Amino acid residue 246 in RF2 is very close to the GGQ motif, four positions towards the N-terminus. In the sequences of all 37 prokaryotic or mitochondrial class 1 RFs currently available in the data banks, this residue is an alanine or a serine, with the single exception of E.coli K12 RF2, where a threonine is found. We show that this exception is not true of E.coli in general, as two E.coli B strains we have sequenced both show alanine at position 246. We suggest that the prfB gene in a common ancestor of the current K12, B and MRE600 strains coded for alanine at this position, as in most other prokaryotic class 1 RFs. Indeed, it cannot be excluded that the Ala246Thr mutation arose as a result of the heavy mutagenesis to which early laboratory K12 strains of E.coli were often subjected (Bachmann, 1987). Here we considerably extend the findings of Uno et al. (1996) who first pointed to the functional importance of this residue in RF2 as critical in explaining the low activity of E.coli RF2 for termination at both UGA and UAA codons in vivo as compared with RF2 in S.typhimurium. This low activity is even more marked in the case of peptidyl-tRNA with very short peptide chains. Both the substitution of Thr246 in place of alanine and the lack of Gln252 N5-methylation introduce a marked dependence on peptide length for the efficiency of termination mediated by RF2. A length dependence in the case of E.coli K12 RF2, in contrast to RF1 from the same strain, was noted previously by Heurgué-Hamard et al. (2000). The effects of residue 246 and of methylation are cumulative; thus, the methylation of glutamine in position 252 increases the termination efficiency of RF2 irrespective of the identity of the amino acid at position 246. Previous findings that overexpressed RF2 has much lower termination activity in simplified in vitro assays than the chromosomally expressed factor (Pavlov et al., 1998) can now be accounted for as resulting from a lack of modification of Gln252.
Finally, the observation that the negative effects on termination efficiency of the substitution of Thr246 in place of alanine and the lack of Gln252 N5-methylation are largely cumulative offers an explanation for the fact that overproduction of E.coli K12 RF2 is lethal to the cell, whereas overproduction of the Ala246 RF2 mutant or the S.typhimurium factor is not (Uno et al., 1996). Overproduction of RF2 appears to saturate the methyltransferase that modifies Gln252 to the extent that no modification of the overproduced protein is visible by MALDI-MS (Figure 4). Thus, whereas the cell can support the decreased termination efficiency associated with one or other defect alone, we propose that the combined effect inhibits termination to an extent incompatible with cell growth.
Materials and methods
Expression vectors for RF1 and RF2
The genes for RF1 and RF2 were cloned into the expression vector pET11a (Stratagene) after amplification by PCR. Pairs of oligonucleotides were used to introduce NdeI and BamHI sites at the initiation codons and just after the termination codons of the genes (for RF1, GCAATCCATATGTTTGAAATTAATCC and TGTTGGATCCTCATAACCCTG, respectively; and for RF2, GCATTTACGCCATATGAAGC and ATATGGATCCATT ATTCC, respectively). PCR was performed on DNA extracted from K12 strain Xac (Coulondre and Miller, 1977), and the product was digested with NdeI and BamHI prior to ligation between the same sites in plasmid pET11a. The frameshift site in prfB (Craigen and Caskey, 1986) was removed and the Thr246Ala change was introduced by a method employing two PCR steps (Good and Nazar, 1992). All cloned PCR products were verified by sequencing on both strands by the dideoxy method of Sanger et al. (1977).
DNA sequence analysis of prfB in E.coli K12 and E.coli B strains
DNA was prepared from the E.coli K12 strains Xac (Coulondre and Miller, 1977) and W3110 (Bachmann, 1987), E.coli B strain BL21 (Studier and Moffatt, 1986) and MRE600 as described by Gay (1984). The prfC region was amplified by PCR and sequenced on both strands by the dideoxy method (Sanger et al., 1977).
Purification of release factors
Chromosomally expressed RF2 was purified from E.coli MRE600 or W3110 cells that were grown in TY2 medium supplemented with glucose and harvested in late log phase. A 280 g aliquot of frozen cell paste was thawed and suspended in buffer A [40 mM Tris–HCl pH 7.5, 10 mM MgCl2, 100 mM KCl, 1 mM dithiothreitol (DTT), 100 µM phenylmethylsulfonyl fluoride (PMSF)]. DNase I was added to the homogenate (1 µg/ml) and cells were disrupted with a French press. Cell debris was removed by centrifugation at 13 000 g for 2 h and applied to a DEAE–Sepharose CL6B column (5 × 12 cm, Amersham Pharmacia Biotech). Protein was eluted with a 2 l linear KCl gradient (100–500 mM) in the same buffer. The fractions were tested for termination activity on release complexes with peptidyl-tRNA in the ribosomal P-site and UGA in the A-site (Freistroffer et al., 1997; Dinçbas et al., 1999). Fractions with RF2 activity were pooled, precipitated with ammonium sulfate (0.35 g/ml) and centrifuged in a GS-3 rotor at 8000 r.p.m. for 40 min. The pellet was dissolved in a minimum volume of buffer B (40 mM Tris–HCl pH 7.5, 10 mM MgCl2, 80 mM KCl, 1 mM DTT, 100 µM PMSF) and applied to a gel filtration column (AcA44 from Sepracor, 3 × 200 cm) equilibrated in the same buffer. Fractions were tested for RF2 activity in termination as above and analysed for purity by 10% SDS–PAGE. Fractions containing RF2 were pooled and precipitated with ammonium sulfate (0.35 g/ml). The pellet was dissolved in a minimal volume of buffer C (50 mM potassium phosphate pH 7.2, 1 mM DTT, 100 µM PMSF) and applied to a DEAE–cellulose column (DE-52, 2 × 12 cm, Whatman) equilibrated in the same buffer. Protein was eluted with a 500 ml linear gradient of potassium phosphate (50 mM potassium phosphate pH 7.2 to 250 mM potassium phosphate pH 6.7) in buffer C. Fractions containing RF2 were identified by SDS–PAGE, precipitated with ammonium sulfate and resubjected to gel filtration on AcA44 as above. Following this last chromatographic step, the protein was 99% pure according to SDS–PAGE. Proteins were stored in polymix buffer at –80°C. Purification of overproduced RF2(oeThr246), RF2(oeAla246), RF2(oeThr246Glu252) and RF2(oeAla246Glu252) proteins was performed as described before (Pavlov et al., 1997).
IEF
IEF was performed with a PhastSystem (Pharmacia, Sweden), using PhastGel IEF media with a pH range 4–6.5 to separate the proteins. After pre-focusing for 75 Vh, 1 µl of each sample (1.5 µg protein/µl) was applied to the gel at 200 V. Focusing was run at 5 mA and 3.5 W for 800 Vh. Coomassie Blue staining was used to detect the proteins.
Primary structure analysis
MALDI-MS analysis was performed with a Kompakt IV spectrometer (Kratos, UK) and MS/MS analysis was done at the department of Plant Biology, Swedish University of Agricultural Sciences, Uppsala, Sweden with the use of a Q-Tof instrument (Micromass, UK). The amino acid sequence was analysed with an ABI 477 A system (PE-Biosystems). A final purification of RF2 prior to tryptic digestion was done by gel filtration on a Superdex 75 column eluted with 0.15 M ammonium bicarbonate using a SMART HPLC system (Amersham Pharmacia Biotech, Sweden). Tryptic digestion of RF2 was done in 0.15 M ammonium bicarbonate with modified trypsin (Promega) at an enzyme:substrate ratio of 1:50 for 15 h at 37°C. Tryptic peptides were separated using a C2/C18 RPC column (Amersham Pharmacia Biotech) eluted with a gradient of acetonitrile in 0.1% trifluoroacetic acid.
N5-methylglutamine was prepared by reacting pyroglutamic acid with methylamine in water at 65°C for several days as described by Lichtenstein (1942) but omitting the purification steps. The derivative was checked by amino acid analysis on an ion-exchange ninhydrin system. The derivative eluted at a position after glutamine.
The normal PTH analysis system used for protein sequencing failed to resolve the PTH derivatives of glutamic acid and N5-methylglutamine. These were therefore analysed on a Nova Pack column (2.1 × 200 mm, Waters) mounted on the PTH analyser and eluted with a gradient of acetonitrile and a buffer composed of 0.1% pre-mix buffer solution (PE-ABI) in water (Klotz et al., 1990). The elution was optimized for separation of PTH-glutamic acid from PTH-N5-methylglutamine and DMPTU. PTH-N5-methylglutamic acid eluted prior to DMPTU, and PTH-N5-methylglutamine eluted following DMPTU. The separation of other PTH derivatives was not complete under these conditions of gradient and temperature.
Amino acid composition analysis was performed at the Center for Amino Acid Analysis at the Department of Biochemistry, University of Uppsala using established procedures.
In vitro assays
GTP and ATP were from Pharmacia, Sweden. Putrescine, spermidine, phosphoenolpyruvate (PEP) and myokinase (MK) were from Sigma (St Louis). Pyruvate kinase (PK) was from Boehringer-Mannheim. [3H]methionine was from Du-Pont NEN. Polymix buffer contained 5 mM magnesium acetate, 5 mM ammonium chloride, 95 mM potassium chloride, 0.5 mM calcium chloride, 8 mM putrescine, 1 mM spermidine, 5 mM potassium phosphate and 1 mM DTT (Jelenc and Kurland, 1979).
Preparation of mRNAs 02.MI2.Oa, 02.MFI2.Oa and 02.MFTI2.Oa with ORF sequences, AUGAUA, AUGUUCAUA and AUGUUCACGAUA, respectively, was done as described before (Dinçbas et al., 1999). Bacterial elongation factors EF-Tu, EF-Ts and EF-G, bulk tRNA and Phe-tRNA synthetase were purified from MRE600 cells according to Ehrenberg et al. (1990). Thr-tRNA synthetase was purified according to Brunel et al. (1993). Initiation factors IF1, IF2 and IF3 were purified from overproducing strains according to Soffientini et al. (1994). Ribosomes were prepared from E.coli 017 cells according to Jelenc (1980). Preparations of Ile-tRNA synthetase and f[3H]Met-tRNAMet were according to Freistroffer et al. (1997). Ribosomal release complexes were prepared with ribosomes programmed with 02.MI2.Oa, 02.MFI2.Oa and 02.MFTI2.Oa mRNAs as described before (Dinçbas et al., 1999).
Measurement of kc. The catalytic rate constant (kc) of hydrolysis of peptidyl-tRNAs in ribosomal complexes paused at the UAA stop codon was measured as single RF2 cycle experiments at 37°C. A factor mix containing (in 50 µl) 0.1 µmol ATP, 1 µmol PEP, 0.1 µmol GTP, 5 µg PK, 0.3 µg MK and 120 pmol RF2 was prepared in polymix buffer. RF2(oeThr246)-specific termination reactions for ribosomal complexes programmed with 02.MI2.Oa and 02.MFI2.Oa mRNAs were done by mixing equal volumes of factor mix and ribosomal release complex in polymix buffer. The extent of hydrolysis of the peptidyl-tRNAs was analysed by withdrawing aliquots at different time points for quenching with an equal volume of 40% ice-cold formic acid. All other single-cycle termination rates were measured with a quench-flow device (Chemical-Quench-Flow Model RQF-3, KinTek Corp.) Reactions (40 µl) were quenched with 90 µl of 30% formic acid. After centrifugation at 22 000 g in an Eppendorf centrifuge for 12 min at 4°C, the radioactivity in the supernatant was counted in Aquasafe 300 plus (Zinsser).
Measurement of kcat/Km. kcat/Km values of RFs for different ribosomal release complexes were measured by varying the RF concentration (0.02–0.7 µM) and keeping the release complex concentration fixed (0.05 µM). Reactions were stopped by adding 20% formic acid at different times that gave hydrolysis of ∼30% of the total peptidyl-tRNA. The quench-flow apparatus was used when the reactions were fast. Eadie–Hofstee plots were used to estimate kcat/Km values. The concentrations of active RFs were obtained by active site titrations (Pavlov et al., 1998).
kcat/Km was estimated also by measuring the extent of hydrolysis of peptidyl-tRNA at a ribosome concentration of 3 × 10–3 µM and RF concentrations of 1 × 10–4 µM (Dinçbas et al., 1999). Under these conditions, the rate of termination is far below kcat, and the free active RF concentration is approximately equal to the total. RF3 (0.1 µM) was added to the reaction to minimize sequestering of RFs by ribosomes (Freistroffer et al., 1997). The two methods gave identical estimates for kcat/Km values.
Relationships between parameters obtained in single and multiple cycle experiments. The Michaelis–Menten parameter kcat/Km is defined as the association rate constant (ka) for factor binding to the ribosome multiplied by the probability that a binding event results in hydrolysis of peptidyl-tRNA. If the dissociation of the factor from the ribosome before hydrolysis is governed by the rate constant kd, then kcat/Km is related to kc, ka and kd through kcat/Km = ka/(1 + kd/kc). In experiments where RFs are present in catalytic amounts and recycle many times (Freistroffer et al., 1997), kcat/Km has the same meaning as here and kcat = kckdiss/(kc + kdiss), where kdiss is the rate constant for factor dissociation after hydrolysis of peptidyl-tRNA.
Acknowledgments
Acknowledgements
We are very grateful to Dr Bo Ek at the Department of Plant Biology, Swedish University of Agricultural Sciences, Uppsala for MS/MS analysis, and the Amino Acid Laboratory at the Department of Biochemistry, Uppsala University for the amino acid analysis. This work was supported by the Swedish Research Council for Engineering Sciences, the Swedish Natural Science Research Council, the Knut and Alice Wallenberg Foundation (WCN2000-UU/SLU-003) and the Centre National pour la Recherche Scientifique (UPR9073).
References
- Adamski F.M., McCaughan,K.K., Jørgensen,F., Kurland,C.G. and Tate,W.P. (1994) The concentration of polypeptide chain release factors 1 and 2 at different growth rates of Escherichia coli. J. Mol. Biol., 238, 302–308. [DOI] [PubMed] [Google Scholar]
- Bachmann B.J. (1987) Derivations and genotypes of some mutant derivatives of Escherichia coli K12. In Neidhardt,F.C., Ingraham,J.L., Brooks Low,K., Magasanik,B., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. American Society for Microbiology, Washington, DC, Vol. 2, pp. 1190–1219. [Google Scholar]
- Brunel C., Romby,P., Moine,H., Caillet,J., Grunberg-Manago,M., Springer,M., Ehresmann,B. and Ehresmann,C. (1993) Translational regulation of the Escherichia coli threonyl-tRNA synthetase gene: structural and functional importance of the thrS operator domains. Biochimie, 75, 1167–1179. [DOI] [PubMed] [Google Scholar]
- Buckingham R.H., Grentzmann,G. and Kisselev,L. (1997) Polypeptide chain release factors. Mol. Microbiol., 24, 449–456. [DOI] [PubMed] [Google Scholar]
- Caskey C.T., Beaudet,A.L., Scolnick,E.M. and Rosman,M. (1971) Peptidyl transferase hydrolysis of fMet-tRNA. Proc. Natl Acad. Sci. USA, 68, 3163–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caskey C.T., Forrester,W.C., Tate,W.P. and Ward,C.D. (1984) Cloning of the Escherichia coli release factor 2 gene. J. Bacteriol., 158, 365–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson C., Lhoest,J. and Urlings,C. (1979) Genetics of ribosomal protein methylation in Escherichia coli. III. Map position of two genes, prmA and prmB, governing methylation of proteins L11 and L3. Mol. Gen. Genet., 169, 245–250. [DOI] [PubMed] [Google Scholar]
- Coulondre C. and Miller,J.H. (1977) Genetic studies of the lac repressor III: additional correlation of mutational sites with specific amino acid residues. J. Mol. Biol., 117, 525–575. [DOI] [PubMed] [Google Scholar]
- Craigen W.J. and Caskey,C.T. (1986) Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature, 322, 273–275. [DOI] [PubMed] [Google Scholar]
- Craigen W.J., Cook,R.G., Tate,W.P. and Caskey,C.T. (1985) Bacterial peptide chain release factors: conserved primary structure and possible frameshifting regulation of release factor 2. Proc. Natl Acad. Sci. USA, 82, 3616–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinçbas V., Heurgué-Hamard,V., Buckingham,R.H., Karimi,R. and Ehrenberg,M. (1999) Shutdown in protein synthesis due to the expression of mini-genes in bacteria. J. Mol. Biol., 291, 745–759. [DOI] [PubMed] [Google Scholar]
- Ehrenberg M., Bilgin,N. and Kurland,C.G. (1990) Design and use of a fast and accurate in vitro translation system. In Spedding,G. (ed.), Ribosomes and Protein Synthesis. IRL Press at Oxford University Press, Oxford, UK, pp. 101–129. [Google Scholar]
- Elliott T. and Wang,X. (1991) Salmonella typhimurium prfA mutants defective in release factor 1. J. Bacteriol., 173, 4144–4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freistroffer D.V., Pavlov,M.Y., MacDougall,J., Buckingham,R.H. and Ehrenberg,M. (1997) Release factor RF3 in E.coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP dependent manner. EMBO J., 16, 4126–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freistroffer D.V., Kwiatkowski,M., Buckingham,R.H. and Ehrenberg,M. (2000) The accuracy of codon recognition by ribosome release factors. Proc. Natl Acad. Sci. USA, 97, 2046–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L. et al. (1994) A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature, 372, 701–703. [DOI] [PubMed] [Google Scholar]
- Frolova L.Y., Tsivkovskii,R.Y., Sivolobova,G.F., Oparina,N.Y., Serpinski,O.I., Blinov,V.M., Tatkov,S.I. and Kisselev,L.L. (1999) Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA, 5, 1014–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay N.J. (1984) Construction and characterization of an Escherichia coli strain with a uncI mutation. J. Bacteriol., 158, 820–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good L. and Nazar,R.N. (1992) An improved thermal cycle for 2-step PCR-based targeted mutagenesis. Nucleic Acids Res., 20, 4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández J., Ontiveros,C., Valadez,C., Buckingham,R.H. and Guarneros,G. (1997) Regulation of protein synthesis by minigene expression. Biochimie, 79, 527–531. [DOI] [PubMed] [Google Scholar]
- Hernández-Sánchez J., Valadez,J.G., Herrera,J.V., Ontiveros,C. and Guarneros,G. (1998) λ bar minigene-mediated inhibition of protein synthesis involves accumulation of peptidyl-tRNA and starvation for tRNA. EMBO J., 17, 3758–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurgué-Hamard V., Dinçbas,V., Buckingham,R.H. and Ehrenberg,M. (2000) Origins of minigene-dependent growth inhibition in bacterial cells. EMBO J., 19, 2710–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Uno,M. and Nakamura,Y. (2000) A tripepeptide anticodon deciphers stop codons in messenger RNA. Nature, 403, 680–684. [DOI] [PubMed] [Google Scholar]
- Jelenc P.C. (1980) Rapid purification of highly active ribosomes from Escherichia coli. Anal. Biochem., 105, 369–374. [DOI] [PubMed] [Google Scholar]
- Jelenc P.C. and Kurland,C.G. (1979) Nucleotide triphosphate regeneration decreases the frequency of translation errors. Proc. Natl Acad. Sci. USA, 76, 3174–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K. and Nakamura,Y. (1990) Autogenous suppression of an opal mutation in the gene encoding peptide chain release factor-2. Proc. Natl Acad. Sci. USA, 87, 8432–8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K., Inada,T. and Nakamura,Y. (1988) Conditionally lethal and recessive UGA-suppressor mutations in the prfB gene encoding peptide chain release factor 2 of Escherichia coli. J. Bacteriol., 170, 5378–5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz A.V., Thomas,B.A., Glazer,A.N. and Blacher,R.W. (1990) Detection of methylated asparagine and glutamine residues in polypeptides. Anal. Biochem., 186, 95–100. [DOI] [PubMed] [Google Scholar]
- Lhoest J. and Colson,C. (1977) Genetics of ribosomal protein methylation in Escherichia coli. II. A mutant lacking a new type of methylated amino acid, N5-methylglutamine, in protein L3. Mol. Gen. Genet., 154, 175–180. [DOI] [PubMed] [Google Scholar]
- Lichtenstein N. (1942) Preparation of γ-alkylamides of glutamic acid. J. Am. Chem. Soc., 64, 1021–1022. [Google Scholar]
- Mikuni O., Kawakami,K. and Nakamura,Y. (1991) Sequence and functional analysis of mutations in the gene encoding peptide-chain-release factor 2 of Escherichia coli. Biochimie, 73, 1509–1516. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Kawakami,K. and Mikuni,O. (1990) Alternative translation and functional diversity of release factor 2 and lysyl-tRNA synthetase. In McCarthy,J.E.G. and Tuite,M.F. (eds), Post-Transcriptional Control of Gene Expression. Springer-Verlag, Berlin, pp. 455–464. [Google Scholar]
- Pape T., Wintermeyer,W. and Rodnina,M. (1999) Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J., 18, 3800–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov M.Y., Freistroffer,D., MacDougall,J., Buckingham,R.H. and Ehrenberg,M. (1997) Fast recycling of E.coli ribosomes requires both ribosome recycling factor (RRF) and release factor RF3. EMBO J., 16, 4134–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov M.Y., Freistroffer,D.V., Dinçbas,V., MacDougall,J., Buckingham,R.H. and Ehrenberg,M. (1998) A direct estimation of the context effect on the efficiency of termination. J. Mol. Biol., 284, 579–590. [DOI] [PubMed] [Google Scholar]
- Rydén S.M. and Isaksson,L.A. (1984) A temperature-sensitive mutant of Escherichia coli that shows enhanced misreading of UAG/A and increased efficiency for some tRNA suppressors. Mol. Gen. Genet., 193, 38–45. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen,S. and Coulson,A.R. (1977) DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA, 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolnick E.M., Tompkins,R., Caskey,C.T. and Nirenberg,M. (1968) Release factors differing in specificity for terminator codons. Proc. Natl Acad. Sci. USA, 61, 768–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soffientini A., Lorenzetti,R., Gastado,L., Spurio,R., La Teana,A. and Khalid,I. (1994) Purification procedure for bacterial initiation factors IF1 and IF2. Expr. Purif., 5, 118–124. [DOI] [PubMed] [Google Scholar]
- Song H., Mugnier,P., Das,A.K., Webb,H.M., Evans,D.R., Tuite,M.F., Hemmings,B.A. and Barford,D. (2000) The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell, 100, 311–321. [DOI] [PubMed] [Google Scholar]
- Studier F.W. and Moffatt,B.A. (1986) Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol., 189, 113–130. [DOI] [PubMed] [Google Scholar]
- Tate W.P. et al. (1993) Translational stop signal: evolution, decoding for protein synthesis and recoding for alternative events. In Nierhaus,K.H., Franceshi,F., Subramanian,A.R., Erdmann,V.A. and Wittmann-Liebold,B. (eds), The Translational Apparatus: Structure, Function, Regulation, Evolution. Plenum Press, New York, NY, pp. 253–261. [Google Scholar]
- Uno M., Ito,K. and Nakamura,Y. (1996) Functional specificity of amino acid at position 246 in the tRNA mimicry domain of bacterial release factor 2. Biochimie, 78, 935–944. [DOI] [PubMed] [Google Scholar]
- Weiss R.B., Murphy,J.P. and Gallant,J.A. (1984) Genetic screen for cloned release factor genes. J. Bacteriol., 158, 362–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. and Cech,T.R. (1997) Peptide bond formation by in vitro selected ribozymes. Nature, 390, 96–100. [DOI] [PubMed] [Google Scholar]