Abstract
Mononuclear iron-containing oxygenases conduct a diverse variety of oxidation functions in biology1,2, including the oxidative demethylation of methylated nucleic acids and histones3,4. E. coli AlkB is the first such enzyme that was discovered to repair methylated nucleic acids (Fig. 1)5,6, which are otherwise cytotoxic and/or mutagenic. AlkB human homologues are known to play pivotal roles in various processes7–11. Presented here is the first structural characterization of oxidation intermediates for these demethylases. Employing a chemical cross-linking strategy12,13, complexes of AlkB-dsDNA containing 1,N6-etheno adenine (εA), N3-methyl thymine (3-meT), and N3-methyl cytosine (3-meC) were stabilized and crystallized, respectively. Exposing these crystals, grown under anaerobic conditions containing iron(II) and α-ketoglutarate (αKG), to dioxygen initiates oxidation in crystallo (Supplementary Fig. 1). A glycol (from εA) and a hemiaminal (from 3-meT) intermediates are captured; a zwitterionic intermediate (from 3-2 meC) is also proposed, based on crystallographic observations and computational analysis. The observation of these unprecedented intermediates provides direct support for the oxidative demethylation mechanism for these demethylases. This study also depicts a general mechanistic view of how a methyl group is oxidatively removed from different biological substrates.
E. coli AlkB is a prototype of iron-containing dioxygenases that catalyze oxidative demethylation of nucleic acids and histones3,4. Its human homologue ABH1 (also known as ALKBH1) demethylates 3-meC in DNA and RNA7, ABH2 (ALKBH2) guards the mammalian genome against N1-methyl adenine (1-meA) damage8, ABH3 (ALKBH3) may repair methylated RNA damage9,10, and FTO, which demethylates 3-meT and N3-methyl uracil (3-meU)14,15, is a key factor in regulating energy homeostasis and obesity11. In addition, within this iron(II)/α-ketoglutarate(αKG)-dependent subfamily, the JHDM proteins are engaged in the human epigenetic regulation by catalyzing the oxidative demethylation of methylated histones4. AlkB is quite versatile in that it can recognize and process three different types of base damages: (1) 3-meC and 1-meA, which formally bear a positive charge on the damaged nitrogen under physiological pH and are the most efficient substrates for AlkB5,6; (2) neutral 3-meT and N1-methylated guanine (1-meG)16–18; and (3) cyclic adducts such as εA and 3,N4-ethenocytosine (Supplementary Fig. 2)19,20. Using a previously developed disulphide cross-linking approach12,13, we crystallized AlkB-dsDNA complexes containing 3-meC, 3-meT, and εA, and solved the structures to high resolutions (Supplementary Table 1 and Supplementary Fig. 3). In these structures, the catalytically essential iron(II) was replaced by manganese(II), which occupies the same binding site, but doesn’t support catalysis21,22.
Comparisons of these structures, including a previously published 1-meA-containing structure (pdb code: 3BIE), provide hints on how AlkB recognizes a diverse range of substrates with different hydrogen-bonding capacities and base-dimensions. Of the four damaged bases, only one direct hydrogen bond is observed, which is between the side chain of Asp 135 and the exocyclic amine of 1-meA or 3-meC (Supplementary Fig. 4). Mutation of this residue yielded mutant proteins (Asp135Ala, Asp135Ser, or Asp135Asn) that display ~5–15% wild-type activity towards 1-meA at pH=7.0 (Supplementary Fig. 5)23. Natural bond orbital (NBO) charge analysis suggests that the exocyclic amine bears a large portion of the delocalized positive charge for 1-meA and 3-meC (Supplementary Fig. 6), which agrees with the observation that charged lesions in category (1) are preferentially recognized by AlkB. For the neutral lesions (3-meT and εA) that instead possess a hydrogen bond acceptor at the equivalent position, such an interaction is replaced by water-mediated hydrogen bonds (Supplementary Fig. 4). On the contrary, 3-meT is a preferred substrate of FTO, and a hydrogen-bonding interaction between O4 of 3-meT and the amide nitrogen of Glu 234 has been observed in the crystal structure of FTO24; the disruption of this hydrogen bond results in the loss of the demethylation activity of FTO towards 3-meT. In the εA-containing structure, the cyclic adduct portion stacks against the side chain of Asp 133, which perhaps contributes to extra binding affinity of the lesioned base to the active site as compared to the repair product adenine (Supplementary Fig. 4a). On the other hand, substrates similar in size overlap well when bound in the active site, regardless of their hydrogen-bonding capacities (Supplementary Fig. 7). The final positions of a damaged purine and pyrimidine are consistently different, and are adjusted by AlkB so that the aberrant alkyl group is positioned towards the metal site for efficient catalysis (Supplementary Fig. 8 and Supplementary Note).
With several substrates covalently locked in the active site of AlkB, we set out to perform oxidation reactions in single crystals with a catalytically active iron centre (Supplementary Fig. 1). Performing enzymatic reactions in protein single crystals has been shown to be very effective in trapping and identifying unstable intermediates as well as elucidating reaction mechanisms25–27. The enzyme active sites provide nanoscale reaction vessels that allow isolation of otherwise transient intermediates.
Covalently linked AlkB-dsDNA complexes were crystallized with cofactors iron(II) and αKG under anaerobic conditions. Then, these crystals were exposed to air to initiate oxidation in crystallo (Supplementary Fig. 1). With the εA-containing DNA a (S,S)-22,23-glycol intermediate 1 was captured (Fig. 2a and Supplementary Fig. 9a). The structure shows two newly formed hydroxyl groups that are involved in multiple hydrogen bonds: O26 forms a water-mediated interaction with Glu 136, and O24 hydrogen bonds directly to Arg 210 and succinate derived from αKG. The N6 atom of 1 forms indirect hydrogen bonds with the side chain of Asp 135 and the backbone amide of Lys 134, as also observed in the εA structure. Since O26 points away from the metal while O24 is within coordination distance to the iron (2.1 Å), we assigned O24 as the oxygen atom derived from O2 oxidation of εA to yield the epoxide intermediate, which then undergoes ring-opening by a water molecule attacking at C23 to afford the trans-glycol 1 (Fig. 2a and Supplementary Fig. 10a). The observation of this glycol intermediate firmly confirms the epoxidation repair mechanism of εA by AlkB. Subsequent protonation at N6 of 1 is anticipated to break N1-C22 and N6-C23 bonds and eventually produce an intact adenine base (Supplementary Fig. 10a). Indeed, species with molecular weight corresponding to epoxide and glycol 1 were observed by mass spectrometry analysis20. Despite considerable efforts, we have yet to capture the epoxide intermediate in these crystals; the epoxide may not be as stable as 1 inside the enzymatic pocket of AlkB.
Figure 2. Intermediates trapped during in crystallo oxidation of εA and 3-meT.
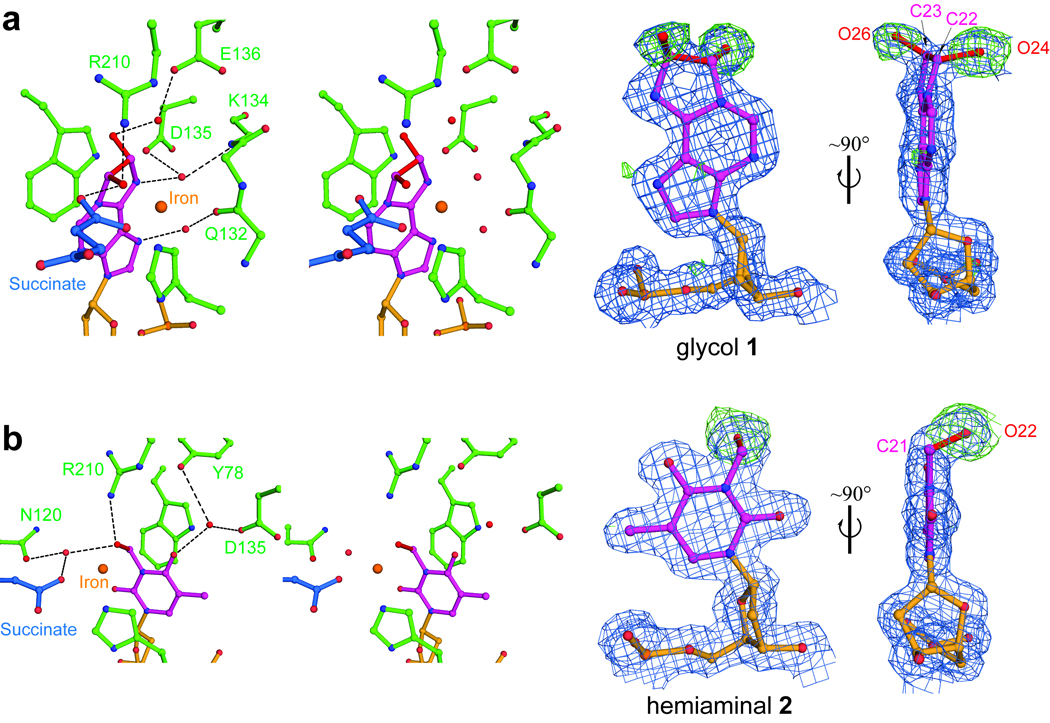
a, Stereo views and electron density maps of the glycol intermediate 1 during εA repair. b, Stereo pairs and density maps of the hemiaminal intermediate 2 during the oxidation demethylation of 3-meT. The blue 2Fobs-Fcal maps are contoured at 1.0σ and the green Fobs-Fcal simulated-annealing omit maps were computed by removing extra atoms of intermediates (compared to the original substrates) and are contoured at 3.0σ. The extra atoms are shown in red. Hydrogen bonds are shown as dotted lines.
When 3-meT-containing crystals were exposed to air, the aberrant methyl group was oxidized to a hydroxymethyl group, which points towards the metal (3.6 Å) and hydrogen-bonds to Arg 210, Asn 120 and succinate (Fig. 2b and Supplementary Fig. 9b). To our knowledge, this hemiaminal intermediate 2 is the first species of this type ever being observed in oxidative demethylation in biology. The mono-dentate succinate shifts away from the newly formed hydroxymethyl group in this hemiaminal structure compared to the glycol structure, which may reflect different recognition of these intermediates in the enzymatic pocket (Supplementary Fig. 11).
We next studied in crystallo oxidation of 3-meC, a preferred substrate of AlkB. Exposing crystals that contain 3-meC with air led to observation of extra electron density on the aberrant methyl group (Supplementary Fig. 12). The best fitting model was obtained after releasing certain geometry restraints on angles and bond lengths of the hydroxymethyl group in the 3-hydroxymethylcytosine (3-hmC) dictionary file during refinement (Supplementary Table 2). In this model 3, C20-O21 distance was refined to ~1.2 Å, which is shorter than a typical C-O single bond (~1.4 Å) (Supplementary Fig. 13a). While protein crystallography at this resolution cannot conclusively assign the exact structure of 3 (Supplementary Fig. 13b), we chemically synthesized a 3-meC analogue, 3-deaza-3-methyl cytosine (3-deazameC), to further investigate the 3-meC demethylation process (Supplementary Fig. 14). Crystals of AlkB-dsDNA containing 3-deazameC were also exposed to air, and in the resulting structure, an oxidized 3-deaza-3-hydroxymethyl cytosine 4 (3-deazahmC) was unambiguously observed (Fig. 3a and Supplementary Fig. 15). Due to C3 substitution, 3-deazameC is neutral and its oxidized product 4 is a stable alcohol. We also demonstrate that 3-deazameC can be converted to 4 by AlkB in vitro, although with a lower activity compared to 3-meC (Supplementary Fig. 16), as expected chemically.
Figure 3. A zwitterionic intermediate 3 is proposed for the demethylation of 3-meC.

a, Oxidation of 3-deazameC in single crystals to yield 3-deazahmC, intermediate 4. The density map and labels are generated and shown as in Fig. 2. b, QM/MM calculated structure of 4. c, Optimized structure of model 3 from the oxidized 3-meC crystal. For both calculated structures, carbon atoms are colored in cyan, nitrogen in blue, oxygen in red, iron in pink, and hydrogen in grey. Red NBO charges are labelled for several base atoms and key distances are marked in black (Å).
We then subjected the crystal structure containing 4 to combined quantum mechanical/molecular mechanical (QM/MM) minimizations (Supplementary Note). With a hydroxide ligand to iron(II) and a hydroxymethyl state of 4, the optimized structure is stable and overlaps well with the crystal structure (Fig. 3b and Supplementary Fig. 17). Replacing the C3 carbon in the optimized 3-deazahmC structure to a nitrogen atom causes spontaneous proton transfer from the hydroxymethyl group of 4 to the hydroxide ligand, resulting in an iron(II)-H2O state and a zwitterionic structure 5 (Supplementary Fig. 18a). In addition, an independent QM/MM minimization starting from the crystal structure with oxidized 3-meC 3 also leads to an iron(II)-H2O state, and the optimized structure agrees with the crystal structure (Fig. 3c and Supplementary Fig. 18b). NBO analysis of the optimized structure containing 3 indicates that O21 bears a large negative partial charge, making the hemiaminal 3 actually a zwitterion (Fig. 3c), which is virtually the same as the computed 5. Moreover, starting from the optimized model of 3, replacing N3 with a carbon atom yields a converged structure that is very similar to the optimized structure containing 3-deazahmC (4), with the key features—iron(II)-OH− and the hydroxymethyl state—reproduced (Supplementary Figs 18c and 19). This pKa difference reflects the intrinsic chemical property of 3-meC, in which the cytosine ring bears a formal positive charge (Supplementary Fig. 20). Taken together, the evidence indicates that such a zwitterionic hemiaminal 3, which explains the shortened C20-O21− distance, is very likely an intermediate of 3-meC oxidative demethylation.
E coli. AlkB and its human homologues ABH2 and ABH3 prefer positively charged 1-meA and 3-meC to neutral 3-meT and 1-meG as substrates. With neutral 3-meT, the intermediate 2 is a relatively “stable” hemiaminal as compared with that derived from 3-meC (Supplementary Fig. 10). Protonation at the O4 atom of 2 initiates bond migration and decomposition of 2 to liberate formaldehyde and yield the final intact thymine base (Supplementary Fig. 10b). In contrast, hydroxylation of 3-meC yields 3-hmC, which readily deprotonates to form the more stable zwitterionic intermediate 3 (Supplementary Fig. 10c). Since the positively charged cytosine base is a much better leaving group than the neutral thymine at physiologic pH, the collapse of zwitterion 3 to cytosine and formaldehyde is expected to have a lower energetic barrier. This may partially explain the much faster repair of 1-meA and 3-meC by AlkB as compared to 1-meG and 3-meT.
The charge-bearing feature of 1-meA and 3-meC is reminiscent of a positively-charged trimethyl-lysine residue, the demethylation process of which and its dedicated enzymes are of great interest (Supplementary Fig. 21)4. Within the AlkB family, FTO exhibits demethylation activity of neutral 3-meT and 3-meU, but not with 1-meA or 3-meC14,15. Depending on the charge state of the substrate, intermediates similar to 2 or 3 could form during the demethylation processes catalyzed by these enzymes. The chemical nature of these intermediates can profoundly affect the reaction mechanism, reaction rate, and substrate specificity (Supplementary Fig. 21).
In summary, AlkB has the ability to work on a diverse range of substrates. Besides the DNA repair function that maintains the integrity of the genome28–30, members from this protein family also play diverse roles in biology9–11. The capture and structural characterization of several different intermediates presented in this study dissect the differences in the mechanism used by these enzymes to oxidatively remove a methyl group from biological substrates. This work also serves as an example of in crystallo reaction that leads to the trapping and characterization of otherwise unstable intermediates to help fully elucidate the reaction mechanism.
METHODS SUMMARY
The disulphide cross-linked protein-DNA complexes were purified using Mono-Q anion exchange chromatography, and crystals were grown using hanging-drop vapour diffusion methods. Diffraction data were collected from cryo-preserved crystals at beamlines 23ID-B, 21ID-D, 19BM-D and 14BM-C at the Advanced Photon Source (APS), Argonne National Laboratory. The structures of AlkB-dsDNA complexes were solved by molecular replacement. Data collection and refinement parameters for all structures are given in Supplementary Table 1. Detailed procedures are presented in Supplementary Information.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Methods
Oligonucleotide synthesis
Oligonucleotides containing a disulfide-tethered cytosine and damaged bases (εA, 3-meT, and 3-meC) were prepared by incorporating the O4-triazolyl-dU-CE phosphoramidite (Glen Research, Inc.), etheno adenosine phosphoramidite, N3-methyl thymidine CED phosphoramidite, and N3-Methyl deoxy cytidine CED phosphoramidite (the latter three are purchased from ChemGenes Corp.) at the desired position during solid-phase synthesis31. All synthetic oligonucleotides were purified with reverse-phase HPLC.
Cross-linking and purification of the AlkB-dsDNA complexes
A truncated AlkB with deletion of the N-terminal 11 amino acids was cloned into a pET30a vector (Novagen) and overexpressed in E. coli BL21(DE3)21. The protein was purified following a previously described procedure32. The Ser 129 to Cys mutation was introduced using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). Synthetic oligonucleotides (5′-TAGGTAAXAC*CGT, where C* is a disulfide-tethered cytosine and X stands for εA, 3-meT or 3-meC, and its complement strand 5′-AACGGTT_TACCT-3′, where “_” stands for an abasic site) were prepared, annealed, and cross-linked to the S129C mutant AlkB as described12. The covalently linked AlkB-dsDNA complexes were purified using Mono-Q anion exchange chromatography (GE Healthcare), after which the buffer was exchanged to 100 mM NaCl, 10 mM Tris-HCl (pH 7.4), and the complexes were concentrated to 12–15 mg/mL before drops were set up.
Crystallization of the AlkB-DNA complexes containing Mn(II) and αKG
For the crystals grown under aerobic conditions, 1.0 mM MnCl2 (Mn(II) is able to occupy the Fe(II) site, but is incapable of catalysis) and 2.0 mM αKG were added to the complexes, and they were mixed in a 1:1 ratio with well solution containing 100 mM NaCl, 25 mM MgCl2, 100 mM cacodylate (pH 6.5) and 20–24% w/v PEG 8K or 4K. Crystals appeared after 2–4 days and were allowed to grow for several weeks. They were transferred into a cryo-protectant solution containing the reservoir solution plus 20% glycerol, and frozen in liquid nitrogen for X-ray data collection.
Conditions for in crystallo oxidation reactions
All buffers and solutions used in crystallization trials were degassed with N2 for 30 min, and then transferred into an anaerobic chamber to equilibrate for at least 16 h before usage. The purified AlkB-dsDNA complexes were buffer-exchanged with degassed buffer at least four times, and then equilibrated in the anaerobic chamber before setting up crystallization trials. To set up drops, 1.0 mM (NH4)2Fe(SO4) and 2.0 mM αKG (the 50mM stock solution of which are always freshly prepared by dissolving (NH4)2Fe(SO4) and αKG solid in pre-equilibrated Milli-Q water in the anaerobic chamber) were added to the complexes (around 0.35~0.45mM), and they were mixed in a 1:1 ratio with well solution containing 100 mM NaCl, 50 mM MgCl2, 100 mM cacodylate (pH 6.5) and 20–24% w/v PEG 8K or 4K. Crystals appeared after 2–4 days, and after crystals appeared they were exposed to air by opening the cover slides periodically, and were then immediately frozen in liquid N2 for X-ray data collection. Different time points of air exposure were sampled (2–4 min, 1–2 h, 5–6 h, 2 d, and 9 d). Since these crystals are sensitive to oxygen, drops were normally set up no more than 2 weeks in advance from the actual date of X-ray data collection.
Air exposure of the AlkB-DNA complex containing 3-meC, Mn(II) and αKG
Mn(II) has previously been shown to occupy the metal-binding site of AlkB, but is catalytically inactive21,22. To be extra cautious, the in crystallo oxidation procedure for the Fe(II)/αKG-containing AlkB-dsDNA complex was performed for the Mn(II)/αKG-containing AlkB-dsDNA complex. The base lesion 3-meC was selected since it’s the most efficient substrate of AlkB among the three damaged bases tested (εA, 3-meT, and 3-meC). Crystallization drops were set up under anaerobic conditions and cover slides were opened to expose these Mn(II)/αKG-containing crystals to air. Two independent batches of such crystals were tested and crystals were picked up after 2 h and 6 h air exposure, respectively. The structures of the 2 h- and 6 h-air-exposed crystals were solved to 1.92 Å and 1.98 Å, with Rwork/Rfree of 17.9/22.5 and 18.9/24.3, respectively. The αKG cofactor has good electron density in both structures, indicating that Mn(II) is incapable of oxidation catalysis; the 3-meC base remains unmodified in both structures as well (Supplementary Fig. 22). Since these structures are the same as the structure 3O1M [(Manganese/αKG)AlkB-DNA: 3-meC], they are not deposited in the PDB database. In summary, these control experiments confirms AlkB-mediated in crystallo oxidation with Fe(II) as the catalytically essential metal ion.
Structure determination and refinement
The AlkB-DNA complex structure was phased by molecular replacement (using Phaser)33, using the previously published AlkB structure as a search model. The model was built by using COOT and refinement was carried out with the program REFMAC5 from the CCP4 suite34,35. Simulated annealing omit maps were generated with Phenix36. Data collection and refinement parameters for all structures are given in Supplementary Table 1. Molecular graphics figures were prepared with PyMOL37.
Ligand refinement and molecular modeling
Regular damaged bases (εA, 3-meT, and 3-meC) were used as the initial model for the crystals obtained from in crystallo oxidation. Based on the resultant Fobs – Fcalc maps, appropriate modifications were modeled into the density to obtain the best fit of the data. Dictionaries for the glycol and hemiaminal intermediates were produced with Monomer Library Sketcher (in ccp4 suite). Occupancy of the additional atoms of the intermediate structures was ~100% based on the average temperature factors (that are similar to those of the rest of the base atoms) and the lack of Fobs–Fcalc density contoured at 3.0σ. Model bias associated with ligand refinement was evaluated by, in addition to simulated annealing omit maps using Phenix, examining omit difference maps. The extra atoms were removed from the final refined models, and resulting models were allowed to refine with at lease 10 cycles of restrained refinement. The resultant positive density in the difference Fobs–Fcalc maps was found to agree with the refined positions and structures of the base intermediates in the full models.
Reproducibility of the observed intermediates
The three intermediate structures reported here were also observed in several additional, independently refined structural datasets obtained from crystals prepared using the same procedure described (data not shown). These observations affirm a physical basis for the observed reactivity in crystalline form.
Synthetic protocols of compound XIII from IV
Compound IV was obtained according to literature methods and was directly isolated from the a-anomer by careful column chromatography38.
4-Amino-1-(2'-deoxy-β-D-erythro-pentofanosyl)-2-pyridone (X)
To a solution of IV (200 mg, 0.418 mmol) in methanol (5 mL) was added 25% sodium methoxide (0.5 mL) dropwise and the resulting solution was stirred at rt for 30 min. Acetic acid was added dropwise to adjust PH=7. Removal of the solvents under reduced pressure and the residue was purified by silica gel chromatography, eluting with 5–15% MeOH in CH2Cl2, to give X (90.4 mg, 90%) as white foam. 1H NMR (500.1 MHz) (CD3OD) δ: 7.64 (d, J=7.5 Hz, 1H), 6.51 (t, J=6.5 Hz, 1H), 6.02 (d, J=7.5 Hz, 1H), 4.36 (m, 1H), 3.94 (m, 1H), 3.80 (dd, J=12.0, 4.0 Hz, 1H), 3.74 (dd, J=12.0, 4.0 Hz, 1H), 2.37 (m, 1H), 2.10 (m, 1H), 1.91 (s, 3H). 13C NMR (125.8 MHz) (CD3OD) δ: 162.8, 154.6, 130.0, 100.3, 87.1, 85.2, 70.8, 61.6, 41.0, 8.0.
4-Phenoxyacetylamido-1-(2'-deoxy-β-D-erythro-pentofanosyl)-2-pyridone (XI)
To a solution of X (82 mg, 0.34 mmol) in pyridine was added trimethylsilyl chloride (10 eq., 0.425 mL) and the mixture was stirred at rt for 2 h. Then phenoxyacetyl chloride (0.236 mL, 5 eq.) was added and the mixture was stirred for another 2 h. 1 mL ammonia hydroxide was added and stirred for 1 h. Removal of the solvents gave a residue, which was purified by silica gel chromatography, eluting with 3–10% MeOH in CH2Cl2, to give XI (100 mg, 78%) as white foam. 1H NMR (500.1 MHz) (CD3Cl+CD3OD) δ: 7.88 (d, J=8.0 Hz, 1H), 7.33 (m, 2H), 7.24 (d, J=8.0 Hz, 1H), 7.03 (m, 1H), 6.97 (d, J=8.5 Hz, 2H), 6.40 (t, J=6.0 Hz, 1H), 4.66 (s, 2H), 4.43 (m, 1H), 3.97 (m, 1H), 3.81 (m, 1H), 3.74 (m, 1H), 2.46 (m, 1H), 2.11 (m, 1H), 2.01 (s, 3H). 13C NMR (125.8 MHz) (CD3Cl+CD3OD) δ: 171.6, 166.7, 160.8, 147.7, 134.3, 133.7, 126.4, 118.5, 106.2, 91.4, 90.4, 74.3, 71.3, 65.4, 45.3, 13.5.
4-Phenoxyacetylamido-1-[(5'-O-(4,4'-dimethoxytrityl)-2'-deoxy-β-D-erythropentofanosyl]-2-pyridone (XII) XI
(90 mg, 0.24 mmol) was dissolved in pyridine (3 mL), and 4,4'-dimethoxytrityl chloride (97.6 mg, 0.288 mmol) was added while stirring the solution. After being stirred overnight at room temperature, the reaction mixture was quenched with MeOH (1 mL), and stirred for an additional 5 min. The reaction mixture was concentrated to dryness under vacuum. Dichloromethane (100 mL) was added and washed with sodium hydrogen carbonate (5%, 50 mL) and brine and then dried over sodium sulfate. After the organic phase was concentrated to dryness, the residue was purified by silica gel chromatography, eluting with 1–3% MeOH in dichloromethane containing 0.2% Et3N, to give XII (145 mg, 91%) as a white foam. 1H NMR (500.1 MHz) (CD3CN) δ: 7.71 (d, J=7.5 Hz, 1H), 7.47 (d, J=7.5 Hz, 2H), 7.34 (m, 9H), 7.25 (m, 1H), 7.05 (m, 2H), 6.92 (d, J=6.5 Hz, 1H), 6.88 (m, 4H), 6.39 (m, 1H), 4.67 (s, 2H), 4.43 (m, 1H), 3.98 (m, 1H), 3.78 (m, 6H), 3.35 (m, 2H), 2.46 (m, 1H), 2.11 (m, 1H), 1.99 (s, 3H). 13C NMR (125.8 MHz) (CD3CN) δ: 162.0, 158.7, 157.3, 149.7, 144.9, 143.2, 135.9, 135.8, 130.08, 130.06, 129.9, 129.8, 128.1, 128.0, 127.0, 123.8, 122.0, 117.3, 116.8, 114.8, 114.2, 113.1, 101.3, 86.4, 85.9, 85.5, 70.2, 67.3, 63.0, 54.9, 41.2, 31.5, 9.6.
4-Phenoxyacetylamido-1-[(5'-O-(4,4'-dimethoxytrityl)- 3'-O-(2-cyanoethyl-N,N-diisopropyl)phosphoramidite-2'-deoxy-β-D-erythro-pentofanosyl]-2-pyridone (XIII) XII
(140 mg, 0.21 mmol) was dissolved in dry CH2Cl2 (5 mL) and 1-methylimidazole (3.40 mg, 41.5 µmol). N,N-diisopropylethylamine (156 mg 0.84 mmol) was added to the stirring solution followed by 2-cyanoethyl N,N-(diisopropylchloro)-phosphoramidite (100 mg, 0.42 mmol). After being stirred at room temperature for 2 h, the reaction mixture was added to dichloromethane (5 mL) and the mixture was washed with 5% aqueous sodium bicarbonate and brine, dried over sodium sulfate and concentrated. The residue was purified by silica gel chromatography, eluting with 10–12% acetone in dichloromethane containing 0.2% Et3N, to give XIII (150 mg, 81%) as a white foam. 31P NMR (202.5 MHz) (CD3CN) δ: 148.3 and 148.4 ppm. HRMS for C49H57N4NaO9P, [MNa]+ 399.3761 (calcd.); 899.3753 (found).
Supplementary Material
Figure 1. Oxidative repair of damaged nucleic acid bases by AlkB.

Oxidative repair of εA, 3-meT, and 3-meC by AlkB with intermediates glycol 1, hemiaminal 2, and zwitterion 3 proposed in this study.
Acknowledgements
This study was supported by National Institutes of Health (GM071440 to C.H.), (GM084028 to Q.C.), and Beamlines 23ID-B (General Medicine and Cancer Institutes Collaborative Access Team [GM/CA-CAT]), 19BM-D (Structual Biology Center [SBC-CAT]), 14BM-C (BioCARS), and 21ID-D (Life Sciences Collaborative Access Team [LS-CAT]) at the Advanced Photon Source at Argonne National Laboratory; National Institutes of Health and the United States Department of Energy. Computational resources from the National Center for Supercomputing Applications at the University of Illinois and the Center of High Throughput Computing at UW-Madison are greatly appreciated. We also thank Drs. X. Yang, Z. Ren, and E. Duguid for crystallographic discussions.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Competing Interest Statement The authors declare they have no competing financial interests.
Author Contributions C.Y., G.J. and C.H. designed the experiments. Experiments were performed by C.Y., G.J., Q.D., W.Z., G.Z., X.J. and C.-G.Y.; computational analyses were performed by G.H. and Q.C. C.Y. and C.H. wrote the paper and G.H., Q.D. and Q.C. contributed to editing the manuscript.
Author Information Atomic coordinates have been deposited in the Protein Data Bank under the accession numbers 3O1M [(Manganese/αKG)AlkB-DNA: 3-meC], 3O1O [(Manganese/αKG)AlkB-DNA: 3-meT], 3O1P [(Manganese/αKG)AlkB-DNA: εA], 3O1R[(Manganese/αKG)AlkB-DNA: 3-deazameC], 3O1S [(iron/succinate)AlkB-DNA: oxidized 3-meC 3], 3O1T [(iron/succinate)AlkB-DNA: hemiaminal 2], 3O1U [(iron/succinate)AlkB-DNA: glycol 1], and 3O1V [(iron/succinate)AlkB-DNA: 3-deazahmC 4].
References
- 1.Kovaleva EG, Lipscomb JD. Versatility of biological non-heme Fe(II) centers in oxygen activation reactions. Nat Chem Biol. 2008;4:186–193. doi: 10.1038/nchembio.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schofield CJ, Zhang Z. Structural and mechanistic studies on 2-oxoglutarate-dependent oxygenases and related enzymes. Curr Opin Struct Biol. 1999;9:722–731. doi: 10.1016/s0959-440x(99)00036-6. [DOI] [PubMed] [Google Scholar]
- 3.Yi C, Yang CG, He C. A non-heme iron-mediated chemical demethylation in DNA and RNA. Acc Chem Res. 2009;42:530–541. doi: 10.1021/ar800178j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 5.Falnes PO, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 6.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 7.Westbye MP, et al. Human AlkB homolog 1 is a mitochondrial protein that demethylates 3-methylcytosine in DNA and RNA. J Biol Chem. 2008;283:25046–25056. doi: 10.1074/jbc.M803776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ringvoll J, et al. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J. 2006;25:2189–2198. doi: 10.1038/sj.emboj.7601109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aas PA, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 10.Sundheim O, et al. Human ABH3 structure and key residues for oxidative demethylation to reverse DNA/RNA damage. EMBO J. 2006;25:3389–3397. doi: 10.1038/sj.emboj.7601219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang CG, et al. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature. 2008;452:961–965. doi: 10.1038/nature06889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi Y, et al. Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme. Nature. 2009;462:762–766. doi: 10.1038/nature08561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerken T, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia G, et al. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008;582:3313–3319. doi: 10.1016/j.febslet.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koivisto P, Robins P, Lindahl T, Sedgwick B. Demethylation of 3-methylthymine in DNA by bacterial and human DNA dioxygenases. J Biol Chem. 2004;279:40470–40474. doi: 10.1074/jbc.M407960200. [DOI] [PubMed] [Google Scholar]
- 17.Falnes PO. Repair of 3-methylthymine and 1-methylguanine lesions by bacterial and human AlkB proteins. Nucleic Acids Res. 2004;32:6260–6267. doi: 10.1093/nar/gkh964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delaney JC, Essigmann JM. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc Natl Acad Sci U S A. 2004;101:14051–14056. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mishina Y, Yang CG, He C. Direct repair of the exocyclic DNA adduct 1,N6-ethenoadenine by the DNA repair AlkB proteins. J Am Chem Soc. 2005;127:14594–14595. doi: 10.1021/ja055957m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delaney JC, et al. AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat Struct Mol Biol. 2005;12:855–860. doi: 10.1038/nsmb996. [DOI] [PubMed] [Google Scholar]
- 21.Yu B, et al. Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB. Nature. 2006;439:879–884. doi: 10.1038/nature04561. [DOI] [PubMed] [Google Scholar]
- 22.Yu B, Hunt JF. Enzymological and structural studies of the mechanism of promiscuous substrate recognition by the oxidative DNA repair enzyme AlkB. Proc Natl Acad Sci U S A. 2009;106:14315–14320. doi: 10.1073/pnas.0812938106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holland PJ, Hollis T. Structural and Mutational Analysis of Escherichia coli AlkB Provides Insight into Substrate Specificity and DNA Damage Searching. PLoS One. 2010;5:e8680. doi: 10.1371/journal.pone.0008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han Z, et al. Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature. 2010;464:1205–1209. doi: 10.1038/nature08921. [DOI] [PubMed] [Google Scholar]
- 25.Schlichting I, et al. The catalytic pathway of cytochrome p450cam at atomic resolution. Science. 2000;287:1615–1622. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]
- 26.Kovaleva EG, Lipscomb JD. Crystal structures of Fe2+ dioxygenase superoxo, alkylperoxo, and bound product intermediates. Science. 2007;316:453–457. doi: 10.1126/science.1134697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burzlaff NI, et al. The reaction cycle of isopenicillin N synthase observed by X-ray diffraction. Nature. 1999;401:721–724. doi: 10.1038/44400. [DOI] [PubMed] [Google Scholar]
- 28.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hitomi K, Iwai S, Tainer JA. The intricate structural chemistry of base excision repair machinery: implications for DNA damage recognition, removal, and repair. DNA Repair (Amst) 2007;6:410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 30.Fromme JC, Banerjee A, Verdine GL. DNA glycosylase recognition and catalysis. Curr Opin Struct Biol. 2004;14:43–49. doi: 10.1016/j.sbi.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Mishina Y, He C. Probing the structure and function of the Escherichia coli DNA alkylation repair AlkB protein through chemical cross-linking. J Am Chem Soc. 2003;125:8730–8731. doi: 10.1021/ja034636c. [DOI] [PubMed] [Google Scholar]
- 32.Mishina Y, Chen LX, He C. Preparation and characterization of the native iron(II)-containing DNA repair AlkB protein directly from Escherichia coli. J Am Chem Soc. 2004;126:16930–16936. doi: 10.1021/ja045066z. [DOI] [PubMed] [Google Scholar]
- 33.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 34.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 37.Delano WLDS. Palo Alto, CA: 2002. [Google Scholar]
- 38.Searls T, McLaughlin LW. Synthesis of the analogue nucleoside 3-deaza-2'-deoxycytidine and its template activity with DNA polymerase. Tetrahedron. 1999;55:11985–11996. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


