Abstract
Increasing evidence indicates the significance of platelet-derived growth factor receptor-β (β-PDGFR) signaling in prostate cancer (PCa). Accordingly, preclinical studies suggest the potential of β-PDGFR as a therapeutic target in metastatic PCa. However, a ligand responsible for β-PDGFR activation in PCa was unknown, and recent clinical trials with imatinib mesylate showed limited success due to normal tissue toxicity. Similarly, in spite of mounting evidence indicating the significance of matriptase in PCa, little is known about its substrates or molecular actions during PCa progression. Here, we identified PDGF-D as a ligand for β-PDGFR in PCa and discovered matriptase as its regulator. Matriptase activates PDGF-D by proteolytic removal of the CUB domain in a two-step process, creating a hemidimer (HD) followed by growth factor domain dimer (GFD-D) generation. Matriptase can deactivate PDGF-D by further proteolytic cleavage within the GFD, revealing its biphasic regulation. Importantly, PDGF-D/matriptase co-localization is accompanied with β-PDGFR phosphorylation in human PCa tissues. This study unveiled a novel signaling axis of matriptase/PDGF-D/β-PDGFR in PCa, providing new insights into functional interplay between serine protease and growth factor signaling networks.
Keywords: Genitourinary cancers, prostate, Protease-inhibitor systems, Growth factors and receptors
INTRODUCTION
Members of the platelet-derived growth factor (PDGF) family are well known inducers of cell migration, proliferation, and transformation [reviewed in (1)]. While the two classic PDGF members, PDGF-A and -B, are secreted as active growth factor homo- or hetero-dimers and readily activate their cognate receptors, α-PDGFR and β-PDGFR, the newly discovered PDGF family members PDGF-C and PDGF-D are secreted as latent homodimers containing the N-terminal CUB domain and the C-terminal growth factor domain (GFD) (2–4). The removal of the CUB domain is required for the GFD dimers (GFD-D) of PDGF-C and -D to activate α-PDGFR and β-PDGFR, respectively (5, 6).
Increasing evidence suggests critical roles for PDGF during cancer progression, especially in prostate cancer (PCa). A majority of prostate tumor tissues at both primary and metastatic sites express β-PDGFR at increased levels as determined by immunohistochemical analysis (7–9). A recent study revealed that β-PDGFR is one of five genes that predict PCa recurrence after prostatectomy (10), and preclinical studies suggested the potential of β-PDGFR as a therapeutic target in metastatic cancers (11–16). However, little research has been conducted to identify PDGF ligand(s) responsible for β-PDGFR activation during PCa progression. PDGF-B, originally thought to be the sole ligand for β-PDGFR, has been difficult to find in PCa patient specimens (12, 17). This raises the possibility that the newly discovered PDGF ligand, PDGF-D, may be responsible for activation of β-PDGFR signaling in human PCa. In order to study the roles of PDGF-D in PCa, we previously established human PCa cell lines engineered to overexpress PDGF-D (6, 18). We showed that PDGF-D accelerates the early onset of prostate tumor growth and drastically enhances PCa cell invasion into surrounding stroma in a mouse model (18).
In this study, we wished to identify a proteolytic activator of PDGF-D in PCa cells and to validate their expression in human PCa tissues. We found that matriptase is a PDGF-D-inducible protease that regulates PDGF-D activity. Matriptase, also referred to as MT-SP1 (membrane type serine protease type 1), is a type II transmembrane serine protease often found in complex with its cognate inhibitor, hepatocyte growth factor activator inhibitor-1 (HAI-1) (19–21). Athough mounting evidence showed increased matritpase expression in human tumors of epithelial origin, including prostate (22, 23), breast (24), cervical (25), ovarian (26), and gastrointestinal tract (27), little is known about its substrates or its molecular actions during cancer progression at present. Here, we report that matriptase activates PDGF-D in a two-step process, creating a hemidimer (HD) containing a 50 kDa full-length monomer and an 18 kDa GFD before producing the active dimer consisting of two 18 kDa GFD peptides. Through site-directed mutagenesis, we identified the matriptase cleavage site of PDGF-D at R247/R249 within the hinge region between the CUB and GFD of the full-length PDGF-D. Interestingly, we found that the GFD-D is susceptible to further cleavage by matriptase, generating a biologically inactive 15 kDa fragment, revealing its biphasic regulation of PDGF-D’s activity. Immunohistochemical analysis of human prostate tumor tissues demonstrated increased expression of both PDGF-D and matriptase. In situ hybridization analysis of PDGF-D mRNA confirmed that PDGF-D is produced predominantly by carcinoma cells. Importantly, confocal microscopic analysis revealed co-localization of PDGF-D and matriptase in human prostate tumor tissues. Consistent with our finding that matriptase is an activator of PDGF-D signaling, increased β-PDGFR phosphorylation was detected in malignant prostate tissues with increased expression of both markers, especially in poorly differentiated prostate carcinoma and the neoplastic glands where perineural invasion occurs. Taken together, the present study identifies matriptase as a regulator of PDGF-D activity in human PCa and provides molecular insight into matriptase-mediated dynamic proteolytic processing of PDGF-D.
MATERIALS AND METHODS
Cell lines
Human prostate carcinoma LNCaP cells over-expressing PDGF-D and vector controlled cells were established previously and maintained in RPMI164 medium with 5% fetal bovine serum (FBS) (18). Mouse fibroblast NIH3T3 cells were purchased from ATCC and maintained in DMEM/F12 medium with 10% bovine serum (Invitrogen). Human prostate fibroblast FTX1532 cells were obtained from Dr. R. Fridman (Wayne State University, Detroit, MI) and maintained in RPMI1640 medium with 10% FBS.
Antibodies, Proteases, and Inhibitors
Generation of custom antibody that recognizes GFD of PDGF-D (8D2) was described previously (18). Anti-matriptase antibodies M32 and S5 were described previously (21). Anti-phosphorylated-β-PDGFR (pY751) antibody was purchased from Cell Signaling Technology. Anti-HAI-1 ectodomain Ab was purchased from R&D Systems. Purification of the serine-protease domain of matriptase (rMatriptase) was described previously (28). HAI-1 construct within a pcDNA3.1 vector was a kind gift from Dr. R. Fridman (Wayne State University, Detroit, MI).
siRNA transfection
LNCaP-PDGF-D cells were transfected with non-targeting or matriptase SMARTPOOL™ siRNA (Dharmacon, Inc.) using Lipofectamine 2000 according to manufacturer’s protocol. After transfection for 5 hours, cells were maintained in RPMI1640 medium with 5% FBS for 48 hours before serum starvation. CM and cell lysates were collected after culturing in serum-free medium for 48 hours. Cell lysates were harvested with RIPA lysis buffer.
Isolation of the PDGF-D dimer species
Using a duplicate non-reducing western blot as a guide, the appropriate bands were cut out of the SDS-PAGE gel at specific molecular weights. Excised gel fragments were minced by passing through syringe followed by eluting with elution buffer (50mM Tris-HCl, 150mM NaCl, and 0.1mM EDTA; pH 7.5) at room temperature overnight. Eluates containing rPDGF-D was analyzed by immunoblotting.
Immunohistochemical analysis of PDGF-D and matriptase
Formalin fixed, paraffin-embedded human PCa specimens were obtained from the Wayne State University Pathology Research Core Facility. Antigen retrieval was performed in sodium citrate buffer. Slides were incubated overnight at 4°C with either anti-PDGF-D Ab (8D2) or anti-matriptase Ab (S-5) and visualized with DAB (Vector Labs). Mayer’s hematoxylin was used to counterstain nuclei. Negative controls were performed without the corresponding primary antibody.
In Situ Hybridization
Digoxigenin-labeled antisense probes were synthesized from pCR2.1-PDGF-D using the DIG RNA Labeling Kit (Roche Applied Science) according to the manufacturer’s instructions. PDGF-D sense probes, created with pcDNA3.1-PDGF-D, were used as a negative control. Slides were prehybridized for 1hr at 65°C followed by incubation with either PDGF-D antisense or sense probes overnight at 65°C. The probe was detected using anti-digoxigenin antibodies and visualized using DAB.
Immunofluorescent double staining of matriptase and PDGF-D in human prostate tissue
Slides were double probed with anti-PDGF-D (8D2) and anti-matriptase antibodies (M32). Texas Red-conjugated-anti-mouse and FITC-conjugated-anti-rabbit secondary antibodies were used to detect matriptase and PDGF-D, respectively (Jackson Immunoresearch Laboratories, West Grove, PA). Confocal immunofluorescence microscopic analysis was performed at the Microscopy, Imaging and Cytometry Resources Core at Wayne State University, School of Medicine, using a Zeiss LSM 510 confocal microscopy system equipped with a C-Apochromat (NA = 1.2) 63× korr objective lens (Carl Zeiss MicroImaging, Inc., Thornwood, NY).
RESULTS
Identification of matriptase as an activator of PDGF-D in human prostate carcinoma LNCaP cells
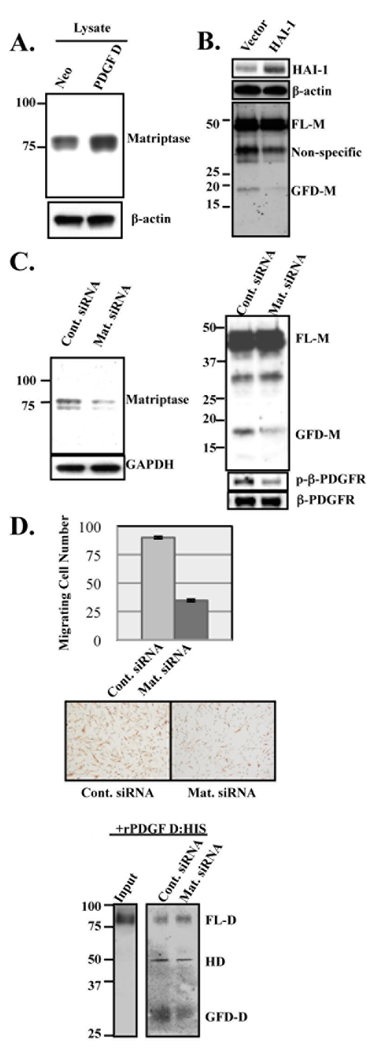
We previously showed that LNCaP cells fail to process PDGF-D into the GFD in the presence of the serine protease inhibitor aprotinin (6). Furthermore, we showed that recombinant PDGF-D (rPDGF-D) undergoes proteolytic processing more effectively in the presence of PDGF-D-transfected LNCaP (LNCaP-PDGF-D) cells compared to control LNCaP-neo cells (18). These results suggest that a PDGF-D-inducible serine protease is responsible for the processing of PDGF-D in LNCaP cells. To identify this enzyme, RNA samples collected from serum-free culture of LNCaP-PDGF-D and LNCaP-neo cells were analyzed via microarray, screening a gene chip containing 12,814 unique human clones (Agilent Technologies). Each sample was run in triplicate against the reference RNA sample, and gene expression levels in LNCaP-PDGF-D were compared to those in LNCaP-neo, with the cut-off fold-change being 2.0 fold (p-value < 0.01). This analysis revealed that the mRNA level of matriptase increased in LNCaP-PDGF-D cells compared to LNCaP-neo by 2.5 fold (p-value = 0.00435). Matriptase, also known as MT-SP1, is an 80 kDa membrane bound serine protease that was originally isolated from breast and PCa cells (20, 29). Increased matriptase expression in LNCaP-PDGF-D cells was confirmed at the protein level (Fig. 1A). To determine whether LNCaP-produced matriptase is critical for PDGF-D processing, LNCaP-PDGF-D cells were transiently transfected with HAI-1, its endogenous inhibitor, or with siRNA. Generation of the GFD of PDGF-D was reduced following HAI-1 transfection or siRNA-mediated matriptase knockdown, suggesting a role for matriptase in the proteolytic processing of PDGF-D in LNCaP cells (Fig. 1B and C). In a separate experiment, we examined the biological consequence of reduced PDGF-D processing on its ability to activate β–PDGFR and induce cell motility in a paracrine manner. To this end, serum-starved fibroblasts were treated with conditioned media (CM) collected from control and matriptase-knockdown LNCaP-PDGF-D cells and analyzed for the status of β–PDGFR activation and for their motility.β–PDGFR was readily activated in NIH3T3 cells following treatment with CM from LNCaP-PDGF-D cells, whereas β–PDGFR activation was far less apparent by treatment with CM from matriptase-knockdown LNCaP-PDGF-D cells. Accordingly, FTX-1532 cell migration was greatly reduced following treatment with CM from matriptase-knockdown LNCaP-PDGF-D cells compared to the control (Fig. 1D, top and middle panels). Lastly, we examined the effects of matriptase knockdown on the ability of LNCaP cells to process exogenous PDGF-D protein. While recombinant full-length PDGF-D dimer (rPDGF-D) was effectively processed into the GFD-D when incubated with LNCaP-PDGF-D cells as previously reported (18), the processing was significantly reduced in the absence of matriptase expression in these cells (Fig. 1D, bottom panel). It should be mentioned that PDGF-D analyzed in Fig. 1D (bottom panel) was almost exclusively rPDGF-D, since the incubation period was short enough not to include a significant amount of PDGF-D secreted from LNCaP cells as we previously demonstrated (18). Taken together, these results demonstrated a critical role for matriptase in the activation of PDGF-D in LNCaP cells.
Fig. 1. Increased matriptase expression in LNCaP-PDGF-D cells and its role in PDGF-D processing.
A. Immunoblot analysis of matriptase in LNCaP-neo and LNCaP-PDGF-D cell lysates using M32 antibody. B. Immunoblot analysis of HAI-1 and β-actin in cell lysates (top and middle panels, respectively) and PDGF-D in CM (bottom panel) from LNCaP-PDGF-D cells transfected with vector control or HAI-1 expression plasmid. C. LNCaP-PDGF-D cells were transfected with non-target siRNA control (Cont. siRNA) or matriptase-targeting siRNA. Equal amount of cell lysates (left panel) or the same volume of CM (top right panel) were analyzed by immunoblot analyses for matriptase and PDGF-D accordingly. Serum-starved NIH3T3 cells were stimulated for 10 minutes with CM from control or matriptase-knockdown cells. Immunoblot analysis of lysates was performed using anti-phosphorylated-β-PDGFR (pY751) (right middle panel) and anti-β-PDGFR antibodies (right bottom panel). D. Top and middle panels: Migration of FTX-1532 co-cultured with control or matriptase-downregulated LNCaP-PDGF-D cells for 12 hours. Data points represent average total number of FTX-1532 migrating to bottom of filter. Experiments were done in quadruplicates. P<0.01 as determined by Student’s t test. Bottom panel: Medium was collected after 4 hr incubation of rPDGF-D with serum-starved control or matriptase-knockdown cells. PDGF-D dimer species were analyzed by non-reducing immunoblot analysis, with input rPDGF-D shown on the left.
Matriptase regulation of PDGF-D activity in a biphasic manner: activation and deactivation
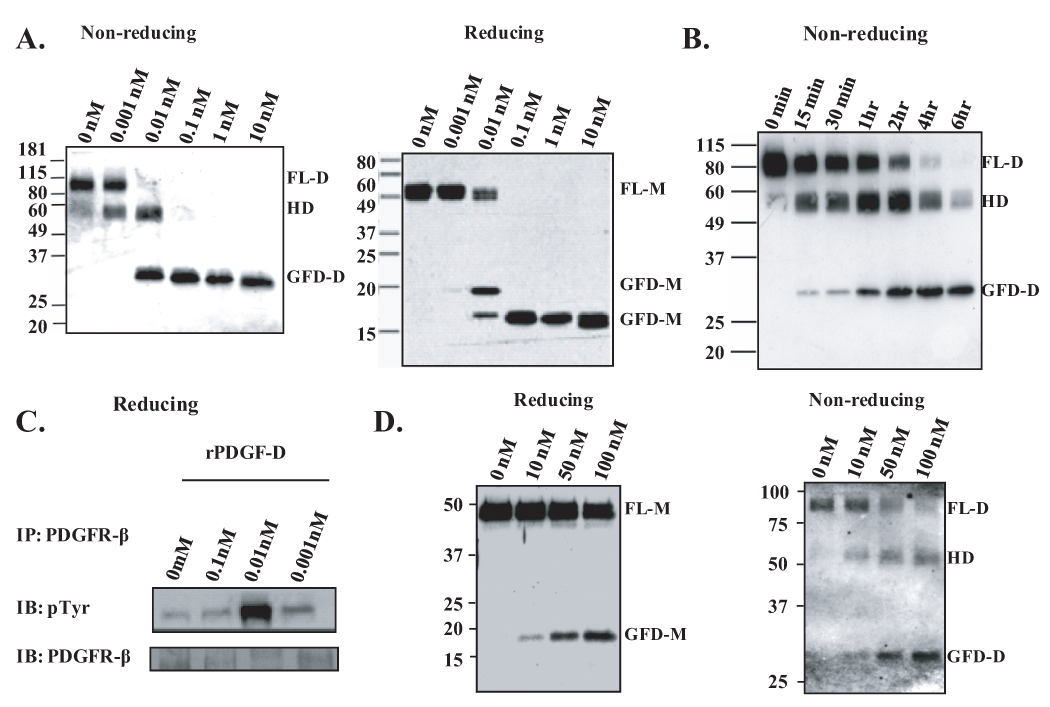
To test whether PDGF-D is a direct substrate for matriptase, rPDGF-D was incubated overnight with increasing concentrations of the purified catalytic domain of matriptase. Immunoblot analysis under non-reducing condition revealed that matriptase can process full-length PDGF-D dimer (FL-D) into GFD-D in a two-step manner, generating a 58 kDa intermediate complex (Fig. 2A). When these samples were analyzed under reducing condition, matriptase-treated rPDGF-D either yielded a 50 kDa full-length monomer (FL-M) or an 18 or 15 kDa GFD monomer (GFD-M), suggesting that the 58 kDa intermediate complex is a hemidimer (HD) containing one FL-M and one GFD-M. We then examined whether two-step, matriptase-mediated processing of PDGF-D into HD and GFD-D can be detected in a time-dependent manner. As shown in Fig. 2B, sequential cleavage of rPDGF-D by 0.1 nM matriptase was detected as the incubation time increases. To examine which of the processed forms of PDGF-D are biologically active, serum-starved NIH3T3 cells were stimulated with matriptase-treated rPDGF-D fragments from panel A. The β-PDGFR protein was immunoprecipitated with an anti-β-PDGFR Ab, and the active form was detected by immunoblot analysis using an anti-phosphotyrosine Ab (Fig. 2C). Consistent with the previous reports that 50 kDa FL-PDGF-D is a latent form, undigested (0 nM) or 0.001 nM matriptase-cleaved rPDGF-D containing mostly 50 kDa FL-PDGF-D failed to activate β-PDGFR. Interestingly, β-PDGFR was readily activated following 10 min incubation with 0.01 nM matriptase-cleaved rPDGF-D containing 18 kDa GFD, but not with 0.1 nM matriptase-cleaved rPDGF-D containing solely 15 kDa GFD. These data indicate that matriptase activates PDGF-D by generating an 18 kDa GFD fragment and deactivates by further proteolytic processing to a 15 kDa fragment.
Fig. 2. Matriptase effectively activates rPDGF-D in a two-step manner and deactivates it by further proteolytic cleavage within the growth factor domain.
A,B. rPDGF-D was incubated at 37°C with increasing concentrations of rMatriptase overnight (A) or with 0.1 nM for the indicated time periods (B). Samples were analyzed by immunoblot analysis using anti-PDGF-D antibody. C. Serum-starved NIH3T3 cells were stimulated for 10 minutes with rPDGF-D processed by indicated concentrations of rMatriptase (as shown in panel A). β-PDGFR was immunoprecipitated and immunoblotted with anti-phosphotyrosine (top panel) and anti-β-PDGFR antibodies (bottom panel). D. rPDGF-D was incubated at 37°C with increasing nanomolar concentrations of recombinant uPA overnight. Samples were analyzed by immunoblot analysis using anti-PDGF-D antibody.
We previously identified uPA as the PC3-expressed protease responsible for PDGF-D activation (6), while the present study identified matriptase as a PDGF-D regulator in LNCaP cells, which produce little to no uPA. We wished to evaluate the relative efficiency between the two serine proteases, matriptase and uPA, for the processing of PDGF-D. rPDGF-D was processed by 1–10 pM matriptase upon overnight incubation (Fig. 2A) or by 0.1 nM matriptase within 15 min (Fig. 2B). In contrast, a significantly higher level of uPA (more than 10 nM) was necessary for the processing of rPDGF-D by overnight incubation at 37°C (Fig. 2D). These data suggest that matriptase may be a more effective activator of PDGF-D compared to uPA.
Identification of the matriptase cleavage site in the hinge region of full-length PDGF-D and characterization of PDGF-D hemidimers
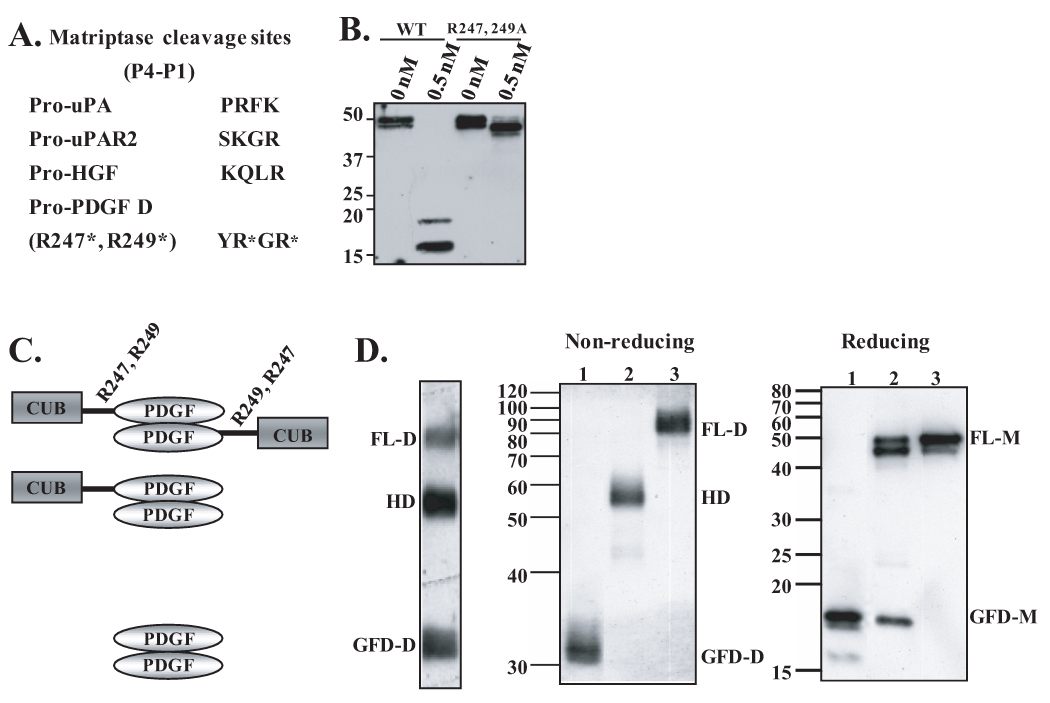
Previous sequence analysis of the amino terminus of the GFD of PDGF-D revealed the proteolytic cleavage site at R247 or R249 within the hinge region by unknown proteases (4). Like most serine proteases, matriptase cleaves at a lysine or an arginine in the P1 position of the substrate, with preference to arginine (21). Further analysis of matriptase cleavage sites using phage display libraries and computer modeling predicted that glycine/phenylalanine/small residue is preferred at the P2 site, and that P3/P4 sites are of a basic/non-basic or vice-versa relationship (30). Comparison of the R247/R249 region of PDGF-D with the known cleavage sites of other matriptase substrates yielded reasonable conservation, as shown in Fig. 3A (31). When these arginine residues were mutated to alanine (PDGF-DR247,249A), matriptase failed to process PDGF-D into GFD, therefore identifying the cleavage site of matriptase within the hinge region of FL-PDGF-D (Fig. 3B).
Fig. 3. Identification of matriptase cleavage site in the hinge region between the CUB and growth factor domains and characterization of PDGF-D dimer species.
A. Comparison of cleavage sites of known matriptase substrates, pro-uPA, pro-uPAR2, and pro-HGF, to the putative matriptase cleavage site within hinge region of full-length PDGF-D. B. Wild-type rPDGF-D and rPDGF-DR247,249A mutant proteins, in which arginine residues at amino acid 247 and 249 are mutated to alanines (6), were incubated with 0.5 nM matriptase overnight at 37°C. Samples were analyzed by immunoblot analysis in reducing condition. C. Schematic diagram of FL-D, HD, and GFD-D. R247 and R249 in the hinge between the CUB and GFD are labeled. D. PDGF-D dimer species (FL-D, HD, and GFD-D) produced by HeLa cells were isolated from a non-reducing SDS-PAGE gel (left panel), resolved on non-reducing or reducing SDS-PAGE gel, then immunoblotted with anti-PDGF-D antibody.
The above results show that the catalytic domain of matriptase activates FL-PDGF-D in a two-step process, with the HD as an intermediate dimer species as depicted by a diagram shown in Fig. 3C. To test whether HD can be generated in HeLa cells, which express matriptase at a high level (32), PDGF-D was expressed in these cells using the vaccinia expression system. Immunoblot analysis in a non-reducing condition revealed three major PDGF-D dimer species, presumably FL-D, HD, and GFD-D (Fig. 3D). To further reveal the nature of these species, these three protein bands were isolated by gel extraction, followed by immunoblot analysis under both non-reducing and reducing conditions. FL-D contained 50 kDa full-length monomers (FL-M) in a reducing condition as expected (Fig. 3D, lane 3). The HD (lane 2) consisted of FL-M and GFD-M, while GFD-D (lane 1) contained GFD-M as well as a smaller GFD-M species. This confirmed that HD contains one FL-M and one GFD-M, and also showed further proteolytic cleavage within GFD-D, generating a 15 kDa fragment.
Increased PDGF-D expression, co-localization of PDGF-D with matriptase, and increased β-PDGFR phosphorylation in human prostate carcinoma
To examine whether PDGF-D and matriptase are expressed in human prostate carcinoma specimens, resulting in β-PDGFR activation, and whether co-localization of PDGF-D and matriptase can be detected, we obtained tissue sections of both benign prostate and prostate carcinoma specimens from our Pathology Research Core. Sixty-six individuals were included in the immunohistochemical analysis, and the distribution of individuals was tabulated by American Joint Committee on Cancer (AJCC) tumor stage and Gleason score (Supplemental Tables 1 and 2). The immunohistochemical staining was evaluated by an Uro-Pathologist, and a blind assessment of the percentage of cells stained for PDGF-D and matriptase was performed for semi-quantitation. Kendall’s tau was used to assess the association of tumor stage and Gleason score with PDGF-D and matriptase staining. PDGF-D staining is strongly associated with both higher tumor stage (p=0.006) and higher Gleason score (p=0.01), while matriptase has a weaker association with both tumor stage (p=0.08) and Gleason score (p=0.11) (Table 1).
Table 1.
Association between tumor stage and Gleason score with PDGF-D and matriptase staining was assessed using Kendall’s tau. P-value shown within parentheses.
| N | Tumor Stage | Gleason Score | |
|---|---|---|---|
| PDGF D | 66 | 0.27 (0.006) | 0.25 (0.01) |
| Matriptase | 62 | 0.18 (0.08) | 0.17 (0.11) |
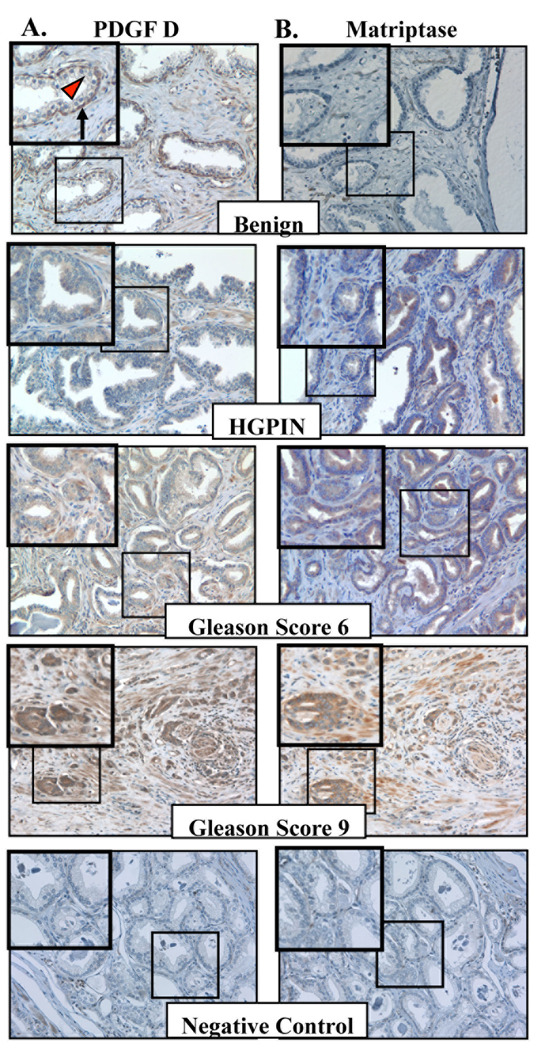
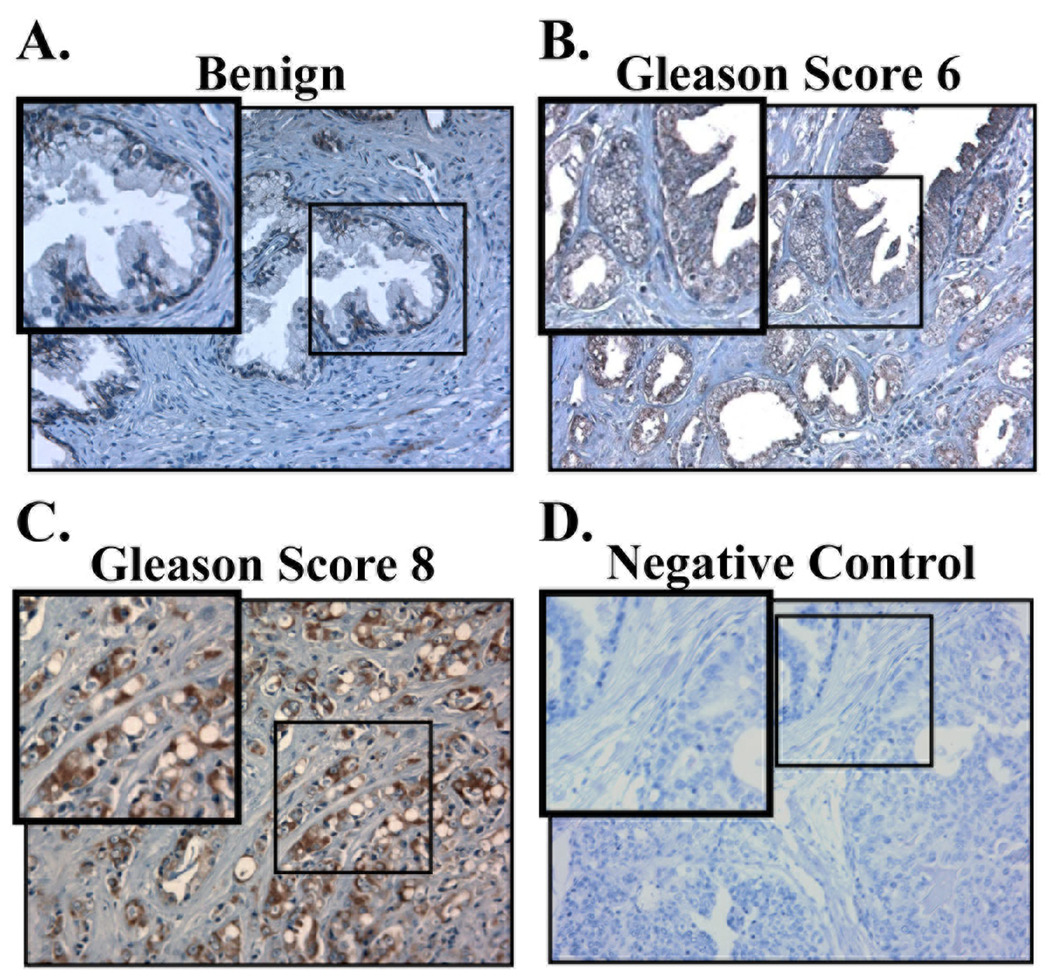
When the staining patterns of PDGF-D and matriptase were analyzed (Fig. 4A and B), expression of PDGF-D was detected in the basal cells but not in luminal cells within benign prostatic glands, while expression of matriptase was barely detected in benign glands. A mild increase of both markers was observed in some of the glands with high-grade prostatic intraepithelial neoplasia (HGPIN). The neoplastic glands in well to moderately differentiated prostate carcinoma (Gleason score 6) largely showed focal and variable staining for PDGF-D and matriptase. Significantly increased staining of both markers was observed in poorly differentiated prostate carcinoma (Gleason score 8 or higher). Unlike matriptase, which is an epithelial cell-specific enzyme, PDGFs can be produced by many different cell types, including epithelium and mesenchyme. To localize the cells that produce PDGF-D in these tissue samples, we performed in situ hybridization analysis of PDGF-D mRNA. PDGF-D mRNA was almost exclusively localized in epithelium (Fig. 5). Consistent with the immunostaining patterns of PDGF-D proteins, PDGF-D mRNA was detected in the basal cells but not in secretory cells within benign prostatic glands, and a marked increase in hybridization signal was observed in poorly differentiated prostate carcinoma cells.
Fig. 4. Expression of PDGF-D and matriptase in human prostate carcinoma.
A,B. Representative images of benign prostate, high-grade prostatic intraepithelial neoplasia (HGPIN), and prostate adenocarcinoma from Gleason scores 6 and 9 immunostained with antibodies against PDGF-D (A) and matriptase (B). Images were taken at 200× magnification. Insert shows enlargement of boxed area. Basal cells and luminal cells in prostate glands are indicated by black arrow and red arrow head, respectively.
Fig. 5. Expression of PDGF-D mRNA in human prostate carcinoma.
In situ hybridization was performed on sections of benign prostate (A) and adenocarcinoma from Gleason scores 6 (B) and 8 (C) using PDGF-D antisense probes. Images were taken at 200× magnification. Insert shows enlargement of boxed area. PDGF-D sense probe was used as a negative control on human prostate carcinoma (D).
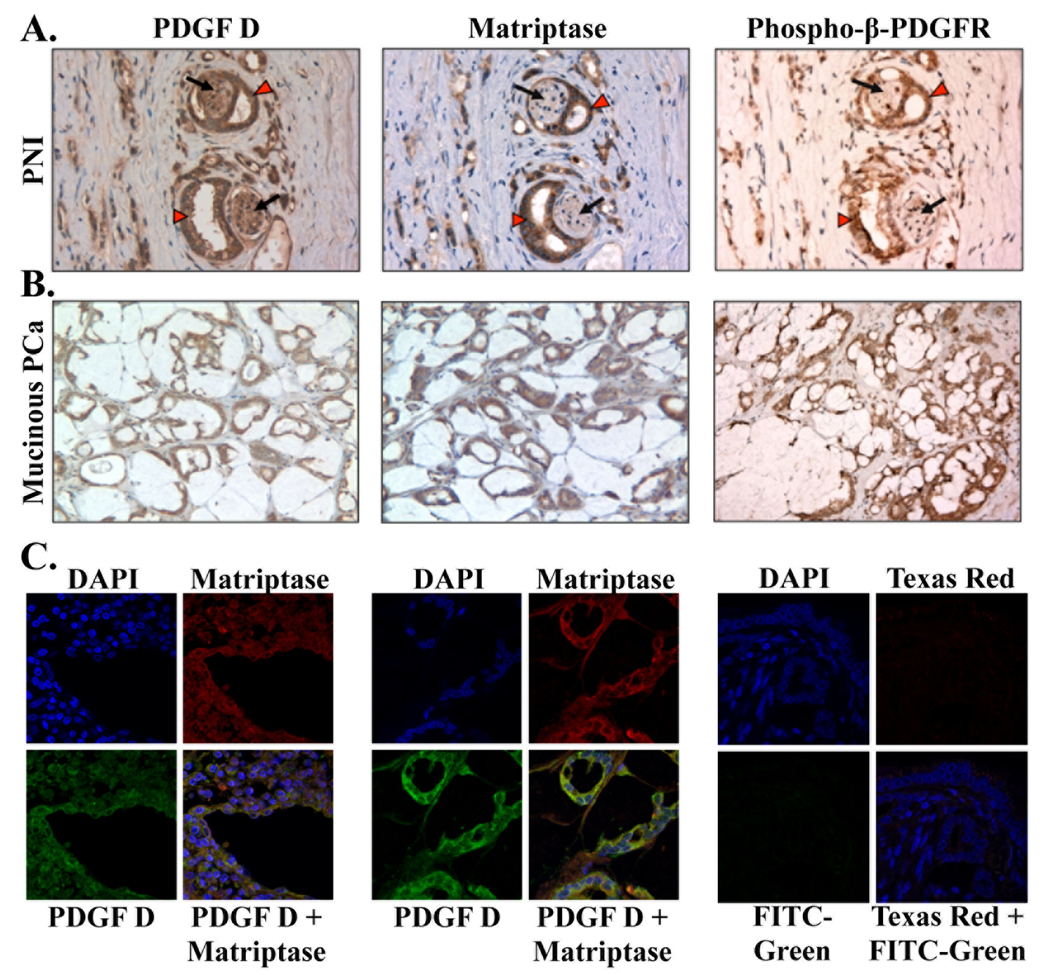
A striking feature of PDGF-D and matriptase expression was the conspicuous over-expression of both markers in the neoplastic glands surrounding nerves, referred to as perineural invasion (Fig. 6A). Consistent with our in vitro finding that matriptase is an activator of PDGF-D/β-PDGFR signaling, phosphorylated β-PDGFR was readily detected in prostate carcinoma cells surrounding nerves. Of note, β-PDGFR phosphorylation was barely detected in nerve cells that express PDGF-D without matriptase. Marked over-expression of both markers as well as β-PDGFR phosphorylation were also detected in the mucinous variant of poorly differentiated prostate carcinoma, typically Gleason score 8 or higher (Fig. 6B). Overall, as the levels of PDGF-D and matriptase increased, phosphorylated β-PDGFR expression was prominent in high-grade prostate carcinoma compared to the benign or low grade prostate carcinoma (Supplemental Fig. 1).
Fig. 6. Expression and co-localization of PDGF-D, matriptase, and phosphorylated-β-PDGFR in PNI and mucinous prostate carcinoma.
A,B. Serial sections of neoplastic glands with perineural invasion (PNI, at 400×) (A) and sections of the mucinous variant of poorly differentiated PCa (mucinous PCa, at 200×) (B) were immunostained with anti-PDGF-D, anti-matriptase, and anti-phospho-β-PDGFR antibodies. Carcinoma and nerve cells are indicated by red arrow heads and black arrows, respectively. C. Immunofluorescence analysis of matriptase (Texas Red) and PDGF-D (FITC-Green) in PCa with Gleason score 8 (left panel) and in a mucinous variant of PCa (middle panel) was performed at 630× magnification. Cell nuclei were stained with DAPI (blue). Yellow in merged panel indicates co-localization of the proteins.
We examined whether co-localization of PDGF-D and matriptase can be detected by confocal immunofluorescence microscopic analysis. As shown in Fig. 6C, co-localization of PDGF-D and matriptase was readily detected in PCa specimens and in the mucinous variant of poorly differentiated prostate carcinoma, where increased expression of both markers was detected by immunohistochemical analysis.
Taken together, the present study unveiled a novel signaling axis of matriptase/PDGF-D/β-PDGFR during human PCa progression.
DISCUSSION
The present study identifies PDGF-D as a substrate for matriptase and provides molecular insight into how matriptase regulates the activities of PDGF-D by its dynamic proteolytic processing. This finding is of particular importance not only for understanding molecular actions of PDGF-D but also unveiling the oncogenic activities of matriptase in human cancer. Matriptase was originally identified from human breast cancer cells (29), and independently from a human PCa cell line (20). Although matriptase was suggested to be involved in the degradation/remodeling of extracellular matrix and activation of growth factor signaling such as hepatocyte growth factor (30, 33), little was known about its substrates or its oncogenic actions at the molecular level. The present study provides an important clue as to how matriptase initiates growth factor signaling networks in PCa, via activation of tumor-derived PDGF-D, which can activate β-PDGFR in tumor cells in an autocrine manner as well as in stromal cells in a paracrine manner. Our finding that matriptase can regulate PDGF-D activity in a biphasic manner (generation of active 18 kDa GFD and further proteolytic cleavage into a biologically inactive 15 kDa fragment) is critical to understanding the molecular basis of PDGF-D actions in relation to serine proteases. This information would be vital in guiding future clinical studies to identify the biologically active PDGF-D dimers in the physiological/pathological conditions. The PDGF/VEGF family is characterized by eight conserved cysteine residues with similar spacing in between, except the GFD of PDGF-D, which lacks the fifth conserved cysteine residue [reviewed in (34)]. It was initially reported that PDGF-D GFD dimer is an approximately three-fold less efficient competitor for β-PDGFR than PDGF-B dimer in a ligand-binding assay. Since the fifth invariant cysteine residue is known to be involved in an intra-chain disulfide bond within the PDGF-A or -B chain, we and others previously hypothesized that missing the fifth invariant cysteine residue in PDGF-D GFD alters the three-dimensional structure, resulting in lower binding affinity to the β-PDGFR (3, 34). In light of our new data, we now speculate that the lower binding affinity of recombinant PDGF-D GFD dimer may be attributed, at least in part, to its unstable nature due to the susceptibility to further proteolytic inactivation.
We reported here, for the first time to our knowledge, the expression pattern of PDGF-D in normal and malignant human prostate tissues. The exclusive expression of PDGF-D in the basal layer, but not the secretory layer, of normal prostate glands is unique among potential oncogenic growth factors examined in human prostate tissues including insulin-like growth factor (IGF), hepatocyte growth factor (HGF), fibroblast growth factor (FGF), and vascular endothelial growth factor (VEGF) (35–37). Interestingly, we noticed prominent expression of PDGF-D in neoplastic secretory cells. It would be of interest to examine whether PDGF-D plays a role in neoplastic transformation of basal cells, considering the current theory, although debatable, that neoplastic secretory cells predominating the tumor glands may stem from neoplastic transformation of the basal cells (38, 39). Most interestingly, the abundant expression of PDGF-D and matriptase was detected in neoplastic glands surrounding the nerve, referred to as perineural invasion (PNI). Although little is known about PNI, and its significance in cancer progression and metastasis is the subject for debate in the field [reviewed in (40)], a statistical link has been reported between PNI and the size of tumor, as well as the risk of tumor recurrence after radiation therapy (41–43). PNI may be associated with a high incidence of metastasis from the primary site, as in vitro studies show mutual beneficial effects between PCa cells and ganglia/nerve in terms of neurite growth, proliferation and migration of PCa cells (44, 45). These studies suggest that the cross-talk between nerve cells and cancer cells supports cell proliferation, and that the axons may provide a pathway for cancer cells to escape from the primary site. A role of PDGF-D in PNI and tumor interactions with the nerve cells remains to be fully investigated.
Recent studies revealed a critical role for β-PDGFR in PCa. A majority of prostate tumor tissues at both primary and metastatic sites in the bone express β-PDGFR, as determined by immunohistochemical analysis (12). Consistent with the potential oncogenic activity of PDGF signaling in PCa, investigators have reported a therapeutic potential of PDGF/PDGFR targeting in clinical trials of hormone refractory PCa (12, 14–16, 46). In animal experiments, imatinib mesylate (also known as STI571 or Gleevec) was shown to inhibit phosphorylation of PDGFR and significantly suppresses experimental human PCa bone metastasis and angiogenesis, especially when administrated in combination with paclitaxel (Taxol) (11) or with paclitaxel and PKI166, an inhibitor of epidermal growth factor receptor (13). However, recent clinical trials with imatinib mesylate showed only limited success with unanticipated side effects such as diarrhea related to inhibition of c-kit in intestines, hypopigmentation in skin, and cardiotoxicity associated with inhibition of c-abl in cardiac myocytes (47–49). Improvement of β-PDGFR targeting specific to PCa would require identification of the prostate-specific PDGF ligand and characterization of its molecular actions. Importantly, the present study identified PDGF-D as a ligand for β-PDGFR in PCa. Our study revealed statistically significant correlation between PDGF-D expression with higher Gleason scores or advanced tumor stages, identifying PDGF-D as one of the very few growth factors with potential prognostic values in PCa. Importantly, the present study also revealed increased expression of matriptase, a regulator of PDGF-D, in human prostate carcinoma, resulting in activation of β-PDGFR signaling. Taken together, our studies unveil a novel signaling axis of matriptase/PDGF-D/β-PDGFR in PCa, providing valuable information for the design of more specific therapeutic approaches for the improvement of the effectiveness with fewer side effects.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH/NCI RO1 Grants CA64139 and CA123362 (H-R. C. K.), as well as the Ruth L. Kirschstein National Research Service Award T32-CA009531 (M.K.C-L.). We are grateful to Dr. R. Fridman and his lab for assistance with the vaccinia virus system, and the WSU Pathology Research Core Facility for assistance with immunohistochemistry assays. Special thanks to Dr. Alan Dombkowski and Amy Diakiw of the WSU Microarray and Bioinformatics Facility Core at the Institute of Environmental Health Sciences for their work with the microarray analysis. Microarray and Bioinformatics Core was supported in part by NIEHS Center Grant P30 ES06639. The Microscopy, Imaging and Cytometry Resources Core is supported, in part, by NIH Center grant P30CA22453 to Karmanos Cancer Institute and the Perinatology Research Branch of the National Institutes of Child Health and Development, WSU.
REFERENCES
- 1.Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22:1276–1312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li X, Ponten A, Aase K, et al. PDGF-C is a new protease-activated ligand for the PDGF alpha-receptor. Nat Cell Biol. 2000;2:302–309. doi: 10.1038/35010579. [DOI] [PubMed] [Google Scholar]
- 3.Bergsten E, Uutela M, Li X, et al. PDGF-D is a specific, protease-activated ligand for the PDGF beta-receptor. Nat Cell Biol. 2001;3:512–516. doi: 10.1038/35074588. [DOI] [PubMed] [Google Scholar]
- 4.LaRochelle WJ, Jeffers M, McDonald WF, et al. PDGF-D, a new protease-activated growth factor. Nat Cell Biol. 2001;3:517–521. doi: 10.1038/35074593. [DOI] [PubMed] [Google Scholar]
- 5.Fredriksson L, Li H, Fieber C, Li X, Eriksson U. Tissue plasminogen activator is a potent activator of PDGF-CC. Embo J. 2004;23:3793–3802. doi: 10.1038/sj.emboj.7600397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ustach CV, Kim HR. Platelet-derived growth factor D is activated by urokinase plasminogen activator in prostate carcinoma cells. Mol Cell Biol. 2005;25:6279–6288. doi: 10.1128/MCB.25.14.6279-6288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fudge K, Wang CY, Stearns ME. Immunohistochemistry analysis of platelet-derived growth factor A and B chains and platelet-derived growth factor alpha and beta receptor expression in benign prostatic hyperplasias and Gleason-graded human prostate adenocarcinomas. Mod Pathol. 1994;7:549–554. [PubMed] [Google Scholar]
- 8.George DJ. Receptor tyrosine kinases as rational targets for prostate cancer treatment: platelet-derived growth factor receptor and imatinib mesylate. Urology. 2002;60:115–121. doi: 10.1016/s0090-4295(02)01589-3. discussion 22. [DOI] [PubMed] [Google Scholar]
- 9.Chott A, Sun Z, Morganstern D, et al. Tyrosine kinases expressed in vivo by human prostate cancer bone marrow metastases and loss of the type 1 insulin-like growth factor receptor. Am J Pathol. 1999;155:1271–1279. doi: 10.1016/S0002-9440(10)65229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh D, Febbo PG, Ross K, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 11.Uehara H, Kim SJ, Karashima T, et al. Effects of blocking platelet-derived growth factor-receptor signaling in a mouse model of experimental prostate cancer bone metastases. J Natl Cancer Inst. 2003;95:458–470. doi: 10.1093/jnci/95.6.458. [DOI] [PubMed] [Google Scholar]
- 12.Ko YJ, Small EJ, Kabbinavar F, et al. A multi-institutional phase ii study of SU101, a platelet-derived growth factor receptor inhibitor, for patients with hormone-refractory prostate cancer. Clin Cancer Res. 2001;7:800–805. [PubMed] [Google Scholar]
- 13.Kim SJ, Uehara H, Yazici S, et al. Simultaneous blockade of platelet-derived growth factor-receptor and epidermal growth factor-receptor signaling and systemic administration of paclitaxel as therapy for human prostate cancer metastasis in bone of nude mice. Cancer Res. 2004;64:4201–4208. doi: 10.1158/0008-5472.CAN-03-3763. [DOI] [PubMed] [Google Scholar]
- 14.van der Poel HG. Smart drugs in prostate cancer. Eur Urol. 2004;45:1–17. doi: 10.1016/j.eururo.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 15.Sintich SM, Lamm ML, Sensibar JA, Lee C. Transforming growth factor-beta1-induced proliferation of the prostate cancer cell line, TSU-Pr1: the role of platelet-derived growth factor. Endocrinology. 1999;140:3411–3415. doi: 10.1210/endo.140.8.6921. [DOI] [PubMed] [Google Scholar]
- 16.Ng SS, MacPherson GR, Gutschow M, Eger K, Figg WD. Antitumor effects of thalidomide analogs in human prostate cancer xenografts implanted in immunodeficient mice. Clin Cancer Res. 2004;10:4192–4197. doi: 10.1158/1078-0432.CCR-03-0700. [DOI] [PubMed] [Google Scholar]
- 17.Fudge K, Bostwick DG, Stearns ME. Platelet-derived growth factor A and B chains and the alpha and beta receptors in prostatic intraepithelial neoplasia. Prostate. 1996;29:282–286. doi: 10.1002/(SICI)1097-0045(199611)29:5<282::AID-PROS2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 18.Ustach CV, Taube ME, Hurst NJ, Jr, et al. A potential oncogenic activity of platelet-derived growth factor d in prostate cancer progression. Cancer Res. 2004;64:1722–1729. doi: 10.1158/0008-5472.can-03-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paoloni-Giacobino A, Chen H, Peitsch MC, Rossier C, Antonarakis SE. Cloning of the TMPRSS2 gene, which encodes a novel serine protease with transmembrane, LDLRA, and SRCR domains and maps to 21q22.3. Genomics. 1997;44:309–320. doi: 10.1006/geno.1997.4845. [DOI] [PubMed] [Google Scholar]
- 20.Takeuchi T, Shuman MA, Craik CS. Reverse biochemistry: use of macromolecular protease inhibitors to dissect complex biological processes and identify a membrane-type serine protease in epithelial cancer and normal tissue. Proc Natl Acad Sci U S A. 1999;96:11054–11061. doi: 10.1073/pnas.96.20.11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin CY, Anders J, Johnson M, Dickson RB. Purification and characterization of a complex containing matriptase and a Kunitz-type serine protease inhibitor from human milk. J Biol Chem. 1999;274:18237–18242. doi: 10.1074/jbc.274.26.18237. [DOI] [PubMed] [Google Scholar]
- 22.Riddick AC, Shukla CJ, Pennington CJ, et al. Identification of degradome components associated with prostate cancer progression by expression analysis of human prostatic tissues. Br J Cancer. 2005;92:2171–2180. doi: 10.1038/sj.bjc.6602630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saleem M, Adhami VM, Zhong W, et al. A novel biomarker for staging human prostate adenocarcinoma: overexpression of matriptase with concomitant loss of its inhibitor, hepatocyte growth factor activator inhibitor-1. Cancer Epidemiol Biomarkers Prev. 2006;15:217–227. doi: 10.1158/1055-9965.EPI-05-0737. [DOI] [PubMed] [Google Scholar]
- 24.Kang JY, Dolled-Filhart M, Ocal IT, et al. Tissue microarray analysis of hepatocyte growth factor/Met pathway components reveals a role for Met, matriptase, and hepatocyte growth factor activator inhibitor 1 in the progression of node-negative breast cancer. Cancer Res. 2003;63:1101–1105. [PubMed] [Google Scholar]
- 25.Lee JW, Yong Song S, Choi JJ, et al. Increased expression of matriptase is associated with histopathologic grades of cervical neoplasia. Hum Pathol. 2005;36:626–633. doi: 10.1016/j.humpath.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Oberst MD, Johnson MD, Dickson RB, et al. Expression of the serine protease matriptase and its inhibitor HAI-1 in epithelial ovarian cancer: correlation with clinical outcome and tumor clinicopathological parameters. Clin Cancer Res. 2002;8:1101–1107. [PubMed] [Google Scholar]
- 27.Zeng L, Cao J, Zhang X. Expression of serine protease SNC19/matriptase and its inhibitor hepatocyte growth factor activator inhibitor type 1 in normal and malignant tissues of gastrointestinal tract. World J Gastroenterol. 2005;11:6202–6207. doi: 10.3748/wjg.v11.i39.6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedrich R, Fuentes-Prior P, Ong E, et al. Catalytic domain structures of MT-SP1/matriptase, a matrix-degrading transmembrane serine proteinase. J Biol Chem. 2002;277:2160–2168. doi: 10.1074/jbc.M109830200. [DOI] [PubMed] [Google Scholar]
- 29.Lin CY, Wang JK, Torri J, Dou L, Sang QA, Dickson RB. Characterization of a novel, membrane-bound, 80-kDa matrix-degrading protease from human breast cancer cells. Monoclonal antibody production, isolation, and localization. J Biol Chem. 1997;272:9147–9152. [PubMed] [Google Scholar]
- 30.Takeuchi T, Harris JL, Huang W, Yan KW, Coughlin SR, Craik CS. Cellular localization of membrane-type serine protease 1 and identification of protease-activated receptor-2 and single-chain urokinase-type plasminogen activator as substrates. J Biol Chem. 2000;275:26333–26342. doi: 10.1074/jbc.M002941200. [DOI] [PubMed] [Google Scholar]
- 31.Uhland K. Matriptase and its putative role in cancer. Cell Mol Life Sci. 2006;63:2968–2978. doi: 10.1007/s00018-006-6298-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santin AD, Cane S, Bellone S, et al. The novel serine protease tumor-associated differentially expressed gene-15 (matriptase/MT-SP1) is highly overexpressed in cervical carcinoma. Cancer. 2003;98:1898–1904. doi: 10.1002/cncr.11753. [DOI] [PubMed] [Google Scholar]
- 33.Lee SL, Dickson RB, Lin CY. Activation of hepatocyte growth factor and urokinase/plasminogen activator by matriptase, an epithelial membrane serine protease. J Biol Chem. 2000;275:36720–36725. doi: 10.1074/jbc.M007802200. [DOI] [PubMed] [Google Scholar]
- 34.Yu J, Ustach C, Kim HR. Platelet-derived growth factor signaling and human cancer. J Biochem Mol Biol. 2003;36:49–59. doi: 10.5483/bmbrep.2003.36.1.049. [DOI] [PubMed] [Google Scholar]
- 35.Ittman M, Mansukhani A. Expression of fibroblast growth factors (FGFs) and FGF receptors in human prostate. J Urol. 1997;157:351–356. [PubMed] [Google Scholar]
- 36.Kambhampati S, Ray G, Sengupta K, Reddy VP, Banerjee SK, Van Veldhuizen PJ. Growth factors involved in prostate carcinogenesis. Front Biosci. 2005;10:1355–1367. doi: 10.2741/1625. [DOI] [PubMed] [Google Scholar]
- 37.Tam NN, Chung SS, Lee DT, Wong YC. Aberrant expression of hepatocyte growth factor and its receptor, c-Met, during sex hormone-induced prostatic carcinogenesis in the Noble rat. Carcinogenesis. 2000;21:2183–2191. doi: 10.1093/carcin/21.12.2183. [DOI] [PubMed] [Google Scholar]
- 38.Algaba F, Trias I, Arce Y. Natural history of prostatic carcinoma: the pathologist's perspective. Recent Results Cancer Res. 2007;175:9–24. doi: 10.1007/978-3-540-40901-4_2. [DOI] [PubMed] [Google Scholar]
- 39.Long RM, Morrissey C, Fitzpatrick JM, Watson RW. Prostate epithelial cell differentiation and its relevance to the understanding of prostate cancer therapies. Clin Sci (Lond) 2005;108:1–11. doi: 10.1042/CS20040241. [DOI] [PubMed] [Google Scholar]
- 40.Harnden P, Shelley MD, Clements H, et al. The prognostic significance of perineural invasion in prostatic cancer biopsies: a systematic review. Cancer. 2007;109:13–24. doi: 10.1002/cncr.22388. [DOI] [PubMed] [Google Scholar]
- 41.Beard C, Schultz D, Loffredo M, et al. Perineural invasion associated with increased cancer-specific mortality after external beam radiation therapy for men with low- and intermediate-risk prostate cancer. Int J Radiat Oncol Biol Phys. 2006;66:403–407. doi: 10.1016/j.ijrobp.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 42.Lee IH, Roberts R, Shah RB, Wojno KJ, Wei JT, Sandler HM. Perineural invasion is a marker for pathologically advanced disease in localized prostate cancer. Int J Radiat Oncol Biol Phys. 2007 doi: 10.1016/j.ijrobp.2007.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Villers A, McNeal JE, Redwine EA, Freiha FS, Stamey TA. The role of perineural space invasion in the local spread of prostatic adenocarcinoma. J Urol. 1989;142:763–768. doi: 10.1016/s0022-5347(17)38881-x. [DOI] [PubMed] [Google Scholar]
- 44.Ayala GE, Wheeler TM, Shine HD, et al. In vitro dorsal root ganglia and human prostate cell line interaction: redefining perineural invasion in prostate cancer. Prostate. 2001;49:213–223. doi: 10.1002/pros.1137. [DOI] [PubMed] [Google Scholar]
- 45.Ayala GE, Dai H, Ittmann M, et al. Growth and survival mechanisms associated with perineural invasion in prostate cancer. Cancer Res. 2004;64:6082–6090. doi: 10.1158/0008-5472.CAN-04-0838. [DOI] [PubMed] [Google Scholar]
- 46.Mathew P, Thall PF, Jones D, et al. Platelet-derived growth factor receptor inhibitor imatinib mesylate and docetaxel: a modular phase I trial in androgen-independent prostate cancer. J Clin Oncol. 2004;22:3323–3329. doi: 10.1200/JCO.2004.10.116. [DOI] [PubMed] [Google Scholar]
- 47.Guilhot F. Indications for imatinib mesylate therapy and clinical management. Oncologist. 2004;9:271–281. doi: 10.1634/theoncologist.9-3-271. [DOI] [PubMed] [Google Scholar]
- 48.Kerkela R, Grazette L, Yacobi R, et al. Cardiotoxicity of the cancer therapeutic agent imatinib mesylate. Nat Med. 2006;12:908–916. doi: 10.1038/nm1446. [DOI] [PubMed] [Google Scholar]
- 49.Deininger MW, O'Brien SG, Ford JM, Druker BJ. Practical management of patients with chronic myeloid leukemia receiving imatinib. J Clin Oncol. 2003;21:1637–1647. doi: 10.1200/JCO.2003.11.143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.