Abstract
Abdominal aortic aneurysm (AAA) is a multifactorial disease with a strong genetic component. Since first candidate gene studies were published 20 years ago, nearly 100 genetic association studies using single nucleotide polymorphisms (SNPs) in biologically relevant genes have been reported on AAA. The studies investigated SNPs in genes of the extracellular matrix, the cardiovascular system, the immune system, and signaling pathways. Very few studies were large enough to draw firm conclusions and very few results could be replicated in another sample set. The more recent unbiased approaches are family-based DNA linkage studies and genome-wide genetic association studies, which have the potential of identifying the genetic basis for AAA, if appropriately powered and well-characterized large AAA cohorts are used. SNPs associated with AAA have already been identified in these large multicenter studies. One significant association was of a variant in a gene called CNTN3 which is located on chromosome 3p12.3. Two follow-up studies, however, could not replicate the association. Two other SNPs, which are located on chromosome 9p21 and 9q33 were replicated in other samples. The two genes with the strongest supporting evidence of contribution to the genetic risk for AAA are the CDKN2BAS gene, also known as ANRIL, which encodes an antisense RNA that regulates expression of the cyclin-dependent kinase inhibitors CDKN2A and CDKN2B, and DAB2IP, which encodes an inhibitor of cell growth and survival. Functional studies are now needed to establish the mechanisms by which these genes contribute to AAA pathogenesis.
INTRODUCTION
Abdominal aortic aneurysm (AAA) is a life threatening disease that affects 10% of Caucasian males over 65 years of age.1 It is the 17th leading cause of death in the United States with about 15,000 deaths per year (WISQARS Leading Causes of Death Reports, 1999–2004.2007). Thanks to improved diagnostic efforts using ultrasonography imaging of the abdominal aorta to detect a dilatation, AAAs are identified earlier and only about 15% of AAA present as ruptured AAA in hospitals of industrialized nations.2
AAA is a complex disease with several recognized risk factors such as smoking, male sex, age over 65 years, and positive family history.1, 3, 4 Protective factors are type 2 diabetes mellitus, African-American ethnicity and female sex.5–9 The familial nature of AAA was first reported in a study published in 1977 where three brothers were all described to have AAA.10 Subsequent studies on families in which at least two relatives had AAA, demonstrated that the risk for developing an AAA is eight-fold higher for a first-degree relative than in the general population.11 It is, therefore, common practise for every vascular surgeon to include questions about family history of AAA when examining a patient.
The first studies on the genetic basis of AAA were reported more than 25 years ago and compared the frequencies of blood groups or HLA-A and HLA-B in AAA patients and controls.12 Subsequent candidate gene studies in single AAA families with several affected individuals identified mutations in connective tissue genes.13–16 These mutations, however, were not seen in sporadic AAA patients. More recently genome-wide approaches were used to provide unbiased ways of identifying genetic risk factors for AAA. These included DNA linkage studies in affected relative pairs from AAA families17, 18 and genome-wide association studies (GWAS)19, 20 with AAA cases and controls.
Another approach to study disease pathogenesis at the molecular level is to carry out a genome-wide expression analysis to identify genes actively transcribed in AAA and non-aneurysmal aortic tissue. Only one such large-scale microarray-based study has been reported for AAA tissue21 and it identified over 3,000 genes with significantly elevated or decreased expression levels in AAA aortic wall tissue compared to control aortic tissue. A follow-up study analyzed the promoter regions of the up- and downregulated genes with an in silico approach and determined the transcription factor (TF) binding sites which were overrepresented in the 5-kb promoter regions of the 3,274 genes. 22 There were only 13 distinct overrepresented transcription factor binding sites on the upregulated genes and eight of them belong to the ETS family. Additionally, NFκB and its subunits p50 and p65 also showed enrichment.22 Another microarray-based expression study investigated the differences between ruptured and unruptured AAA samples.23 A severe limitation of these studies is the fact that tissue specimens can be obtained only from the end-stage disease, when the patient undergoes repair operation. In most cases the architecture of the aortic wall has changed considerably. Expression profiles of peripheral blood have also been generated for AAA patients24 with the hope of being able to study different stages of the AAA development, and identify biomarkers for AAA.
Studies on early stages of AAA development are currently feasible only in animal models. Three mouse models have been established but none of them simulate all the features of human AAAs.4 For example, the widely-used model in which aneurysms are induced with angiotensin II-treatment in mice lacking the apolipoprotein E gene (APOE) produces suprarenal AAAs with dissection, which is only in rare cases a feature of the human infrarenal AAAs.
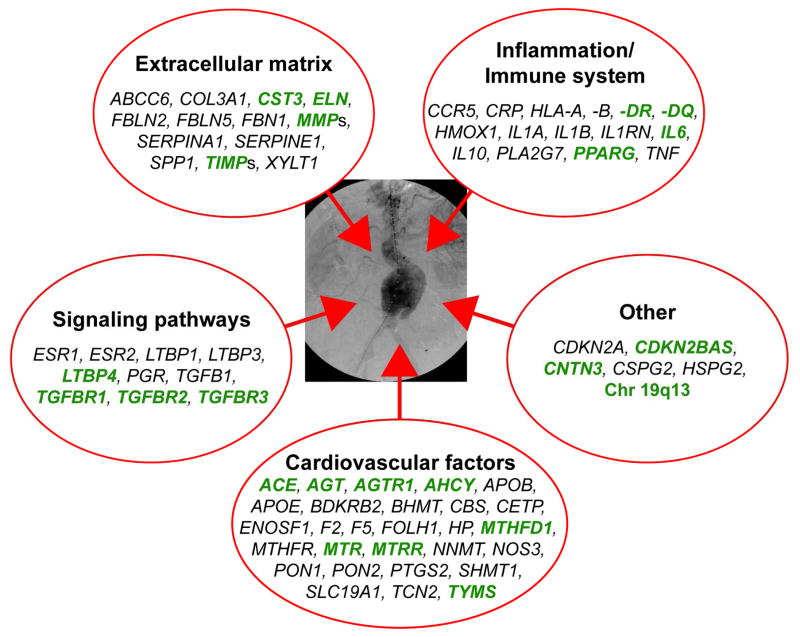
In this review we summarize the results of nearly 100 genetic studies on AAA. The individual studies and their results are summarized in Tables I-VI. To help the reader evaluate these studies we have separated the studies showing genetic association from those with no association. Furthermore, we provide an indicator of the robustness of the result based on the number of AAA patients and controls studied. Finally, for those readers of the Annals who just want a quick summary, Figure 1 provides the genes studied with associations indicated in green type. Most of these findings still need to be replicated in additional sample sets. In addition, the biological mechanisms need to be elucidated as to how the variation in each of the replicated genes contribute to increased risk for AAA. The ultimate goals of genetic research on AAA are to 1) predict who is at increased risk for developing AAA, and 2) identify the biological pathways involved in AAA pathogenesis and subsequently design ways to interfere their function to slow growth or prevent rupture of AAAs. To date these goals have not yet been attained, but some progress has been made.
Table I.
AAA candidate genes investigated by DNA sequencing
| Gene | Sequence variant | Number of AAA patients/controls |
|
|---|---|---|---|
| with mutations | without mutations | ||
| COL3A1 | G619R | 1 Family15 | |
| G136R, T501P | 1 Family16 | 49 Families16 | |
| 607 C>T | 113 | 39/2913 | |
| G+1 IVS20 | 1 Family14 | ||
| FBLN2 | 1203 G>A, 1295 C>T, 500/502 G>A, 1115 G>A, 1150 A>G, 1229 A>C, 1278 A>G, 1779 C>T, 2574 G>A, 2629 G>A, 2754 C>T, 3408 G>A, 3540 T>C | 11/228 | |
| MMP2 | IVS1+31 C>G, IVS7−18 G>A, IVS10+26 C>T, 124 G>A, 1368 C>T, 1860 C>T, −1586 C>T, −1070 T>G, −61 G>C, 228 G>A, 678 G>C, 750 C>T, IVS512 C>T, C1149 C>T, 1380 G>A, rs243834, rs11541998, rs10775332 | 51/4832 | |
| TIMP1 | 434 C>T | 6/036 | 64/29 (only males)37 |
| 20/22 (only females)37 | |||
| rs4898 | 50/4438 | 96/8938 | |
| rs1043428, 328+16 C>T | 50/4438 | ||
| 323 C>T | 84/5137 | ||
| TIMP2 | 573 G>A | 64/29 (only males)37 | 20/22 (only females)37 |
| 306 C>T | 84/5137 | ||
| −621 C>T, −596 A>C, rs8179096, −261 G>A, 231+23 C>T, rs2277698 | 50/4141 | ||
| CETP | 4 microsatellite loci | 82/7942 | |
| Exon 9 nt 170 A>T, Intron 12 nt 312 C>T, Exon 14 nt 16 G>A, Intron 15 nt 151 G>A | 85/3443 | ||
Gene symbols available from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) were used.
Table VI.
Family-based DNA linkage studies, genome-wide association studies (GWAS) and follow-up studies
| Locus | Polymorphisms | Number of studies* |
|
|---|---|---|---|
| Associated | Not associated | ||
|
19q1317, 18 | |||
| 608 SNPs | 1 (1)203 | ||
| rs16968029, rs281407, rs1075453, rs6509496, rs1530878, rs285676, rs576556, rs4802552 | 1 (1)203 | 1 (1)203 | |
|
9p21208 | |||
| rs10116277, rs1333040, rs2383207 | 2 (2)20 | ||
| rs7025486 | 2 (2)20 | ||
| CDKN2BAS | rs10757278-G | 5 (5)208 | 4 (4)208, 210 |
| CDKN2A | rs10811661-T | 7 (7)208 | |
| HSPG2 | rs2270701, rs3767137, rs7556412, rs7355045, rs2305562, rs12404444, rs2454290, rs11810496, rs2501255, rs6698486, rs4654773, rs12081298 | 1 (1)211 | |
| CSPG2 | rs2652106 | 1 (1)211 | 1 (1)211 |
| rs28899, rs962066, rs11746859, rs33599, rs6452540, rs160380, rs13686, rs17348129, rs188703, rs2445874, rs647873, rs309596, rs309590, rs309587, rs12653308, rs17205986, rs160187, rs160280, rs309584, rs309578, rs11726 | 1 (1)211 | ||
|
3p12.319 | |||
| rs9876789, rs6549604, rs4076052 | 1 (1)207 | ||
| CNTN3 | rs7635818 (upstream to CNTN3) | 2 (2)19 | 3 (3)20, 207 |
|
9p3320 | |||
| rs7025486 | 4 (4)20 | 6 (6)20 | |
Number in parenthesis indicates the number of studies with a sample size of at least 150 cases and 150 controls.
If multiple sample sets were used in one publication, they were treated as independent studies.
Gene symbols available from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) were used.
Fig. 1.
Summary on the biological pathways thought to be involved in AAA pathogenesis. The genes investigated for conferring genetic risk for AAA can be divided into five different functional groups. Genes with best evidence for contributing to AAA pathogenesis are shown in green. Studies on these genes had sufficient numbers of cases and controls making it likely that the results are not false positive associations. Details on the results of each gene are provided in Tables II through VI. To make it easier to find the genes in each functional group we have alphabetized the gene symbols, which are available from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/).
CANDIDATE GENE STUDIES (Tables I–V)
Table V.
Genes of signaling pathways
| Gene | Polymorphisms | Number of studies* |
|
|---|---|---|---|
| Associated | Not associated | ||
| TGFB1 | Bsu361 RFLPs | 1 (0)56 | |
| −1347 G>A, 7440 A>C, 11089 G>A, 13077 A>G, 20743 G>A, 25782 C>T, 26718 G>A | 3 (3)199 AAA expansion | ||
| −509 T>C | 1 (1) 48 | ||
| TGFBR1 | rs10819634 | 2 (2)200 | |
| rs1571590 | 1 (1)200 | 1 (1)200 | |
| rs1626340 | 1 (1)200 | 1 (1)200 | |
| 9A/6A | 1 (1)105 | ||
| TGFBR2 | rs1078985 | 2 (2)201 | |
| rs764522, rs3087465, rs13075948, rs9831477, rs1346907, rs9843143 | 1 (1)200 | 1 (1)200 | |
| rs1036095, rs4522809 | 2 (2)200 | ||
| rs304839 | 2 (2)200 | ||
| TGFB3 | −614 G>A | 1 (1)199 AAA expansion | 2 (2)199 AAA expansion |
| 715 T>G, 2469 T>C, 7602 T>C, 17368 A>G, 19456 A>T, 23653 G>A | 3 (3)199 AAA expansion | ||
| LTBP1 | −256 G>C | 3 (3)199 AAA expansion | |
| LTBP3 | 4598 A>G, 9048 T>C, 22537 G>A | 3 (3)199 AAA expansion | |
| LTBP4 | −4234 A>G, 10384 G>A, 21011 A>T, 25859 C>T, 32603 C>G | 1 (1)199 AAA expansion | 2 (2)199 AAA expansion |
| 1334C>G, 573 G>A, 20757 C>T | 3 (3)199 AAA expansion | ||
| ESR1 | −397 T>C, −351 A>G | 1 (0)56 | |
| ESR2 | 1730 A>G | 1 (0)56 | |
| PGR | TaqI RFLPs | 1 (0)56 | |
Number in parenthesis indicates the number of studies with a sample size of at least 150 cases and 150 controls.
If multiple sample sets were used in one publication, they were treated as independent studies. AAA, abdominal aortic aneurysm
Gene symbols available from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) were used.
Candidate gene study-approach has been used widely and has been the most common form of study design for the genetics of AAA. It requires selection of candidate genes based on their biological function and plausibility as genes with potential role in disease pathogenesis. One approach is to sequence the complete coding region, adjacent parts of the introns and the promoter of a biologically relevant candidate gene in the patient’s DNA to find new sequence variants. Follow-up studies determine the variant’s frequency in the general population and in larger patient groups. Most sequencing studies analysed only a small number of individuals (Table I). The other approach is to use known genetic markers called polymorphisms and carry out a genetic association study with cases and controls (Tables II–V). Most studies use SNPs as markers since they are easy to detect with high-throughput techniques. Blood samples are collected from cases and controls and DNA isolated from the blood. Only small quantities of genomic DNA are needed from the study subjects to perform these studies using PCR-based genotyping assays.
Table II.
Genes of the extracellular matrix (ECM) – structure and remodeling of the aortic wall
| Gene | Polymorphisms | Number of studies* |
|
|---|---|---|---|
| Associated | Not associated | ||
| COL3A1 | G619R | 3 (0)44 in 28 AAA-families, 45, 46 | |
| A531T | 3 (0)44, 45, 47 one study on AAA+PAA | ||
| 581 T>C | 1 (1)48 | ||
| Ava II rare allele | 1 (0)45 | ||
| FBN1 | 4 allele repeat | 1 (0)47 AAA vs AAA+PAA | |
| ELN | 422 G>A | 1 (1)48 in males with family history | 2 (1)48, 56 |
| ABCC6 | 31 mutations, c.1964 A>G, c.2171 G>A, c.2175 A>T, c.2224 A>T, c.4254 G>A | 1 (1)57 | |
| FBLN5 | rs2498834, rs2430366, rs2254320 | 1 (1)65 | |
| MMP1 | −1670 G/GG | 1 (1)48 | |
| MMP2 | −955 A>C | 1 (1)48 | |
| −1360 C>T | 2 (2)67, 68 AAA expansion | ||
| rs243865, rs2053605 | 1 (1)216 | ||
| MMP3 | −1612 5A/6A | 2 (2)48 in males with family history,70 | 3 (2)48, 67, 71 one study on AAA expansion |
| −1171 5A/6A | 1 (1)68 AAA expansion | ||
| MMP9 | rs3918242 | 1 (1)74 AAA vs PVD | 8 (6)48, 67, 68, 71, 75, 76 two studies on AAA expansion |
| −131 to −90 (CA repeat) | 2 (0)76, 77 | ||
| MMP10 | 180 A>G | 1 (1)48 | |
| MMP12 | −82 A>G | 3 (3)48, 67, 68 two studies on AAA expansion | |
| MMP13 | −77 A>G | 1 (1)48 | |
| TIMP1 | 434 C>T | 1 (1)48 males without family history | 1 (1)48 males with family history |
| −372 C>T | 1 (1)68 AAA expansion | ||
| rs2070584 | 1 (1)48 males without family history | 1 (1)48 males with family history | |
| TIMP2 | rs2009196 | 1 (1)48 | |
| TIMP3 | −1296 T>C | 1 (1)48 | |
| SERPINA1 | α1-AT P1 deficiency | 1 (0)82 | 1 (0)81 AAA vs IA |
| SERPINE1 | −675 4G/5G | 1 (0)217 AAA families | 3 (3)67, 68, 71, 86 three studies on AAA expansion |
| −847 A>G | 1 (1)68 AAA expansion | ||
| SPP1 | rs1126772, rs9138, rs4754, rs1126616, rs11730582 | 1 (1)91 | |
| XYLT1 | 343 G>T | 1 (0)92 | |
| CST3 | −148 G>A | 1 (1)96 AAA expansion | |
| −82 G>C | 1 (1)96 AAA expansion | ||
Number in parenthesis indicates the number of studies with a sample size of at least 150 cases and 150 controls.
If multiple sample sets were used in one publication, they were treated as independent studies.
AAA, abdominal aortic aneurysm; PAA, popliteal artery aneurysm; IA, intracranial aneurysm; PVD: peripheral vascular disease
Gene symbols available from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) were used.
Analysis of Candidate Genes by DNA Sequencing (Table I)
In only a few studies the entire candidate gene was sequenced in a family or a group of AAA patients (Table I). Since mutations in fibrillin I (FBN1) gene were found to cause the Marfan syndrome,25 it was plausible to consider a role for the extracellular matrix (ECM) genes also in AAA, since these molecules are the main structural components of the aortic wall. Thus the genes for the α1 chain of type III collagen (COL3A1), elastin (ELN), fibulin 2 (FBLN2), FBN1 and the genes of matrix degrading proteases and their inhibitors were of great interest in AAA research.
The α1 chain of type III collagen (COL3A1)
Collagens are major components of the ECM, and provide support and tensile strength to the aorta.26 The loss of collagen plays a role in aneurysm rupture.26 Mutations in this gene are responsible for the rare genetic disorder Ehlers-Danlos syndrome type IV, also known as the vascular type of the Ehlers-Danlos syndrome.27 COL3A1 was the first candidate gene to be investigated by DNA sequencing in patients with aortic aneurysms. Sequencing revealed a G619R mutation in a family with three generations of individuals with aortic aneurysms15 and an RNA splicing mutation G+1 IVS20 in another family.14 Detailed clinical evaluation showed, however, that both of these families likely had the Ehlers-Danlos syndrome type IV even though diagnosis had not been made before the sequencing results were revealed.
In subsequent studies the COL3A1 gene was sequenced in 50 different families with at least one additional blood relative with AAA.16 Two mutations were detected (G136R and T501P), but they were present in only 2% of AAA patients.16 Another study examined the COL3A1 sequences in 40 AAA patients with 22.5% having positive family history and 29 patients with aortic occlusive disease.13 Only one patient among the 40 AAA patients was found with a mutation (607C>T, which replaces a leucine with phenylalanine) in the coding region of COL3A1.13 In conclusion, mutations in the COL3A1 can be found in only a small fraction of AAA patients, most likely in patients who have an undiagnosed Ehlers-Danlos syndrome type IV.
Fibulin 2 (FBLN2)
FBLN2 is also a protein of the ECM. A candidate gene study in which the cDNA of FBLN2 was sequenced in eleven AAA patients and two controls found no mutations. A total of 14 SNPs were detected, but these variants did not segregate with AAA in families.28
Matrix metallopeptidase 2 (MMP2)
Matrix metallopeptidase 2 (MMP2) is one of the most abundant collagen-degrading proteinases in the ECM. Increased mRNA and protein levels in the wall of AAA have been shown in several studies.29–31 The coding region (13 exons with adjacent parts of the introns), 5′-untranslated region (UTR) and selected parts of the promoter region of the MMP2 gene were analyzed from genomic DNA in 51 AAA patients and 48 controls. No mutations were found. Eighteen SNPs were described, ten of which were located in the coding region, three SNPs in the promoter region and 5′-UTR, but none of the SNPs showed an association with the AAA phenotype.32
Tissue inhibitor of metallopeptidase 1 and 2 (TIMP1 and 2)
Metallopeptidases and their inhibitors, the TIMPs have a dynamic balance in the ECM. Imbalance of their activity or expression levels can cause degradation of the ECM. Decreased mRNA levels of TIMPs have been reported in the aneurysmal aortic wall.33, 34 In a mouse model, elastase perfusion led to a development of larger aneurysms in Timp1-deficient mice than wild-type mice.35 In humans, TIMP1 is localized on the X chromosome and is, therefore, of special interest in AAA, which is a disease with male predominance.
The first DNA sequencing analysis of TIMP1 was carried out in six AAA patients and revealed a silent polymorphism in two of the six patients.36 A subsequent sequencing study found a SNP (C>T) at nucleotide (nt) position 434.37 The investigators showed a significant association with the C-allele in female AAA patients.37 Due to the small number (n = 20) of patients analyzed, the results should be interpreted with caution. Another sequence analysis of the coding region of TIMP1 gene (six exons) in 50 patients did not detect any mutations and the three SNPs that were found were not associated with AAA.38
Serum levels of TIMP2 are decreased in AAA patients.39 Additionally the ratio of MMP2/TIMP2 mRNA levels is elevated in AAA tissue compared to control aortic tissue40 suggesting a role for TIMP2 in AAA development. One DNA sequencing study of the TIMP2 gene revealed a SNP at nt 573G>A. The G allele was significantly more frequent in males with AAA (n = 64) than in male controls (n = 29).37 Another complete sequence analysis of the TIMP2 gene (50 AAA patients and 41 controls) identified six polymorphisms, but no mutations and did not find an association between the SNP nt 573G>A with AAA phenotype.41
Cholesteryl ester transfer protein (CETP)
After a promising, although small (44 AAA cases and 32 controls), genetic association study published in 1990 in which a polymorphism at the CETP locus was associated with AAA,42 a candidate gene study of the 16 exons in the CETP gene was performed by single stranded conformation polymorphism analysis, followed by a restriction enzyme analysis and DNA sequencing in 85 AAA patients and 34 controls. The study revealed four polymorphisms (two in exons 9 and 14, and two in introns 12 and 15). The polymorphism in exon 9 was an A>T transversion which produced a stop codon in the CETP protein and could potentially result in an inactive protein. The polymorphism in exon 14 was a G>A transition which resulted in a substitution of isoleucine for valine. The surprising finding was that none of the polymorphisms differed in frequency between the cases and the controls.43
Genetic Association Studies Using Known Polymorphisms in Candidate Genes (Tables II–V)
Most genetic studies on AAA have been genetic association studies which focused on specific known SNPs or other genetic variants (see below) in biologically plausible candidate genes. Taken together the results from these various genetic studies on AAA suggest that mutations in one single gene cannot account for the liability to AAA. Altogether 83 candidate genes with more than 1,000 SNPs or other gene alterations have been studied. Most of the early genetic association studies were substantially underpowered with fewer than 150 cases and 150 controls. The number of samples required for a genetic association study is dependent on allele frequency, genotypic relative risk (penetrance), linkage disequilibrium (marker to effective variant) and can be much larger than 150.
The candidate genes can be divided into four different groups4 according to their potential role in the AAA pathophysiology (Fig. 1; Tables II through VI). We discuss these categories separately in this review. All studies published by July 2010 and identified in a PubMed search were included in this review.
Group I: Genes of the extracellular matrix (ECM) – Structure and Remodelling of the Aorta (Table II)
Type III Collagen (COL3A1)
As mentioned above COL3A1 gene was the first ECM gene analyzed by DNA sequencing (Table I). Using genetic association study approach, five reports have been published investigating the potential role of COL3A1 polymorphisms in AAA pathogenesis,44–48 but no evidence of association was shown in any of these studies.
Fibrillin I (FBN1)
A large number of different mutations have been identified in FBN1 in patients with the Marfan syndrome.49–51 Concerning AAA, one study, which compared patients with AAA to those with AAA and popliteal artery aneurysms (PAA), showed a significant difference in the frequency of a repeat polymorphism in the FBN1 gene between the two study groups, but the number of samples investigated was too small to draw any conclusions about the findings.47
Elastin (ELN)
Elastin is also a component of the aortic ECM, accounting for 30% of the aorta by weight.52 Its function is to maintain the shape of the vessel,26 and loss of elastin in the aortic wall causes dilatation without rupture.26 Elastin deposition in the vessel wall does not continue in adulthood. Mutations in the ELN gene cause supravalvular aortic stenosis and Williams-Beuren syndrome (different cardiac valve pathologies with neurobehavioral features and mental retardation).53, 54 Mutations in the ELN gene are also found in patients with another connective tissue disease, cutis laxa.55 Two association studies investigated a SNP located in the ELN gene (nt 422 G>A) in AAA patients and controls.48, 56 No association was shown in sporadic AAA cases,48, 56 but a nominally significant association was shown in a subgroup of patients with a positive family history.48
ATP-binding cassette transporter, subfamily C (CFTR/MRP), member 6 (ABCC6)
Mutations in the ABCC6 gene are known to cause pseudoxanthoma elasticum (PXE), a hereditary connective tissue disorder characterized by fragmentation of elastic fibers. It is, therefore, plausible that this gene might play a role also in AAA.57 One study investigated ABCC6 polymorphisms in 133 AAA patients and 910 controls, detected only five genetic variants (missense or silent amino acid variants) in the 133 AAA cases, and no significant associations.57
Fibulin 5 (FBLN5)
Fibulin 5 is expressed in the great vessels during embryogenesis and in many adult tissues including the aorta.58–60 It is an elastin-binding protein which localizes to the surface of elastic fibers in vivo.58 Mice lacking Fbln5 gene have fragmented elastin.61 The protein acts as a bridging peptide between blood vessel wall cell surface integrins and elastin fibers.62 Mutations in FBLN5 gene cause cutis laxa and might contribute to age-related macular degeneration.63, 64 Thus FBLN5 is of interest also in the AAA pathogenesis. One recent genetic association study on AAA used three SNPs in the FBLN5 gene in a total of 230 AAA cases and 278 controls and showed no association with AAA.65
Family of Matrix Metallopeptidases (MMPs)
Several studies have focussed on the MMP genes as candidate genes since elevated levels of MMPs, especially MMP9 and MMP2 have been measured in the serum and aortic wall of AAA patients.29–31, 40, 66 The list of MMP genes investigated for their role in AAA includes MMP1, MMP2, MMP3, MMP9, MMP10, MMP12 and MMP13. SNPs in genes of MMP1, MMP2, MMP10, MMP12, and MMP13 showed no associations with AAA.48 One of the MMP2 SNPs (rs243865) was also investigated with regard to rate of AAA growth, but no association was shown.67, 68
MMP3 is a known activator of MMP9 and its mRNA levels are significantly increased in AAA tissue.69 The promoter region of MMP3 which has a SNP (5A/6A) at nt –1621 has gained special interest. One study with 405 AAA patients and controls showed a significant association with the 5A allele.70 Other studies, including AAA growth studies, could not detect this association.48, 67, 68, 71 A meta analysis of this MMP3 polymorphism found 5A allele associated with a dominant model giving the best fit (OR, 1.4; 95% CI, 1.12 to 1.76; p = 0.003).72
The T allele of C>T SNP (rs3918242) in the promoter region of MMP9 results in increased transcriptional activity compared to the C-allele.73 The frequency distribution of the genotypes for this SNP in 414 AAA cases was significantly different from 172 cases with peripheral vascular disease (PVD) (p = 0.0013), but not if compared to 203 healthy controls (p = 0.334).74 Subsequent studies with larger sample sizes showed no association.48, 75 The results were also negative when using aneurysm growth rate as the phenotype.67, 68 Two smaller studies showed no association with AAA when studying a CA-repeat in the MMP9 promoter region.76, 77 Two meta analyses found contradictory results in that, one meta analysis published in 2008 demonstrated a significant allelic dose effect for rs3918242,78 whereas a more recent meta analysis which included an additional non-significant association study from Smallwood and co-workers75 with 678 AAA patients and 659 controls, found no association.72
Tissue Inhibitor of Metallopeptidase 1, 2 and 3 (TIMP1, -2 and -3)
DNA sequence analysis detected a SNP in the TIMP1 gene (nt 434 C>T) which was associated (p = 0.0047) with AAA in 235 male AAA patients without family history and 425 controls.48 A meta analysis of this TIMP1 SNP, however, found no association.78 Another SNP (rs2070584) in the same gene was also associated with AAA in male patients without family history.48 In the same study SNPs in TIMP2 and TIMP3 genes showed no association to AAA phenotype.48
Serine peptidase inhibitor, clade A, member 1 (SERPINA1)
This peptidase inhibitor is better known under the name of α1-antitrypsin (α1-AT), the deficiency of which leads to a severe hereditary disease, characterized by lung emphysema, which is aggravated by smoking. α1-AT levels are influenced by variation at the protease inhibitor (PI) locus. The SERPINA1 locus was previously known as the PI locus and was studied by isoelectric focusing, which identified over 50 different alleles. 79, 80 The so called PI*S- and PI*Z alleles lead to lower serum levels of α1-AT than the more frequent PI*M1, PI*M2, PI*M3 and PI*M4 alleles. Two studies measured the PI*S and PI*Z α1-AT-deficiency alleles in aneurysm patients by isoelectric focusing in the plasma, and both observed no excess in the PI deficiency alleles compared to controls.81, 82 Another study detected fewer PI*MM phenotypes (p = 0.0003) and significantly more PI*MV phenotypes (p = 0.0001) in AAA patients.82 The functional role of the PI*MV allele was not investigated.
Serine peptidase inhibitor, clade E, member 1 (SERPINE1)
Plasminogen activator inhibitor-1 (PAI-1), now formally known as SERPINE1, can inhibit MMPs and also fibrin degradation by repressing plasmin.83 The amount of SERPINE1 is decreased in cultured smooth vessel cells isolated from the aortic wall of AAAs compared to those from non-aneurysmal aorta.84 Most studies have focused on a functional repeat variant (4G/5G) located at nt –675 in the promoter region. Individuals who are homozygous for the 4G allele (4G/4G genotype) have increased SERPINE1 expression.85 A study with 39 AAA cases with strong family history and 163 controls showed a significantly higher frequency of the 5G allele in the AAA group,85 but a previous study could not detect an association when analyzing 47 AAA patients without family history and 173 controls.71 Other studies investigated the AAA expansion rate related to the 4G/5G allele distribution with no association.67, 68, 86 A meta analysis of this SERPINE1 variant concluded that there is no association with AAA.72
Secreted phosphoprotein 1 (SPP1)
Experimental studies support a role for SPP1, also known as osteopontin (OPN), in promoting inflammation, proteolysis, and atherosclerosis.87–89 SPP1 has been linked to development of AAA in an animal model, in which double knockout (Apoe−/−Opn−/−) mice developed smaller aneurysms after angiotensin II-infusion with decreased MMP2 and MMP9 activity compared to Apoe−/−Opn+/+ mice (p < 0.005).90 In human AAA patients, SPP1 concentrations in the serum and aortic tissue are significantly increased particularly in aortic wall specimens of small AAA with a diameter from 3.0 to 5.0 cm, but no genetic association has been shown between five polymorphisms in the SPP1 gene and AAA.91
Xylosyltransferase 1 (XYLT1)
XYLT1 is the first enzyme in the biosynthesis of the proteoglycan-linked glycosaminoglycan chains. Proteoglycan content and biosynthesis are altered in AAA and thus make XYLT1 a plausible candidate gene to study in AAA.92 A genetic association study using 129 AAA cases and 129 controls demonstrated that individuals with the T-allele of SNP G>T at nt 343 had a 5-fold risk of developing AAA (p = 0.011).92 These intriguing preliminary findings need replication in larger samples.
Cystatin C (CST3)
Cystatin C is a natural inhibitor of elastolytic cysteine peptidase such as the cathepsin family of peptidases. Cathepsin inhibitors are potentially relevant since cathepsins S and K are overexpressed at sites of arterial elastin damage and in vascular smooth muscle cells.93 Decreased cystatin C levels measured from serum have been correlated with the presence of AAA and with the growth rate of aneurysms.94, 95 A British-Swedish study examined 424 AAA patients with two SNPs located in the CST3 gene. The number of homozygote individuals with the C-allele (CC genotype) for one of the SNPs (nt -82G>C) was low, and individuals who were homozygous for the A-allele (AA genotype) for another SNP (nt 148G>A) had a slower growth rate of AAA.96
Group II: Genes of the Cardiovascular System (Table III)
Table III.
Genes of the cardiovascular system
| Gene | Polymorphisms | Number of studies* |
|
|---|---|---|---|
| Associated | Not associated | ||
| ACE | insertion/deletion intron 16 (rs4646994) | 4 (2)103–106 | 7 (4)101–103, 107,108§ |
| AGT | rs699 | 1 (1)107 | 2 (2)107 |
| AGTR1 | rs5186 | 2 (2)107 | 3 (3)105, 107, 109 |
| BDKRB2 | rs5223 | 3 (3)107 | |
| APOB | ApoB (Xbal) | 1 (0)47 AAA vs AAA+PAA | |
| APOE | APO E3E3 and E3E4 | 1 (0)113 AAA expansion | 1 (1)114 AAA expansion |
| rs405509, rs429358, rs4420638, rs7412, rs8106922 | 1 (1)114 | ||
| CETP | TaqIA RFLP | 1 (0)42 | |
| F2 | FII G20210A | 1 (1)115 | |
| F5 | Factor V Leiden | 1 (1)115 | |
| MTHFR | rs1801133 | 4 (0)120–123 | 6 (3)115, 118, 123–126 |
| rs1801131 | 1 (0)126 male and non-smoker | 2 (1)125, 126 | |
| rs2274976, rs48846049 | 1 (1)125 | ||
| MTHFD1 | rs8003379 | 1 (1)125 | |
| rs2357481, rs1076991, rs3783732, rs1950902, rs4902283, rs2236225 | 1 (1)125 | ||
| MTR | rs2853523 | 1 (1)125 | |
| rs4659725, rs1805087, rs2275566, rs6676866 | 1 (1)125 | ||
| MTRR | rs326118 | 1 (1)125 | |
| rs1801394, rs1532268, rs2303080, rs10064631, rs16879334, rs8659 | 1 (1)125 | ||
| TYMS | rs16430 | 1 (1)125 | |
| rs502396 | 1 (1)125 | ||
| ENOSF1 | rs8423 | 1 (1)125 | |
| AHCY | rs819149, | 1 (1)125 | |
| rs7271501 | 1 (1)125 | ||
| FOLH1 | rs202680 | 1 (0)125 subgroup AAA diameter | 1 (1)125 |
| rs202676 | 1 (1)125 | ||
| BHMT | rs651852, rs567754, rs3733890, rs585800 | 1 (1)125 | |
| BHMT2 | rs644191, rs682985 | 1 (1)125 | |
| CBS | 8344 C>T, 1542 C>T, 3753 C>T, 3758 G>A, rs1051319 | 1 (1)125 | |
| NNMT | rs566775, rs10891639, rs4646335, rs3819100 | 1 (1)125 | |
| PON1 | −108 C>T | 1 (0)126 only > 65 years and smoker | 1 (0)126 |
| rs854560, rs662, rs3917594 | 1 (1)125 | ||
| PON2 | rs12026 | 1 (1)125 | |
| SLC19A1 | rs3788205, rs3177999, rs1051266 | 1 (1)125 | |
| SHMT1 | rs638416, rs1979277 | 1 (1)125 | |
| TCN2 | rs5749131, rs1801198, rs10418 | 1 (1)125 | |
| NOS3 | Intron 4 27–bp repeat (4a/4b) | 2 (1)135, 141 | |
| 894 G>T | 2 (1)135, 136 | ||
| −786 T>C | 1 (1)135 | ||
| PTGS2 | rs20417, rs4648307 | 1 (1)146 | |
| HP | Variant α-chains α1/α2 | 2 (0)42, 148 one study on AAA expansion | 2 (0)44, 147 in 28 AAA families |
|
Blood groups | |||
| ABO | A | 1 (0)12 117/59862 |
|
| RHD/d | Rh- | 1 (0)12 | |
| KEL | positive | 1 (0)12 | |
| MNS | MNss | 1 (0)12 | |
Number in parenthesis indicates the number of studies with a sample size of at least 150 cases and 150 controls.
Study with 1,000 cases and controls
If multiple sample sets were used in one publication, they were treated as independent studies. AAA, abdominal aortic aneurysm; PAA, popliteal artery aneurysm
Gene symbols available from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) were used.
Genes of the cardiovascular system constitute another group of genes investigated for their role in AAA. Proteins regulating blood pressure, and factors involved in blood cell development and coagulation or genes known to be associated with other cardiovascular diseases serve as candidate genes (Table III).
Genes involved in the renin-angiotensin-aldosterone system
The renin-angiotensin-aldosterone system plays a critical role in blood pressure regulation and angiotensin converting enzyme inhibitors are one of the most frequently prescribed antihypertensive medications. Clinical studies on AAA patients have demonstrated that ACE-inhibitors augment collagen synthesis, reduce stiffness of the aortic wall,97 and reduce the risk of rupture of AAA.98 Thus, different genes of the renin-angiotensin-aldosterone system have been investigated for their role in AAA pathogenesis.
Angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 (ACE)
ACE itself has been investigated for its role in AAA pathogenesis. An insertion(I)/deletion(D) polymorphism in an Alu repeat in intron 16 (rs4646994) correlates with plasma levels of ACE, and homozygotes with the deletion allele have higher ACE levels.99, 100 A small study (125 AAA cases and 153 controls) showed no association with aneurysm.101 Another study investigated the polymorphism in AAA growth in 58 AAA patients, detected 14 patients with D/D, 29 with D/I and 15 with I/I genotype, but no association to aneurysm growth rate (median AAA growth rate was 0.28 cm/year) was shown.102 The small numbers in the subgroups, however, make conclusions difficult.
Another small study (133 AAA cases and 152 controls) showed an association only when comparing subgroups of hypertensive AAA patients (n = 81) with normotensive AAA patients (n = 43).103 In another study Pola and co-workers showed an association with the deletion allele and AAA in 56 normotensive AAA when compared to 122 controls.104 Since all of the studies were small, the results should be interpreted with caution. Three other larger studies detected an association with the deletion allele.105–107 Two meta analyses observed an association between AAA and this ACE polymorphism with the best fit in a dominant model,72, 78 but significant inter-study heterogeneity.72 Recently, another study tried to replicate the association in a large British case-control set of 1,155 AAA patients and 996 controls with an estimated power of 90%. No association was shown.108
Angiotensinogen (AGT)
In a large genetic association study the T-allele of rs699 in exon 2 of AGT was associated with AAA in a patient cohort (576 AAA cases and 472 controls) from New Zealand. The association, however, could not be replicated in cohorts from the United Kingdom (298 AAA cases and 912 controls) and Australia (352 AAA cases and 339 controls).107
Angiotensinogen II receptor, type 1 (AGTR1)
The most significant genetic association in the angiotensin-renin-system was shown for a SNP (1166A>C; rs5186) in AGTR1.107 The odds ratio (OR) was 1.60 (95% CI: 1.32–1.93; p = 1.1×10−6).107 Two other large studies, however, were not able to replicate this association,105, 109 but a meta analysis for the same SNP found an association in a dominant genetic model.72
Bradykinin receptor 2 (BDKRB2)
Bradykinin is a vaso-depressor which works in the renin-angiotensin-system as a functional antagonist of angiotensin II, and it can be inactivated by ACE. No association between a polymorphism (rs5223; I/D of 9 bp, in exon 1 and the 5′-UTR) in BDKRB2 and the presence of AAA, or AAA growth rate, was shown.107
Genes involved in lipid metabolism
Because of the crucial role of lipid metabolism in atherosclerosis, the genes involved in this pathway are also of interest in AAA pathogenesis.
Apolipoprotein B (including Ag(x) antigen) (APOB)
APOB is a marker for triglyceride-rich lipoproteins and the Xbal polymorphism in this gene is correlated with plasma levels of triglycerides.110 One small study investigated the Xbal polymorphism in AAA patients without (n = 124) and with (n = 24) additional PAA and no differences in allele frequencies were seen.47
Apolipoprotein E (APOE)
APOE polymorphism has long been known to be associated with risk for atherosclerosis.111 In mice Apoe deficiency predisposes to AAA formation and atherosclerotic changes after angiotensin II-infusion.112 The APOE*2, -*3, and *4 alleles were investigated in patients with small AAAs (n = 57), and the AAA expansion rates were followed over 2 to 4.5 years.113 Compared with the E3/E3 genotype, individuals with the E3/E4 genotype had a significantly lower AAA expansion rate (p = 0.001).113 These findings, however, were not replicated in another larger study (640 men with small AAA).114
Cholesteryl ester transfer protein, plasma (CETP)
The first small study on CETP and AAA was published in 1990 and it investigated the TaqIA polymorphism at the CETP locus in AAA patients (n = 44) and controls (n = 32).42 The frequency of the rare 9.0 kb allele was significantly increased in AAA patients (p < 0.01).42 Another later study in which the entire gene was sequenced revealed no mutations or associations with AAA.43 In both studies the sample numbers were too small to draw conclusions.
Coagulation factor II (thrombin; F2) and coagulation factor V (proaccelerin, labile factor; F5)
Since the aneurysm sac is nearly always filled with mural thrombus, a role for thrombophilic risk factors in AAA pathogenesis has been suggested.115 Two genetic variants predisposing to thrombosis, the so called Leiden mutation of F5 and the G20210A variant of F2 were investigated in 438 AAA cases and 438 controls, but no associations were shown.115
Genes involved in the methionine metabolism
Hyperhomocysteinemia is a risk factor for thrombosis and atherosclerosis and in animal models hyperhomocysteinemia induced an ECM remodelling of the arterial wall.116, 117 Two studies demonstrated a correlation between AAA and mild hyperhomocysteinemia.115, 118 Homocysteine is an intermediate of the methionine metabolism. Several polymorphisms in 17 genes coding for enzymes involved in the methionine metabolism have been tested for association with hyperhomocysteinemia, which might play a role in the development of AAA.
5,10-Methylenetetrahydrofolate reductase (MTHFR)
MTHFR is a cardiovascular candidate gene, located on chromosome 1p36.3 and is involved in homocysteine metabolism. Studies have focused on the SNP (rs1801133; nt 677 C>T), which results in an alanine to valine substitution and in 70% reduction in enzymatic activity in vitro.119 Three small studies showed that the T-allele was associated with AAA.120–122 Another small study restricted the analysis to normotensive AAA patients and showed an association with AAA.123 Recent, larger studies, however, were not able to confirm these associations.115, 118, 124, 125 When combining all the studies published to date, two meta analyses revealed an association between the T allele and AAA72, 78 in a dominant model. Analysis of three additional polymorphisms, two exonic SNPs with amino acid substitutions (rs1801131: A1298C and rs2274976: C1793T) and one in the 3′-UTR region (rs48846049) showed no association with AAA.125 A small subgroup analysis in male, non-smoking AAA patients (20 vs 33) showed an association with the A1298C SNP.126
Methylenetetrahydrofolate dehydrogenase 1 (MTHFD1)
One large study (423 AAA cases and 423 controls) which evaluated 56 polymorphisms in 17 genes coding for enzymes in the methionine metabolism demonstrated an association between one SNP (rs8003379) in MTHFD1 and AAA, whereas six additional SNPs in the same gene were not associated.125 Of interest is that smaller aortic diameter in AAA patients was associated with SNP rs8003379, which is located in an intron and no functional information is known about it.125
5-Methyltetrahydrofolate-homocysteine methyltransferase (MTR)
The MTR enzyme catalyses the final step in methionine biosynthesis.125 Five polymorphisms were investigated in the MTR gene, and one of them (rs2853523) showed an association with AAA for the C-allele, with individuals having the A-allele showing smaller aortic diameter.125 This SNP is localized in the 3′-UTR which can have a role in the control of gene expression.125
5-Methyltetrahydrofolate-homocysteine methyltransferase reductase (MTRR)
In the methionine metabolism MTR enzyme eventually becomes inactive due to the oxidation of its cob(I)alamin cofactor. The protein encoded by MTRR gene regenerates a functional MTR enzyme via reductive methylation. A study investigating the MTRR gene showed that one SNP (rs326118) out of seven polymorphisms, was associated with AAA (A-allele).125 This variant is located in the 5′-UTR of the gene.125
Thymidylate synthetase (TYMS)
There is a SNP (rs16430) in the 3′-UTR of the TYMS gene which influences the expression and stability of TYMS mRNA,127 and is associated with higher red blood cell folate and homocysteine concentrations.128 The C-allele of this variant was associated with AAA in one study, while another SNP (rs502396) in the same gene was not associated.125 The naturally occurring antisense transcript to the human thymidylate synthase (TYMS) gene is enolase superfamily member 1 (ENOSF1). One SNP (rs8423) in the ENOSF1 gene has been investigated in one study, and no association was shown with AAA.125
Adenosylhomocysteinase (AHCY)
AHCY regulates the intracellular S-adenosylhomocysteine concentration and is important in transmethylation reactions.125 Deficiency in the protein is one of the causes of hypermethioninemia. The promoter region of AHCY contains a SNP (rs819146) near the start codon which might have regulatory role on gene expression.125 Interestingly, this SNP was associated with AAA in one study.125
Folate hydrolase 1 (FOLH1)
Folic acid play a role in intestinal absorption of dietary folates, and therefore, a genetic variation could lead to an altered absorption followed by low blood folate levels and hyperhomocysteinemia. A SNP (rs202676; Q75X), which results in a stop codon was studied, but no association with AAA was shown.125 Another SNP (rs202680) was associated with larger aortic diameter in AAAs, but increased levels of homocysteine were not detected.125
Several other SNPs in genes involved in the methionine metabolism have been investigated in AAA, mostly in only one study and all results were negative for an association with AAA: Four polymorphisms in the betaine-homocysteine methyltransferase (BHMT) gene have been investigated for their role in AAA pathogenesis with no associations identified.125 In betaine-homocysteine methyltransferase 2 (BHMT2) two SNPs have been studied, but no associations were shown with AAA.125 Five SNPs in the cystathione-beta-synthase (homocysteine; CBS), three of them leading to amino acid substitutions, and one located in the 3′-UTR were tested without evidence for association with AAA.125 Nicotinamide-N-methyltransferase (NNMT) is another important enzyme in homocysteine metabolism.129 Four SNPs have been investigated, two in the 5′ region, one in 5′-UTR and one intronic, but no associations were shown with AAA.125 The enzyme activity of paraoxonase 1 (PON1) is predictive of vascular disease,125 but polymorphisms found in this gene (rs8545560, rs662 and rs3917594) cannot explain this variation in activity.125 One small study detected an association between a SNP (nt –108 C>T) in the PON1 gene and AAA in a subgroup analysis of a small number of AAA patients who were more than 65 years of age and smokers.126 In paraoxonase 2 (PON2) one amino acid changing SNP (rs12026; A148G) has been investigated, but no association with AAA was shown.125 Three SNPs have been analyzed in the gene called solute carrier family 19 (folate transporter), member 1 (SLC19A1), all located in the 5′ end of the gene or 5′-UTR, but they were not associated with AAA.125 Serine hydroxymethyl-transferase, cytosolic (SHMT1) has two SNPs, one in 5′-UTR, the other in the exon with amino acid change L474F. Both of these SNPs were analysed in one study, but no evidence for association with AAA was detected.125 And three SNPs (rs1801198: P259R; rs5749131 and rs10418) in transcobalamin II (TCN2) were studied, but no association with AAA was shown.125
Endothelial constitutive nitric oxide synthase (NOS3)
NOS3 is a modulator of vascular disease and maintains endothelial function and antithrombotic intravascular environment. A SNP (G894T in exon 7) is associated with reduced basal nitric oxide production.130 The minor, less common T-allele leads to an amino acid substitution Q298N. This SNP is associated with coronary artery disease131 and carotid atherosclerosis.132 Studies on mice lacking this gene gave evidence for the role of nitric oxide in controlling vessel wall geometry.133 Thus NOS3 is a plausible candidate gene also for AAA.134 A study published in 2005, analyzed 250 AAA patients and 250 controls and showed an association between the G894T SNP and AAA.135 The association remained significant in multivariate analysis adjusted for other vascular risk factors and other atherosclerotic localizations.135 An association was also shown in a smaller study (62 AAA cases and 62 controls).136
A SNP located in the promoter region (nt –786T>C) of NOS3 gene influences promoter activity with the T-allele showing 50% more transcriptional activity than the C-allele.137 No association, however, has been detected between this SNP and AAA.135 Another study focused on a variable number tandem repeat (VNTR), called 4a/4b polymorphism, located in intron 4, which is correlated with altered nitric oxide plasma levels.138 The 4a/4b polymorphism is associated with hypertension139 and coronary lesions in smokers.140 In 58 AAA patients and 410 controls no association was shown with this VNTR, although a subgroup analysis showed an association with the need of surgery in AAA patients, but the sample numbers were small (34 surgical and 24 non-surgical AAA patients).141 A second study with more samples did not observe any association between the VNTR and AAA.135 A meta analysis of these two studies concluded that there was no association.78
Prostaglandin-endoperoxide synthase 2 (PTGS2)
The biological activity of PTGS2 (known also as cyclooxygenase 2) is critical in the inflammatory response, modulates cardiovascular diseases and promotes atherosclerotic plaque progression and instability.142, 143 A SNP located in the promoter region (nt –756G>C; rs20417) is functionally important, and the C-allele produces less PTGS2 mRNA than the G-allele, and correlates with significantly reduced serum levels of C-reactive protein in myocardial infarction or stroke patients.144 This SNP may act as a protective factor against myocardial infarction and stroke.144 It has been speculated that a reduced PTGS2 expression and the consequent suppression of local and systemic inflammation may also be protective in AAA development and rupture.145 One study investigated PTGS2 in AAA and showed no association with the SNPs in this gene with AAA.146
Haptoglobin (HP)
HP was one of the first genes investigated in AAA patients. In a small sample group the α1 chain was seen more frequently than the α2 chain,42 but in a follow-up study of 28 AAA-families the HP variants were equally distributed.44 Another study investigated the HP phenotypes by starch gel electrophoresis in 52 AAA, 37 PVD patients, and a control group of 37 without atherosclerosis. The 3 different HP phenotypes were equally distributed in the 3 study groups.147 Recently a study assessed the relationship between AAA expansion and HP phenotypes. They determined three major HP phenotypes (1-1, 2-1, and 2-2) in 83 AAA patients. Rescanning at 6- to 12-month intervals up to a period of 2 to 7 years, the mean yearly rate of growth of the AAA was calculated from the largest transverse diameter for each group of HP patients and HP 2-1 patients had a significantly higher growth rate (3.69 [2.40] mm/y) compared with patients with HP 2-2 (1.24 [0.79], p < 0.00001) and HP 1-1 (1.45 [0.68], p = 0.00004). This association remained significant in the multivariate analysis. Elastase serum activity and CRP serum levels were also higher in AAA patients with HP 2-1. Larger follow-up studies are needed to confirm if the HP 2-1 phenotype is useful as independent predictor of AAA expansion rate.148
Blood groups
A Swedish study investigated the ABO blood groups and Rh-system in 117 AAA patients and 59,862 controls.149 Blood group A of the ABO-system was relatively common in AAA patients but the difference between AAA patients and controls did not reach statistical significance.149 A statistically lower frequency of the Rh-negative blood group of the Rh-system was observed in the AAA group.149 The MNS blood group was studied in 54 AAA cases and 2,164 controls, and the frequencies of the MNss type and the number of MN heterozygotes were significantly increased in AAA patients.149 The KELL blood group was investigated in 54 AAA cases and 287 controls, and the AAA group had significantly more KELL positive individuals than the control group.149
Group III: Genes of the Immune System (Table IV)
Table IV.
Genes of the immune system
| Gene | Polymorphisms | Number of studies* |
|
|---|---|---|---|
| Associated | Not associated | ||
| HLA | -A, -B | 1 (0)12 | |
| -DRB1*15 | 1 (0)155 | ||
| -DQB1, -DRB1*0404 | 1 (0)155 | ||
| -DRB1*04, -DRB1*02 | 2 (0)156, 157 | ||
| -DQA*0120, -DQA1, -DRB1, -DRB1,3-5 | 1 (1)162 in male Belgians | 1 (1)162 in male Canadians | |
| -DR2(15) | 1 (0)158 | ||
| -DQ3 | 1 (0)159 | ||
| -DRB1*0401 | 1 (0)160 | ||
| -DQA1*0102 | 1 (1)218 | ||
| -A, -A2, -B, -B61, -DRB1*, -B1*1502 | 1 (0)161 | ||
| -A1, -A3, -A11, -A24, -A26, -A30, -A31, -A33; -B7, -B13, -B62, -B75, -B71, -B35, -B37, -B38, -B39, -B60, -B40, -B44, -B47, -B46, -B48, -B51, -B52, -B54, -B55, -B56, -B58, -B59, -B67; -DRB1*0101, -1501, -1602, -0301, -0302, -0401, -0404, .0405, -0410, -0403, -0406, -0407, -1101, -1102, -1201, -1202, -1301, -1302, -1401, -1402, -1405, -1407, -1403, -1406, -0701, -0803, -0801, -0802, -0804, -0811, -0901, -1001 | 1 (0)161 | ||
| -A*11, -B*08, -B1*11 | 1 (1)163 | ||
| CCR5 | −308 (32 pair deletion) | 1 (0)170 in subgroups AAA vs PVD, AAA vs carotid stenosis, rAAA vs eAAA | 2 (1)170, 171 |
| IL1A | −889 C>T, 4845 G>T | 1 (0)176 | |
| IL1B | −511 C>T, −31 C>T | 1 (0)176 | |
| 3953 C>T | 2 (0)176, 177 | ||
| IL1RN | 2018 C>T | 1 (0)176 | |
| IL6 | rs1800796 | 1 (1)179 recessive model | 1 (1)179 |
| rs1800795 | 3 (2)177–179 | ||
| rs1800797 | 1 (1)179 | ||
| IL10 | −1082 G>A | 1 (0)177 | 1 (1)182 |
| −592 C>A | 1 (0)177 | ||
| PLA2G7 | G994T | 1 (0)183 | |
| TNF | −308 G>A | 1 (0)177 | |
| PPARG | rs1801282 | 1 (1)187 | |
| rs3856806 | 1 (1)187 | ||
| CRP | rs3091244 | 1 (1)193 | |
| HMOX1 | −258 to −195 GT length polymorphism | 1 (0)197 | |
Number in parenthesis indicates the number of studies with a sample size of at least 150 cases and 150 controls.
If multiple sample sets were used in one publication, they were treated as independent studies. AAA, abdominal aortic aneurysm; rAAA, ruptured AAA; eAAA, elective repair of AAA; PVD, peripheral vascular disease
Gene symbols available from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) were used.
AAA disease typically involves inflammation with leukocyte infiltrates and increased amounts of various cytokines.150–153,152–154 A large number of different immune-related genes, especially the human leukocyte antigen (HLA) class II genes, interleukins and the tumor necrosis factor (TNF) gene, have been investigated for their potential role in AAA pathogenesis (Table IV).
Human leukocyte antigen locus (HLA)
The first study concerning the role of HLA-A and –B in AAA was performed 26 years ago in Sweden. Serotyping of patients for 18 HLA-A and –B antigens detected no significant differences between the AAA group (n = 48) and the control group (n = 368). More than ten years later studies using genetic approaches found that HLA-DRB1*15, HLA-DR B1*04 and –B1*02 were associated with AAA but the sample size was rather small.155–157 Several follow-up studies concerning HLA-DR and HLA-B subgroups were also small and their findings should be interpreted with caution.158–161 A larger study showed an association with HLA-DQA1*0102,162 but yet another study showed no evidence of an association between HLA-A*11, HLA-B*08 or HLA-B1*11 and AAA.163
Chemokine (C-C motif) receptor 5 (CCR5)
Chemokines are cytokines that promote migration of leukocytes into inflammatory sites.164 CCR5 is expressed in macrophages, T-cells, aortic smooth muscle cells and coronary artery endothelial cells165, 166 and CCR5 ligands have been detected in atherosclerotic plaques.167 Genetic screening studies showed that CCR5 deficiency protects from severe coronary artery disease.168 A loss of CCR5 receptors on lymphoid cell surfaces is caused by a 32-bp deletion, called delta 32, in the CCR5 gene.169 Investigation of 70 AAA patients, 76 patients with PVD and 62 carotid artery stenosis patients showed that AAA patients had a higher frequency of the deletion allele. Comparing to 172 controls it was not an independent risk factor. The study concluded that the presence of at least one deletion allele provided a 4-fold higher risk for AAA rupture.170 A follow-up study with larger sample sizes (285 AAA cases and 273 controls) showed the same frequency of the deletion allele in both groups.171 To interpret the role of the delta 32 deletion in the development and rupture of AAA, larger sample sizes are necessary.
Interleukin 1α (IL1A), interleukin 1β (IL1B) and interleukin 1 receptor antagonist (IL1RN)
IL1 is an important regulator of the inflammatory cascade and it is present in aneurysmal tissue. AAA patients have significantly higher plasma levels of IL1B than patients with coronary artery disease without AAA.150, 172, 173 IL1 induces MMPs and promotes vascular smooth muscle cell apoptosis.174, 175 Some polymorphisms in the IL1 gene cluster modulate cytokine expression and contribute to a wide range of inflammatory conditions.176 Six SNPs in IL1A, IL1B and IL1RN were investigated in 135 AAA cases and 270 controls, but no associations between the SNPs and AAA were detected.176 One SNP in the IL1B gene (nt 3953 C>T) was investigated in two different small case-control studies and no association was shown.176, 177 A meta analysis of the two studies found no association with AAA.78
Interleukin 6 (interferon β2; IL6)
IL6 is a pro-inflammatory cytokine, and elevated levels of circulating IL6 have been measured in AAA patients178 and IL6 has increased expressed in the wall of AAA and popliteal aneurysms153. In a large well-designed case-control study (677 AAA and 656 controls) three SNPs in the IL6 gene promoter were examined. One SNP (rs1800796; –572 G>C), which was present in only 1.5% of cases, was identified as an independent risk factor for AAA in an analysis using a recessive model, and no association was seen in an additive or dominant model.179 Two other SNPs (rs1800795 and rs1800796) showed no association.177, 178 A meta analysis of one SNP (rs1800795; IL6 G174C) found no evidence of association.72
Interleukin 10 (IL10)
IL10 is a predominantly anti-inflammatory cytokine. In a small study increased levels of IL10 were measured in the aortic wall of patients with AAA,180 but in another study mRNA levels did not differ between AAA tissue and control tissue.153 The presence of an A-allele at nt -1082 instead of a G-allele resulted in 25% reduction in IL10 production by lymphocytes.181 This SNP has been associated with chronic inflammatory diseases.182 The same SNP was first tested in a small study on AAA and an association with AAA was seen,177 but a confirmation study with larger sample groups (389 AAA cases and 404 controls) showed that after a multivariate regression analysis the association was not independent from age, smoking, hypertension or history of PVD.182 A meta analysis of the two studies, however, found that the best fit for an association with AAA was the dominant genetic model.72
Phospholipase A2, group VII (PLA2G7)
PLA2G7 catalyzes platelet activating factor (PAF) and inactivates its inflammatory function. Deficiency of the PLA2G7 gene is caused by a missense mutation G994T, which has been described only in the Japanese population. The T-allele had a significantly higher frequency (independent from other risk factors) in the AAA group (n = 131) than in the controls (n = 106) in a Japanese study.183
Tumor necrosis factor (TNF superfamily, member 2; TNF)
TNFα is a pro-inflammatory cytokine produced by macrophages. The A-allele of a SNP located at nt –308 produces six to nine times more TNFα mRNA in human B-cells than the G-allele.184. A small association study in AAA patients and controls could not show any association between the A-allele and AAA patients.177
Peroxisome proliferator-activated receptor gamma (PPARG)
PPARG is a target receptor for TNFRSF11B (tumor necrosis factor receptor superfamily, member 11 B), which is also called osteoprotegerin. High concentrations of TNFRSF11B were detected in human AAA tissue and serum levels of TNFRSF11B were elevated in AAA patients.185 Based on the known influence of PPARG on TNFRSF11B,186 two SNPs were examined in 689 AAA patients and 3,538 controls.187 For one SNP (rs1801282; c.34 G>C), which causes an amino acid substitution P12A, a weak association with AAA was shown in a dominant model, and for another SNP (rs3856806; c.1347 C>T), which is a silent SNP, only the subgroup with faster AAA progression had significantly higher frequency of the TT genotype.187
C-reactive protein (CRP)
CRP is an acute phase protein produced mainly in hepatocytes and lymphocytes and its production is stimulated by cytokines.188, 189 Higher than control serum levels of CRP have been detected in AAA patients.190, 191 One study tested a SNP (rs3091244)192 for an association with AAA and if the serum concentrations of CRP were influenced by this SNP.193 Serum levels of CRP were significantly higher in the AAA group than in the control group and especially in patients with large AAAs, but the SNP was not associated with AAA nor with elevated CRP serum levels.193
Heme oxygenase (decycling) 1 (HMOX1)
HMOX1 is a vascular anti-inflammatory factor with antioxidant capacity and is produced by vascular smooth muscle cells.194 HMOX1 is one of the stress proteins expressed in atheroclerotic plaque lesions in the aorta.195 A dinucleotide repeat present in the promoter region of this gene modulates the level of transcription and shorter repeats correlate with increased HMOX1 levels.196 A small case-control study (70 AAA cases and 61 controls) showed that fewer AAA patients than controls had the shorter repeats.197
Group IV: Genes of Signaling Pathways (Table V)
Transforming growth factor β (TGFB) plays a role in growth and ECM protein expression in vascular smooth muscle cells, and TGFB can be induced by angiotensin II.105 Variants in the TGFB1, −2, and −3 genes can inhibit development of vascular smooth muscle cells, and mutations in the TGFB-receptor genes cause syndromic forms of thoracic aortic aneurysms.198 It is, therefore, plausible that the TGFB-pathway with its receptors and binding proteins is involved in AAA development (Table V). Other candidate genes belonging to this group are the estrogen and progesterone receptors.
Transforming growth factor β1 (TGFB1) and Transforming growth factor β1-receptor (TGFBR1)
Two studies have investigated the role of SNPs in the TGFB1 gene,48, 199 but no associations with AAA were seen. One study analyzed a repeat variant in the TGFBR1 gene in 201 AAA cases and 252 controls, and showed no association.105 A Dutch group examined three SNPs in the TGFBR1 gene with two independent large case-control sets, but showed no associations.200
Transforming growth factor β2-receptor (TGFBR2)
SNPs located in the TGFBR2 gene have also been investigated for their potential role in AAA. Study populations from Australia (640 AAA cases and 1071 controls) and New Zealand (654 AAA cases and 389 controls) were examined for 49 SNPs in the TGFBR2 gene, only one of which (rs1078985) showed a weak association with AAA, which did not withstand correction for multiple testing.201 A Dutch study investigated nine different SNPs with two large case-control sets, and two of the SNPs (rs1036095 and rs4522809) showed an association with AAA in both case-control sets.200
Transforming growth factor β3 (TGFB3)
A SNP located in the TGFB3 gene (rs11466414) was examined for its role in the growth of AAAs, but no association was shown.199
Latent TGF-β binding protein 1, 3 and 4 (LTBP1, LTBP3 and LTBP4)
Selected SNPs in the LTBP1 and LTBP3 genes were investigated in four geographically different case-control sets (altogether 1,890 AAA cases and 3,785 controls) but no association with the presence of AAA nor growth rate of AAA was detected.199 LTBP4 gene is located on chromosome 19q13, in the candidate interval identified in a DNA linkage study.17 Several SNPs (nt –4234A>G, 10384 G>A, 21011A>T, 25859C>T and 32603C>G) in the LTBP4 gene showed an association with AAA growth rates.199
Estrogen receptors 1 (ESR1), and 2 (ESR2), and Progesterone receptor (PGR)
Based on the fact that AAAs are more common in males and that the prevalence of AAA increases by age in women, it is plausible to hypothesize that female sex hormones play a role in AAA development. Most biological effects of steroids are mediated by their nuclear receptors and genetic variants in these receptor genes might contribute to the disease.56 Two SNPs in the ESR1 gene, one in the ESR2 gene, and one in the PGR gene were investigated in one study with 99 AAA patients and 225 controls. Only the SNP located in the ESR2 gene (nt 1730A>G) showed an association with AAA.56
DNA LINKAGE STUDIES IN AAA FAMILIES
DNA linkage studies are used to find regions on the human chromosomes that harbor sequence changes contributing to the disease. They require samples from several family members. The identified regions are called susceptibility loci. The first DNA linkage analysis for AAA was performed in 48 AAA families with DNA markers located in only three candidate genes: 1) BHMT (betaine-homocysteine methyltransferase), 2) COL1A2 (the α2 chain of type I collagen) and 3) CTSH (cathepsin H) gene. None of the selected candidate genes showed linkage to AAA.202 The first genome-wide DNA linkage study on AAA used an affected-relative-pair design because it is difficult to find large families for AAA which is a late-age-at-onset and deadly disease.17 A set of 235 affected relative-pairs from 119 families showed a strong linkage to chromosomes 19q13 and 4q31. These genomic regions have now been designated as the AAA1 and AAA2 susceptibility loci for AAA.17 Another DNA linkage study carried out on Dutch AAA families also found evidence for linkage on chromosome 19q13.18
A recent genetic association study followed-up on the linkage findings by using 1,615 sequence variants on chromosome 19q13 in two independent case control sets.203 Altogether eight SNPs (rs16968029, rs281407, rs1075453, rs6509496, rs1530878, rs285676, rs576556, rs4802552) had a significant association (p = 0.015) in the first case-control-set, but the association could not be confirmed in the second case-control set (Table VI).203 These results suggest that the genetic basis of familial and sporadic AAA might be different.
GENOME-WIDE ASSOCIATION STUDIES
Another unbiased genetic approach to study complex diseases without prior hypotheses on candidate genes is a GWAS.204 Two GWASs on AAA have so far been reported.19, 20 It is expected that more such studies are in progress from at least one large consortium.205
The first GWAS for AAA was carried out using pooled DNA samples from 123 AAA cases and 112 controls on a SNP array (Affymetrix 500K) which can measure half a million SNPs simultaneously. A SNP (rs763518) located on chromosome 3p12.3, a region near contactin-3 (CNTN3) gene, was associated with AAA.19 The association was confirmed by individual genotype assays on 502 AAA cases and 736 controls, and by a replication on a second independent set (448 cases and 410 controls). By analysing subgroups the association with rs763518 was even stronger in the subset of smokers. The function of this SNP is unclear and it is considered a marker for a functional genetic variant most likely located upstream in the regulatory region of CNTC3 gene.19 The CNTC3 protein is a lipid-anchored cell adhesion molecule known to be expressed in nervous tissue206 and aortic wall.19 A group from New Zealand followed-up these findings by genotyping rs763518 in 567 cases and 552 controls but could not confirm the association. The group also tested three SNPs in intron 2 of CNTC3 and found a weak association with AAA.207
The second GWAS on AAA was carried out with 1,292 AAA patients and 30,503 controls from Iceland and the Netherlands, with a follow-up of most significant findings in up to 3,267 AAA patients and 7,451 controls. The A allele of a SNP (rs7025486) on 9q33 was found to associate with AAA, with an OR of 1.21 and p = 4.6 × 10−10. The same SNP was also found to be associated with early onset myocardial infarction (OR = 1.18, p = 3.1 × 10−5), peripheral artery disease (OR = 1.14, p = 3.9 × 10−5) and pulmonary embolism (OR = 1.20, p = 0.00030), but not with intracranial aneurysm or ischemic stroke. No association was observed between this SNP and common risk factors for arterial and venous diseases such as smoking, lipid levels, obesity, type 2 diabetes or hypertension. This SNP is located within DAB2IP, which encodes an inhibitor of cell growth and survival.20
Altogether 24 additional SNPs with p < 5.5×10−5 on 13 chromosomes were detected in the combined GWA analysis of Icelandic and Dutch samples, but none of them were replicated in other sample sets.20
OTHER FINDINGS
Several studies have investigated the possibility that AAA could share genetic risk factors with other vascular diseases. In one such study, five different vascular phenotypes, AAA, intracranial aneurysms, coronary artery disease, peripheral artery disease and atherosclerotic stroke, were analyzed for SNPs found to be associated with myocardial infarction and type 2 diabetes mellitus.208 The AAA study samples were from seven different geographical regions and included 2,836 AAA cases and 16,732 controls. The SNP previously associated with myocardial infarction and located in the CDKN2BAS gene, previously known as ANRIL, (which encodes an antisense RNA that regulates expression of cyclin-dependent kinase inhibitor 2B) at locus 9p21 was strongly associated with AAA, whereas the near-by SNP associated with diabetes was not associated with AAA (Table VI).208 The latter finding is in agreement with clinical findings that type 2 diabetes is not a risk factor for AAA.209
The G allele of SNP rs10757278 on CDKN2BAS gene was associated with AAA in five of the seven geographically different (Iceland, Belgium, Canada, US-Pennsylvania, the Netherlands, UK and New Zealand) AAA study populations. Belgian and Canadian study populations did not reach statistical significance. In a meta-analysis of all seven populations the association was highly significant (Table VI).208 The same SNP has also been investigated in other case-control sets from UK and Australia. A significant association with AAA phenotype was confirmed, but no correlation to aneurysm growth rate was found.210
In another study perlecan (officially known as heparan sulfate proteoglycan of basement membrane, HSPG2) and versican (known as chondroitin sulfate proteoglycan 2, CSPG2), which are susceptibility genes for intracranial aneurysms and located at 9p21, were investigated as potential risk loci for AAA.211 The study examined 12 SNPs in the HSPG2 gene and 22 in the CSPG2 gene. No associations were found (Table VI).211
Another recently published study measured the telomere length of chromosomes in leukocytes in blood samples collected from 190 AAA patients and 183 age-matched controls and observed significantly shorter telomeres in the AAA group.212 It is not immediately obvious how this relates to the disease pathogenesis, and it might just be an indication of vascular aging, since the same phenomena have been seen in other cardiovascular diseases.
CONCLUSIONS
AAA is a multifactorial disease, with both genetic and environmental risk factors contributing to the underlying pathobiology.22, 152, 213, 214 The mutations responsible for the phenotype in syndromic forms of aortic aneurysms such as the Marfan syndrome are different from the common form of AAA. Many methodological challenges of genetic association studies,204 such as the requirement of a large number of patients and controls, have slowed down the progress of identifying genetic variants associated with AAA. Approximately half of the genetic association studies discussed in this review do not have the number of samples required to get reliable results, which are likely to be replicated (see Tables II–VI; Fig. 1). In addition to genome-wide studies, nearly 100 candidate genes have been investigated for potential role in the AAA pathogenesis, and approximately quarter of them have been found to be associated with AAA (Fig. 1). These genes cover a wide range of functional groups and biological pathways.
Genetic heterogeneity is likely to play a role in AAA and there might also be differences in different populations. From the clinical point of view, vascular surgeons have observed that AAA patients are a heterogeneous group. Most AAAs have some degree of atherosclerotic occlusive disease. A large number of AAA patients also have hernias suggesting that their condition might be due to a more generalized connective tissue disease.215 Additional differences are that some AAAs grow faster than others, some lead to rupture, and some have large thrombi. Some AAAs have clinically obvious inflammatory reaction in the aortic wall, whereas in other cases the wall is only thinner without signs of inflammation. Some AAA patients have additional arterial aneurysms, most often in the popliteal artery. These variations in the clinical presentation suggest different subtypes and could help us determine which subgroups have better long-term outcomes in endovascular and open repair. In future genetic studies it will be useful to focus on well-defined subgroups. Here the interest and clinical expertise of vascular surgeons is needed to collect all the appropriate and relevant data. Future studies will include replication studies as well as detailed characterization of the genes and their potential role in AAA development.
Acknowledgments
Grant support: Dr. Hinterseher is a recipient of a Fellowship from the Deutsche Forschungsgemeinschaft.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alcorn HG, Wolfson SK, Jr, Sutton-Tyrrell K, et al. Risk factors for abdominal aortic aneurysms in older adults enrolled in The Cardiovascular Health Study. Arterioscler Thromb Vasc Biol. 1996;16(8):963–70. doi: 10.1161/01.atv.16.8.963. [DOI] [PubMed] [Google Scholar]
- 2.Cowan JA, Jr, Dimick JB, Henke PK, et al. Epidemiology of aortic aneurysm repair in the United States from 1993 to 2003. Ann N Y Acad Sci. 2006;1085:1–10. doi: 10.1196/annals.1383.030. [DOI] [PubMed] [Google Scholar]
- 3.Wahlgren CM, Larsson E, Magnusson PK, et al. Genetic and environmental contributions to abdominal aortic aneurysm development in a twin population. J Vasc Surg. 2009 doi: 10.1016/j.jvs.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 4.Nischan J, Lenk GM, Boddy AM, et al. Abdominal Aortic Aneurysms - A Complex Genetic Disease. In: Morel ALaE., editor. Aneurysms Types, Risks, Formation and Treatment. Nova Science Publishers, Inc; 2009. pp. 35–93. [Google Scholar]
- 5.Shantikumar S, Ajjan R, Porter KE, et al. Diabetes and the abdominal aortic aneurysm. Eur J Vasc Endovasc Surg. 2010;39(2):200–7. doi: 10.1016/j.ejvs.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 6.LaMorte WW, Scott TE, Menzoian JO. Racial differences in the incidence of femoral bypass and abdominal aortic aneurysmectomy in Massachusetts: relationship to cardiovascular risk factors. J Vasc Surg. 1995;21(3):422–31. doi: 10.1016/s0741-5214(95)70284-9. [DOI] [PubMed] [Google Scholar]
- 7.Derubertis BG, Trocciola SM, Ryer EJ, et al. Abdominal aortic aneurysm in women: prevalence, risk factors, and implications for screening. J Vasc Surg. 2007;46(4):630–635. doi: 10.1016/j.jvs.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 8.Lederle FA, Johnson GR, Wilson SE. Abdominal aortic aneurysm in women. J Vasc Surg. 2001;34(1):122–6. doi: 10.1067/mva.2001.115275. [DOI] [PubMed] [Google Scholar]
- 9.Lederle FA, Johnson GR, Wilson SE, et al. The aneurysm detection and management study screening program: validation cohort and final results. Aneurysm Detection and Management Veterans Affairs Cooperative Study Investigators. Arch Intern Med. 2000;160(10):1425–30. doi: 10.1001/archinte.160.10.1425. [DOI] [PubMed] [Google Scholar]
- 10.Clifton MA. Familial abdominal aortic aneurysms. Br J Surg. 1977;64(11):765–6. doi: 10.1002/bjs.1800641102. [DOI] [PubMed] [Google Scholar]
- 11.Ogata T, MacKean GL, Cole CW, et al. The lifetime prevalence of abdominal aortic aneurysms among siblings of aneurysm patients is eightfold higher than among siblings of spouses: an analysis of 187 aneurysm families in Nova Scotia, Canada. J Vasc Surg. 2005;42(5):891–7. doi: 10.1016/j.jvs.2005.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norrgard O, Cedergren B, Angquist KA, et al. Blood groups and HLA antigens in patients with abdominal aortic aneurysms. Hum Hered. 1984;34(1):9–13. doi: 10.1159/000153411. [DOI] [PubMed] [Google Scholar]
- 13.Anderson DW, Edwards TK, Ricketts MH, et al. Multiple defects in type III collagen synthesis are associated with the pathogenesis of abdominal aortic aneurysms. Ann N Y Acad Sci. 1996;800:216–28. doi: 10.1111/j.1749-6632.1996.tb33312.x. [DOI] [PubMed] [Google Scholar]
- 14.Kontusaari S, Tromp G, Kuivaniemi H, et al. Inheritance of an RNA splicing mutation (G+ 1 IVS20) in the type III procollagen gene (COL3A1) in a family having aortic aneurysms and easy bruisability: phenotypic overlap between familial arterial aneurysms and Ehlers-Danlos syndrome type IV. Am J Hum Genet. 1990;47(1):112–20. [PMC free article] [PubMed] [Google Scholar]
- 15.Kontusaari S, Tromp G, Kuivaniemi H, et al. A mutation in the gene for type III procollagen (COL3A1) in a family with aortic aneurysms. J Clin Invest. 1990;86(5):1465–73. doi: 10.1172/JCI114863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tromp G, Wu Y, Prockop DJ, et al. Sequencing of cDNA from 50 unrelated patients reveals that mutations in the triple-helical domain of type III procollagen are an infrequent cause of aortic aneurysms. J Clin Invest. 1993;91(6):2539–45. doi: 10.1172/JCI116490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shibamura H, Olson JM, van Vlijmen-Van Keulen C, et al. Genome scan for familial abdominal aortic aneurysm using sex and family history as covariates suggests genetic heterogeneity and identifies linkage to chromosome 19q13. Circulation. 2004;109(17):2103–8. doi: 10.1161/01.CIR.0000127857.77161.A1. [DOI] [PubMed] [Google Scholar]
- 18.Van Vlijmen-Van Keulen CJ, Rauwerda JA, Pals G. Genome-wide linkage in three Dutch families maps a locus for abdominal aortic aneurysms to chromosome 19q13.3. Eur J Vasc Endovasc Surg. 2005;30(1):29–35. doi: 10.1016/j.ejvs.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 19.Elmore JR, Obmann MA, Kuivaniemi H, et al. Identification of a genetic variant associated with abdominal aortic aneurysms on chromosome 3p12.3 by genome wide association. J Vasc Surg. 2009;49(6):1525–31. doi: 10.1016/j.jvs.2009.01.041. [DOI] [PubMed] [Google Scholar]
- 20.Gretarsdottir S, Baas AF, Thorleifsson G, et al. Genome-wide association study identifies a sequence variant within the DAB2IP gene conferring susceptibility to abdominal aortic aneurysm. Nat Genet. 2010 doi: 10.1038/ng.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lenk GM, Tromp G, Weinsheimer S, et al. Whole genome expression profiling reveals a significant role for immune function in human abdominal aortic aneurysms. BMC Genomics. 2007;8:237. doi: 10.1186/1471-2164-8-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nischan J, Gatalica Z, Curtis M, et al. Binding sites for ETS family of transcription factors dominate the promoter regions of differentially expressed genes in abdominal aortic aneurysms. Circ Cardiovasc Genet. 2009;2(6):565–72. doi: 10.1161/CIRCGENETICS.108.843854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choke E, Cockerill GW, Laing K, et al. Whole genome-expression profiling reveals a role for immune and inflammatory response in abdominal aortic aneurysm rupture. Eur J Vasc Endovasc Surg. 2009;37(3):305–10. doi: 10.1016/j.ejvs.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 24.Giusti B, Rossi L, Lapini I, et al. Gene expression profiling of peripheral blood in patients with abdominal aortic aneurysm. Eur J Vasc Endovasc Surg. 2009;38(1):104–12. doi: 10.1016/j.ejvs.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 25.McKusick VA. The defect in Marfan syndrome. Nature. 1991;352(6333):279–81. doi: 10.1038/352279a0. [DOI] [PubMed] [Google Scholar]
- 26.Dobrin PB, Baker WH, Gley WC. Elastolytic and collagenolytic studies of arteries. Implications for the mechanical properties of aneurysms. Arch Surg. 1984;119(4):405–9. doi: 10.1001/archsurg.1984.01390160041009. [DOI] [PubMed] [Google Scholar]
- 27.Superti-Furga A, Steinmann B, Ramirez F, et al. Molecular defects of type III procollagen in Ehlers-Danlos syndrome type IV. Hum Genet. 1989;82:104–108. doi: 10.1007/BF00284038. [DOI] [PubMed] [Google Scholar]
- 28.Kuivaniemi H, Marshall A, Ganguly A, et al. Fibulin-2 exhibits high degree of variability, but no structural changes concordant with abdominal aortic aneurysms. Eur J Hum Genet. 1998;6(6):642–6. doi: 10.1038/sj.ejhg.5200245. [DOI] [PubMed] [Google Scholar]
- 29.Davis V, Persidskaia R, Baca-Regen L, et al. Matrix metalloproteinase-2 production and its binding to the matrix are increased in abdominal aortic aneurysms. Arterioscler Thromb Vasc Biol. 1998;18(10):1625–33. doi: 10.1161/01.atv.18.10.1625. [DOI] [PubMed] [Google Scholar]
- 30.McMillan WD, Patterson BK, Keen RR, et al. In situ localization and quantification of seventy-two-kilodalton type IV collagenase in aneurysmal, occlusive, and normal aorta. J Vasc Surg. 1995;22(3):295–305. doi: 10.1016/s0741-5214(95)70144-3. [DOI] [PubMed] [Google Scholar]
- 31.Sakalihasan N, Delvenne P, Nusgens BV, et al. Activated forms of MMP2 and MMP9 in abdominal aortic aneurysms. J Vasc Surg. 1996;24(1):127–33. doi: 10.1016/s0741-5214(96)70153-2. [DOI] [PubMed] [Google Scholar]
- 32.Hinterseher I, Bergert H, Kuhlisch E, et al. Matrix metalloproteinase 2 polymorphisms in a caucasian population with abdominal aortic aneurysm. J Surg Res. 2006;133(2):121–8. doi: 10.1016/j.jss.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Brophy CM, Marks WH, Reilly JM, et al. Decreased tissue inhibitor of metalloproteinases (TIMP) in abdominal aortic aneurysm tissue: a preliminary report. J Surg Res. 1991;50(6):653–7. doi: 10.1016/0022-4804(91)90058-t. [DOI] [PubMed] [Google Scholar]
- 34.Defawe OD, Colige A, Lambert CA, et al. TIMP-2 and PAI-1 mRNA levels are lower in aneurysmal as compared to athero-occlusive abdominal aortas. Cardiovasc Res. 2003;60(1):205–13. doi: 10.1016/s0008-6363(03)00513-3. [DOI] [PubMed] [Google Scholar]
- 35.Eskandari MK, Vijungco JD, Flores A, et al. Enhanced abdominal aortic aneurysm in TIMP-1-deficient mice. J Surg Res. 2005;123(2):289–93. doi: 10.1016/j.jss.2004.07.247. [DOI] [PubMed] [Google Scholar]
- 36.Tilson MD, Reilly JM, Brophy CM, et al. Expression and sequence of the gene for tissue inhibitor of metalloproteinases in patients with abdominal aortic aneurysms. J Vasc Surg. 1993;18(2):266–70. [PubMed] [Google Scholar]
- 37.Wang X, Tromp G, Cole CW, et al. Analysis of coding sequences for tissue inhibitor of metalloproteinases 1 (TIMP1) and 2 (TIMP2) in patients with aneurysms. Matrix Biol. 1999;18(2):121–4. doi: 10.1016/s0945-053x(99)00008-6. [DOI] [PubMed] [Google Scholar]
- 38.Hinterseher I, Krex D, Kuhlisch E, et al. Tissue inhibitor of metalloproteinase-1 (TIMP-1) polymorphisms in a Caucasian population with abdominal aortic aneurysm. World J Surg. 2007;31(11):2248–54. doi: 10.1007/s00268-007-9209-x. [DOI] [PubMed] [Google Scholar]
- 39.Annabi B, Shedid D, Ghosn P, et al. Differential regulation of matrix metalloproteinase activities in abdominal aortic aneurysms. J Vasc Surg. 2002;35(3):539–46. doi: 10.1067/mva.2002.121124. [DOI] [PubMed] [Google Scholar]
- 40.Tamarina NA, McMillan WD, Shively VP, et al. Expression of matrix metalloproteinases and their inhibitors in aneurysms and normal aorta. Surgery. 1997;122(2):264–71. doi: 10.1016/s0039-6060(97)90017-9. discussion 271–2. [DOI] [PubMed] [Google Scholar]
- 41.Hinterseher I, Krex D, Kuhlisch E, et al. Analysis of tissue inhibitor of metalloproteinase-2 gene polymorphisms in a caucasian population with abdominal aortic aneurysms. Zentralbl Chir. 2008;133(4):332–7. doi: 10.1055/s-2008-1076862. [DOI] [PubMed] [Google Scholar]
- 42.Powell JT, Bashir A, Dawson S, et al. Genetic variation on chromosome 16 is associated with abdominal aortic aneurysm. Clin Sci (Lond) 1990;78(1):13–6. doi: 10.1042/cs0780013. [DOI] [PubMed] [Google Scholar]
- 43.Ramsbottom D, O’Neill A, Sexton DM, et al. The cholesteryl ester transfer protein (CETP) locus as a candidate gene in abdominal aortic aneurysm. Clin Genet. 1997;51(4):241–5. doi: 10.1111/j.1399-0004.1997.tb02462.x. [DOI] [PubMed] [Google Scholar]
- 44.Adamson J, Powell JT, Greenhalgh RM. Selection for screening for familial aortic aneurysms. Br J Surg. 1992;79(9):897–8. doi: 10.1002/bjs.1800790914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Powell JT, Adamson J, MacSweeney ST, et al. Influence of type III collagen genotype on aortic diameter and disease. Br J Surg. 1993;80(10):1246–8. doi: 10.1002/bjs.1800801008. [DOI] [PubMed] [Google Scholar]
- 46.Powell JT, Adamson J, MacSweeney ST, et al. Genetic variants of collagen III and abdominal aortic aneurysm. Eur J Vasc Surg. 1991;5(2):145–8. doi: 10.1016/s0950-821x(05)80679-6. [DOI] [PubMed] [Google Scholar]
- 47.MacSweeney ST, Skidmore C, Turner RJ, et al. Unravelling the familial tendency to aneurysmal disease: popliteal aneurysm, hypertension and fibrillin genotype. Eur J Vasc Endovasc Surg. 1996;12(2):162–6. doi: 10.1016/s1078-5884(96)80101-8. [DOI] [PubMed] [Google Scholar]
- 48.Ogata T, Shibamura H, Tromp G, et al. Genetic analysis of polymorphisms in biologically relevant candidate genes in patients with abdominal aortic aneurysms. J Vasc Surg. 2005;41(6):1036–42. doi: 10.1016/j.jvs.2005.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee B, Godfrey M, Vitale E, et al. Linkage of Marfan syndrome and a phenotypically related disorder to two different fibrillin genes. Nature. 1991;352(6333):330–4. doi: 10.1038/352330a0. [DOI] [PubMed] [Google Scholar]
- 50.Ramirez F, Lee B, Vitale E. Clinical and genetic associations in Marfan syndrome and related disorders. Mt Sinai J Med. 1992;59(4):350–6. [PubMed] [Google Scholar]
- 51.Caglayan AO, Dundar M. Inherited diseases and syndromes leading to aortic aneurysms and dissections. Eur J Cardiothorac Surg. 2009;35(6):931–40. doi: 10.1016/j.ejcts.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 52.Thompson RW, Geraghty PJ, Lee JK. Abdominal aortic aneurysms: basic mechanisms and clinical implications. Curr Probl Surg. 2002;39(2):110–230. doi: 10.1067/msg.2002.121421. [DOI] [PubMed] [Google Scholar]
- 53.Ewart AK, Morris CA, Atkinson D, et al. Hemizygosity at the elastin locus in a developmental disorder, Williams syndrome. Nat Genet. 1993;5(1):11–6. doi: 10.1038/ng0993-11. [DOI] [PubMed] [Google Scholar]
- 54.Ewart AK, Morris CA, Ensing GJ, et al. A human vascular disorder, supravalvular aortic stenosis, maps to chromosome 7. Proc Natl Acad Sci U S A. 1993;90(8):3226–30. doi: 10.1073/pnas.90.8.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang MC, He L, Giro M, et al. Cutis laxa arising from frameshift mutations in exon 30 of the elastin gene (ELN) J Biol Chem. 1999;274(2):981–6. doi: 10.1074/jbc.274.2.981. [DOI] [PubMed] [Google Scholar]
- 56.Massart F, Marini F, Menegato A, et al. Allelic genes involved in artery compliance and susceptibility to sporadic abdominal aortic aneurysm. J Steroid Biochem Mol Biol. 2004;92(5):413–8. doi: 10.1016/j.jsbmb.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 57.Schulz V, Hendig D, Schillinger M, et al. Analysis of sequence variations in the ABCC6 gene among patients with abdominal aortic aneurysm and pseudoxanthoma elasticum. J Vasc Res. 2005;42(5):424–32. doi: 10.1159/000087900. [DOI] [PubMed] [Google Scholar]
- 58.Yanagisawa H, Davis EC, Starcher BC, et al. Fibulin-5 is an elastin-binding protein essential for elastic fibre development in vivo. Nature. 2002;415(6868):168–71. doi: 10.1038/415168a. [DOI] [PubMed] [Google Scholar]
- 59.Nakamura T, Ruiz-Lozano P, Lindner V, et al. DANCE, a novel secreted RGD protein expressed in developing, atherosclerotic, and balloon-injured arteries. J Biol Chem. 1999;274(32):22476–83. doi: 10.1074/jbc.274.32.22476. [DOI] [PubMed] [Google Scholar]
- 60.Chu ML, Tsuda T. Fibulins in development and heritable disease. Birth Defects Res C Embryo Today. 2004;72(1):25–36. doi: 10.1002/bdrc.20003. [DOI] [PubMed] [Google Scholar]
- 61.Nakamura T, Lozano PR, Ikeda Y, et al. Fibulin-5/DANCE is essential for elastogenesis in vivo. Nature. 2002;415(6868):171–5. doi: 10.1038/415171a. [DOI] [PubMed] [Google Scholar]
- 62.Spencer JA, Hacker SL, Davis EC, et al. Altered vascular remodeling in fibulin-5-deficient mice reveals a role of fibulin-5 in smooth muscle cell proliferation and migration. Proc Natl Acad Sci U S A. 2005;102(8):2946–51. doi: 10.1073/pnas.0500058102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loeys B, Van Maldergem L, Mortier G, et al. Homozygosity for a missense mutation in fibulin-5 (FBLN5) results in a severe form of cutis laxa. Hum Mol Genet. 2002;11(18):2113–8. doi: 10.1093/hmg/11.18.2113. [DOI] [PubMed] [Google Scholar]
- 64.Stone EM, Braun TA, Russell SR, et al. Missense variations in the fibulin 5 gene and age-related macular degeneration. N Engl J Med. 2004;351(4):346–53. doi: 10.1056/NEJMoa040833. [DOI] [PubMed] [Google Scholar]
- 65.Badger SA, Soong CV, O’Donnell ME, et al. Common polymorphisms of Fibulin-5 and the risk of abdominal aortic aneurysm development. Vasc Med. 2009;15(2):113–7. doi: 10.1177/1358863X09355667. [DOI] [PubMed] [Google Scholar]
- 66.McMillan WD, Pearce WH. Increased plasma levels of metalloproteinase-9 are associated with abdominal aortic aneurysms. J Vasc Surg. 1999;29(1):122–7. doi: 10.1016/s0741-5214(99)70363-0. discussion 127–9. [DOI] [PubMed] [Google Scholar]
- 67.Eriksson P, Jormsjo-Pettersson S, Brady AR, et al. Genotype-phenotype relationships in an investigation of the role of proteases in abdominal aortic aneurysm expansion. Br J Surg. 2005;92(11):1372–6. doi: 10.1002/bjs.5126. [DOI] [PubMed] [Google Scholar]
- 68.Powell JT. Genes predisposing to rapid aneurysm growth. Ann N Y Acad Sci. 2006;1085:236–41. doi: 10.1196/annals.1383.042. [DOI] [PubMed] [Google Scholar]
- 69.Carrell TW, Burnand KG, Wells GM, et al. Stromelysin-1 (matrix metalloproteinase-3) and tissue inhibitor of metalloproteinase-3 are overexpressed in the wall of abdominal aortic aneurysms. Circulation. 2002;105(4):477–82. doi: 10.1161/hc0402.102621. [DOI] [PubMed] [Google Scholar]
- 70.Deguara J, Burnand KG, Berg J, et al. An increased frequency of the 5A allele in the promoter region of the MMP3 gene is associated with abdominal aortic aneurysms. Hum Mol Genet. 2007;16(24):3002–7. doi: 10.1093/hmg/ddm258. [DOI] [PubMed] [Google Scholar]
- 71.Yoon S, Tromp G, Vongpunsawad S, et al. Genetic analysis of MMP3, MMP9, and PAI-1 in Finnish patients with abdominal aortic or intracranial aneurysms. Biochem Biophys Res Commun. 1999;265(2):563–8. doi: 10.1006/bbrc.1999.1721. [DOI] [PubMed] [Google Scholar]
- 72.McColgan P, Peck GE, Greenhalgh RM, et al. The genetics of abdominal aortic aneurysms: a comprehensive meta-analysis involving eight candidate genes in over 16,700 patients. Int Surg. 2009;94(4):350–8. [PubMed] [Google Scholar]
- 73.Zhang B, Ye S, Herrmann SM, et al. Functional polymorphism in the regulatory region of gelatinase B gene in relation to severity of coronary atherosclerosis. Circulation. 1999;99(14):1788–94. doi: 10.1161/01.cir.99.14.1788. [DOI] [PubMed] [Google Scholar]
- 74.Jones GT, Phillips VL, Harris EL, et al. Functional matrix metalloproteinase-9 polymorphism (C-1562T) associated with abdominal aortic aneurysm. J Vasc Surg. 2003;38(6):1363–7. doi: 10.1016/s0741-5214(03)01027-9. [DOI] [PubMed] [Google Scholar]
- 75.Smallwood L, Allcock R, van Bockxmeer F, et al. Polymorphisms of the matrix metalloproteinase 9 gene and abdominal aortic aneurysm. Br J Surg. 2008;95(10):1239–44. doi: 10.1002/bjs.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Armani C, Curcio M, Barsotti MC, et al. Polymorphic analysis of the matrix metalloproteinase-9 gene and susceptibility to sporadic abdominal aortic aneurysm. Biomed Pharmacother. 2007;61(5):268–71. doi: 10.1016/j.biopha.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 77.St Jean PL, Zhang XC, Hart BK, et al. Characterization of a dinucleotide repeat in the 92 kDa type IV collagenase gene (CLG4B), localization of CLG4B to chromosome 20 and the role of CLG4B in aortic aneurysmal disease. Ann Hum Genet. 1995;59(Pt 1):17–24. doi: 10.1111/j.1469-1809.1995.tb01602.x. [DOI] [PubMed] [Google Scholar]
- 78.Thompson AR, Drenos F, Hafez H, et al. Candidate gene association studies in abdominal aortic aneurysm disease: a review and meta-analysis. Eur J Vasc Endovasc Surg. 2008;35(1):19–30. doi: 10.1016/j.ejvs.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 79.Cox D. α1-Antitrypsin deficiency. In: Royce P, Steinman B, editors. Connective Tissue and its Heritable Disoerders. New York: Wiley-Liss; 1993. pp. 549–561. [Google Scholar]
- 80.Fagerhol MK. Quantitative studies on the inherited variants of serum alpha-1-antitrypsin. Scand J Clin Lab Invest. 1969;23(1):97–103. doi: 10.3109/00365516909078092. [DOI] [PubMed] [Google Scholar]
- 81.St Jean P, Hart B, Webster M, et al. Alpha-1-antitrypsin deficiency in aneurysmal disease. Hum Hered. 1996;46(2):92–7. doi: 10.1159/000154333. [DOI] [PubMed] [Google Scholar]
- 82.Schardey HM, Hernandez-Richter T, Klueppelberg U, et al. Alleles of the alpha-1-antitrypsin phenotype in patients with aortic aneurysms. J Cardiovasc Surg (Torino) 1998;39(5):535–9. [PubMed] [Google Scholar]
- 83.Werb Z. ECM and cell surface proteolysis: regulating cellular ecology. Cell. 1997;91(4):439–42. doi: 10.1016/s0092-8674(00)80429-8. [DOI] [PubMed] [Google Scholar]
- 84.Shireman PK, McCarthy WJ, Pearce WH, et al. Elevations of tissue-type plasminogen activator and differential expression of urokinase-type plasminogen activator in diseased aorta. J Vasc Surg. 1997;25(1):157–64. doi: 10.1016/s0741-5214(97)70333-1. [DOI] [PubMed] [Google Scholar]
- 85.Eriksson P, Kallin B, van ‘t Hooft FM, et al. Allele-specific increase in basal transcription of the plasminogen-activator inhibitor 1 gene is associated with myocardial infarction. Proc Natl Acad Sci U S A. 1995;92(6):1851–5. doi: 10.1073/pnas.92.6.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jones K, Powell J, Brown L, et al. The influence of 4G/5G polymorphism in the plasminogen activator inhibitor-1 gene promoter on the incidence, growth and operative risk of abdominal aortic aneurysm. Eur J Vasc Endovasc Surg. 2002;23(5):421–5. doi: 10.1053/ejvs.2002.1633. [DOI] [PubMed] [Google Scholar]
- 87.Golledge J, McCann M, Mangan S, et al. Osteoprotegerin and osteopontin are expressed at high concentrations within symptomatic carotid atherosclerosis. Stroke. 2004;35(7):1636–41. doi: 10.1161/01.STR.0000129790.00318.a3. [DOI] [PubMed] [Google Scholar]
- 88.Denhardt DT, Noda M, O’Regan AW, et al. Osteopontin as a means to cope with environmental insults: regulation of inflammation, tissue remodeling, and cell survival. J Clin Invest. 2001;107(9):1055–61. doi: 10.1172/JCI12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.O’Regan A, Berman JS. Osteopontin: a key cytokine in cell-mediated and granulomatous inflammation. Int J Exp Pathol. 2000;81(6):373–90. doi: 10.1046/j.1365-2613.2000.00163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bruemmer D, Collins AR, Noh G, et al. Angiotensin II-accelerated atherosclerosis and aneurysm formation is attenuated in osteopontin-deficient mice. J Clin Invest. 2003;112(9):1318–31. doi: 10.1172/JCI18141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Golledge J, Muller J, Shephard N, et al. Association between osteopontin and human abdominal aortic aneurysm. Arterioscler Thromb Vasc Biol. 2007;27(3):655–60. doi: 10.1161/01.ATV.0000255560.49503.4e. [DOI] [PubMed] [Google Scholar]
- 92.Gotting C, Prante C, Schillinger M, et al. Xylosyltransferase I variants and their impact on abdominal aortic aneurysms. Clin Chim Acta. 2008;391(1–2):41–5. doi: 10.1016/j.cca.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 93.Sukhova GK, Shi GP, Simon DI, et al. Expression of the elastolytic cathepsins S and K in human atheroma and regulation of their production in smooth muscle cells. J Clin Invest. 1998;102(3):576–83. doi: 10.1172/JCI181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lindholt JS, Erlandsen EJ, Henneberg EW. Cystatin C deficiency is associated with the progression of small abdominal aortic aneurysms. Br J Surg. 2001;88(11):1472–5. doi: 10.1046/j.0007-1323.2001.01911.x. [DOI] [PubMed] [Google Scholar]
- 95.Shi GP, Sukhova GK, Grubb A, et al. Cystatin C deficiency in human atherosclerosis and aortic aneurysms. J Clin Invest. 1999;104(9):1191–7. doi: 10.1172/JCI7709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Eriksson P, Jones KG, Brown LC, et al. Genetic approach to the role of cysteine proteases in the expansion of abdominal aortic aneurysms. Br J Surg. 2004;91(1):86–9. doi: 10.1002/bjs.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Claridge MW, Hobbs SD, Quick CR, et al. ACE inhibitors increase type III collagen synthesis: a potential explanation for reduction in acute vascular events by ACE inhibitors. Eur J Vasc Endovasc Surg. 2004;28(1):67–70. doi: 10.1016/j.ejvs.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 98.Hackam DG, Thiruchelvam D, Redelmeier DA. Angiotensin-converting enzyme inhibitors and aortic rupture: a population-based case-control study. Lancet. 2006;368(9536):659–65. doi: 10.1016/S0140-6736(06)69250-7. [DOI] [PubMed] [Google Scholar]
- 99.Tiret L, Rigat B, Visvikis S, et al. Evidence, from combined segregation and linkage analysis, that a variant of the angiotensin I-converting enzyme (ACE) gene controls plasma ACE levels. Am J Hum Genet. 1992;51(1):197–205. [PMC free article] [PubMed] [Google Scholar]
- 100.Rigat B, Hubert C, Alhenc-Gelas F, et al. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86(4):1343–6. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hamano K, Ohishi M, Ueda M, et al. Deletion polymorphism in the gene for angiotensin-converting enzyme is not a risk factor predisposing to abdominal aortic aneurysm. Eur J Vasc Endovasc Surg. 1999;18(2):158–61. doi: 10.1053/ejvs.1999.0873. [DOI] [PubMed] [Google Scholar]
- 102.Yeung JM, Heeley M, Gray S, et al. Does the angiotensin-converting enzyme (ACE) gene polymorphism affect rate of abdominal aortic aneurysm expansion? Eur J Vasc Endovasc Surg. 2002;24(1):69–71. doi: 10.1053/ejvs.2002.1693. [DOI] [PubMed] [Google Scholar]
- 103.Korcz A, Mikolajczyk-Stecyna J, Gabriel M, et al. Angiotensin-converting enzyme (ACE, I/D) gene polymorphism and susceptibility to abdominal aortic aneurysm or aortoiliac occlusive disease. J Surg Res. 2009;153(1):76–82. doi: 10.1016/j.jss.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 104.Pola R, Gaetani E, Santoliquido A, et al. Abdominal aortic aneurysm in normotensive patients: association with angiotensin-converting enzyme gene polymorphism. Eur J Vasc Endovasc Surg. 2001;21(5):445–9. doi: 10.1053/ejvs.2001.1339. [DOI] [PubMed] [Google Scholar]
- 105.Lucarini L, Sticchi E, Sofi F, et al. ACE and TGFBR1 genes interact in influencing the susceptibility to abdominal aortic aneurysm. Atherosclerosis. 2009;202(1):205–10. doi: 10.1016/j.atherosclerosis.2008.04.038. [DOI] [PubMed] [Google Scholar]
- 106.Fatini C, Pratesi G, Sofi F, et al. ACE DD genotype: a predisposing factor for abdominal aortic aneurysm. Eur J Vasc Endovasc Surg. 2005;29(3):227–32. doi: 10.1016/j.ejvs.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 107.Jones GT, Thompson AR, van Bockxmeer FM, et al. Angiotensin II type 1 receptor 1166C polymorphism is associated with abdominal aortic aneurysm in three independent cohorts. Arterioscler Thromb Vasc Biol. 2008;28(4):764–70. doi: 10.1161/ATVBAHA.107.155564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Obukofe B, Sayers RD, Thompson J, et al. Is There an Association between Angiotensin Converting Enzyme (ACE) Genotypes and Abdominal Aortic Aneurysm? Eur J Vasc Endovasc Surg. 2010 doi: 10.1016/j.ejvs.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 109.Elmore JR, Obmann MA, Kuivaniemi H, et al. Reply to Comment Regarding “Identification of a genetic variant associated with abdominal aortic aneurysms on chromosome 3p12.3 by genome wide association”. J Vasc Surg. 2009;50(5):1246–7. doi: 10.1016/j.jvs.2009.07.098. author reply 1247. [DOI] [PubMed] [Google Scholar]
- 110.Humphries SE, Peacock R, Dunning A, et al. Identification of genetic variation that determines levels of plasma triglycerides and hypercoagulability. Clin Genet. 1994;46(1 Spec):19–31. doi: 10.1111/j.1399-0004.1994.tb04198.x. [DOI] [PubMed] [Google Scholar]
- 111.Davignon J, Gregg RE, Sing CF. Apolipoprotein E polymorphism and atherosclerosis. Arteriosclerosis. 1988;8(1):1–21. doi: 10.1161/01.atv.8.1.1. [DOI] [PubMed] [Google Scholar]
- 112.Daugherty A, Manning MW, Cassis LA. Angiotensin II promotes atherosclerotic lesions and aneurysms in apolipoprotein E-deficient mice. J Clin Invest. 2000;105(11):1605–12. doi: 10.1172/JCI7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gerdes LU, Lindholt JS, Vammen S, et al. Apolipoprotein E genotype is associated with differential expansion rates of small abdominal aortic aneurysms. Br J Surg. 2000;87(6):760–5. doi: 10.1046/j.1365-2168.2000.01486.x. [DOI] [PubMed] [Google Scholar]
- 114.Golledge J, Biros E, Cooper M, et al. Apolipoprotein E genotype is associated with serum C-reactive protein but not abdominal aortic aneurysm. Atherosclerosis. 2010;209(2):487–91. doi: 10.1016/j.atherosclerosis.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sofi F, Marcucci R, Giusti B, et al. High levels of homocysteine, lipoprotein (a) and plasminogen activator inhibitor-1 are present in patients with abdominal aortic aneurysm. Thromb Haemost. 2005;94(5):1094–8. [PubMed] [Google Scholar]
- 116.Jourdheuil-Rahmani D, Rolland PH, Rosset E, et al. Homocysteine induces synthesis of a serine elastase in arterial smooth muscle cells from multi-organ donors. Cardiovasc Res. 1997;34(3):597–602. doi: 10.1016/s0008-6363(97)00048-5. [DOI] [PubMed] [Google Scholar]
- 117.Giusti B, Marcucci R, Lapini I, et al. Role of hyperhomocysteinemia in aortic disease. Cell Mol Biol (Noisy-le-grand) 2004;50(8):945–52. [PubMed] [Google Scholar]
- 118.Brunelli T, Prisco D, Fedi S, et al. High prevalence of mild hyperhomocysteinemia in patients with abdominal aortic aneurysm. J Vasc Surg. 2000;32(3):531–6. doi: 10.1067/mva.2000.107563. [DOI] [PubMed] [Google Scholar]
- 119.Sharp L, Little J. Polymorphisms in genes involved in folate metabolism and colorectal neoplasia: a HuGE review. Am J Epidemiol. 2004;159(5):423–43. doi: 10.1093/aje/kwh066. [DOI] [PubMed] [Google Scholar]
- 120.Peeters AC, van Landeghem BA, Graafsma SJ, et al. Low vitamin B6, and not plasma homocysteine concentration, as risk factor for abdominal aortic aneurysm: a retrospective case-control study. J Vasc Surg. 2007;45(4):701–5. doi: 10.1016/j.jvs.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 121.Ferrara F, Novo S, Grimaudo S, et al. Methylenetetrahydrofolate reductase mutation in subjects with abdominal aortic aneurysm subdivided for age. Clin Hemorheol Microcirc. 2006;34(3):421–6. [PubMed] [Google Scholar]
- 122.Strauss E, Waliszewski K, Gabriel M, et al. Increased risk of the abdominal aortic aneurysm in carriers of the MTHFR 677T allele. J Appl Genet. 2003;44(1):85–93. [PubMed] [Google Scholar]
- 123.Strauss E, Waliszewski K, Pawlak AL. The normotensive carriers of the MTHFR 677T allele, displaying the increased risk of development of the abdominal aortic aneurysm (AAA), occur at the highest frequency among the smoking patients. Przegl Lek. 2004;61(10):1086–9. [PubMed] [Google Scholar]
- 124.Jones GT, Harris EL, Phillips LV, et al. The methylenetetrahydrofolate reductase C677T polymorphism does not associate with susceptibility to abdominal aortic aneurysm. Eur J Vasc Endovasc Surg. 2005;30(2):137–42. doi: 10.1016/j.ejvs.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 125.Giusti B, Saracini C, Bolli P, et al. Genetic analysis of 56 polymorphisms in 17 genes involved in methionine metabolism in patients with abdominal aortic aneurysm. J Med Genet. 2008;45(11):721–30. doi: 10.1136/jmg.2008.057851. [DOI] [PubMed] [Google Scholar]
- 126.Strauss E, Waliszewski K, Pawlak AL. The different genotypes of MTHFR 1298A>C and PON1 -108C>T polymorphisms confer the increased risk of the abdominal aortic aneurysm in the smoking and nonsmoking persons. Przegl Lek. 2005;62(10):1023–30. [PubMed] [Google Scholar]
- 127.Mandola MV, Stoehlmacher J, Zhang W, et al. A 6 bp polymorphism in the thymidylate synthase gene causes message instability and is associated with decreased intratumoral TS mRNA levels. Pharmacogenetics. 2004;14(5):319–27. doi: 10.1097/00008571-200405000-00007. [DOI] [PubMed] [Google Scholar]
- 128.Kealey C, Brown KS, Woodside JV, et al. A common insertion/deletion polymorphism of the thymidylate synthase (TYMS) gene is a determinant of red blood cell folate and homocysteine concentrations. Hum Genet. 2005;116(5):347–53. doi: 10.1007/s00439-004-1243-2. [DOI] [PubMed] [Google Scholar]
- 129.Souto JC, Blanco-Vaca F, Soria JM, et al. A genomewide exploration suggests a new candidate gene at chromosome 11q23 as the major determinant of plasma homocysteine levels: results from the GAIT project. Am J Hum Genet. 2005;76(6):925–33. doi: 10.1086/430409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Veldman BA, Spiering W, Doevendans PA, et al. The Glu298Asp polymorphism of the NOS 3 gene as a determinant of the baseline production of nitric oxide. J Hypertens. 2002;20(10):2023–7. doi: 10.1097/00004872-200210000-00022. [DOI] [PubMed] [Google Scholar]
- 131.Hingorani AD, Liang CF, Fatibene J, et al. A common variant of the endothelial nitric oxide synthase (Glu298-->Asp) is a major risk factor for coronary artery disease in the UK. Circulation. 1999;100(14):1515–20. doi: 10.1161/01.cir.100.14.1515. [DOI] [PubMed] [Google Scholar]
- 132.Lembo G, De Luca N, Battagli C, et al. A common variant of endothelial nitric oxide synthase (Glu298Asp) is an independent risk factor for carotid atherosclerosis. Stroke. 2001;32(3):735–40. doi: 10.1161/01.str.32.3.735. [DOI] [PubMed] [Google Scholar]
- 133.Rudic RD, Shesely EG, Maeda N, et al. Direct evidence for the importance of endothelium-derived nitric oxide in vascular remodeling. J Clin Invest. 1998;101(4):731–6. doi: 10.1172/JCI1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Radomski MW, Palmer RM, Moncada S. Endogenous nitric oxide inhibits human platelet adhesion to vascular endothelium. Lancet. 1987;2(8567):1057–8. doi: 10.1016/s0140-6736(87)91481-4. [DOI] [PubMed] [Google Scholar]
- 135.Fatini C, Sofi F, Sticchi E, et al. eNOS G894T polymorphism as a mild predisposing factor for abdominal aortic aneurysm. J Vasc Surg. 2005;42(3):415–9. doi: 10.1016/j.jvs.2005.05.044. [DOI] [PubMed] [Google Scholar]
- 136.Atli FH, Manduz S, Katrancioglu N, et al. eNOS G894T Polymorphism and Abdominal Aortic Aneurysms. Angiology. 2009 doi: 10.1177/0003319709339589. [DOI] [PubMed] [Google Scholar]
- 137.Nakayama M, Yasue H, Yoshimura M, et al. T-786-->C mutation in the 5′-flanking region of the endothelial nitric oxide synthase gene is associated with coronary spasm. Circulation. 1999;99(22):2864–70. doi: 10.1161/01.cir.99.22.2864. [DOI] [PubMed] [Google Scholar]
- 138.Tsukada T, Yokoyama K, Arai T, et al. Evidence of association of the ecNOS gene polymorphism with plasma NO metabolite levels in humans. Biochem Biophys Res Commun. 1998;245(1):190–3. doi: 10.1006/bbrc.1998.8267. [DOI] [PubMed] [Google Scholar]
- 139.Uwabo J, Soma M, Nakayama T, et al. Association of a variable number of tandem repeats in the endothelial constitutive nitric oxide synthase gene with essential hypertension in Japanese. Am J Hypertens. 1998;11(1 Pt 1):125–8. doi: 10.1016/s0895-7061(97)00419-6. [DOI] [PubMed] [Google Scholar]
- 140.Wang XL, Sim AS, Badenhop RF, et al. A smoking-dependent risk of coronary artery disease associated with a polymorphism of the endothelial nitric oxide synthase gene. Nat Med. 1996;2(1):41–5. doi: 10.1038/nm0196-41. [DOI] [PubMed] [Google Scholar]
- 141.Kotani K, Shimomura T, Murakami F, et al. Allele frequency of human endothelial nitric oxide synthase gene polymorphism in abdominal aortic aneurysm. Intern Med. 2000;39(7):537–9. doi: 10.2169/internalmedicine.39.537. [DOI] [PubMed] [Google Scholar]
- 142.Cipollone F, Prontera C, Pini B, et al. Overexpression of functionally coupled cyclooxygenase-2 and prostaglandin E synthase in symptomatic atherosclerotic plaques as a basis of prostaglandin E(2)-dependent plaque instability. Circulation. 2001;104(8):921–7. doi: 10.1161/hc3401.093152. [DOI] [PubMed] [Google Scholar]
- 143.Cipollone F, Rocca B, Patrono C. Cyclooxygenase-2 expression and inhibition in atherothrombosis. Arterioscler Thromb Vasc Biol. 2004;24(2):246–55. doi: 10.1161/01.ATV.0000104005.92603.f2. [DOI] [PubMed] [Google Scholar]
- 144.Cipollone F, Toniato E, Martinotti S, et al. A polymorphism in the cyclooxygenase 2 gene as an inherited protective factor against myocardial infarction and stroke. Jama. 2004;291(18):2221–8. doi: 10.1001/jama.291.18.2221. [DOI] [PubMed] [Google Scholar]
- 145.Xu Q, Ji YS, Schmedtje JF., Jr Sp1 increases expression of cyclooxygenase-2 in hypoxic vascular endothelium. Implications for the mechanisms of aortic aneurysm and heart failure. J Biol Chem. 2000;275(32):24583–9. doi: 10.1074/jbc.M003894200. [DOI] [PubMed] [Google Scholar]
- 146.Badger SA, Soong CV, Young IS, et al. The influence of COX-2 single nucleotide polymorphisms on abdominal aortic aneurysm development and the associated inflammation. Angiology. 2009;60(5):576–81. doi: 10.1177/0003319709335027. [DOI] [PubMed] [Google Scholar]
- 147.Wiernicki I, Gutowski P, Ciechanowski K, et al. Abdominal aortic aneurysm: association between haptoglobin phenotypes, elastase activity, and neutrophil count in the peripheral blood. Vasc Surg. 2001;35(5):345–50. doi: 10.1177/153857440103500503. discussion 351. [DOI] [PubMed] [Google Scholar]
- 148.Wiernicki I, Safranow K, Baranowska-Bosiacka I, et al. Haptoglobin 2-1 phenotype predicts rapid growth of abdominal aortic aneurysms. J Vasc Surg. 2010 doi: 10.1016/j.jvs.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 149.Norrgard O, Frohlander N, Beckman G, et al. Association between haptoglobin groups and aortic abdominal aneurysms. Hum Hered. 1984;34(3):166–9. doi: 10.1159/000153455. [DOI] [PubMed] [Google Scholar]
- 150.Freestone T, Turner RJ, Coady A, et al. Inflammation and matrix metalloproteinases in the enlarging abdominal aortic aneurysm. Arterioscler Thromb Vasc Biol. 1995;15(8):1145–51. doi: 10.1161/01.atv.15.8.1145. [DOI] [PubMed] [Google Scholar]
- 151.MacSweeney ST, Powell JT, Greenhalgh RM. Pathogenesis of abdominal aortic aneurysm. Br J Surg. 1994;81(7):935–41. doi: 10.1002/bjs.1800810704. [DOI] [PubMed] [Google Scholar]
- 152.Kuivaniemi H, Platsoucas CD, Tilson MD., 3rd Aortic aneurysms: an immune disease with a strong genetic component. Circulation. 2008;117(2):242–52. doi: 10.1161/CIRCULATIONAHA.107.690982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Abdul-Hussien H, Hanemaaijer R, Kleemann R, et al. The pathophysiology of abdominal aortic aneurysm growth: corresponding and discordant inflammatory and proteolytic processes in abdominal aortic and popliteal artery aneurysms. J Vasc Surg. 2010;51(6):1479–87. doi: 10.1016/j.jvs.2010.01.057. [DOI] [PubMed] [Google Scholar]
- 154.Koch AE, Haines GK, Rizzo RJ, et al. Human abdominal aortic aneurysms. Immunophenotypic analysis suggesting an immune-mediated response. Am J Pathol. 1990;137(5):1199–213. [PMC free article] [PubMed] [Google Scholar]
- 155.Rasmussen TE, Hallett JW, Jr, Metzger RL, et al. Genetic risk factors in inflammatory abdominal aortic aneurysms: polymorphic residue 70 in the HLA-DR B1 gene as a key genetic element. J Vasc Surg. 1997;25(2):356–64. doi: 10.1016/s0741-5214(97)70358-6. [DOI] [PubMed] [Google Scholar]
- 156.Rasmussen TE, Hallett JW, Jr, Schulte S, et al. Genetic similarity in inflammatory and degenerative abdominal aortic aneurysms: a study of human leukocyte antigen class II disease risk genes. J Vasc Surg. 2001;34(1):84–9. doi: 10.1067/mva.2001.115603. [DOI] [PubMed] [Google Scholar]
- 157.Rasmussen TE, Hallett JW, Jr, Tazelaar HD, et al. Human leukocyte antigen class II immune response genes, female gender, and cigarette smoking as risk and modulating factors in abdominal aortic aneurysms. J Vasc Surg. 2002;35(5):988–93. doi: 10.1067/mva.2002.121753. [DOI] [PubMed] [Google Scholar]
- 158.Hirose H, Takagi M, Miyagawa N, et al. Genetic risk factor for abdominal aortic aneurysm: HLA-DR2(15), a Japanese study. J Vasc Surg. 1998;27(3):500–3. doi: 10.1016/s0741-5214(98)70324-6. [DOI] [PubMed] [Google Scholar]
- 159.Hirose H, Tilson MD. Negative genetic risk factor for abdominal aortic aneurysm: HLA-DQ3, a Japanese study. J Vasc Surg. 1999;30(5):959–60. doi: 10.1016/s0741-5214(99)70025-x. [DOI] [PubMed] [Google Scholar]
- 160.Monux G, Serrano FJ, Vigil P, et al. Role of HLA-DR in the pathogenesis of abdominal aortic aneurysm. Eur J Vasc Endovasc Surg. 2003;26(2):211–4. doi: 10.1053/ejvs.2002.1897. [DOI] [PubMed] [Google Scholar]
- 161.Sugimoto T, Sada M, Miyamoto T, et al. Genetic analysis on HLA loci in Japanese patients with abdominal aortic aneurysm. Eur J Vasc Endovasc Surg. 2003;26(2):215–8. doi: 10.1053/ejvs.2002.1912. [DOI] [PubMed] [Google Scholar]
- 162.Ogata T, Gregoire L, Goddard KA, et al. Evidence for association between the HLA-DQA locus and abdominal aortic aneurysms in the Belgian population: a case control study. BMC Med Genet. 2006;7:67. doi: 10.1186/1471-2350-7-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Badger SA, Soong CV, O’Donnell ME, et al. The role of human leukocyte antigen genes in the formation of abdominal aortic aneurysms. J Vasc Surg. 2007;45(3):475–80. doi: 10.1016/j.jvs.2006.09.067. [DOI] [PubMed] [Google Scholar]
- 164.Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392(6676):565–8. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- 165.Rottman JB, Ganley KP, Williams K, et al. Cellular localization of the chemokine receptor CCR5. Correlation to cellular targets of HIV-1 infection. Am J Pathol. 1997;151(5):1341–51. [PMC free article] [PubMed] [Google Scholar]
- 166.Schecter AD, Calderon TM, Berman AB, et al. Human vascular smooth muscle cells possess functional CCR5. J Biol Chem. 2000;275(8):5466–71. doi: 10.1074/jbc.275.8.5466. [DOI] [PubMed] [Google Scholar]
- 167.Wilcox JN, Nelken NA, Coughlin SR, et al. Local expression of inflammatory cytokines in human atherosclerotic plaques. J Atheroscler Thromb. 1994;1 (Suppl 1):S10–3. doi: 10.5551/jat1994.1.supplemment1_s10. [DOI] [PubMed] [Google Scholar]
- 168.Szalai C, Duba J, Prohaszka Z, et al. Involvement of polymorphisms in the chemokine system in the susceptibility for coronary artery disease (CAD). Coincidence of elevated Lp(a) and MCP-1 -2518 G/G genotype in CAD patients. Atherosclerosis. 2001;158(1):233–9. doi: 10.1016/s0021-9150(01)00423-3. [DOI] [PubMed] [Google Scholar]
- 169.Smith MW, Dean M, Carrington M, et al. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science. 1997;277(5328):959–65. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- 170.Ghilardi G, Biondi ML, Battaglioli L, et al. Genetic risk factor characterizes abdominal aortic aneurysm from arterial occlusive disease in human beings: CCR5 Delta 32 deletion. J Vasc Surg. 2004;40(5):995–1000. doi: 10.1016/j.jvs.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 171.Sandford B, Bown M, London N, et al. The role of the CCR5 Delta32 polymorphism in abdominal aortic aneurysms. Int J Immunogenet. 2009;36(4):199–205. doi: 10.1111/j.1744-313X.2009.00845.x. [DOI] [PubMed] [Google Scholar]
- 172.Juvonen J, Surcel HM, Satta J, et al. Elevated circulating levels of inflammatory cytokines in patients with abdominal aortic aneurysm. Arterioscler Thromb Vasc Biol. 1997;17(11):2843–7. doi: 10.1161/01.atv.17.11.2843. [DOI] [PubMed] [Google Scholar]
- 173.Shah PK. Inflammation, metalloproteinases, and increased proteolysis: an emerging pathophysiological paradigm in aortic aneurysm. Circulation. 1997;96(7):2115–7. doi: 10.1161/01.cir.96.7.2115. [DOI] [PubMed] [Google Scholar]
- 174.Geng YJ, Wu Q, Muszynski M, et al. Apoptosis of vascular smooth muscle cells induced by in vitro stimulation with interferon-gamma, tumor necrosis factor-alpha, and interleukin-1 beta. Arterioscler Thromb Vasc Biol. 1996;16(1):19–27. doi: 10.1161/01.atv.16.1.19. [DOI] [PubMed] [Google Scholar]
- 175.Keen RR, Nolan KD, Cipollone M, et al. Interleukin-1 beta induces differential gene expression in aortic smooth muscle cells. J Vasc Surg. 1994;20(5):774–84. doi: 10.1016/s0741-5214(94)70165-2. discussion 784–6. [DOI] [PubMed] [Google Scholar]
- 176.Marculescu R, Sodeck G, Domanovits H, et al. Interleukin-1 gene cluster variants and abdominal aortic aneurysms. Thromb Haemost. 2005;94(3):646–50. [PubMed] [Google Scholar]
- 177.Bown MJ, Burton PR, Horsburgh T, et al. The role of cytokine gene polymorphisms in the pathogenesis of abdominal aortic aneurysms: a case-control study. J Vasc Surg. 2003;37(5):999–1005. doi: 10.1067/mva.2003.174. [DOI] [PubMed] [Google Scholar]
- 178.Jones KG, Brull DJ, Brown LC, et al. Interleukin-6 (IL-6) and the prognosis of abdominal aortic aneurysms. Circulation. 2001;103(18):2260–5. doi: 10.1161/01.cir.103.18.2260. [DOI] [PubMed] [Google Scholar]
- 179.Smallwood L, Allcock R, van Bockxmeer F, et al. Polymorphisms of the interleukin-6 gene promoter and abdominal aortic aneurysm. Eur J Vasc Endovasc Surg. 2008;35(1):31–6. doi: 10.1016/j.ejvs.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 180.Davis VA, Persidskaia RN, Baca-Regen LM, et al. Cytokine pattern in aneurysmal and occlusive disease of the aorta. J Surg Res. 2001;101(2):152–6. doi: 10.1006/jsre.2001.6281. [DOI] [PubMed] [Google Scholar]
- 181.Turner DM, Williams DM, Sankaran D, et al. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet. 1997;24(1):1–8. doi: 10.1111/j.1365-2370.1997.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 182.Bown MJ, Lloyd GM, Sandford RM, et al. The interleukin-10-1082 ‘A’ allele and abdominal aortic aneurysms. J Vasc Surg. 2007;46(4):687–93. doi: 10.1016/j.jvs.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 183.Unno N, Nakamura T, Mitsuoka H, et al. Association of a G994 -->T missense mutation in the plasma platelet-activating factor acetylhydrolase gene with risk of abdominal aortic aneurysm in Japanese. Ann Surg. 2002;235(2):297–302. doi: 10.1097/00000658-200202000-00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wilson AG, Symons JA, McDowell TL, et al. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci U S A. 1997;94(7):3195–9. doi: 10.1073/pnas.94.7.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Moran CS, McCann M, Karan M, et al. Association of osteoprotegerin with human abdominal aortic aneurysm progression. Circulation. 2005;111(23):3119–25. doi: 10.1161/CIRCULATIONAHA.104.464727. [DOI] [PubMed] [Google Scholar]
- 186.Moran CS, Cullen B, Campbell JH, et al. Interaction between angiotensin II, osteoprotegerin, and peroxisome proliferator-activated receptor-gamma in abdominal aortic aneurysm. J Vasc Res. 2009;46(3):209–17. doi: 10.1159/000163019. [DOI] [PubMed] [Google Scholar]
- 187.Moran CS, Clancy P, Biros E, et al. Association of PPARgamma allelic variation, osteoprotegerin and abdominal aortic aneurysm. Clin Endocrinol (Oxf) 2010;72(1):128–32. doi: 10.1111/j.1365-2265.2009.03615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Baumann H, Jahreis GP, Morella KK, et al. Transcriptional regulation through cytokine and glucocorticoid response elements of rat acute phase plasma protein genes by C/EBP and JunB. J Biol Chem. 1991;266(30):20390–9. [PubMed] [Google Scholar]
- 189.Kuta AE, Baum LL. C-reactive protein is produced by a small number of normal human peripheral blood lymphocytes. J Exp Med. 1986;164(1):321–6. doi: 10.1084/jem.164.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Domanovits H, Schillinger M, Mullner M, et al. Acute phase reactants in patients with abdominal aortic aneurysm. Atherosclerosis. 2002;163(2):297–302. doi: 10.1016/s0021-9150(02)00006-0. [DOI] [PubMed] [Google Scholar]
- 191.Powell JT, Muller BR, Greenhalgh RM. Acute phase proteins in patients with abdominal aortic aneurysms. J Cardiovasc Surg (Torino) 1987;28(5):528–30. [PubMed] [Google Scholar]
- 192.Carlson CS, Aldred SF, Lee PK, et al. Polymorphisms within the C-reactive protein (CRP) promoter region are associated with plasma CRP levels. Am J Hum Genet. 2005;77(1):64–77. doi: 10.1086/431366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Badger SA, Soong CV, O’Donnell ME, et al. C-reactive protein (CRP) elevation in patients with abdominal aortic aneurysm is independent of the most important CRP genetic polymorphism. J Vasc Surg. 2009;49(1):178–84. doi: 10.1016/j.jvs.2008.07.081. [DOI] [PubMed] [Google Scholar]
- 194.Otterbein LE, Bach FH, Alam J, et al. Carbon monoxide has anti-inflammatory effects involving the mitogen-activated protein kinase pathway. Nat Med. 2000;6(4):422–8. doi: 10.1038/74680. [DOI] [PubMed] [Google Scholar]
- 195.Wang LJ, Lee TS, Lee FY, et al. Expression of heme oxygenase-1 in atherosclerotic lesions. Am J Pathol. 1998;152(3):711–20. [PMC free article] [PubMed] [Google Scholar]
- 196.Yamada N, Yamaya M, Okinaga S, et al. Microsatellite polymorphism in the heme oxygenase-1 gene promoter is associated with susceptibility to emphysema. Am J Hum Genet. 2000;66(1):187–95. doi: 10.1086/302729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Schillinger M, Exner M, Mlekusch W, et al. Heme oxygenase-1 gene promoter polymorphism is associated with abdominal aortic aneurysm. Thromb Res. 2002;106(2):131–6. doi: 10.1016/s0049-3848(02)00100-7. [DOI] [PubMed] [Google Scholar]
- 198.Loeys BL, Schwarze U, Holm T, et al. Aneurysm syndromes caused by mutations in the TGF-beta receptor. N Engl J Med. 2006;355(8):788–98. doi: 10.1056/NEJMoa055695. [DOI] [PubMed] [Google Scholar]
- 199.Thompson AR, Cooper JA, Jones GT, et al. Assessment of the association between genetic polymorphisms in transforming growth factor beta, and its binding protein (LTBP), and the presence, and expansion, of Abdominal Aortic Aneurysm. Atherosclerosis. 2010;209(2):367–73. doi: 10.1016/j.atherosclerosis.2009.09.073. [DOI] [PubMed] [Google Scholar]
- 200.Baas AF, Medic J, van ‘t Slot R, et al. Association of the TGF-beta receptor genes with abdominal aortic aneurysm. Eur J Hum Genet. 2009 doi: 10.1038/ejhg.2009.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Golledge J, Clancy P, Jones GT, et al. Possible association between genetic polymorphisms in transforming growth factor beta receptors, serum transforming growth factor beta1 concentration and abdominal aortic aneurysm. Br J Surg. 2009;96(6):628–32. doi: 10.1002/bjs.6633. [DOI] [PubMed] [Google Scholar]
- 202.van Vlijmen-van Keulen CJ, Vahl AC, Hennekam RC, et al. Genetic linkage of candidate genes in families with abdominal aortic aneurysms? Eur J Vasc Endovasc Surg. 2003;26(2):205–10. doi: 10.1053/ejvs.2002.1874. [DOI] [PubMed] [Google Scholar]
- 203.Baas AF, Medic J, van ‘t Slot R, et al. Association Study of Single Nucleotide Polymorphisms on Chromosome 19q13 With Abdominal Aortic Aneurysm. Angiology. 2010 doi: 10.1177/0003319709354752. [DOI] [PubMed] [Google Scholar]
- 204.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363(2):166–76. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 205.Genome Wide Association Studies: identifying the genes that determine the risk of abdominal aortic aneurysm. Eur J Vasc Endovasc Surg. 2008;36(4):395–6. doi: 10.1016/j.ejvs.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Yoshihara Y, Kawasaki M, Tamada A, et al. Overlapping and differential expression of BIG-2, BIG-1, TAG-1, and F3: four members of an axon-associated cell adhesion molecule subgroup of the immunoglobulin superfamily. J Neurobiol. 1995;28(1):51–69. doi: 10.1002/neu.480280106. [DOI] [PubMed] [Google Scholar]
- 207.Jones GT, van Rij AM. Regarding “Identification of a genetic variant associated with abdominal aortic aneurysms on chromosome 3p12.3 by genome wide association”. J Vasc Surg. 2009;50(5):1246–7. doi: 10.1016/j.jvs.2009.07.098. author reply 1247. [DOI] [PubMed] [Google Scholar]
- 208.Helgadottir A, Thorleifsson G, Magnusson KP, et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40(2):217–24. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 209.Chaikof EL, Brewster DC, Dalman RL, et al. SVS practice guidelines for the care of patients with an abdominal aortic aneurysm: executive summary. J Vasc Surg. 2009;50(4):880–96. doi: 10.1016/j.jvs.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 210.Thompson AR, Golledge J, Cooper JA, et al. Sequence variant on 9p21 is associated with the presence of abdominal aortic aneurysm disease but does not have an impact on aneurysmal expansion. Eur J Hum Genet. 2009;17(3):391–4. doi: 10.1038/ejhg.2008.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Baas AF, Medic J, van ‘t Slot R, et al. The Intracranial Aneurysm Susceptibility Genes HSPG2 and CSPG2 Are Not Associated With Abdominal Aortic Aneurysm. Angiology. 2010 doi: 10.1177/0003319709354751. [DOI] [PubMed] [Google Scholar]
- 212.Atturu G, Brouilette S, Samani NJ, et al. Short leukocyte telomere length is associated with abdominal aortic aneurysm (AAA) Eur J Vasc Endovasc Surg. 2010;39(5):559–64. doi: 10.1016/j.ejvs.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 213.Tromp G, Kuivaniemi H. Developments in genomics to improve understanding, diagnosis and management of aneurysms and peripheral artery disease. Eur J Vasc Endovasc Surg. 2009;38(6):676–82. doi: 10.1016/j.ejvs.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 214.Tromp G, Kuivaniemi H, Hinterseher I, et al. Novel genetic mechanisms for aortic aneurysms. Curr Atheroscler Rep. 2010;12(4):259–66. doi: 10.1007/s11883-010-0111-x. [DOI] [PubMed] [Google Scholar]
- 215.Takagi H, Sugimoto M, Kato T, et al. Postoperative incision hernia in patients with abdominal aortic aneurysm and aortoiliac occlusive disease: a systematic review. Eur J Vasc Endovasc Surg. 2007;33(2):177–81. doi: 10.1016/j.ejvs.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 216.Smallwood L, Warrington N, Allcock R, et al. Matrix metalloproteinase-2 gene variants and abdominal aortic aneurysm. Eur J Vasc Endovasc Surg. 2009;38(2):169–71. doi: 10.1016/j.ejvs.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 217.Rossaak JI, Van Rij AM, Jones GT, et al. Association of the 4G/5G polymorphism in the promoter region of plasminogen activator inhibitor-1 with abdominal aortic aneurysms. J Vasc Surg. 2000;31(5):1026–32. doi: 10.1067/mva.2000.104589. [DOI] [PubMed] [Google Scholar]
- 218.Tromp G, Ogata T, Gregoire L, et al. HLA-DQA is associated with abdominal aortic aneurysms in the Belgian population. Ann N Y Acad Sci. 2006;1085:392–5. doi: 10.1196/annals.1383.045. [DOI] [PubMed] [Google Scholar]