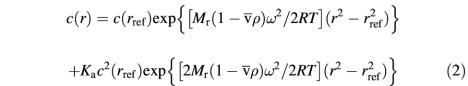Abstract
The cDNA encoding rice methionyl-tRNA synthetase was isolated. The protein exhibited a C-terminal polypeptide appended to a classical MetRS domain. This supplementary domain is related to endothelial monocyte activating polypeptide II (EMAPII), a cytokine produced in mammals after cleavage of p43, a component of the multisynthetase complex. It is also related to Arc1p and Trbp111, two tRNA binding proteins. We expressed rice MetRS and a derivative with a deletion of its EMAPII-like domain. Band-shift analysis showed that this extra-domain provides MetRS with non-specific tRNA binding properties. The EMAPII-like domain contributed a 10-fold decrease in KM for tRNA in the aminoacylation reaction catalyzed by the native enzyme, as compared with the C-terminally truncated MetRS. Consequently, the EMAPII domain provides MetRS with a better catalytic efficiency at the free tRNA concentration prevailing in vivo. This domain binds the acceptor minihelix of tRNAMet and facilitates its aminoacylation. These results suggest that the EMAPII module could be a relic of an ancient tRNA binding domain that was incorporated into primordial synthetases for aminoacylation of RNA minihelices taken as the ancestor of modern tRNA.
Keywords: EMAPII/methionyl-tRNA synthetase/protein–RNA interactions/RNA binding domain
Introduction
Aminoacyl-tRNA synthetases (aaRS) catalyze the aminoacylation of tRNA molecules in a two-step reaction: in the first step, the amino acid is activated in the form of an enzyme-bound aminoacyl adenylate intermediate; the second step involves the transfer of the amino acid to the 3′-end of the tRNA molecule to give the aminoacyl-tRNA product. Protein–RNA interactions play a central role in this essential process.
Aminoacyl-tRNA synthetases bind their cognate tRNA isoacceptors through a set of specific (amino acid side chains and tRNA bases) or non-specific (amino acid side chains and tRNA phosphate–sugar backbone) interactions. The amino acid acceptor helical arm of tRNA is directed to the active site crevice of the synthetase via RNA–protein interactions involving the catalytic domain, but specific contacts between RNA binding modules of the synthetase and other domains of the tRNA molecule also contribute to its proper positioning. Whereas only two structurally distinct catalytic domains have been described, built around a parallel or antiparallel β-sheet, the RNA binding domains show more structural diversity. The anticodon of tRNA interacts with an α-helical cage in GluRS (Nureki et al., 1995), an α-helix bundle in MetRS (Mechulam et al., 1999; Sugiura et al., 2000), ArgRS (Cavarelli et al., 1998), IleRS (Nureki et al., 1998) and LeuRS (Cusack et al., 2000), a β-barrel domain in GlnRS (Rould et al., 1991), AspRS (Cavarelli et al., 1993), LysRS (Cusack et al., 1996) and AsnRS (Berthet-Colominas et al., 1998), and a mixed α–β domain in HisRS (Arnez et al., 1995), GlyRS (Logan et al., 1995), ProRS (Cusack et al., 1998) and ThrRS (Sankaranarayanan et al., 1999). A long coiled-coil domain interacts with the D loop and TΨC loop of tRNA in SerRS (Biou et al., 1994) and PheRS (Goldgur et al., 1997). Basically, this two-domain architecture represents the elementary synthetase throughout evolution. Additional non-specific RNA binding modules appended to aminoacyl-tRNA synthetases have been described (Simos et al., 1996; Whelihan and Schimmel, 1997; Wang and Schimmel, 1999; Cahuzac et al., 2000; Frugier et al., 2000). They also contribute to the aminoacylation activity by enhancing tRNA binding.
A recurrent RNA binding domain associated in cis or in trans with various aminoacyl-tRNA synthetases has been described in the three kingdoms of life. The minimum domain corresponds to a 111 amino acid residue protein, Trbp111, described in Escherichia coli or in the extreme thermophilic bacterium Aquifex aeolicus (Morales et al., 1999). A Trbp-like domain associated in cis to several eubacterial and archaeal MetRSs was shown to be responsible for the dimerization of the enzyme (Cassio and Waller, 1971; Kohda et al., 1987; Mellot et al., 1989). In addition, the dissociation constants of the complexes of MetRS with tRNAfMet or with a microhelix mimicking the acceptor stem of tRNAfMet were found to be ∼10-fold higher with a C-terminally truncated derivative of the E.coli enzyme as compared with wild-type MetRS (Blanquet et al., 1973; Gale et al., 1996). In eubacterial PheRSs, a Trbp-like domain is recovered at the N-terminus of the large β-subunit of the α2β2 tetramer. It forms an oligonucleotide binding (OB)-fold (Goldgur et al., 1997). A homologous domain with an additional 60 amino acid residue C-terminal polypeptide was first identified as the putative cytokine endothelial monocyte activating polypeptide II (EMAPII) (Kao et al., 1994) and was shown to be the C-terminal domain of the p43 component of the mammalian multisynthetase complex (Quevillon et al., 1997). This protein could modulate ArgRS activity (Park et al., 1999). EMAPII-like domains have been recovered associated in cis with Caenorhabditis elegans MetRS or with human (Kleeman et al., 1997) or bovine (Levanets et al., 1997) TyrRS. In TyrRS, this domain was shown to be dispensable for aminoacylation (Wakasugi and Schimmel, 1999). In the yeast Saccharomyces cerevisiae, the EMAPII-like domain of the protein Arc1p is associated in trans with MetRS and GluRS, and increases their aminoacylation efficiency (Simos et al., 1998).
In this work, we isolated the cDNA encoding MetRS from the plant Oryza sativa. We found that the rice enzyme possesses an EMAPII-like C-terminal appended domain. We expressed the full-length and a C-terminally truncated MetRS, and investigated their oligomeric structure and their non-specific tRNA binding properties. Since the EMAPII-like domain of plant MetRS proved to have general tRNA binding capacities, we addressed the problem of the function of this appended domain on the efficiency of aminoacylation catalyzed by plant MetRS. Given the early emergence of related EMAPII-like domains in evolution, we considered the possibility that this RNA binding domain could interact with the amino acid acceptor helical arm of tRNA, which potentially mimics early tRNA substrates, and could direct it to the active site of the enzyme.
Results
Molecular cloning of cytoplasmic methionyl-tRNA synthetase from rice and comparison with other MetRSs
A search in the Protein Information Resource database identified a partial cDNA clone from O.sativa (C2054) encoding an EMAPII-like domain. This rice cDNA clone was obtained from the National Institute of Agrobiological Resources (Japan) and the 1.05 kbp cDNA insert was used to isolate the corresponding full-length cDNA. A cDNA of 2850 nucleotides encoding a bona fide MetRS from the plant O.sativa was isolated by screening a cDNA library with cDNA probes, as previously described (Deniziak et al., 1998).
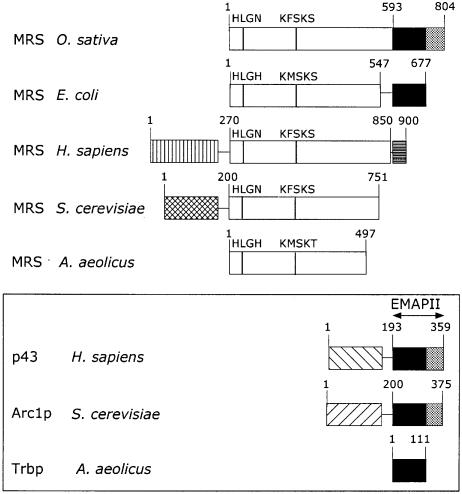
The encoded protein contained 804 amino acid residues, with an apparent molecular mass of 89.7 kDa. A schematic alignment of rice MetRS with MetRSs from other species is shown Figure 1. MetRSs can be divided into five structural groups. Only one member of each group is shown Figure 1. (i) The minimum core enzyme corresponds to the 497-residue protein found in the eubacterium A.aeolicus, but also in some Gram-positive bacteria from the subgroups of actinobacteria (Mycobacterium tuberculosis) or bacillus (Mycoplasma pneumoniae and Mycoplasma genitalium), in mitochondria (S.cerevisiae, Schizosaccharomyces pombe and Arabidopsis thaliana) and in the cytoplasm of some eukaryotes (S.pombe). (ii) Cytoplasmic MetRS from S.cerevisiae possesses a 200-residue N-terminal polypeptide extension that associates with the N-terminal moiety of Arc1p, which provides in trans an EMAPII-like domain. (iii) Human MetRS has a large N-terminal extension that is involved in its association with eight other aminoacyl-tRNA synthetases, which form a multienzyme complex. The human enzyme possesses a short C-terminal appended domain of 50 amino acid residues that contributes an RNA binding domain distinct from EMAPII, also recovered in other eukaryotic synthetases [GlyRS, HisRS, TrpRS, GluRS and ProRS (Berthonneau and Mirande, 2000)]. (iv) In most of the eubacteria [from the β (Neisseria meningitidis), γ (E.coli and Haemophilus influenzae, Vibrio cholerae, Xylella fastidiosa) and ε (Helicobacter pylori and Campylobacter jejuni) subdivisions of proteobacteria, from the bacillus subgroup of Gram-positive bacteria (Bacillus subtilis and Bacillus stearothermophilus), from the Thermus/Deinococcus group (T.thermophilus and D.radiodurans), or from thermotogales (Thermotoga maritima)], a C-terminal Trbp-like domain related to EMAPII, comprising ∼120 amino acid residues, is adjoined to the minimum core MetRS found in Aquifex. MetRSs from the Euryarchaeota kingdom of archaea (Methanobacterium thermoautotrophicum, Methanococcus jannaschii, Archaeoglobus fulgidus or Pyrococcus horikoshii) display a similar structural organization, whereas MetRSs from the kingdom Crenarchaeota (Aeropyrum pernix or Sulfolobus solfataricus) lack this C-terminal domain. (v) Finally, MetRSs from O.sativa as well as from several other eukaryotic origins (A.thaliana or C.elegans) display an EMAPII-like C-terminal appended domain comprising a Trbp-like domain (black boxes in Figure 1) with an additional 60 amino acid residue C-terminal polypeptide (gray boxes in Figure 1). Eubacteria from the spirochete family (Borrelia burgdorferi or Treponema pallidum) also display an EMAPII-like domain.
Fig. 1. The five groups of MetRS and alignment of the EMAPII related RNA binding domains. Only one member of each group is indicated. A more detailed list of MetRSs related to these groups is included in the main text. The open boxes indicate the conserved minimal domain of MetRS, as found in the eubacterium A.aeolicus. MetRS from O.sativa displays an additional C-terminal domain related to the EMAPII moiety of Homo sapiens p43, also recovered in S.cerevisiae Arc1p. The first part of this domain, indicated as a black box, is appended to E.coli MetRS. It corresponds to a dimeric tRNA binding protein (Trbp111) recovered in A.aeolicus. MetRSs from H.sapiens and S.cerevisiae possess N-terminal polypeptide extensions involved in their association with the multisynthetase complex or Arc1p, respectively. The human enzyme has an additional C-terminal polypeptide unrelated to EMAPII or Trbp111 motifs, corresponding to a RNA binding motif found as repeated units in the multifunctional GluProRS. The two conserved functional motifs related to HIGH and KMSKS sequences of MetRSs are indicated. Numbers refer to amino acid residues.
Plant MetRS is a monomer in solution
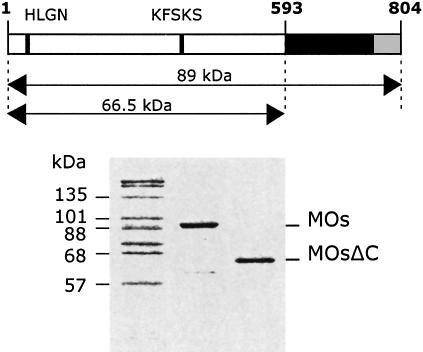
To probe the functional role of the appended C-terminal domain of plant MetRS, the native enzyme (MetRS-Os; 89 kDa) and a derivative with a deletion from residue 594 to 804 (MetRSΔC-Os; 66.5 kDa) were cloned into the pET28b expression plasmid. A stretch of seven proline residues located at positions 4–10 was also removed in both constructs to ensure a better expression in E.coli (the expression was 10-fold lower in the presence of the proline stretch). The two proteins were purified to homogeneity (Figure 2) after fractionation on a Q Sepharose FF column, followed by a second fractionation step on a SOURCE 15S (MetRS-Os) or a SOURCE 15Q column (MetRSΔC-Os).
Fig. 2. Expression of plant MetRS. The full-length MetRS from O.sativa (MOs; 89 kDa) and a derivative with a deletion of the EMAPII-like C-terminal domain (MOsΔC; 66.5 kDa), expressed in E.coli and purified to homogeneity, were analyzed (1 µg of protein) by SDS–PAGE on a 10% polyacrylamide gel and visualized by Coomassie Blue staining. The multisynthetase complex from rabbit (2 µg) is shown on the left as a size marker.
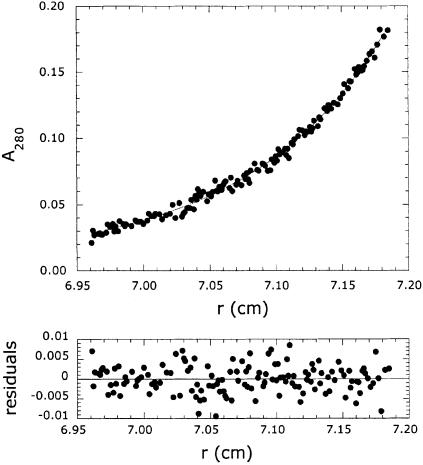
The oligomeric structure of MetRS-Os and MetRSΔC-Os was determined by gel filtration on a Superose 12 HR 10/30 column equilibrated in 200 mM potassium phosphate buffer pH 7.0, 10% glycerol and 1 mM dithiothreitol (DTT), conditions that prevented protein–matrix interaction. MetRS-Os and MetRSΔC-Os were eluted as symmetrical peaks with apparent molecular masses of 110 and 62 kDa, respectively, corresponding to monomers in solution (not shown). To confirm the monomeric structure of plant MetRS, MetRS-Os and MetRSΔC-Os were analyzed by sedimentation equilibrium. As shown in Figure 3, when MetRS-Os was subjected to centrifugation equilibrium at an initial protein concentration of 1 µM, experimental data could be fitted to a monomer–dimer equilibrium with a dissociation constant KD = 2.5 ± 0.8 µM. MetRSΔC-Os behaved exclusively as a monomer (not shown). This analysis clearly showed that plant MetRS is a monomer at physiological protein concentrations, but could form a dimer in vitro at high protein concentration.
Fig. 3. Quaternary structure of plant MetRS. The purified full-length MetRS from O.sativa (MetRS-Os) was analyzed by equilibrium sedimentation at 10 000 r.p.m. in 50 mM potassium phosphate pH 7.5, 10% glycerol, 1 mM DTT at 4°C. Experimental values (closed circles) were fitted (curves) to a monomer–dimer equilibrium with the mass of the monomer equal to 89 kDa and a dissociation constant KD = 2.5 ± 0.8 µM. The residuals are indicated.
The EMAPII-like domain provides rice MetRS with RNA binding properties
Because the EMAPII domain of the p43 component of the multisynthetase complex displays a general tRNA binding ability (Quevillon et al., 1997), we analyzed the ability of full-length and C-terminally truncated plant MetRS to form complexes with various tRNA molecules. Radiolabeled in vitro transcribed tRNAiMet from S.cerevisiae was incubated with increasing amounts of MetRS-Os (30–2000 nM), and free and bound tRNA species were analyzed by a gel-retardation assay (Figure 4). Two band shifts were observed. They most likely corresponded to 1:1 and 2:1 protein:tRNA complexes. The apparent dissociation constant was 80 ± 20 nM, taking into account the monomeric status of MetRS-Os. This binding capacity is much higher than that determined for the isolated human EMAPII domain (∼20 µM; Quevillon et al., 1997), or for the isolated C-terminal domain of MetRS-Os (15 µM; results not shown). As determined above, MetRS-Os formed a dimer at high protein concentration. This observation suggested that the 2:1 complex corresponded to association of dimeric MetRS to one tRNA molecule. However, the possibility that two monomers of MetRS could bind simultaneously to a single tRNA molecule could not be excluded. As shown in Figure 4, the removal of the EMAPII-like domain of MetRS-Os was accompanied by the loss of its tRNA binding capacity. MetRSΔC-Os at 2 µM (also assayed up to 36 µM) was unable to form a stable complex with tRNAiMet. Surprisingly, MetRS-Os also formed stable complexes with non-cognate tRNAs. As shown in Figure 4, MetRS-Os contributed a stable complex with in vitro transcribed yeast tRNAAsp (apparent KD = 120 ± 20 nM), whereas MetRSΔC-Os did not. Essentially identical results were obtained with other non-cognate tRNAs [yeast tRNAPhe or human tRNALys (not shown)], exemplifying the general tRNA binding capacity provided by the EMAPII-like domain of plant MetRS.
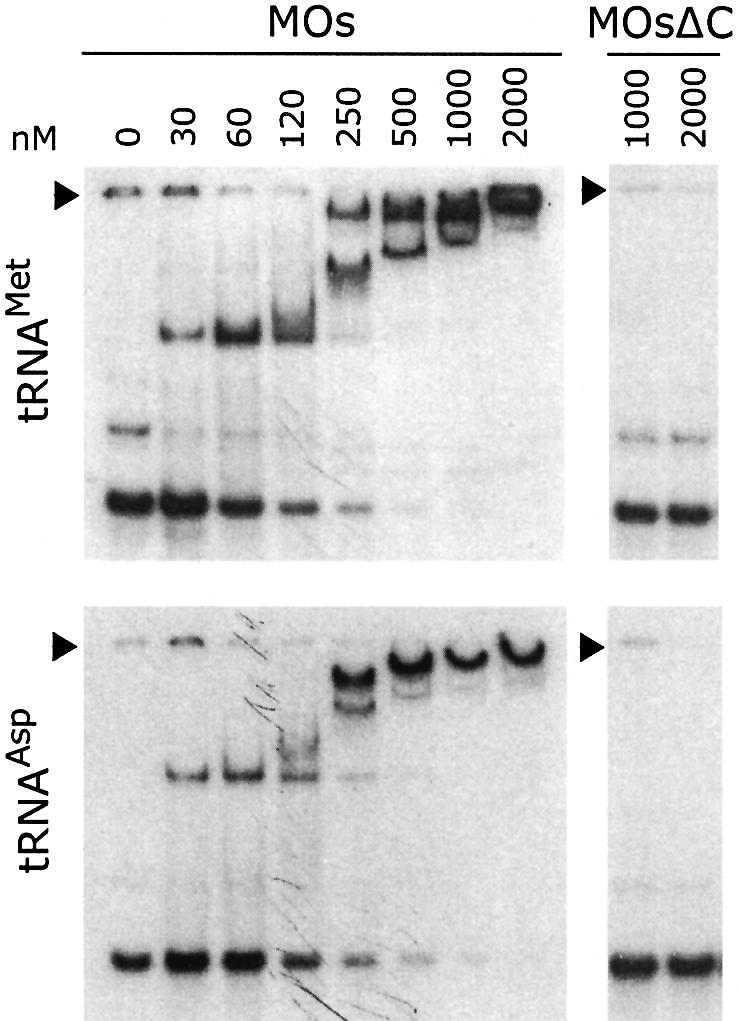
Fig. 4. The EMAPII-like domain confers on plant MetRS a general tRNA binding property. 32P-labeled in vitro transcribed tRNAMet(upper panel) or tRNAAsp (lower panel) was incubated with native (MOs) or C-terminally truncated (MOsΔC) plant MetRS at different concentrations (0–2000 nM). After electrophoresis at 4°C on a 6% native polyacrylamide gel, the mobility shift of tRNA was visualized by autoradiography.
Since tRNAMet and tRNAAsp exhibited similar affinities for MetRS-Os, we examined further the possibility that these two tRNAs could bind MetRS at identical, distinct or partially overlapping sites. To distinguish between these three possibilities, we performed a competition experiment. MetRS-Os was incubated with 32P-labeled tRNAMet at a protein concentration (1 µM) at which all tRNA formed a complex with the protein (Figure 5, lane 0), with the additional presence of increasing amounts of either unlabeled tRNAMet or tRNAAsp. As shown in Figure 5, tRNAAsp also displaced labeled tRNAMet from the synthetase, but less efficiently than did tRNAMet. Thus, although the RNA binding property of MetRS-Os is not specific for the cognate tRNA, tRNAMet and tRNAAsp binding sites are only partially overlapping. This suggested that tRNAAsp interaction with MetRS-Os, which mainly occurs through its appended C-terminal domain, did not form a productive complex for aminoacylation.
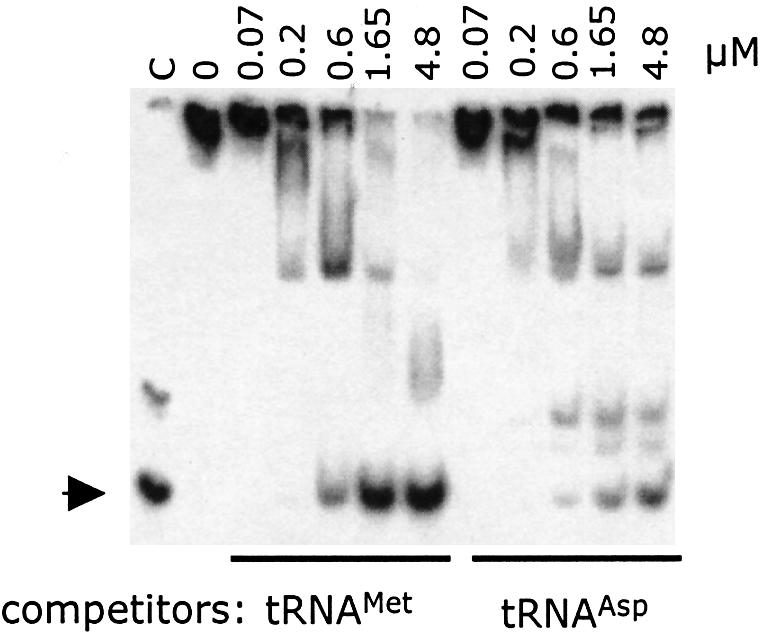
Fig. 5. The tRNAMet and tRNAAsp binding sites are partially overlapping. 32P-labeled tRNAMet was incubated with MetRS-Os (1 µM) to form a tRNA–protein complex in the absence (lane 0) or in the presence of increasing amounts of unlabeled in vitro transcribed tRNAMet or tRNAAsp, at concentrations ranging from 0.07 to 4.8 µM. At equilibrium, free tRNA and tRNA–protein complexes were visualized after electrophoresis at 4°C on a 6% polyacrylamide geland autoradiography. Free tRNA in lane C is indicated by an arrow.
The tRNA binding domain of plant MetRS acts as a cis-activating factor for aminoacylation
Total yeast tRNA (methionine acceptance of 5.9 pmol/A260) was used to assess the specificity of MetRSΔC-Os for tRNAMet aminoacylation as compared with the wild-type enzyme. The same plateau values of aminoacylation were obtained with MetRS-Os and MetRSΔC-Os in the presence of saturating amounts of enzyme, thus showing that, in vitro, both enzymes remained specific for their cognate methionine-acceptor tRNA. MetRS-Os and MetRSΔC-Os displayed indistinguishable specific activities of 340 and 375 U/mg of protein, respectively. The impact of the deletion of the EMAPII-like C-terminal domain on the steady-state kinetic parameters for the aminoacylation reaction was examined with total yeast tRNA (Table I). The catalytic efficiency (kcat/KM) of the deletion mutant MetRSΔC-Os was only 1.9-fold lower as compared with MetRS-Os, resulting from an identical KM for tRNA and a 2-fold decrease in kcat.
Table I. Aminoacylation kinetics of plant MetRS with total yeast tRNA (Y-tRNA) or with homogeneous in vitro transcribed tRNA (tRNAiMet).
| Enzyme | KM (µM) | kcat (s–1) | (kcat/KM) (µM–1 s–1) |
|---|---|---|---|
| MetRS-Os | |||
| Y-tRNA | 0.70 ± 0.09 | 0.53 ± 0.08 | 0.76 |
| tRNAiMet | 0.75 ± 0.07 | 0.29 ± 0.02 | 0.39 |
| MetRSΔC-Os | |||
| Y-tRNA | 0.66 ± 0.07 | 0.26 ± 0.02 | 0.39 |
| tRNAiMet | 6.5 ± 0.8 | 0.36 ± 0.08 | 0.055 |
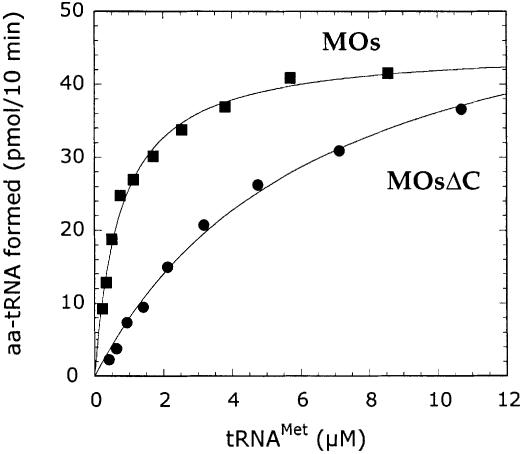
Given the low amount of tRNAMet in the total yeast tRNA fraction used in that study, we considered the possibility that the large excess of non-cognate tRNA added in the assay at tRNAMet saturation could neutralize the appended RNA binding domain of MetRS-Os. To test this hypothesis, we produced homogeneous yeast tRNAiMet by in vitro transcription of the cloned tRNA gene with T7 RNA polymerase. Using pure tRNAiMet, no change in catalytic efficiency was observed for the wild-type enzyme. In contrast, MetRSΔC-Os showed a dramatic 10-fold decrease in KM, resulting in a lower catalytic efficiency (Table I; Figure 6). However, MetRS-Os and MetRSΔC-Os displayed similar kcat values. Therefore, at the concentrations of free, non-aminoacylated tRNA prevailing within the cell (∼0.1 µM), the tRNA binding ability of the EMAPII-like domain significantly improves the catalytic potency of plant MetRS.
Fig. 6. The removal of the EMAPII-like domain affects KM but not kcat in the aminoacylation reaction. The tRNA saturation kinetics in the tRNAMet aminoacylation reaction was determined with pure in vitro transcribed tRNAMet (methionine acceptance of 1150 pmol/A260 unit) in the presence of 2.5 nM MetRS-Os (MOs) or MetRSΔC-Os (MOsΔC). Experimental values (closed symbols) were fitted to the Michaelis–Menten equation (curves).
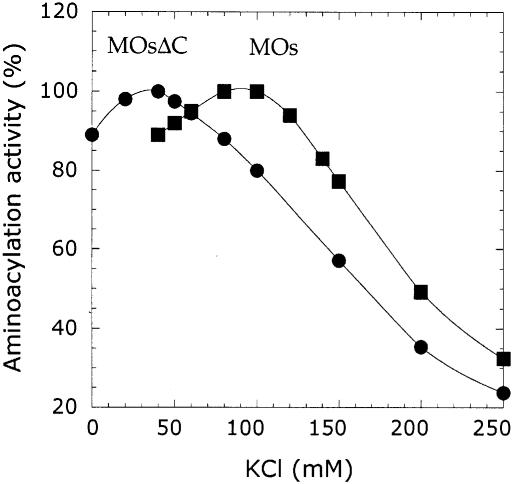
Since the appended domain of MetRS-Os contributed an additional non-specific binding site for tRNA, we surmised that this supplementary interaction could involve electrostatic interactions between positively charged side chains from the appended domain and the phosphate–sugar backbone of tRNA. Since ionic interactions are salt sensitive, we reasoned that there may be differences in the relative aminoacylation activities of MetRS-Os and MetRSΔC-Os at various concentrations of salt. To address this issue, we compared the aminoacylation activities catalyzed by MetRS-Os and MetRSΔC-Os in the presence of increasing concentrations of KCl (Figure 7). We found that after removal of the EMAPII-like domain, the optimum KCl concentration for aminoacylation of either total yeast tRNA or homogeneous tRNAiMet was lowered from 90 mM for MetRS-Os to 35 mM for MetRSΔC-Os. In contrast, MetRS-Os and MetRSΔC-Os displayed identical KCl dependency in the ATP–PPi exchange reaction. Thus, the appended RNA binding domain of MetRS-Os is likely to interact with tRNA during the aminoacylation step.
Fig. 7. Salt dependency of the aminoacylation reaction. Initial velocity in the in vitro transcribed tRNAMet aminoacylation reaction was determined for MetRS-Os (MOs) and MetRSΔC-Os (MOsΔC) as a function of KCl concentration in the assay. The results are plotted as percentage of maximum activity obtained at 35 and 90 mM KCl for MetRSΔC-Os and MetRS-Os, respectively.
The EMAPII-like domain of MetRS-Os interacts with the acceptor stem of tRNA and contributes to its aminoacylation
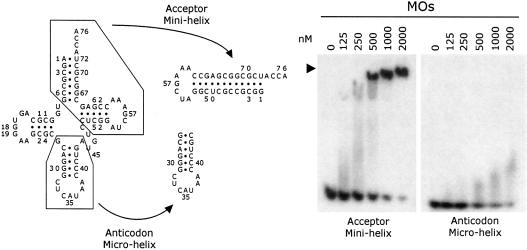
The two primary interaction loci for synthetase–tRNA complexes are the acceptor stem and the anticodon stem–loop structures of tRNA. To probe the interaction of tRNAMet with the appended domain of MetRS-Os, the two major domains of tRNAiMet were synthesized in vitro. Radiolabeled amino acid acceptor minihelix and anticodon microhelix were used in a gel-mobility shift assay to examine their association with full-length plant MetRS (Figure 8). The acceptor minihelix formed a stable complex with MetRS-Os with an apparent KD of ∼500 nM. Essentially identical results were obtained with a minihelix mimicking the acceptor stem of yeast tRNAAsp (not shown). This dissociation constant of minihelixMet is only 6-fold higher than that observed with the complete tRNAiMet. Although the anticodon microhelix also interacted with MetRS-Os, as judged from the disappearance of the band corresponding to the free RNA species (Figure 8), in accordance with the general RNA binding properties of the EMAPII-like domain of MetRS-Os, no stable complex could be recovered, suggesting that dissociation occurred during electrophoresis (the labeled RNA distributed as a smear visible after overexposure of the autoradiography film). We concluded that the appended RNA binding domain of MetRS-Os provides a binding site for the amino acid acceptor stem minihelix of tRNA.
Fig. 8. The amino acid acceptor arm of tRNAMet binds to plant MetRS. Left: sequence and cloverleaf structure of S.cerevisiae initiator tRNAMet and sequence and hairpin structures of acceptor minihelix and anticodon microhelix. Right: band shift assay performed with 32P-labeled minihelix and microhelix in the presence of 125–2000 nM MetRS-Os (MOs), as described in the legend of Figure 4.
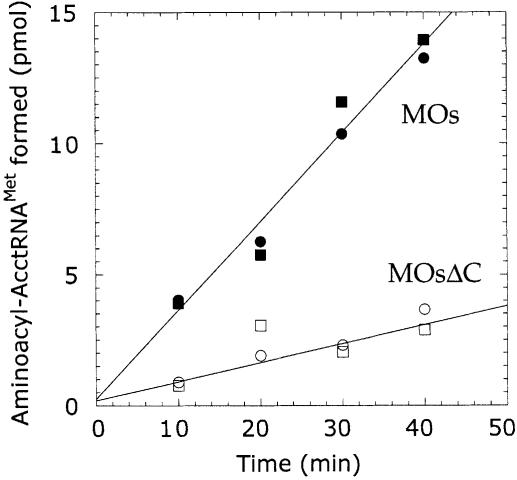
MetRS from E.coli (Martinis and Schimmel, 1992) or S.cerevisiae (Senger et al., 1995) can aminoacylate RNA substrates mimicking the acceptor minihelix of tRNA. Given the robust interaction of MetRS-Os with the amino acid acceptor minihelix of tRNAMet via association with the EMAPII-like domain, we investigated the possibility that minihelix aminoacylation could be strengthened in MetRS-Os as compared with MetRSΔC-Os. As expected, the rate of tRNAiMet-minihelix aminoacylation by plant MetRS was increased by the presence of the RNA binding domain (Figure 9). The linear time course of methionine incorporation into minihelix of tRNAiMet was significantly higher (4- to 5-fold) with MetRS-Os as compared with MetRSΔC-Os. This stimulation of minihelix aminoacylation when the appended RNA binding domain is present is consistent with the involvement of this domain in binding the acceptor stem of tRNA and presenting the 3′-terminal adenosine in a conformation suited for aminoacylation. We also tested the possibility that binding of anticodon microhelix of tRNAiMet on MetRS-Os or MetRSΔC-Os could stimulate charging of acceptor minihelix through interdomain communication. As observed for MetRS from E.coli (Alexander et al., 1998) or S.cerevisiae (Senger et al., 1995), neither enhancement nor inhibition of minihelix aminoacylation occurred upon its addition (Figure 9).
Fig. 9. Time course of aminoacylation of the amino acid acceptor minihelix of tRNAiMet by plant MetRS. Aminoacylation of the amino acid acceptor minihelix of tRNAMet was conducted at 25°C, with 50 µM RNA and 8 µM MetRS-Os (MOs, closed symbols) or MetRSΔC-Os (MOsΔC, open symbols). In one set of experiments, the anticodon microhelix of tRNAiMet was also added at 8 µM in the reaction mixture (squares).
Discussion
Methionyl-tRNA synthetase from O.sativa displays a C-terminal polypeptide extension related to EMAPII. Human EMAPII is a pro-inflammatory cytokine that stimulates chemotactic migration of polymorphonuclear leukocytes and mononuclear phagocytes, and induces tissue factor activity on endothelial cells (Kao et al., 1992, 1994). The EMAPII cytokine is produced after cleavage of a pro-cytokine precursor identified as the p43 component of the mammalian multisynthetase complex (Quevillon et al., 1997). Caspase 7 was shown to be able to process in vitro translated proEMAPII (Behrensdorf et al., 2000). However, EMAPII-like domains are likely to be primarily involved in protein biosynthesis. We reported that EMAPII is a monomer and has a general RNA binding property. Similarly, we showed here that the EMAPII-like domain of MetRS-Os displays a weak non-specific RNA binding capacity (KD ∼15 µM). No stable tRNA–protein complex could be observed with a C-terminally truncated derivative of MetRS-Os. In contrast, the native monomeric enzyme bound tRNA with an apparent KD of 80 nM. Therefore, when the core catalytic domain MetRSΔC-Os and the EMAPII-like domain are fused, the synergy of several weak contributions provides MetRS-Os with substantial tRNA binding capacity. Likewise, the EMAPII-like domain of Arc1p, the yeast homolog of p43, required additional N-terminal sequences for efficient tRNA binding (Simos et al., 1998).
As shown in Figure 1, MetRSs across evolution display three types of C-terminally appended RNA binding domains: a repeated unit similar to that isolated in GluProRS, a Trbp111-like domain or an EMAPII-like domain. Whereas the repeated units display no sequence similarity with EMAPII and constitute a distinct family of RNA binding motifs with a helix–loop–helix conformation (Cahuzac et al., 2000), Trbp111 is a subdomain related to EMAPII. The EMAPII-like RNA binding domain of MetRS-Os can be divided into two parts: an N-terminal moiety of ∼110 amino acid residues related to Trbp111 (30–40% identities) to which is adjoined a C-terminal polypeptide of ∼60 amino acid residues. The findings that EMAPII and MetRSs carrying an EMAPII-like domain are monomers, and that Trbp111 and proteins containing a C-terminal Trbp-like domain without the C-terminal extension found in EMAPII (i.e. E.coli MetRS) are dimers suggest that the dimeric interface of Trbp111 is masked by the extra C-terminal 60 amino acid residues found in EMAPII.
In contrast to EMAPII, Trbp111 is a dimer and has a strong affinity for tRNA (KD ∼30 nM). It binds one tRNA per dimer. The weak tRNA binding displayed by monomeric EMAPII-like domains as compared with dimeric Trbp111 could be due to their different oligomeric structures, Trbp111 contributing potentially two RNA binding sites. However, the mode of tRNA binding by Trbp111, EMAPII and related domains is not known. It should be argued that from a functional point of view, the modes of tRNA binding are essentially different between Trbp111 and proteins carrying EMAPII-like domains (i.e. MetRS-Os) or degenerated Trbp-like domains (i.e. E.coli MetRS). Whereas Trbp111 binds tRNA but does not bind to an aminoacyl-tRNA synthetase, EMAPII-like domains are invariably associated with aminoacyl-tRNA synthetase domains, either in cis (MetRS-Os) or in trans via protein–protein interactions (MetRS and GluRS from S.cerevisiae with Arc1p; synthetases from the multisynthetase complex with p43). Morales et al. (1999) suggested that Trbp111 could stabilize the L-shaped structure of tRNA and act as a tRNA-specific chaperone. Although it is conceivable that Trbp111 binding to tRNA might not inhibit the tRNA aminoacylation reaction, a strong and quasi-irreversible binding of tRNA to an aminoacyl-tRNA synthetase via a non-specific RNA binding domain would greatly impair dissociation of the aminoacylated tRNA. Therefore, the relatively weak RNA binding property of EMAPII-like domains could be an intrinsic feature associated with their amino acid composition, related to the requirement for efficient tRNA turnover on the synthetase.
In this work, we established that the EMAPII-like C-terminal appended domain of rice MetRS (MetRS-Os) contributes a non-specific tRNA binding domain. The removal of this domain (MetRSΔC-Os) is accompanied by a 10-fold decrease in the catalytic efficiency for aminoacylation of tRNAMet. Cognate and non-cognate tRNAs were found to bind to MetRS with similar affinities, but only tRNAMet was aminoacylated. Therefore, the general RNA binding capacity of the EMAPII-like domain did not interfere with the tRNA aminoacylation selectivity of MetRS-Os. Interestingly, the increased catalytic efficiency could not be observed in vitro when total tRNA was used in the aminoacylation assay. However, when homogeneous tRNAMet was used, MetRS with a deletion of its C-terminal domain showed a 10-fold increase in KM and a similar kcat compared with wild type. This suggests that under conditions of saturating tRNA concentrations, the large excess of non-cognate tRNA could bind and neutralize the appended domain of MetRS. In mammalian cells, the in vivo concentration of individual tRNAs has been estimated to be in the 1 µM range, corresponding to a total tRNA concentration of ∼10–20 µM (Smith, 1975). The active species of tRNA for ribosomal protein synthesis is in the form of the ternary complex EF-1α–GTP–aminoacyl-tRNA. The concentration of elongation factor EF-1α has been calculated to be ∼30 µM (Browning et al., 1990), in excess as compared with total tRNA, and the in vivo aminoacylation level of tRNAMet from exponentially growing cells has been shown to be >95% (Lazard et al., 1987). Accordingly, the in vivo concentration of free non-aminoacylated individual tRNAs is likely to be low, <0.1 µM, a value much lower than the KM for tRNAMet determined for MetRS-Os. Therefore, in the conditions of a non-saturating concentration of free tRNA prevailing within the cell, the improvement in catalytic efficiency provided by the C-terminal appended domain of MetRS-Os is likely to contribute to an efficient capture of tRNA by the synthetase. This could be a means to ensure effective tRNA channeling in eukaryotic cells (Negrutskii and Deutscher, 1991). Similarly, the EMAPII-like domain of yeast Arc1p has been shown to be involved in effective recruitment of tRNAs for MetRS and GluRS (Simos et al., 1998), and was able to confer on E.coli GlnRS the ability to complement a disrupted allele of its yeast homolog (Wang and Schimmel, 1999).
The EMAPII-like domain of plant MetRS is able to produce a stable complex with the amino acid acceptor minihelix of tRNA. Similarly, the EMAPII-like domain of Arc1p, a trans-acting cofactor of yeast MetRS and GluRS, has been shown to bind preferentially the acceptor minihelix of tRNA (Simos et al., 1996). In that connection, according to the crystal structure of PheRS from T.thermophilus complexed with tRNAPhe, the Trbp-like domain of bacterial PheRS adopts an OB-fold topology characteristic of various RNA binding proteins, and is located adjacent to the acceptor stem of tRNA (Goldgur et al., 1997). Moreover, in the crystal structure of E.coli or T.thermophilus MetRS, obtained for the monomeric enzymes with a deletion of their C-terminal Trbp-like domain (Mechulam et al., 1999; Sugiura et al., 2000), the C-terminus folds back to the catalytic domain close to the entrance of the active site crevice. Therefore, in the native enzymes, the EMAPII/Trbp-like domains of MetRSs are likely to be located near the acceptor stem of tRNA.
Interestingly, EMAPII/Trbp-like proteins are widespread among all living organisms, from eubacteria, archaea or eukarya. This suggests that this recurrent RNA binding domain is a primitive protein that arose early in evolution. Basically, aminoacyl-tRNA synthetases are built around a common architecture: a catalytic domain that interacts with the acceptor stem of tRNA; an anticodon binding domain. Numerous aminoacyl-tRNA synthetases are able to aminoacylate minihelices derived from the acceptor-TΨC stem–loop of their corresponding tRNAs (Beuning and Musier-Forsyth, 1999). Assuming that the very primordial synthetases were solely made of the catalytic domain and catalyzed aminoacylation of acceptor minihelices of tRNA (Schimmel and de Pouplana, 1995), it is tempting to speculate that the interaction between the EMAPII-like domain of plant MetRS and the acceptor stem of tRNAiMet recapitulates a second stage primitive enzyme with an appended general RNA binding domain that might have improved the proper positioning of the acceptor minihelix in the active site. In this connection, a 368 amino acid residue N-terminal fragment of E.coli AlaRS that is fully competent for alanyl adenylate formation could regain its ability to aminoacylate RNA substrates following addition of the EMAPII-like non-specific RNA binding domain of Arc1p (Chihade and Schimmel, 1999). However, the finding that EMAPII/Trbp domains are generally appended at the C-terminus of the enzyme, separated from the catalytic domain by the anticodon binding domain, seems to be at odds with this prediction. This structural arrangement would rather suggest that the anticodon binding domains were appended to the catalytic domains prior to the EMAPII/Trbp-like domains. However, the possibility that anticodon binding domains were late insertions between the catalytic and RNA binding domains of primordial synthetases cannot be dismissed. The EMAPII/Trbp domains of the resulting fusion proteins would gain more flexibility to bind accessible parts of tRNAs. According to the ensuing topology, the general tRNA binding properties of EMAPII/Trbp domains could stabilize tRNA–protein interactions either through the binding of the amino acid acceptor (probably in the case of MetRS) or anticodon domain of tRNA.
Materials and methods
cDNA cloning
Plasmid containing the rice cDNA clone C2054 was obtained from Dr Yoshiaki Nagamura (Rice Genome Research Program, Japan). The cDNA insert was radiolabeled by random oligonucleotide priming and used to screen a λgt11 cDNA library made from total poly(A)+ mRNA from O.sativa (Clontech), as previously described (Deniziak et al., 1998). The nucleotide sequence has been deposited in the DDBJ/EMBL/GenBank databank under accession No. AF040700.
Protein overexpression and purification
The NcoI–XhoI cDNA fragment encoding full-length rice MetRS, starting with the ATG translation initiation codon and containing the TAG stop codon, was inserted into the NcoI–XhoI sites of the bacterial expression vector pET-28b (Novagen). Deletion of the seven proline residues located at positions 4–10 was performed by replacing the original 5′-terminal NcoI–NcoI fragment by the mutated fragment obtained by PCR using oligonucleotides MRS07 (5′-CCCCCATGGCGTCGCCGAAGCTGCCGGTCCCC-3′) and MRS08 (5′-ATGGACAGCATGATACTTGT-3′) to give pET/MOs. To obtain a C-terminal truncated variant of MetRS (MOsΔC) we replaced the wild-type 832 bp HindIII–XhoI fragment bearing the 3′-coding region of cDNA by a 185 bp PCR product amplified between oligonucleotides MRS05 (5′-CCCCTCGAGCTACTCAGCTTGGCTGCCTG-3′) and MRS06 (5′-CATGCCTTCATTTTCCAATG-3′), which introduced a TAG stop codon after the Glu residue at position 593, followed by an XhoI restriction site. All constructs were verified by DNA sequencing.
The proteins encoded by the recombinant plasmids were expressed in the E.coli BL21(DE3) host strain grown in LB medium supplemented with kanamycin. Cultures (15 l) were grown at 37°C to an A600 of 0.25, transferred at 28°C, and expression was induced at A600 = 0.5 by addition of 1 mM isopropyl-β-d-thiogalactopyranoside for 12 h. Cells were washed with ice-cold extraction buffer (5 mM potassium phosphate pH 7.0, 2 mM DTT, 10% glycerol) supplemented with protease inhibitors (1 mM di-isopropyl fluorophosphate, 1 mM phenylmethylsulfonyl fluoride, and 2 µg/ml leupeptin, chymostatin and pepstatin) and homogenized with an Eaton press. All purification steps were conducted at 4°C. The clear lysate obtained after centrifugation at 25 000 g for 30 min was loaded onto a Q Sepharose Fast Flow column (5 × 23.5 cm) equilibrated in extraction buffer and developed with a 20 column vol. gradient of potassium phosphate from 5 to 275 mM. MetRS-Os and MetRSΔC-Os were eluted at phosphate concentrations of 80 and 165 mM, respectively.
Fractions containing MetRS-Os or MetRSΔC-Os were applied to a 2.0 × 9.5 cm column of SOURCE 15S or 15Q (Amersham Pharmacia Biotech.) equilibrated in extraction buffer. Loading on the column was accomplished by mixing 1 vol. of the protein solution with 2 vols (MetRS-Os) or 4 vols (MetRSΔC-Os) of equilibration buffer. After washing with the same buffer, MetRSs were eluted by a 50 column vol. gradient of potassium phosphate from 5 to 275 mM. Fractions containing MetRS-Os (eluted at 80 mM) or MetRSΔC-Os (110 mM) were pooled, concentrated by vacuum dialysis, dialyzed against 25 mM potassium phosphate pH 7.0, 2 mM DTT, 55% glycerol, and stored at –20°C. Protein concentrations were determined by using calculated absorption coefficients of 1.345 and 1.666 A280 units/mg/cm2 for MetRS-Os and MetRSΔC-Os, respectively.
Sedimentation equilibrium
Ultracentrifugation experiments were conducted as described previously (Agou et al., 1996) in a Beckman Optima XL-A analytical ultracentrifuge, using an An 60 Ti rotor and a double-sector cell of 12 mm path length. Equilibrium was verified from the superimposition of duplicate scans recorded at 4 h intervals.
The experimental sedimentation equilibrium data were fitted to a model for a single homogeneous species following the equation:
where c(r) is the protein concentration at radial position r, c(rref) is the concentration of the protein at an arbitrary reference radial distance rref, Mr is the molecular weight (89 000 and 66 500 for MetRS-Os and MetRSΔC-Os, respectively), ![]() the partial specific volume (0.729 and 0.7288 at 4°C for MetRS-Os and MetRSΔC-Os, respectively) of the solute, ρ is the density of the solvent, ω is the angular velocity of the rotor, and R and T are the molar gas constant and the absolute temperature, respectively.
the partial specific volume (0.729 and 0.7288 at 4°C for MetRS-Os and MetRSΔC-Os, respectively) of the solute, ρ is the density of the solvent, ω is the angular velocity of the rotor, and R and T are the molar gas constant and the absolute temperature, respectively.
Where experimental data could not be fitted to a model for a single species, a monomer–dimer equilibrium was considered, according to equation (2):
where Ka is the association constant of the dimer.
Gel-retardation assay
Protein–tRNA interactions were assayed using a band shift assay as described previously (Quevillon et al., 1997). Plasmids carrying the tRNA genes were linearized and subjected to in vitro transcrip tion. T7 RNA polymerase was purified from strain BL21/pAR1219, generously provided by Professor W.Studier (Brookhaven National Laboratory). 32P-labeled tRNAs were synthesized in a reaction mixture (50 µl) containing 2 µg of template DNA, 40 mM Tris–HCl pH 7.9, 6 mM MgCl2, 2 mM spermidine, 10 mM DTT, 0.01% Triton X-100, 1 mM each CTP, UTP and GTP, 10 µM [α-32P]ATP (200 Ci/mmol), 4000 U/ml T7 RNA polymerase. After incubation at 37°C for 4 h, the transcripts were purified by electrophoresis on a denaturing 12% polyacrylamide gel (mono:bis, 19:1), recovered from the gel by electroelution in a Bio-Trap apparatus (Schleicher & Schuell), precipitated with ethanol and resuspended in 20 mM Tris–HCl pH 7.5, 10 mM MgCl2.
Homogeneous MetRS-Os or MetRSΔC-Os was incubated at increasing concentrations with radiolabeled tRNA (100 000 c.p.m. Cerenkov per point) in an 11 µl volume containing 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 10 mM MgCl2, 10 mM 2-mercaptoethanol, 10% glycerol and bovine serum albumin (BSA) at 0.1 mg/ml. After incubation at 25°C for 20 min, the mixture was placed on ice and loaded onto a 6% polyacrylamide gel (mono:bis, 29:1) containing 5% glycerol in 0.5× TBE at 4°C. After electrophoresis, the gel was fixed, dried and subjected to autoradiography. Free and bound tRNA were quantified by densitometry analysis.
Aminoacylation assay
Initial rates of tRNA aminoacylation were measured at 25°C in 0.1 ml of 20 mM imidazole–HCl buffer pH 7.5, 100 mM KCl, 0.5 mM DTT, 7 mM MgCl2, 2 mM ATP, 52 µM 14C-labeled methionine (NEN; 58 Ci/mol) and saturating amounts of tRNA, as described previously (Mirande et al., 1983). Total brewer’s yeast tRNA (Roche; methionine acceptance of 5.9 pmol/A260) or homogeneous yeast tRNAiMet obtained by in vitro transcription (methionine acceptance of 1150 pmol/A260) was used as tRNA substrate. The incubation mixture contained catalytic amounts (1–10 nM) of enzymes appropriately diluted in 10 mM Tris–HCl pH 7.5, 10 mM 2-mercaptoethanol, containing BSA at 4 mg/ml. One unit of activity is the amount of enzyme producing 1 nmol of methionine-tRNAMet/min at 25°C. For the determination of KM values for tRNA, tRNAMet concentrations of 0.05–10 µM were used. Michaelian parameters were obtained by non-linear regression of the theoretical Michaelis–Menten equation to the experimental curve using KaleidaGraph 3.0.8 software (Abelbeck Software).
The time course of aminoacylation of tRNAiMet minihelix was conducted at 25°C as described above in the presence of 50 µM RNA substrate and 2–10 µM enzyme. At different intervals, aliquots were withdrawn and quenched on 3MM Whatman papers pre-soaked with ice-cold 5% trichloroacetic acid (TCA) and 1 mM [12C]methionine. After a 1 h incubation on ice, filters were washed five times for 10 min in ice-cold 5% TCA containing 1 mM methionine, once in 95% ethanol, dried and processed for liquid scintillation counting.
For large-scale synthesis of RNA substrates, in vitro transcription was conducted on 1 mg of linearized template DNA, in a final volume of 10 ml containing 40 mM Tris–HCl pH 7.9, 22 mM MgCl2, 1 mM spermidine, 5 mM DTT, 0.01% Triton X-100, 4 mM each CTP, UTP, GTP and ATP, 16 mM GMP, 1000 U/ml T7 RNA polymerase. After a 1 h incubation at 37°C, 200 U of inorganic pyrophosphatase (BioLabs) were added, and synthesis was resumed for a 4 h period. After phenol–chloroform extraction, transcripts were purified on a 1.6 mm thick 12% polyacrylamide–urea gel (15% for tRNA minihelix or microhelix), electroeluted, ethanol precipitated, resuspended in 20 mM Tris–HCl pH 7.5, 10 mM MgCl2, and renatured by heating for 15 min at 60°C and cooling for 20 min at room temperature.
Acknowledgments
Acknowledgements
We thank Y.Nagamura for the gift of the rice cDNA clone C2054 and W.Studier for the BL21/pAR1219 strain. The excellent technical assistance of Françoise Triniolles and the involvement of Pierre-Yves Lozach in some parts of this work are gratefully acknowledged. This work was supported by grants from the ‘Programme Physique et Chimie du Vivant’ from CNRS, the ‘Association pour la Recherche sur le Cancer’ and ‘La Ligue’.
References
- Agou F., Waller,J.P. and Mirande,M. (1996) Expression of rat aspartyl-tRNA synthetase in Saccharomyces cerevisiae—role of the NH2-terminal polypeptide extension on enzyme activity and stability. J. Biol. Chem., 271, 29295–29303. [DOI] [PubMed] [Google Scholar]
- Alexander R.W., Nordin,B.E. and Schimmel,P. (1998) Activation of microhelix charging by localized helix destabilization. Proc. Natl Acad. Sci. USA, 95, 12214–12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnez J.G., Harris,D.C., Mitschler,A., Rees,B., Francklyn,C.S. and Moras,D. (1995) Crystal structure of histidyl-tRNA synthetase from Escherichia coli complexed with histidyl-adenylate. EMBO J., 14, 4143–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrensdorf H.A., van de Craen,M., Knies,U.E., Vandenabeele,P. and Clauss,M. (2000) The endothelial monocyte-activating polypeptide II (EMAP II) is a substrate for caspase-7. FEBS Lett., 466, 143–147. [DOI] [PubMed] [Google Scholar]
- Berthet-Colominas C., Seignovert,L., Härtlein,M., Grotli,M., Cusack,S. and Leberman,R. (1998) The crystal structure of asparaginyl-tRNA synthetase from Thermus thermophilus and its complexes with ATP and asparaginyl-adenylate: the mechanism of discrimination between asparagine and aspartic acid. EMBO J., 17, 2947–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthonneau E. and Mirande,M. (2000) A gene fusion event in the evolution of aminoacyl-tRNA synthetases. FEBS Lett., 470, 300–304. [DOI] [PubMed] [Google Scholar]
- Beuning P.J. and Musier-Forsyth,K. (1999) Transfer RNA recognition by aminoacyl-tRNA synthetases. Biopolymers, 52, 1–28. [DOI] [PubMed] [Google Scholar]
- Biou V., Yaremchuk,A., Tukalo,M. and Cusack,S. (1994) The 2.9 Å crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNASer. Science, 263, 1404–1410. [DOI] [PubMed] [Google Scholar]
- Blanquet S., Iwatsubo,M. and Waller,J.P. (1973) The mechanism of action of methionyl-tRNA synthetase from Escherichia coli. 1. Fluorescence studies on tRNAMet binding as a function of ligands, ions and pH. Eur. J. Biochem., 36, 213–226. [DOI] [PubMed] [Google Scholar]
- Browning K.S., Humphreys,J., Hobbs,W., Smith,G.B. and Ravel,J.M. (1990) Determination of the amounts of the protein synthesis initiation and elongation factors in wheat germ. J. Biol. Chem., 265, 17967–17973. [PubMed] [Google Scholar]
- Cahuzac B., Berthonneau,E., Birlirakis,N., Guittet,E. and Mirande,M. (2000) A recurrent RNA-binding domain is appended to eukaryotic aminoacyl-tRNA synthetases. EMBO J., 19, 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassio D. and Waller,J.P. (1971) Modification of E.coli methionyl-tRNA synthetase by proteolytic cleavage and properties of the trypsin modified enzyme. Eur. J. Biochem., 20, 283–300. [DOI] [PubMed] [Google Scholar]
- Cavarelli J., Rees,B., Ruff,M., Thierry,J.C. and Moras,D. (1993) Yeast tRNAAsp recognition by its cognate class-II aminoacyl-tRNA synthetase. Nature, 362, 181–184. [DOI] [PubMed] [Google Scholar]
- Cavarelli J., Delagoutte,B., Eriani,G., Gangloff,J. and Moras,D. (1998) l-arginine recognition by yeast arginyl-tRNA synthetase. EMBO J., 17, 5438–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chihade J.W. and Schimmel,P. (1999) Assembly of a catalytic unit for RNA microhelix aminoacylation using nonspecific RNA binding domains. Proc. Natl Acad. Sci. USA, 96, 12316–12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack S., Yaremchuk,A. and Tukalo,M. (1996) The crystal structures of T.thermophilus lysyl-tRNA synthetase complexed with E.coli tRNALys and a T.thermophilus tRNALys transcript: anticodon recognition and conformational changes upon binding of a lysyl-adenylate analogue. EMBO J., 15, 6321–6334. [PMC free article] [PubMed] [Google Scholar]
- Cusack S., Yaremchuk,A., Krikliviy,I. and Tukalo,M. (1998) tRNAPro anticodon recognition by Thermus thermophilus prolyl-tRNA synthetase. Structure, 6, 101–108. [DOI] [PubMed] [Google Scholar]
- Cusack S., Yaremchuk,A. and Tukalo,M. (2000) The 2 Å crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J., 19, 2351–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deniziak M., Mirande,M. and Barciszewski,J. (1998) Cloning and sequencing of cDNA encoding the rice methionyl-tRNA synthetase. Acta Biochim. Pol., 45, 669–676. [PubMed] [Google Scholar]
- Frugier M., Moulinier,L. and Giegé,R. (2000) A domain in the N-terminal extension of class IIb eukaryotic aminoacyl-tRNA synthetases is important for tRNA binding. EMBO J., 19, 2371–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale A.J., Shi,J.P. and Schimmel,P. (1996) Evidence that specificity of microhelix charging by a class I tRNA synthetase occurs in the transition state of catalysis. Biochemistry, 35, 608–615. [DOI] [PubMed] [Google Scholar]
- Goldgur Y., Mosyak,L., Reshetnikova,L., Ankilova,V., Lavrik,O., Khodyreva,S. and Safro,M. (1997) The crystal structure of phenylalanyl-tRNA synthetase from Thermus thermophilus complexed with cognate tRNAPhe. Structure, 5, 59–68. [DOI] [PubMed] [Google Scholar]
- Kao J. et al. (1992) Endothelial monocyte-activating polypeptide II. A novel tumor-derived polypeptide that activates host-response mechanisms. J. Biol. Chem., 267, 20239–20247. [PubMed] [Google Scholar]
- Kao J. et al. (1994) Characterization of a novel tumor-derived cytokine. Endothelial monocyte activating polypeptide II. J. Biol. Chem., 269, 25106–25119. [PubMed] [Google Scholar]
- Kleeman T.A., Wei,D.B., Simpson,K.L. and First,E.A. (1997) Human tyrosyl-tRNA synthetase shares amino acid sequence homology with a putative cytokine. J. Biol. Chem., 272, 14420–14425. [DOI] [PubMed] [Google Scholar]
- Kohda D., Yokoyama,S. and Miyazawa,T. (1987) Functions of isolated domains of methionyl-tRNA synthetase from an extreme thermophile, Thermus thermophilus HB8. J. Biol. Chem., 262, 558–563. [PubMed] [Google Scholar]
- Lazard M., Mirande,M. and Waller,J.P. (1987) Expression of the aminoacyl-tRNA synthetase complex in cultured Chinese hamster ovary cells. Specific derepression of the methionyl-tRNA synthetase component upon methionine restriction. J. Biol. Chem., 262, 3982–3987. [PubMed] [Google Scholar]
- Levanets O.V., Naidenov,V.G., Odynets,K.A., Woodmaska,M.I., Matsuka,G. and Kornelyuk,A.I. (1997) Homology of C-terminal non-catalytic domain of mammalian tyrosyl-tRNA synthetase with cytokine EMAPII and non-catalytic domains of methionyl-and phenylalanyl-tRNA synthetases. Biopolymers and Cell (Kiev), 13, 474–478. [Google Scholar]
- Logan D.T., Mazauric,M.H., Kern,D. and Moras,D. (1995) Crystal structure of glycyl-tRNA synthetase from Thermus thermophilus. EMBO J., 14, 4156–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinis S.A. and Schimmel,P. (1992) Enzymatic aminoacylation of sequence-specific RNA minihelices and hybrid duplexes with methionine. Proc. Natl Acad. Sci. USA, 89, 65–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechulam Y., Schmitt,E., Maveyraud,L., Zelwer,C., Nureki,O., Yokoyama,S., Konno,M. and Blanquet,S. (1999) Crystal structure of Escherichia coli methionyl-tRNA synthetase highlights species-specific features. J. Mol. Biol., 294, 1287–1297. [DOI] [PubMed] [Google Scholar]
- Mellot P., Mechulam,Y., Le Corre,D., Blanquet,S. and Fayat,G. (1989) Identification of an amino acid region supporting specific methionyl-tRNA synthetase:tRNA recognition. J. Mol. Biol., 208, 429–443. [DOI] [PubMed] [Google Scholar]
- Mirande M., Cirakoglu,B. and Waller,J.P. (1983) Seven mammalian aminoacyl-tRNA synthetases associated within the same complex are functionally independent. Eur. J. Biochem., 131, 163–170. [DOI] [PubMed] [Google Scholar]
- Morales A.J., Swairjo,M.A. and Schimmel,P. (1999) Structure-specific tRNA-binding protein from the extreme thermophile Aquifex aeolicus. EMBO J., 18, 3475–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrutskii B.S. and Deutscher,M.P. (1991) Channeling of aminoacyl-tRNA for protein synthesis in vivo. Proc. Natl Acad. Sci. USA, 88, 4991–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nureki O., Vassylyev,D.G., Katayanagi,K., Shimizu,T., Sekine,S., Kigawa,T., Miyazawa,T., Yokoyama,S. and Morikawa,K. (1995) Architectures of class-defining and specific domains of glutamyl-tRNA synthetase. Science, 267, 1958–1965. [DOI] [PubMed] [Google Scholar]
- Nureki O. et al. (1998) Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science, 280, 578–582. [DOI] [PubMed] [Google Scholar]
- Park S.G., Jung,K.H., Lee,J.S., Jo,Y.J., Motegi,H., Kim,S. and Shiba,K. (1999) Precursor of pro-apoptotic cytokine modulates aminoacylation activity of tRNA synthetase. J. Biol. Chem., 274, 16673–16676. [DOI] [PubMed] [Google Scholar]
- Quevillon S., Agou,F., Robinson,J.C. and Mirande,M. (1997) The p43 component of the mammalian multi-synthetase complex is likely to be the precursor of the endothelial monocyte-activating polypeptide II cytokine. J. Biol. Chem., 272, 32573–32579. [DOI] [PubMed] [Google Scholar]
- Rould M.A., Perona,J.J. and Steitz,T.A. (1991) Structural basis of anticodon loop recognition by glutaminyl-tRNA synthetase. Nature, 352, 213–218. [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan R., Dock-Bregeon,A.C., Romby,P., Caillet,J., Springer,M., Rees,B., Ehresmann,C., Ehresmann,B. and Moras,D. (1999) The structure of threonyl-tRNA synthetase-tRNAThr complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell, 97, 371–381. [DOI] [PubMed] [Google Scholar]
- Schimmel P. and de Pouplana,L.R. (1995) Transfer RNA: from minihelix to genetic code. Cell, 81, 983–986. [DOI] [PubMed] [Google Scholar]
- Senger B., Aphasizhev,R., Walter,P. and Fasiolo,F. (1995) The presence of a D-stem but not a T-stem is essential for triggering aminoacylation upon anticondon binding in yeast methionine tRNA. J. Mol. Biol., 249, 45–58. [DOI] [PubMed] [Google Scholar]
- Simos G., Segref,A., Fasiolo,F., Hellmuth,K., Shevchenko,A., Mann,M. and Hurt,E.C. (1996) The yeast protein Arc1p binds to tRNA and functions as a cofactor for the methionyl- and glutamyl-tRNA synthetases. EMBO J., 15, 5437–5448. [PMC free article] [PubMed] [Google Scholar]
- Simos G., Sauer,A., Fasiolo,F. and Hurt,E.C. (1998) A conserved domain within Arc1p delivers tRNA to aminoacyl-tRNA synthetases. Mol. Cell, 1, 235–242. [DOI] [PubMed] [Google Scholar]
- Smith D.W. (1975) Reticulocyte transfer RNA and hemoglobin synthesis. Science, 190, 529–535. [DOI] [PubMed] [Google Scholar]
- Sugiura I. et al. (2000) The 2.0 Å crystal structure of Thermus thermophilus methionyl-tRNA synthetase reveals two RNA-binding modules. Structure Fold Des., 8, 197–208. [DOI] [PubMed] [Google Scholar]
- Wakasugi K. and Schimmel,P. (1999) Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science, 284, 147–151. [DOI] [PubMed] [Google Scholar]
- Wang C.C. and Schimmel,P. (1999) Species barrier to RNA recognition overcome with nonspecific RNA binding domains. J. Biol. Chem., 274, 16508–16512. [DOI] [PubMed] [Google Scholar]
- Whelihan E.F. and Schimmel,P. (1997) Rescuing an essential enzyme RNA complex with a non-essential appended domain. EMBO J., 16, 2968–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]