Abstract
The cytosolic sulfotransferase hSULT2A1 is the major hydroxysteroid (alcohol) sulfotransferase in human liver, and it catalyzes the 3′-phosphoadenosine-5′-phosphosulfate (PAPS)-dependent sulfation of various endogenous hydroxysteroids as well as many xenobiotics that contain alcohol and phenol functional groups. The hSULT2A1 often displays substrate inhibition, and we have hypothesized that a key element in this response to increasing substrate concentration is the formation of non-productive ternary dead-end enzyme complexes involving the nucleotide product, adenosine 3′,5′-diphosphate (PAP). One of these substrates for hSULT2A1 is dehydroepiandrosterone (DHEA), a major circulating steroid hormone in humans that serves as precursor to both androgens and estrogens. We have utilized DHEA in both initial velocity studies and equilibrium binding experiments in order to evaluate the potential role of ternary complexes in substrate inhibition of the enzyme. Our results indicate that hSULT2A1 forms non-productive ternary complexes that involve either DHEA or dehydroepiandrosterone sulfate, and the formation of these ternary complexes displays negative cooperativity in the binding of DHEA.
Keywords: sulfotransferase, hydroxysteroid, hSULT2A1, substrate inhibition, negative cooperativity, non-productive enzyme complexes
Introduction
The mammalian cytosolic sulfotransferases (SULTs) comprise a group of enzymes that have important physiological functions in the metabolism of endogenous regulatory molecules as well as many xenobiotics [1-6]. For example, the major hydroxysteroid (alcohol) sulfotransferase in human liver, now designated as hSULT2A1 in the current nomenclature for the superfamily of cytosolic sulfotransferases [7], functions in the metabolism of several physiological steroids and also in the detoxication or metabolic activation of many hydroxyl-containing drugs, environmental chemicals, and other xenobiotics. As seen with all SULTs, the enzyme catalyzes the transfer of a sulfuryl group from 3′-phosphoadenosine-5′-phosphosulfate (PAPS) to an acceptor molecule to form a sulfuric acid ester (or sulfate ester) and adenosine-3′,5′-diphosphate (PAP). As is true of most SULTs, the catalytically active enzyme is isolated from tissues as a homodimer [8].
Among the physiologically important reactions catalyzed by hSULT2A1 is the sulfation of dehydroepiandrosterone (DHEA) to form dehydroepiandrosterone sulfate (DHEAS) [8]. DHEA and DHEAS serve as precursors to both androgens and estrogens, and they are among the most abundant circulating steroid hormones in humans. Circulating levels of DHEA and DHEAS are regulated by sulfation of DHEA and desulfation of DHEAS, alteration of the rates of adrenal secretion or hepatic release into the blood, and changes in renal clearance [9]. Moreover, DHEAS can serve as a transport form of DHEA, with the hydrolysis of DHEAS in peripheral tissues catalyzed by sulfatases [10]. In addition, the sulfation of pregnenolone, catalyzed by hSULT2A1 in the adrenal gland of the fetus, is important in the fetoplacental synthesis of estrogens that is required in pregnancy [11]. Although two additional members of SULT family 2, hSULT2B1 and hSULT2B2, also catalyze the sulfation of pregnenolone, the catalytic efficiency (kcat/Km) has been reported to be 3-8 fold higher with hSULT2A1 [12]. Recent evidence also points to a potentially important role of hSULT2A1 in steroid homeostasis through sulfation of androsterone, an abundant circulating 5α-reduced androgen in human serum and a major androgen metabolite in urine [13].
In addition to metabolic functions in the metabolism of these steroid hormones, sulfation reactions catalyzed by hSULT2A1 also have a key role in bile acid metabolism [14-16]. Sulfation of bile acids catalyzed by hSULT2A1 and their excretion as sulfate conjugates provide significant contributions to the detoxication of these endogenous molecules. Indeed, decreased expression of hSULT2A1 in human liver has been associated with the development of chronic liver disease related to the accumulation of hepatic bile acids [17, 18].
hSULT2A1 has also received substantial attention due to its role in the metabolism of drugs, carcinogens, and other xenobiotics. In general, the sulfation of xenobiotics catalyzed by hSULT2A1 is often associated with detoxication of these molecules. However, sulfation of certain benzylic alcohols, allylic alcohols, and N-hydroxyarylamines can lead to the formation of electrophilic sulfuric acid ester products capable of reacting with the nucleophilic sites on DNA, RNA, and protein [19-23].
Studies on the catalytic mechanism of cytosolic sulfotransferases have been primarily focused on the SULT1 family (previously known as either as phenol or aryl sulfotransferases). Kinetic and crystallographic studies on these enzymes have elucidated several general principles regarding the chemical mechanism of sulfuryl transfer from PAPS to an acceptor substrate [1, 24-31], that, based on three-dimensional structural homologies, may be applicable to members of other SULT families. Details of the kinetic mechanisms for individual SULTs, however, continue to provide information on the substrate-dependent differences in sulfation reactions that define the specificities and functions of these enzymes.
One of these substrate-dependent characteristics is the extent to which a SULT-catalyzed reaction exhibits substrate inhibition. One basis for substrate inhibition in reactions catalyzed by SULTs is the formation of non-productive enzyme-product-substrate complexes that inhibit the reaction. Such effects have been seen with SULT1A isoforms [29, 32, 33] and SULT1E isoforms [31]. Another potential mechanism for substrate inhibition, the binding of multiple substrate molecules to the active site as in the case of 4-nitrophenol binding to hSULT1A1, has been observed crystallographically and studied kinetically [25]. Other studies with substrates such as estradiol indicate that, for larger molecules, the formation of enzyme-product-substrate complexes with hSULT1A1 is more important [33]. Crystal structures of hSULT2A1 have provided information on the binding of DHEA and PAP to this enzyme [13, 34-36]. While there is the potential for two different binding modes for DHEA [35], there is no crystallographic evidence for multiple DHEA molecules simultaneously bound at a single active site. However, it has been proposed that alternate orientations for binding of steroid substrates to hSULT2A1 may play a role in substrate inhibition seen with this isoform [37].
Thus, we hypothesized that, similar to previously studied SULT1A enzymes, the formation of non-productive ternary complexes is a key element in the substrate inhibition exhibited by hSULT2A1. Moreover, substrate- or product-induced alterations in binding of PAPS, sulfuryl acceptor, product sulfuric acid ester, or PAP may contribute to these effects. When these characteristics are considered along with substrate inhibition and limitations in the solubility of DHEA and other sulfuryl acceptors for this enzyme, standard analyses of initial rates of reaction at varied concentrations of substrates and products have important limitations in determination of inhibitory complexes. Therefore, we have investigated both the kinetic characteristics of sulfation of DHEA catalyzed by hSULT2A1 as well as the binding of substrates and products to the enzyme and to relevant enzyme-substrate and enzyme-product complexes.
Materials and Methods
Materials
DHEA, PAPS, PAP, potassium phosphate, 2-mercaptoethanol, and methylene blue were purchased from Sigma Chemical Co. (St. Louis, MO). PAPS was further purified using a previously described procedure [38] to a purity of at least 99% by HPLC. Chloroform and anhydrous sodium sulfate were from Fisher Scientific (Pittsburgh, PA). Econo-Safe biodegradable scintillation cocktail was obtained from RPI (Mt. Prospect, IL). DE52 was purchased from Whatman (Fairfield, IL). Hydroxyapatite (Bio-Gel HT) was from Bio-Rad Laboratories (Hercules, CA). 3H-DHEA (94.5 Ci/mmol), and 3H-DHEAS (63 Ci/mmol) were from Perkin Elmer Life and Analytical Sciences (Boston, MA).
Purification of recombinant hSULT2A1
Recombinant hSULT2A1 was expressed in Escherichia coli BL21 (DE3) cells [39]. The extraction and purification of hSULT2A1 from these cells were carried out as previously described [40] Briefly, following isolation of the cell extract, three chromatographic steps (i.e., DE52 anion exchange followed by two hydroxyapatite columns) were used to purify recombinant hSULT2A1. At each purification step, hSULT2A1-containing fractions were identified and quantitated with a previously described methylene blue assay using dehydroepiandrosterone as substrate [41, 42]. Protein content was determined by the modified Lowry procedure with bovine serum albumin as standard [43]. SDS-PAGE with Coomassie blue staining was used to monitor the purification progress and verify the homogeneity of the protein after the final step. The subunit relative molecular mass of the homogeneous protein was found to be 34 kDa, and the kinetic behavior of the protein with DHEA as substrate was completely consistent with the previously reported data for the native enzyme isolated from human liver [8].
Determination of DHEA solubility
In order to assess the limit of solubility of DHEA under the experimental conditions, a previously described light scattering method [44] was employed using a Perkin-Elmer LS55 Luminescence Spectrometer. Both the excitation and the emission wavelengths were set at 400 nm, with entrance and exit slit widths of 12 nm and 2.5 nm, respectively. As the concentration of DHEA was increased, light scattering was first observed at concentrations greater than 80 μM under the conditions utilized for kinetic and binding studies. Therefore, 80 μM was the highest concentration of DHEA utilized for subsequent experiments.
Kinetics of DHEA sulfation catalyzed by hSULT2A1
DHEA and DHEAS differentially partition between chloroform and water, and these differences allowed us to develop a rapid and quantitative radiometric assay for the DHEAS formed in reactions catalyzed by hSULT2A1. DHEA solutions were prepared containing the appropriate specific radioactivity of 3H-DHEA in order to achieve the sensitivity needed to determine the initial velocity of each enzyme-catalyzed reaction. Each reaction mixture (final volume of 200 μL) contained 0.25 M potassium phosphate buffer (pH 7.0), appropriate concentrations of DHEA and PAPS, and 7.5 mM 2-mercaptoethanol. This solution was preincubated for 2 min at 37 °C, and reaction was then initiated by the addition of 25 ng of hSULT2A1 (i.e., a final hSULT2A1 concentration of 1.8 nM based on the dimeric Mr of the protein). After incubation for 10 min at 37 °C, 0.8 mL of 50 mM potassium hydroxide and 0.5 mL of chloroform were added to stop the reaction. Preliminary experiments indicated that the addition of 0.8 mL of 50 mM potassium hydroxide increased the pH to approximately 11.3, and it is known that hSULT2A1 has no catalytic activity at this pH [45]. Following termination of the reaction, each reaction tube was subjected to vortex mixing for 10 sec followed by centrifugation for 5 min at 3500 rpm. A 100 μL aliquot from the resulting aqueous phase was mixed with 10 mL of liquid scintillation cocktail and the radioactivity determined with a Perkin Elmer Tri-Carb 2900TR liquid scintillation analyzer. The mean ± standard deviation of six replicates was determined for each assay. The appropriate use of a 10 min assay to determine initial velocities was verified by examining the time-dependence of the formation of DHEAS at one of the lower concentrations of DHEA utilized (e.g., 0.5 μM). Under these conditions, product formation was linear with respect to incubation time for over 10 min (Fig. S1, Supplementary Material).
Control experiments conducted under the conditions of the enzyme assays revealed that the recovery of DHEAS in the aqueous phase following extraction with chloroform was 85 ± 2% (n=3 at each concentration) over a concentration range of 0.025-5.0 μM. This extraction efficiency was used in calculation of the rates of sulfation of DHEA in hSULT2A1-catalyzed reactions. The partition of DHEA from the aqueous phase into chloroform was also examined. The concentration of DHEA remaining in the aqueous phase following the addition of DHEA to all assay components except PAPS and subsequent extraction with chloroform was determined for each concentration of DHEA utilized in reaction mixtures, and the amount of radioactivity remaining in the aqueous layer was corrected for the residual 3H-DHEA present. In all cases, the amount of DHEA remaining in the aqueous phase was less than 2% of the original concentration of DHEA in the assay mixture.
Ligand-binding studies
The intrinsic fluorescence of tryptophan residues in hSULT2A1 was used to monitor the conformational change of the protein in interaction with its substrates and products. This has been shown to be a sensitive method for determination of the binding of ligands to proteins [46, 47]. These binding studies were conducted in 0.25 M potassium phosphate buffer (pH 7.0) and 7.5 mM 2-mercaptoethanol using a Perkin Elmer LS55 luminescence spectrometer, with an excitation wavelength of 290 nm, emission wavelength of 347 nm, and both entrance and exit slit widths of 10 nm. Consistent with published spectral data on purine nucleotides [48], the absorption of PAPS and PAP at 290 nm at the highest concentrations utilized in these experiments was observed to be less than 1.1% of that determined at 259 nm. No absorption due to PAPS or PAP was observed at 347 nm. As a result, no corrections for potential inner filter effects were necessary. All solutions were filtered with a Millex-GS 0.22 μm filter prior to use. Aliquots of ligands were added to 3.0 mL of the above buffer containing 5.1 μg hSULT2A1 (i.e., a 25 nM concentration of hSULT2A1 based on the Mr of the dimeric protein). This solution was allowed to equilibrate for 15 min before the titrations with ligands. Aliquots of a ligand (0.5 – 3.0 μL volumes) were added successively, with fluorescence measurements taken after each addition. The solutions in the fluorescence cells were stirred with a magnetic stirrer during these titrations. Fluorescence titrations were performed at 37 ± 2 °C and the fluorimeter shutters were closed unless spectra were being recorded to minimize UV exposure time. Control experiments in the absence of ligand indicated that no significant change in the fluorescence of the protein occurred under these conditions. The data obtained were corrected for the small dilution occurring when ligands were added. The absolute value of the decrease in the fluorescence (ΔF) was plotted against the concentration of ligand (L), and these data were fit to the following equations: hyperbolic single site binding equation (Equation 1), the Hill equation (Equation 2), and the Adair equation (Equation 3) [49], and a two-site saturable binding equation (Equation 4).
| Eq (1) |
| Eq (2) |
| Eq (3) |
| Eq (4) |
Binding parameters were calculated on the basis of the best fit to these equations. For non-cooperative ligand binding, Kd values were obtained by fitting the results to the hyperbolic single site binding equation. For cooperative cases, the Hill and the Adair equations were used [50, 51]. In the Hill equation, “n” (the Hill coefficient) was determined to indicate the type of cooperativity. When cooperativity was indicated, the Adair equation (i.e., Eq 3) was utilized to determine the dissociation constants for ligand-binding to the enzyme. In Eq 3, x̄ is the total number of moles of ligand bound per total number of moles of enzyme, and this is calculated by multiplying the fractional saturation at each concentration by the number of possible binding sites (i.e., two for all ligands of SULT2A1). β1 and β2 represent the Adair association constants, with β1 equal to the first association constant, Ka1, of a ligand, and β2 equal to the product of the first and second association constants (Ka1 × Ka2). Following a fit of the data to Eq 3, the dissociation constants, Kd1 and Kd2 for each ligand were determined as the reciprocals of the corresponding association constants.
In the first set of experiments, binding properties of DHEA, DHEAS, PAPS, and PAP were analyzed in individual titrations. In the second set of experiments, depending on the results obtained with the individual ligand titrations, appropriate concentrations of ligands were utilized for pretreatment of the enzyme in order to analyze the effect of one ligand on the binding of a second one. Thus, 80 μM DHEA, 80 μM DHEAS, and 100 μM PAP were used to examine the effects of DHEA, DHEAS, and PAP, respectively. Each set of experiments was performed in triplicate, and the mean ± standard deviation was calculated.
Results
Kinetic studies on hSULT2A1-catalyzed sulfation of DHEA
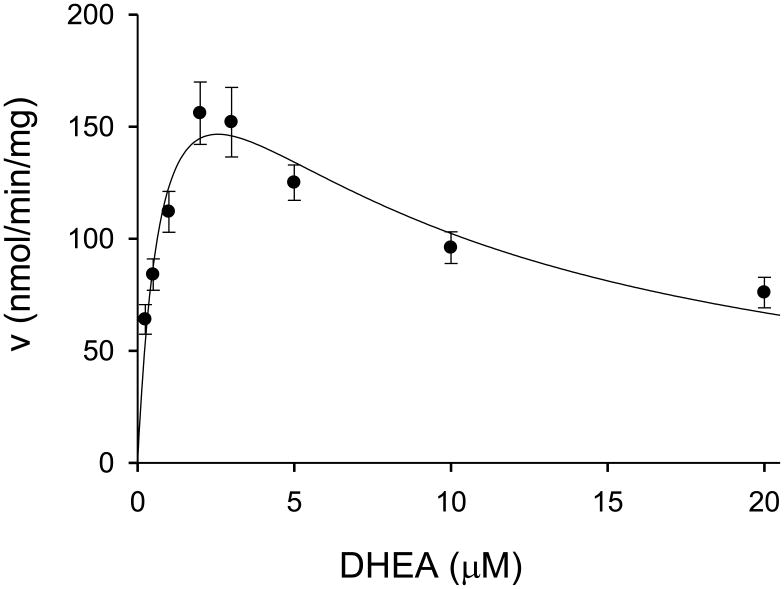
DHEA sulfation was initially examined using a relatively broad concentration range for DHEA (0.25-20 μM) and a single PAPS concentration (200 μM). As seen in Fig. 1, there was an increase in the rate of sulfation at concentrations of DHEA up to approximately 2.0 μM, with a subsequent decrease in the reaction velocity at higher DHEA concentrations. These data for hSULT2A1-catalyzed sulfation of DHEA were fit to the following substrate inhibition equation:
Fig. 1.
Initial velocities of hSULT2A1-catalyzed DHEA sulfation at 0.25-20 μM DHEA and 200 μM PAPS. Data are the means ± standard deviation of 6 replicates.
| Eq (5) |
In this equation, v is the initial velocity of the reaction, Vmax is the maximal velocity, Km is the Michaelis constant, [S] is the substrate (DHEA) concentration, and Kis is the substrate inhibition constant. The values for Vmax, Km, and Kis were calculated as 245 ± 40 nmol/min/mg, 0.8 ± 0.3 μM, and 8 ± 3 μM, respectively. It is noteworthy that the kinetic data in Fig. 1 utilizing homogenous recombinant hSULT2A1 are comparable to the results obtained by Falany and co-workers [8] for DHEA sulfation catalyzed by hSULT2A1 that had been purified directly from human liver homogenates.
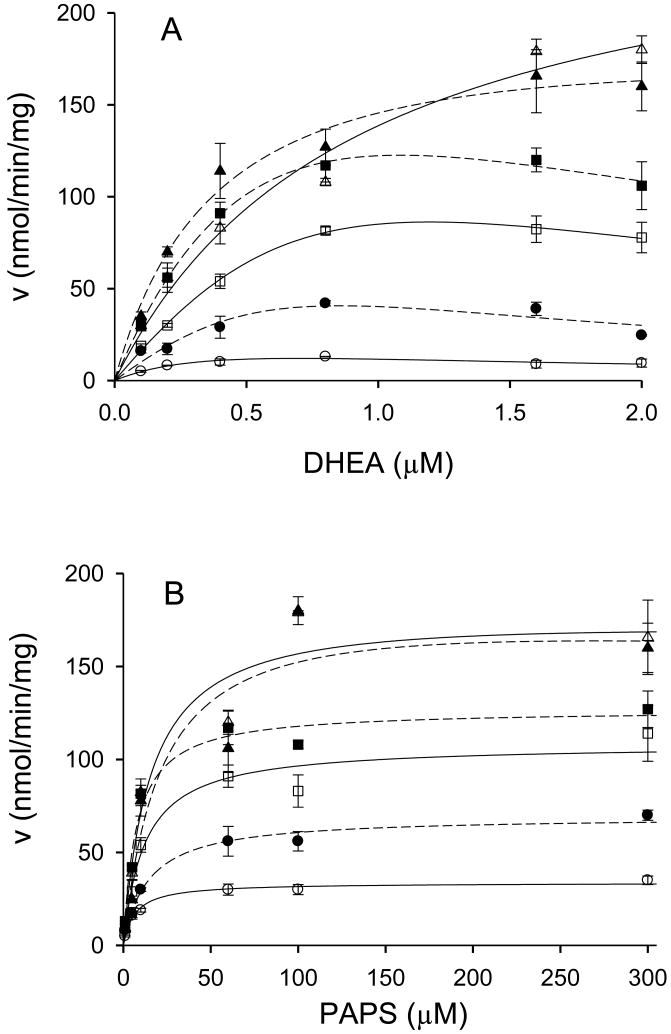
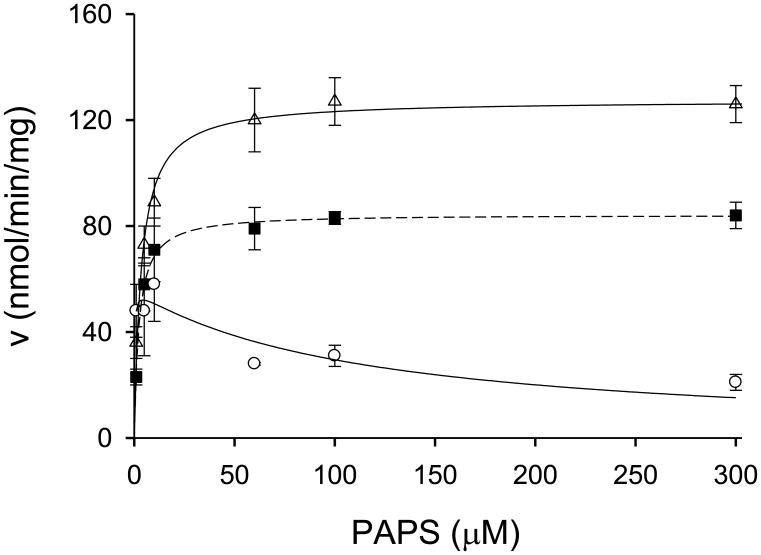
Following these preliminary studies, we investigated initial velocities of DHEA sulfation catalyzed by hSULT2A1 at varied concentrations of DHEA and PAPS. Thus, we analyzed the kinetics of DHEA sulfation at low concentrations of DHEA, where minimal substrate inhibition occurs, and at higher concentrations of DHEA, where significantly more substrate inhibition occurs. In the first set of experiments, the PAPS concentrations utilized were 1.0, 5.0, 10, 60, 100, and 300 μM, and the DHEA concentrations were 0.1, 0.2, 0.4, 0.8, 1.6, and 2.0 μM. As seen in Fig. 2A, substrate inhibition was observed for DHEA at intermediate concentrations of PAPS (i.e., 5.0 – 60 μM), however at the lowest (1.0 μM) and highest (100 and 300 μM) concentrations of PAPS, substrate inhibition was not evident over this concentration range of DHEA. When the concentration of PAPS was varied at relatively low concentrations of DHEA (0.1 μM - 2.0 μM), there was no indication that PAPS displayed substrate inhibition (Fig. 2B). Moreover, as seen in Fig. 3, PAPS also did not exhibit substrate inhibition at DHEA concentrations of 5.0 μM or 20 μM, although an overall reduction in maximal velocity was seen at 20 μM DHEA. Such a decrease in Vmax was expected due to the substrate inhibition by DHEA as seen in Fig. 1. However, at 80 μM DHEA, PAPS displayed significant substrate inhibition. Thus, these studies indicated a relatively complex kinetic pattern wherein substrate inhibition observed with each substrate may be regulated by the other substrate. Since such kinetic behavior could be the result of cooperativity in binding of substrates and/or products, we examined the binding characteristics of both substrates and products of the hSULT2A1-catalyzed sulfation of DHEA.
Fig. 2.
Rates of DHEA sulfation catalyzed by hSULT2A1 at low DHEA concentrations. A, the effect of various PAPS concentrations on the sulfation of 0.1-2.0 μM DHEA. Open circles (○) with a solid line represent sulfation rates at 1 μM PAPS, closed circles (●) with a dashed line represent sulfation rates at 5 μM PAPS, open squares (□) with a solid line represent sulfation rates at 10 μM PAPS, closed squares (■) with a dashed line represent sulfation rates at 60 μM PAPS, open triangles (△) with a solid line represent sulfation rates at 100 μM PAPS, closed triangles (▲) with a dashed line represent sulfation rates at 300 μM PAPS. B, the effect of various DHEA concentrations on the rate of reaction at 1.0 - 300 μM PAPS. Open circles (○) with a solid line represent sulfation rates at 0.1 μM DHEA, closed circles (●) with a dashed line represent sulfation rates at 0.2 μM DHEA, open squares (□) with a solid line represent sulfation rates at 0.4 μM DHEA, closed square (■) with a dotted line represent sulfation rates at 0.8 μM DHEA, open triangles (△) with a solid line represent sulfation rates at 1.6 μM DHEA, closed triangles (▲) with a dashed line represent sulfation rates at 2.0 μM DHEA.
Fig. 3.
Effects of 5.0, 20, and 80 μM DHEA concentrations on the initial velocity of the hSULT2A1-catalyzed reaction at concentrations of PAPS from 1.0 μM to 300 μM. Open triangles (△) with a dashed line represent sulfation rates at 5 μM DHEA, closed squares (■) with a dashed line represent sulfation rates at 20 μM DHEA, and open circles (o) with a solid line represent sulfation rates at 80 μM DHEA.
Binding of substrates and products to hSULT2A1
Binding properties of the substrates and products of DHEA sulfation reaction catalyzed by hSULT2A1 were analyzed in two groups of experiments by determining changes in the intrinsic fluorescence of tryptophan residues in hSULT2A1 upon ligand binding. In the first group, the binding of DHEA, DHEAS, PAPS, and PAP to hSULT2A1 were individually determined. In the second set of titrations, we examined the binding of these molecules to enzyme-substrate or enzyme-product complexes, as appropriate.
Binding of DHEA and DHEAS
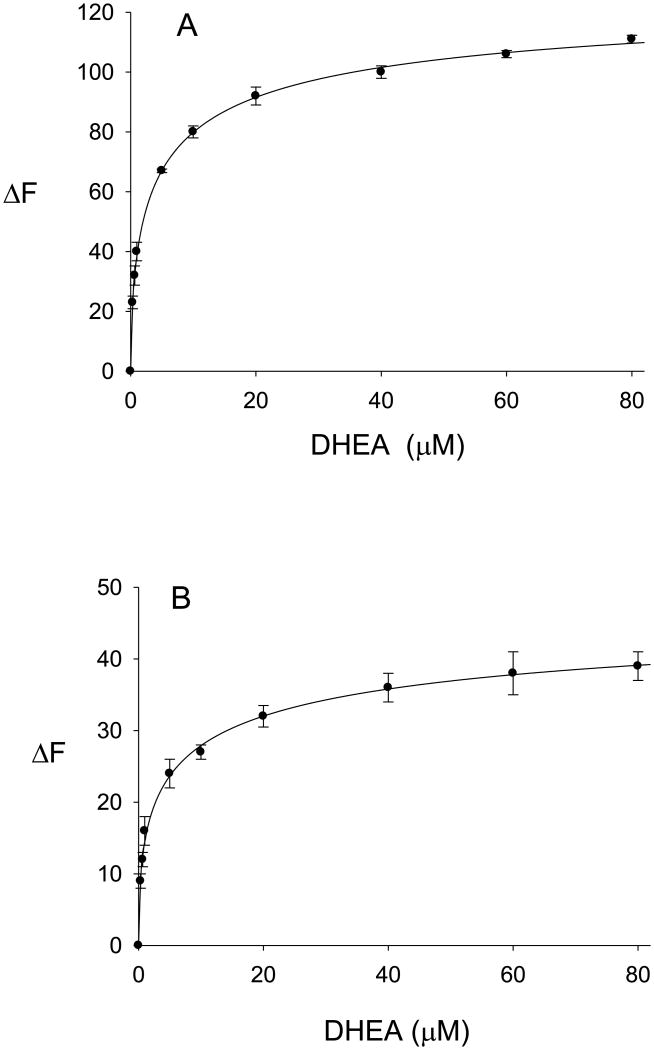
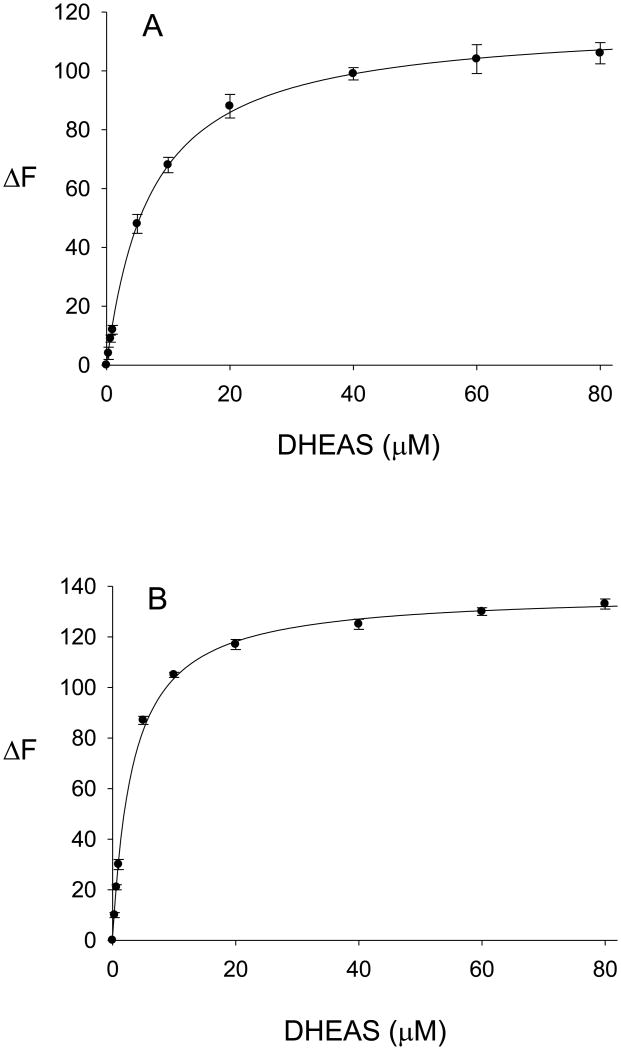
The binding properties of DHEA (0.33 μM – 80 μM) with the ligand-free enzyme and the enzyme-PAP complex are shown in Fig. 4A and Fig. 4B, respectively. DHEA-binding to both hSULT2A1 and PAP-bound hSULT2A1 exhibited negative cooperativity. As seen in Fig. 4, the absolute values of the changes in the intrinsic fluorescence of the enzyme upon titration with DHEA were fit to the Hill equation (Equation 2), and the calculated Hill coefficients were 0.57 (Fig. 4A) and 0.48 (Fig. 4B), indicating negative cooperativity in the binding of DHEA to both the free enzyme (Fig. 4A) and the enzyme-PAP complex (Fig. 4B). Attempts to fit the data for binding of DHEA to hSULT2A1 (i.e., data in Fig. 4A) and the hSULT2A1-PAP complex (i.e., data in Fig. 4B) to a hyperbolic single site model (Equation 1) resulted in lower values of r2 (0.9745 and 0.9593, respectively). We then utilized the Adair equation (Equation 3) to calculate the dissociation constants for DHEA. The Kd1 and Kd2 values for the binding of DHEA to the free enzyme were 0.6 ± 0.1 μM and 12.4 ± 2.5 μM, respectively, and the Kd1(app) and Kd2(app) values for the binding of DHEA to the enzyme in the presence of 100 μM PAP were 2.4 ± 0.6 μM and 22.0 ± 3.2 μM, respectively. Kd1 and Kd2 were labeled as apparent dissociation constants, since the binding to the enzyme-PAP complex was determined at a single concentration of PAP. The observed differences between Kd1 and Kd2 values for DHEA-binding to the free enzyme, and between Kd1(app) and Kd2(app) for DHEA-binding to the enzyme-PAP complex, were consistent with the results of the analyses by the Hill equation indicating negative cooperativity.
Fig. 4.
Binding of DHEA to hSULT2A1 and to the hSULT2A1-PAP complex. Values are the mean ± standard deviation (triplicate determinations) of the absolute value of the change in intrinsic fluorescence of the protein. The curves represent the fit of the data to the Hill equation. A, binding of DHEA to ligand-free hSULT2A1 (r2=0.9994). B, binding of DHEA to the enzyme-PAP complex (r2=0.9976).
The binding of the product DHEAS (0.33 μM - 80 μM) to hSULT2A1 was also studied with both the ligand-free enzyme and the enzyme-PAP complex. As seen in Fig. 5A, the titration of hSULT2A1 with DHEAS fit well to a classical hyperbolic curve (Equation 1) with no evidence of cooperativity in binding. This absence of cooperativity for binding of DHEAS was significantly different from the negative cooperativity observed for the binding of DHEA to the enzyme. The dissociation constant calculated for DHEAS was 7.2 ± 0.3 μM. Upon investigation of the binding of DHEAS to hSULT2A1 in the presence of 100 μM PAP, we found that the binding was also consistent with the absence of any cooperative effects (Fig. 5B). The Kd(app) value for DHEAS binding to the enzyme-PAP complex was calculated to be 3.3 ± 0.2 μM. This lack of cooperativity was confirmed by a fit of the data to the Hill equation model, where the calculated Hill coefficient was 1.097 for the binding of DHEA to hSULT2A1 in either the presence or absence of 100 μM PAP (r2 values of 0.9998 and 0.9993, respectively).
Fig. 5.
Binding of DHEAS to hSULT2A1 and to the hSULT2A1-PAP complex. Values are the mean ± standard deviation (triplicate determinations) of the absolute value of the change in intrinsic fluorescence of the protein. The curves represent the fit of the data to a classical hyperbolic binding curve (i.e., no cooperativity). A, binding of DHEAS to ligand-free hSULT2A1 (r2=0.9993). B, binding of DHEAS to the enzyme-PAP complex (r2=0.9985).
Binding of PAPS and PAP
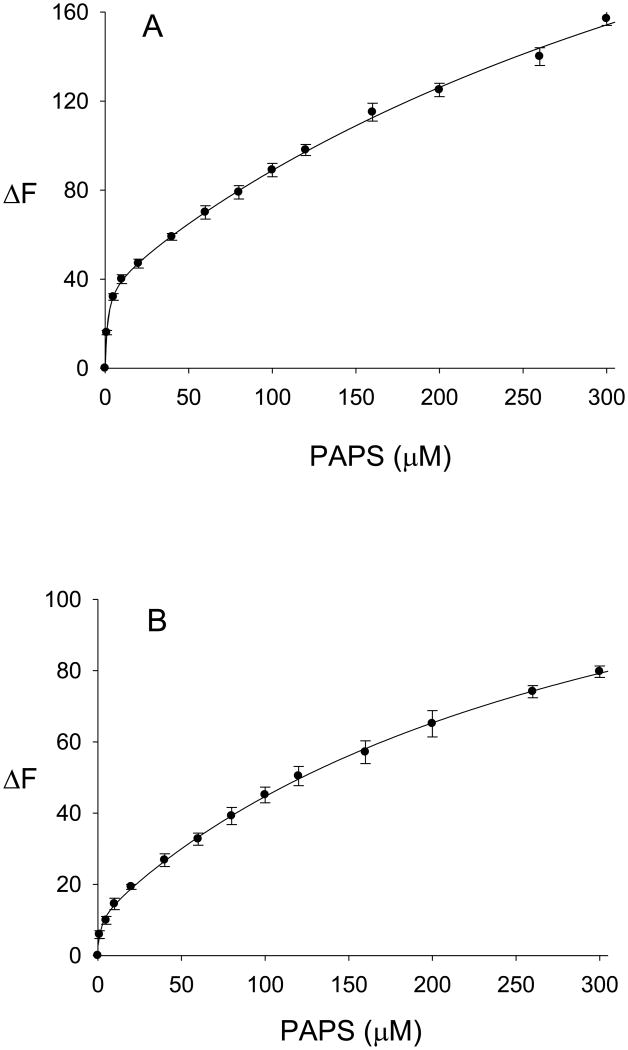
Studies on the binding of PAPS and PAP, at a range of concentrations between 1.0 μM and 300 μM, were carried out with ligand-free hSULT2A1 and with the binary complexes hSULT2A1-DHEA and hSULT2A1-DHEAS. As shown in Fig. 6A, the binding of PAPS to hSULT2A1 was well-modeled by a two-site saturable binding equation. This result was indicative of two non-equivalent sites for the binding of PAPS, the dissociation constants for PAPS, as determined using equation 4, were calculated to be Kd1 = 1.5 ± 0.4 μM and Kd2 = 518 ± 91 μM. Thus, there was a greater than 300-fold difference between the first and second dissociation constants for PAPS.
Fig. 6.
Binding of PAPS to hSULT2A1 and to the hSULT2A1-DHEAS complex. Values are the mean ± standard deviation (triplicate determinations) of the absolute value of the change in intrinsic fluorescence of the protein. The curves represent the fit of the data to a two-site saturable binding equation. A, binding of PAPS to ligand-free hSULT2A1 (r2=0.9989). B, binding of PAPS to the enzyme-DHEAS complex (r2=0.9995).
The formation of an enzyme-DHEAS-PAPS ternary complex was analyzed by titration with PAPS (1.0 μM – 300 μM) after pretreatment of the enzyme with 80 μM DHEAS. Analysis of the formation of this ternary complex using a two-site saturable binding model (equation 4) was consistent with two non-equivalent binding sites for PAPS-interaction with he enzyme-DHEAS binary complex (Fig. 6B), and Kd1(app) and Kd2(app) values were calculated as 1.3 ± 0.4 μM and 315 ± 27 μM, respectively. Thus, the Kd1(app) value for PAPS-binding to the enzyme-DHEAS complex was approximately equal to the Kd1 for binding of PAPS to the ligand-free enzyme. Although the Kd2(app) for PAPS-binding to the enzyme-DHEAS complex was slightly lower than the corresponding second dissociation constant for PAPS with the free enzyme, the Kd2(app) for the hSULT2A1-DHEAS-PAPS complex was still over 200-fold greater than Kd1(app) for the hSULT2A1-DHEAS-PAPS complex.
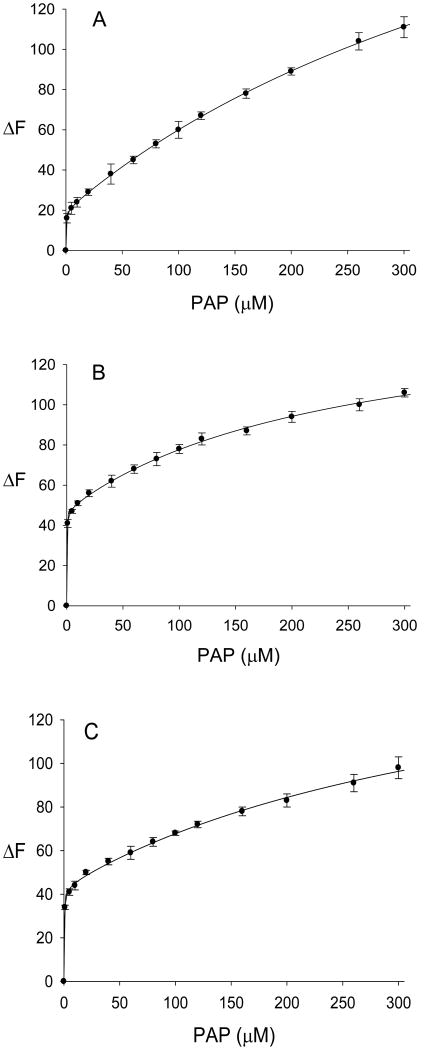
Results for the binding of PAP to ligand-free hSULT2A1 were similar to those obtained for the analogous binding of PAPS to the enzyme. The binding of PAP to ligand-free hSULT2A1 fit well to the saturable binding equation for two non-equivalent sites (Fig. 7A). Upon analysis by this model, Kd1 was 0.3 ± 0.05 μM, and Kd2 was 536 ± 26 μM. We then evaluated the binding of PAP to the enzyme in the presence of either DHEA or DHEAS. In the first group of experiments, we analyzed the effects of DHEA on the binding of PAP to hSULT2A1. In order to provide the enzyme-DHEA binary complex, we pretreated the hSULT2A1 with 80 μM DHEA. This concentration of DHEA was at the limit of solubility in the assay. As seen in Fig. 7B, the results fit well to the two-site saturable binding equation. Dissociation constants for the hSULT2A1-DHEA-PAP complex, as calculated by equation 4, were Kd1(app) = 0.2 ± 0.04 μM and Kd2(app) = 243 ± 27 μM. Thus, the Kd1 (app) calculated for the binding of PAP to the hSULT2A1-DHEA complex was similar to the Kd1 for the binding of PAP to the ligand-free hSULT2A1.
Fig. 7.
Binding of PAP to hSULT2A1 and to the hSULT2A1-DHEA and hSULT2A1 DHEAS complexes. Values are the mean ± standard deviation (triplicate determinations) of the absolute value of the change in intrinsic fluorescence of the protein. The curves represent the fit of the data to a two-site saturable binding equation. A, binding of PAP to ligand-free hSULT2A1 (r2=0.9999). B, binding of PAP to the enzyme-DHEA complex (r2=0.9988). C, binding of PAP to the enzyme-DHEAS complex (r2=0.9985).
The binding of PAP to hSULT2A1 in the presence of DHEAS was also investigated. In order to provide the enzyme-DHEAS binary complex, hSULT2A1 was pretreated with 80 μM DHEAS, and the binding of PAP (concentrations from 1.0 μM to 300 μM) was determined by monitoring the change in intrinsic fluorescence of the protein. Analysis of the results with a model for two non-equivalent binding sites (equation 4) provided dissociation constants for PAP in the enzyme-PAP-DHEAS complex, and Kd1(app) and Kd2(app) were calculated as 0.3 ± 0.06 μM and 415 ± 75 μM, respectively. Thus, there was little effect of DHEAS on the dissociation constants for binding of PAP to the enzyme. The binding parameters for all of the substrates and products of hSULT2A1 that were studied are summarized in Tables 1 and 2.
Table 1. Dissociation constants calculated for binding of substrates and products to hSULT2A1.
| Substrate or Product | ||||
|---|---|---|---|---|
| DHEAa | DHEAS | PAPS | PAP | |
| Kd1 (μM) | 0.6 ± 0.1 | 7.2 ± 0.3 | 1.5 ± 0.4 | 0.3 ± 0.05 |
| Kd2 (μM) | 12.4 ± 2.5 | 7.2 ± 0.3 | 518 ± 91 | 536 ± 26 |
The binding of DHEA to hSULT2A1exhibited negative cooperativity with a Hill coefficient of 0.57.
Table 2. Apparent dissociation constants for the formation of various hSULT2A1-ternary complexes at the specified concentration of one substrate or product and varied concentrations of the second substrate or product.
| Substrate or Product | |||||
|---|---|---|---|---|---|
| DHEAa binding at 0.1 mM PAP | DHEAS binding at 0.1 mM PAP | PAP binding at 0.08 mM DHEA | PAP binding at 0.08 mM DHEAS | PAPS binding at 0.08 mM DHEAS | |
| Kd1(app) (μM) | 2.4 ± 0.6 | 3.3 ± 0.2 | 0.2 ± 0.04 | 0.3 ± 0.06 | 1.3 ± 0.4 |
| Kd2(app) (μM) | 22 ± 3.2 | 3.3 ± 0.2 | 243 ± 27 | 415 ± 75 | 315 ± 27 |
The binding of DHEA to hSULT2A1 in the presence of PAP exhibited negative cooperativity with a Hill coefficient of 0.48.
Discussion
When substrate inhibition data are analyzed along with equilibrium binding data for substrates and products, our results indicate the ability of hSULT2A1 to form non-productive ternary complexes that involve either DHEA or DHEAS. The formation of these ternary complexes is subject to negative cooperativity in the binding of DHEA. Moreover, the binding of both PAPS and PAP to the enzyme is best described by a model with two saturable binding sites, with Kd1 values for PAPS and PAP in the 0.3 – 1.5 μM range and Kd2 values 200-300 fold higher. Since hepatic concentrations of PAPS have been measured as approximately 70 nmoles/g tissue in the rat [52], it could be reasonably assumed that the second binding site for PAPS is not highly occupied under physiological conditions (i.e., at approximately 70-80 μM PAPS). Thus, the hSULT2A1 exhibits behavior approaching that described for other enzymes as a “half-of-the-sites reactivity”[53]. However, the smaller differences between Kd1 and Kd2 for binding of DHEA to the free enzyme and to the enzyme-PAP complex suggest that the concentration of DHEA may play a more subtle regulatory role in catalysis and in its physiological sulfation. In addition, the high affinity of DHEAS for the enzyme and the lack of cooperativity in its interaction with hSULT2A1 are of interest. The affinity of DHEAS for both the free enzyme and the enzyme-PAP complex suggests that such product inhibition might serve as another component in the regulation of intracellular concentrations of DHEA and DHEAS.
A proposed model for roles of non-productive complexes in the function of hSULT2A1
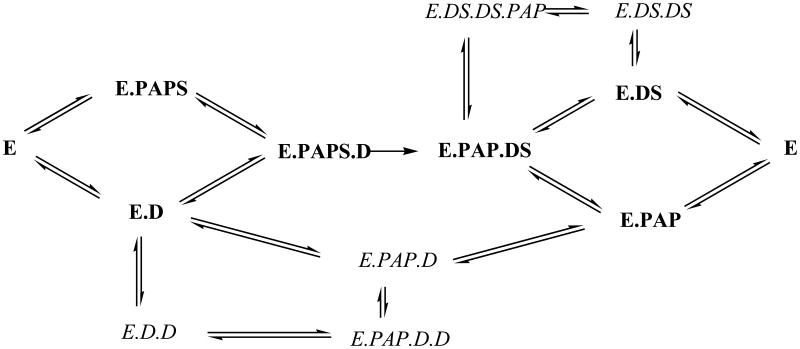
Recent studies combining molecular modeling with kinetic analyses are consistent with a random bi bi mechanism for hSULT2A1 with DHEA as substrate [54]. Our current results on both the kinetics of DHEA-sulfation and the roles of substrate and product complexes with hSULT2A1 are summarized in Figure 8. According to this model, an enzyme-DHEAS-PAP ternary complex occurs after the binding of DHEA and PAPS and catalysis of the sulfuryl transfer reaction. Since there is no evidence for a sulfuryl transfer from DHEAS to PAP catalyzed by hSULT2A1, this ternary complex can dissociate into either enzyme-PAP or enzyme-DHEAS. Dissociation of either of these binary complexes would yield the free enzyme, which is available for the next catalytic turnover. However, since DHEA and PAPS are available in the assay system, it is still possible to observe ternary complex formation from either an enzyme-DHEAS or an enzyme-PAP binary complex. The first possibility is the formation of enzyme-PAP-DHEA complex from the enzyme-PAP binary complex. This complex is non-productive, and the dissociation constant for PAP in this complex is similar to the dissociation constant for PAP from its binary complex with the enzyme. Moreover, at higher concentrations of DHEA, the formation of this enzyme-PAP-DHEA complex from the enzyme-PAP complex is favored. Interestingly, the DHEA concentration at the maximum rate of sulfation is around 2-3 μM. This concentration of DHEA yields an almost fully occupied first site and a partially occupied second DHEA-binding site, with increasing concentrations of DHEA resulting in reduced reaction velocity. Therefore, this model identifies the enzyme-PAP-DHEA ternary complex as a major component of substrate inhibition with DHEA.
Fig. 8.
A proposed model describing major pathways for the formation of dead-end complexes important for substrate and product inhibition in the hSULT2A1-catalyzed sulfation of DHEA. Abbreviations used for the enzyme complexes are: E: hSULT2A1, D: DHEA, DS: DHEAS.
The possibility of a significant contribution of an enzyme-DHEAS-PAPS ternary complex at a sufficiently high concentration of DHEAS could not be ruled out by the binding experiments. Under the initial velocity conditions utilized, however, the lack of substrate inhibition by PAPS makes this an unlikely possibility. Likewise, depending on the amount of PAP and DHEAS present, an enzyme-PAP-DHEAS ternary complex or an enzyme-PAP-DHEAS-DHEAS complex may provide product inhibition. PAP has a high affinity for the enzyme-DHEAS complex and DHEAS has a high affinity for the enzyme-PAP complex, however such product inhibition by PAP or DHEAS would require the accumulation of both products. Intracellular PAP can be hydrolyzed by action of a (2′)3′,5′-bisphosphate nucleotidase [55, 56], and DHEAS is subject to intracellular hydrolysis catalyzed by steroid sulfatase [57] and to alteration of its cellular concentration by anionic transporters [58]. Regulation of these mechanisms for cellular concentrations of DHEAS and PAP might, therefore, have effects on the intracellular rate of hSULT2A1-catalyzed sulfation.
Thus, the concentrations of both DHEAS and DHEA relative to the catalytic formation of PAP are important components in inhibition of the enzyme, with a role for DHEAS in product inhibition and DHEA involved in substrate inhibition. These results share similarities with the previous findings on substrate inhibition seen with other cytosolic sulfotransferases, since studies on family 1 cytosolic sulfotransferases (i.e., rSULT1A1, hSULT1A1, hSULT1E1) also indicated that enzyme-product-substrate type ternary enzyme complexes are responsible for substrate inhibition [29, 31-33].
Our studies on the kinetics of the hSULT2A1-catalyzed sulfation of DHEA and the binding of subtrates and products to the enzyme were also evaluated in conjunction with the several published studies on the crystal structures of hSULT2A1. Pederson and co-workers reported the first crystal structure of hSULT2A1 with PAP present [34]. In the crystal structure, they found PAP-containing monomers in asymmetric units and they concluded that the protein interaction in the crystal structure revealed a possible dimer-directed conformational alteration that may regulate the enzyme activity. Zhou and co-workers published additional crystallographic studies on hSULT2A1 [36], in which they examined the ligand-free enzyme, DHEA-bound enzyme, and both DHEA- and PAP-bound enzyme. According to their analysis, all of the crystal forms of the enzyme had different dimer arrangements. They further noted that the PAP-containing crystal form was not found in either the DHEA-containing or apocrystal forms. Moreover, they also reported that the crystal packing of the hSULT2A1 crystallized in the presence of DHEA was significantly different from crystals obtained in the presence of DHEA and PAP. One of the critical observations that they made was on the effect of PAP on the DHEA-bound crystals of the enzyme: the hSULT2A1 crystals obtained through the co-crystallization with DHEA were immediately destroyed when exposed to PAP. According to their conclusion, this was suggestive of a possible conformational change upon binding of the PAP yielding the decomposition of the crystals. Such significant conformational changes upon binding of substrate and product ligands would be consistent with the PAP-dependent changes in the dissociation constants for DHEA that are seen in our binding experiments. Moreover, this would point to the possibility of communication of conformational changes between subunits as a basis for the negative cooperativity and substrate inhibition.
Subsequent to the above crystallographic studies, Rehse and co-workers also published the crystal structure of hSULT2A1 with its substrate DHEA [35]. In this research, they demonstrated two different binding orientations for DHEA. They stated that the substrate might show rotations of up to 30°, and there would be corresponding rearrangements of the protein loops contributing to the active site of the enzyme. Our results indicating negative cooperativity in binding of DHEA to hSULT2A1 are consistent with the possibility of different binding orientations for DHEA at the two active sites of the homodimer. An alternate explanation for our results might be the binding of DHEA at an allosteric site in the same subunit. While computational analyses of hSULT2A1 have suggested a possible allosteric binding site for celecoxib and nimesulide, allosteric binding of DHEA to a single subunit was not considered in that study [59]. However, the crystal structures of hSULT2A1 solved to date have not revealed any example of two molecules of DHEA simultaneously bound to a single subunit of the homodimeric protein. Therefore, either the proper crystallization conditions have not yet been found to observe this, the conformational changes resulting from the allosteric interactions within a subunit yield populations of conformations that are too heterogeneous to be amenable to crystallization, or such allosteric binding sites for a second molecule of DHEA are not present within a single subunit of the dimer. It is clear that further studies will be necessary to determine the specific roles that conformational changes within the subunits, and/or those communicated between the subunits may play in the negative cooperativity and substrate inhibition observed with hSULT2A1.
Supplementary Material
Acknowledgments
The authors express their appreciation to Xiaoyan Qin for assistance with the experiments in Figure S1. This study was supported by the National Institutes of Health through research grants R01 CA038683 from the National Cancer Institute and P42 ES013661 from the National Institute of Environmental Health Sciences. Support from the NIEHS through P30 ES05605 is also acknowledged. The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
The abbreviations used are
- DE52
diethylaminoethyl cellulose anion exchanger
- DHEA
dehydroepiandrosterone (5-androsten-3β-ol-17-one)
- DHEAS
dehydroepiandrosterone sulfate
- hSULT2A1
human hydroxysteroid sulfotransferase 2A1
- PAP
adenosine 3′,5′-diphosphate
- PAPS
3′-phosphoadenosine 5′-phosphosulfate
- SULT
cytosolic sulfotransferase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Duffel MW, Marshall AD, McPhie P, Sharma V, Jakoby WB. Drug Metab Rev. 2001;33:369–395. doi: 10.1081/dmr-120001394. [DOI] [PubMed] [Google Scholar]
- 2.Pacifici GM, Coughtrie MW, editors. Human cytosolic sulfotransferases. CRC Press; Boca Raton, FL: 2005. [Google Scholar]
- 3.Falany CN. FASEB J. 1997;11:206–216. doi: 10.1096/fasebj.11.4.9068609. [DOI] [PubMed] [Google Scholar]
- 4.Gamage N, Barnett A, Hempel N, Duggleby RG, Windmill KF, Martin JL, McManus ME. Toxicol Sci. 2006;90:5–22. doi: 10.1093/toxsci/kfj061. [DOI] [PubMed] [Google Scholar]
- 5.Glatt H. Chem Biol Interact. 2000;129:141–170. doi: 10.1016/s0009-2797(00)00202-7. [DOI] [PubMed] [Google Scholar]
- 6.Jakoby WB, Sekura RD, Lyon ES, Markus CJ, Wang JL. In: Enzymatic Basis of Detoxication. Jakoby WB, editor. Vol. 2. Academic Press; New York: 1980. pp. 199–228. [Google Scholar]
- 7.Blanchard RL, Freimuth RR, Buck J, Weinshilboum RM, Coughtrie MW. Pharmacogenetics. 2004;14:199–211. doi: 10.1097/00008571-200403000-00009. [DOI] [PubMed] [Google Scholar]
- 8.Falany CN, Vazquez ME, Kalb JM. Biochem J. 1989;260:641–646. doi: 10.1042/bj2600641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Longcope C. Journal of Endocrinology. 1996;150:S125–127. [PubMed] [Google Scholar]
- 10.Bird CE, Masters V, Clark AF. Clin Invest Med. 1984;7:119–122. [PubMed] [Google Scholar]
- 11.Kallen CB. Obstet Gynecol Clin North Am. 2004;31:795–816. x. doi: 10.1016/j.ogc.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Strott CA. In: Human cytosolic sulfotransferases. Pacifici GM, Coughtrie MWH, editors. CRC Press; Boca Raton, FL: 2005. pp. 231–253. [Google Scholar]
- 13.Chang HJ, Shi R, Rehse P, Lin SX. J Biol Chem. 2004;279:2689–2696. doi: 10.1074/jbc.M310446200. [DOI] [PubMed] [Google Scholar]
- 14.Aksoy IA, Otterness DM, Weinshilboum RM. Drug Metab Dispos. 1993;21:268–276. [PubMed] [Google Scholar]
- 15.Comer KA, Falany JL, Falany CN. Biochem J. 1993;289:233–240. doi: 10.1042/bj2890233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radominska A, Comer KA, Zimniak P, Falany J, Iscan M, Falany CN. Biochem J. 1990;272:597–604. doi: 10.1042/bj2720597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elekima OT, Mills CO, Ahmad A, Skinner GR, Ramsden DB, Bown J, Young TW, Elias E. Liver. 2000;20:45–50. doi: 10.1034/j.1600-0676.2000.020001045.x. [DOI] [PubMed] [Google Scholar]
- 18.Iqbal S, Vickers C, Elias E. J Hepatol. 1990;11:37–42. doi: 10.1016/0168-8278(90)90269-w. [DOI] [PubMed] [Google Scholar]
- 19.Chou HC, Lang NP, Kadlubar FF. Cancer Res. 1995;55:525–529. [PubMed] [Google Scholar]
- 20.Glatt H, Seidel A, Harvey RG, Coughtrie MW. Mutagenesis. 1994;9:553–557. doi: 10.1093/mutage/9.6.553. [DOI] [PubMed] [Google Scholar]
- 21.Okuda H, Nojima H, Miwa K, Watanabe N. Chem Res Toxicol. 1989;2:15–22. doi: 10.1021/tx00007a003. [DOI] [PubMed] [Google Scholar]
- 22.Surh YJ. Chemico-Biol Interact. 1998;109:221–235. doi: 10.1016/s0009-2797(97)00134-8. [DOI] [PubMed] [Google Scholar]
- 23.Watabe T, Ishizuka T, Isobe M, Ozawa N. Science. 1982;215:403–405. doi: 10.1126/science.6800033. [DOI] [PubMed] [Google Scholar]
- 24.Chapman E, Bryan MC, Wong CH. Proc Natl Acad Sci U S A. 2003;100:910–915. doi: 10.1073/pnas.0337638100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gamage NU, Duggleby RG, Barnett AC, Tresillian M, Latham CF, Liyou NE, McManus ME, Martin JL. J Biol Chem. 2003;278:7655–7662. doi: 10.1074/jbc.M207246200. [DOI] [PubMed] [Google Scholar]
- 26.Hoff RH, Czyryca PG, Sun M, Leyh TS, Hengge AC. J Biol Chem. 2006;281:30645–30649. doi: 10.1074/jbc.M604205200. [DOI] [PubMed] [Google Scholar]
- 27.Kakuta Y, Petrotchenko EV, Pedersen LC, Negishi M. J Biol Chem. 1998;273:27325–27330. doi: 10.1074/jbc.273.42.27325. [DOI] [PubMed] [Google Scholar]
- 28.Marshall AD, Darbyshire JF, Hunter AP, McPhie P, Jakoby WB. J Biol Chem. 1997;272:9153–9160. doi: 10.1074/jbc.272.14.9153. [DOI] [PubMed] [Google Scholar]
- 29.Marshall AD, McPhie P, Jakoby WB. Arch Biochem Biophys. 2000;382:95–104. doi: 10.1006/abbi.2000.2020. [DOI] [PubMed] [Google Scholar]
- 30.Teramoto T, Sakakibara Y, Liu MC, Suiko M, Kimura M, Kakuta Y. Biochem Biophys Res Commun. 2009;383:83–87. doi: 10.1016/j.bbrc.2009.03.146. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H, Varlamova O, Vargas FM, Falany CN, Leyh TS. J Biol Chem. 1998;273:10888–10892. doi: 10.1074/jbc.273.18.10888. [DOI] [PubMed] [Google Scholar]
- 32.Duffel MW, Jakoby WB. J Biol Chem. 1981;256:11123–11127. [PubMed] [Google Scholar]
- 33.Gamage NU, Tsvetanov S, Duggleby RG, McManus ME, Martin JL. J Biol Chem. 2005;280:41482–41486. doi: 10.1074/jbc.M508289200. [DOI] [PubMed] [Google Scholar]
- 34.Pedersen LC, Petrotchenko EV, Negishi M. FEBS Lett. 2000;475:61–64. doi: 10.1016/s0014-5793(00)01479-4. [DOI] [PubMed] [Google Scholar]
- 35.Rehse PH, Zhou M, Lin SX. Biochem J. 2002;364:165–171. doi: 10.1042/bj3640165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou M, Rehse P, Chang HJ, Luu-The V, Lin SX. Acta Crystallogr D Biol Crystallogr. 2001;57:1630–1633. doi: 10.1107/s0907444901010964. [DOI] [PubMed] [Google Scholar]
- 37.Lu LY, Hsieh YC, Liu MY, Lin YH, Chen CJ, Yang YS. Mol Pharmacol. 2008;73:660–668. doi: 10.1124/mol.107.041038. [DOI] [PubMed] [Google Scholar]
- 38.Sekura RD. Methods Enzymol. 1981;77:413–415. doi: 10.1016/s0076-6879(81)77026-5. [DOI] [PubMed] [Google Scholar]
- 39.Sheng JJ, Duffel MW. Drug Metab Dispos. 2003;31:697–700. doi: 10.1124/dmd.31.6.697. [DOI] [PubMed] [Google Scholar]
- 40.Gulcan HO, Liu Y, Duffel MW. Chem Res Toxicol. 2008;21:1503–1508. doi: 10.1021/tx800133d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nose Y, Lipmann F. J Biol Chem. 1958;233:1348–1351. [PubMed] [Google Scholar]
- 42.Sheng J, Sharma V, Duffel MW. Current Protocols in Toxicology. John Wiley & Sons, Inc.; New York: 2001. pp. 4.5.1–4.5.9. [DOI] [PubMed] [Google Scholar]
- 43.Bensadoun A, Weinstein D. Anal Biochem. 1976;70:241–250. doi: 10.1016/s0003-2697(76)80064-4. [DOI] [PubMed] [Google Scholar]
- 44.Blomquist CH, Kotts CE, Hakanson EY. Anal Biochem. 1978;87:631–635. doi: 10.1016/0003-2697(78)90714-5. [DOI] [PubMed] [Google Scholar]
- 45.Chang HJ, Zhou M, Lin SX. J Steroid Biochem Mol Biol. 2001;77:159–165. doi: 10.1016/s0960-0760(01)00048-6. [DOI] [PubMed] [Google Scholar]
- 46.Beechem JM, Brand L. Annu Rev Biochem. 1985;54:43–71. doi: 10.1146/annurev.bi.54.070185.000355. [DOI] [PubMed] [Google Scholar]
- 47.Vivian JT, Callis PR. Biophys J. 2001;80:2093–2109. doi: 10.1016/S0006-3495(01)76183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cavaluzzi MJ, Borer PN. Nucleic Acids Res. 2004;32:e13. doi: 10.1093/nar/gnh015. 1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neet KN. In: Contemporary enzyme kinetics and mechanism. Purich DL, editor. Academic Press, INC; New Tork: 1983. pp. 267–320. [Google Scholar]
- 50.Adair GS. J Biol Chem. 1925;63:529–545. [Google Scholar]
- 51.Hill AV. Biochem J. 1921;15:577–586. doi: 10.1042/bj0150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brzeznicka EA, Hazelton GA, Klaassen CD. Drug Metab Dispos. 1987;15:133–135. [PubMed] [Google Scholar]
- 53.Koshland DE., Jr Current Opinion in Structural Biology. 1996;6:757–761. doi: 10.1016/s0959-440x(96)80004-2. [DOI] [PubMed] [Google Scholar]
- 54.Cook IT, Leyh TS, Kadlubar SA, Falany CN. Horm Mol Biol Clin Invest. 2010;1:81–87. doi: 10.1515/HMBCI.2010.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meireles Ribeiro J, Ruiz A, Sillero A, Gunther Sillero MA. Biochimie. 1990;72:227–234. doi: 10.1016/0300-9084(90)90077-t. [DOI] [PubMed] [Google Scholar]
- 56.Ramaswamy SG, Jakoby WB. J Biol Chem. 1987;262:10044–10047. [PubMed] [Google Scholar]
- 57.Ghosh D. Cell Mol Life Sci. 2007;64:2013–2022. doi: 10.1007/s00018-007-7175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hagenbuch B, Gui C. Xenobiotica. 2008;38:778–801. doi: 10.1080/00498250801986951. [DOI] [PubMed] [Google Scholar]
- 59.Yalcin EB, Struzik SM, King RS. Drug metabolism letters. 2008;2:198–204. doi: 10.2174/187231208785425755. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.