Abstract
Hybrids from crosses between populations of the flour beetle, Tribolium castaneum, express varying degrees of inviability and morphological abnormalities. The proportion of allopatric population hybrids exhibiting these negative hybrid phenotypes varies widely, from 3% to 100%, depending upon the pair of populations crossed. We crossed three populations and measured two fitness components, fertility and adult offspring numbers from successful crosses, to determine how genes segregating within populations interact in inter-population hybrids to cause the negative phenotypes. With data from crosses of 40 sires from each of three populations to groups of 5 dams from their own and two divergent populations, we estimated the genetic variance and covariance for breeding value of fitness between the intra- and inter-population backgrounds and the sire × dam-population interaction variance. The latter component of the variance in breeding values estimates the change in genic effects between backgrounds owing to epistasis. Interacting genes with a positive effect, prior to fixation, in the sympatric background but a negative effect in the hybrid background cause reproductive incompatibility in the Dobzhansky-Muller speciation model. Thus, the sire × dam-population interaction provides a way to measure the progress toward speciation of genetically differentiating populations on a trait by trait basis using inter-population hybrids.
Introduction
In the Dobzhansky-Muller model, the genetic basis of speciation rests on the development of incompatibilities (negative epistasis) between alleles fixed by a combination of drift and selection in different populations descended from a common ancestor (reviewed in Coyne and Orr, 2004). Specifically, the genetic basis for hybrid inviability and infertility lies with those genes, prior to fixation, which increase (or do not affect) fitness in the sympatric genetic background but diminish fitness in a hybrid background. It is generally believed that such genetic differences accumulate gradually but at an accelerating pace over time (Orr, 1995; Palmer & Feldman, 2009), as genes are acquired and fixed in daughter populations isolated from one another by geographic barriers to gene flow.
Structured populations permit another phenomenon to contribute to reproductive isolation. Wright, in his shifting balance theory of evolution, suggested that “adaptive gene complexes” arise in such populations (Wright, 1977). An adaptive gene complex is a set of genes that confer high fitness together, but are of much lower fitness when combined with other genes. Such co-adapted complexes arise when there are non-additive interactions between loci. For example, allele ‘A’ may increase fitness in combination with allele ‘b’ but decrease fitness in combination with allele ‘B’ (Wade 2001). Because the effect of A on fitness is dependent on the frequency of b in the population, populations with large differences in the frequency of ‘b’ will produce hybrids of reduced fitness proportional to the frequency difference at the B locus. In a large randomly mating population, the positive and negative effects of these allele combinations across loci may cancel one another with no net affect on fitness. Population genetic structure, however, allows for the sorting of existing genetic variation into locally favorable combinations which may be detrimental to fitness when brought together by hybridization (Mayr, 1963; Carson, 1975; Templeton, 1980). Herein, we test for the existence of background specific allele by comparing the breeding values for fitness within and between populations of the red flour beetle, Tribolium castaneum. This theory of speciation predicts that sires will exhibit heterogeneity of breeding values when crossed to dams from different genetic backgrounds and that epistatic gene interactions will be the cause of the variation in breeding values.
Because interaction is necessary for the effect of an allele to change with genetic background at other loci, additive gene action alone (i.e., the absence of dominance , epistasis, and genotypeby-environment interactions), only allows two populations to become different in trait means; they cannot differ in the average effects of alleles (Goodnight, 2000). With epistasis, two populations can have the same trait means, but nevertheless differ in the underlying genetic basis of those traits (Goodnight, 1995; 2000b; Palmer & Feldman, 2009). The magnitude of the among-population variation in average effects depends not only on the differential fixation of genes in genetically isolated populations descended from a common ancestor but also upon the amounts of epistatic relative to additive genetic variance segregating in the ancestral population from which they descended (Wade, 1991; Goodnight, 2000a; 2004). The greater the ratio of epistatic to additive variance in the ancestor, the greater can be the variance in average effects from one population to another. Variation in genic effects owing to epistatic interactions can be estimated as the contribution of the sire × dam-population interaction to the variance in local breeding values among sires (Damon et al., 1961; Roger et al., 1975; Elzo, 1990; Wade & James, 1994; Goodnight & Wade, 2000; Wade, 2001). When the inter-population traits are hybrid fertility and viability, the relative magnitude of this component of variance provides a way to measure the progress toward speciation of genetically differentiating populations. Whereas breeders concerned with exploiting gene interaction and heterosis are interested the selection of purebreds for enhanced crossbred performance (e.g., Ibanez-Escriche et al., (2009)), speciation geneticists are interested in the opposite phenomenon: selection within populations and its correlated effects on diminished hybrid performance. Nevertheless, the methods developed by breeders to measure the possible gains in hybrids (crossbreds) from non-additive gene action can be employed to measure the possible losses to hybrid fitness from epistasis, as we show below.
We investigated the variance in local breeding values for fertility and offspring numbers using three sets of sires, each crossed to groups of dams from three populations of T. castaneum. Two of the populations were collected from the wild, one from India (Bhopal) and one from Africa (Dar es Salaam, Tanzania). These two populations exhibit a low frequency of failed matings when crossed to one another, but a high frequency of antennal and limb deformities among the F1 hybrid and Backcross populations, and substantial epistasis for these traits in diallele crosses (Demuth & Wade, 2007). Our study is similar in experimental design to the studies by Robinson et al. (1994), Wade and Johnson (1994) and Wade et al. (1994) of interspecific hybrids between T. castaneum and T. freeman. However, the taxonomic scale of the present study is much smaller, involving crosses between populations of the same species, T. castaneum, as opposed to the earlier work on crosses between species.
The FST between the Indian and African populations, estimated from microsatellites, is 0.196 and both populations exhibit substantial heterozygosity (see Tables 3 and 1, respectively, in Drury, Siniard et al. (2009)). Whereas the diallele study of Demuth and Wade (2007) emphasized fixed genetic differences between populations, our study is focused on how genetic variation segregating within populations contributes to the negative hybrid phenotypes. The third population, c-SM, is an outbred laboratory strain used in several studies of the genetic differentiation among demes within artificial metapopulations (Wade, 1977; Mccauley & Wade, 1981; Wade & Mccauley, 1984) and in a study of Wright's Shifting Balance Theory (Wade & Goodnight, 1991), which implicated epistasis in the selection response.
Table 3.
Variance components of inter- and intra-population fecundity
| Sire | Dam | σ s2 | SEM | 95% CI | σ P2 | SEM | 95% CI | ||
|---|---|---|---|---|---|---|---|---|---|
| Bhopal | Bhopal | 251 | ± | 168 | -87, 589 | 1755 | ± | 173 | 1409, 2101 |
| cSM | 645 | ± | 249 | 158, 1132 | 3046 | ± | 199 | 2657, 3436 | |
| Dar es salaam | 151 | ± | 125 | -95, 396 | 1997 | ± | 259 | 1490, 2505 | |
| cSM | Bhopal | 284 | ± | 178 | -65, 633 | 2122 | ± | 167 | 1795, 2450 |
| cSM | 208 | ± | 94 | 24, 393 | 1446 | ± | 119 | 1212, 1680 | |
| Dar es salaam | 893 | ± | 267 | 370, 1416 | 2941 | ± | 185 | 2578, 3305 | |
| Dar es salaam | Bhopal | 323 | ± | 157 | 15, 632 | 1485 | ± | 172 | 1147, 1822 |
| cSM | 268 | ± | 151 | -23, 560 | 1215 | ± | 158 | 911, 1520 | |
| Dar es salaam | 141 | ± | 124 | -102, 384 | 1185 | ± | 200 | 794, 1576 |
Values in bold face signify sire component of variance is significantly greater than 0
Table 1.
Analysis of variance of offspring number as a function of sire population, dam population and their interaction.
| Source | d.f. | MS | F | P-value | Prand |
|---|---|---|---|---|---|
| Sire population | 2 | 15804 | 8.2538 | < 0.001 | 0.0005 |
| Dam population | 2 | 33804 | 17.6547 | < 0.001 | 0.0001 |
| Sire population × Dam population | 4 | 7606 | 3.9724 | 0.003 | 0.0030 |
| Residuals | 1515 | 1915 |
Prand refers to probabilities generated from 10 000 randomizations (see text)
For each set of sires, we estimated the heritability of two fitness components, fertility and offspring numbers, within their population of origin and on each of two different population backgrounds, the sire × dam-population interaction, and the genetic correlations between intra-population and inter-population half-sib family means. Like sire × dam-population interactions, variation among backgrounds in heritabilities estimated from the same set of sires in a common environment occurs when there are among-population differences in interacting genes (Damon et al. 1961; Roger et al. 1975; Elzo 1990). Similarly, the genetic correlation between intra- and inter-population half-sib family means should equal one in the absence of epistasis but be significantly less than one in its presence (For a detailed explanation see materials and methods of the current manuscript as well as Damon et al. (1961), Roger et al. (1975), Elzo (1990), Wade and James (1994), Goodnight (2000), Goodnight and Wade (2000), and Wade (2001). The experimental design thus has three different but related features useful for detecting the contribution of genetic interactions to the hybrid fitness traits.
Materials and Methods
Species Description
The red flour beetle, Tribolium castaneum, is a globally distributed human commensal. Tribolium beetles are poor migrators, unable to establish persistent populations on uncracked grain (Sokoloff, 1972). The combination of near obligate commensalism and poor dispersal results in a globally distributed species composed of a multitude of genetically differentiated populations with an average FST of 0.180 (Drury et al., 2009). The FST of 0.196 between the two wild populations used in this study is very close to the average observed by Drury, Siniard and Wade (2009), although both are more genetically different from the laboratory strain (c-SM vs Dar es salaam: 0.371; c-CM vs Bhopal: 0.303).
The individual beetles used in the present study were harvested from three populations. Two of which were collected from Bhopal, India, and Dar es Salaam, Tanzania, and have been maintained in the laboratory since 1989. The third population, c-SM, is an outbred laboratory population (for a detailed description of the stock's origin see Wade (1976)). Both wild caught collections originated from more than 50 adults. All three stocks were established and maintained at a population size of > 200 individuals on standard medium (20:1, flour:brewer's yeast, by weight) and in 24 h darkness.
Experimental design
For each half-sib cross, 40 males from one population were each mated to 15 virgin females, 5 from each of the three populations (40 sires × 3 populations × 15 dams per sire = 1,800 families total). Sire-by-population crosses occurred sequentially, with each sire being exposed to a set of five dams for two days. After two days, all beetles were removed by sifting, mated dams were separated into individual vials containing 8 g of medium, and sires were passaged to a subsequent set of dams. This process was repeated over the three dam types with every sire from each population. Each dam was allowed to lay for two weeks at 34 °C. After that time, the dam was removed, and offspring were allowed to mature to adulthood. We use the number of adult offspring produced as a measure of intra- and inter-population fecundity. To control for the possible effects of sperm depletion, the order in which sires were mated to each set of dams was randomized. Number of offspring produced was not significantly affected by cross order (Kruskal-Wallis rank sum test; df = 2, p-value = 0.3725).
Statistical methods: Variance Components and Heritabilities
To test for the effects of sire population, dam population and their interaction on fitness we performed a two-way ANOVA. All effects were estimated and the significance of each was calculated using the least-squares method. However, due to a high rate of cross incompatibility, our response variable, offspring number, did not conform to the parametric assumptions of the ANOVA. Thus, to confirm the ANOVA results, we performed a randomization (details below) with 10,000 replicates and report the probabilities of both analyses.
The additive genetic variance, heritability of offspring numbers and associated standard errors were estimated from the half-sib breeding design as follows. Let the observational components of the phenotypic variance, σ2P, equal, σ2P= σ2S + σE where σ2S is the variance among sires and σ2E is the residual variance, which includes the dominance variance, variance among mothers within demes, higher order interactions, and environmental effects. The genetic component of the phenotypic variance was obtained using the relationship, σ2S = 1/4VA, where VA is the additive genetic variance. We estimated narrow-sense heritabilities as h2 = (4σ2S) /σ2P. We calculated the heritabilities from variance components estimated using restricted maximum likelihood (REML), followed by a delete-one-sire jackknife. The jackknife has been shown to perform well for estimating variance components, heritabilities and genetic correlations (Knapp et al., 1989; Simons & Roff, 1994). In the delete-one jackknife, one family of paternal half sibs is deleted and a ‘pseudo-value’ of the statistic is calculated. This process is repeated, until a pseudo-value has been created for each family. The estimate of the statistic and its standard error are calculated from the mean and standard error of the full set of pseudo-values (Roff, 2006).
Statistical methods: Genetic Correlations Among Backgrounds
When organisms are reared in two different environments, genetically identical individuals can produce markedly different phenotypes in each environment (Falconer, 1952). Some of the difference between environments is the result of genotype-by-environment interaction. The genetic correlation across environments is expected to be one in the absence of genotype-by-environment interaction but significantly lower than one when a significant genotype-by-environment interaction exists (Falconer, 1952; Banos & Shook, 1990). Analogous to the way genetic correlations among environments detect genotype-by-environment interactions, the genetic correlations across population backgrounds in a common environment provide an estimate of allele-by-allele interaction at a single locus (dominance) or by interaction among multiple loci (epistasis). When there is heritable variation among sires and an absence of non-additive variation, the genetic correlation across backgrounds is expected to be one, while, with interactions (non-additive variation), it is expected to be less than one ((Elzo, 1990; Poivey et al., 2001). When divergent local selection is involved in the genetic diversification of populations, ,the expected genetic correlation is less than one (Wade et al. 1994; Johnson and Wade 1996). We calculated the genetic correlations between offspring produced on the intra- and inter-population backgrounds to estimate the effects of gene interaction in terms of the deviation from additive expectation. To calculate the genetic correlation between intra- and inter-population backgrounds (rgintra.inter), we used the paternal–half-sib family means and variances, i.e., covintra.inter /(VinterxVinter)1/2. As above, standard errors and confidence intervals for the correlation were generated using pseudo-values from a delete-one-sire jackknife.
Statistical methods: Genetic Threshold of Reproductive Isolation
In our data set, there were a number of failed crosses, where paired sires and dams produced no progeny at all, as well as crosses that produced offspring which did not develop to adults. Both are components of genetic incompatibility between populations in addition to reduced adult hybrid offspring numbers. We considered the genetic incompatibility arising from a failure to produce adult offspring as a threshold trait, dependent upon an underlying trait termed ‘liability.’ Liabilities below the threshold produce viable offspring (Roff, 1996; 1997), while liabilities beyond the threshold do not (reproductive incompatibility). To measure the liability of reproductive incompatibility, fertility data were transformed to a binary [0,1] scale. A cross was deemed a success if a single offspring reached adulthood, otherwise the cross was considered a failure. To test the effects of sire population, dam population, and their interaction on cross failure rate, we performed a two-way analysis of variance on cross failure rate. The ANOVA results were confirmed by randomization with 10,000 replicates (Manly, 1991). We used REML to determine if heritable variation for the liability of reproductive incompatibility was present in the between population crosses using variance components estimated from the 0,1 scale (Roff, 1997; 2006). Heritabilities were calculated using a delete-one-sire jackknife and transformed to the underlying scale. Standard errors and confidence intervals were estimated from the pseudo-values created from jackknifing. Heritabilities with 95% CI's not overlapping 0 are considered significant.
Results
Offspring Numbers
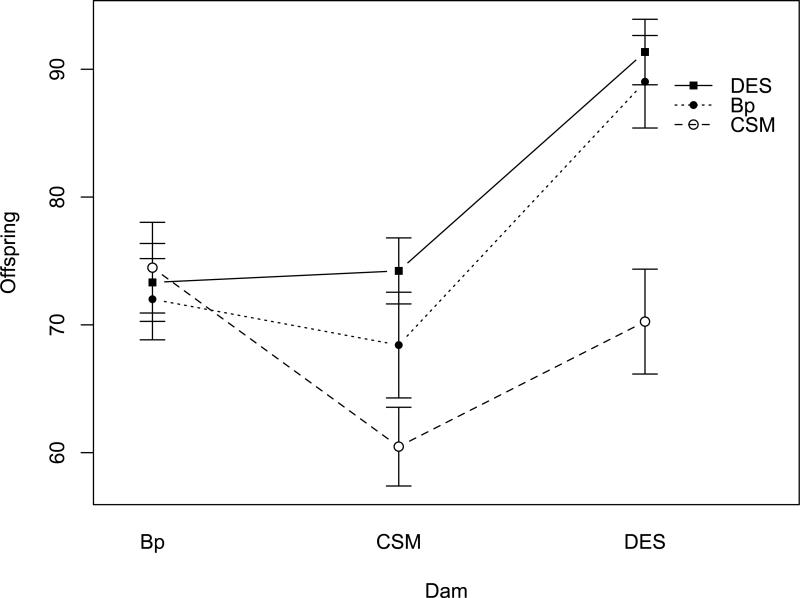
The forty males from each population were each crossed to fifteen dams, for a total of 600 dams. The ANOVA of the total offspring data presented in Table 1 reveals significant sire (P < 0.001), dam (P < 0.001) and sire-by-dam interactions (P < 0.005). Dar es Salaam males had significantly lower mean productivity when crossed with inter-population females than with intra-population females (Dar es Salaam females: 91.34 ± 2.56 offspring; cSM females: 74.22 ± 2.58 offspring; Bhopal females: 73.32 ± 3.05 offspring; Figure 1). In contrast, cSM males exhibited significant heterosis when outcrossed to Dar es Salaam or Bhopal females. Bhopal males were intermediate, displaying heterosis when crossed to Dar es Salaam females but reduced productivity when mated to cSM females. The significant sire-by-dam population interaction for offspring numbers is evidence that epistatic interactions are affecting this component of hybrid fitness (Table 1) and the hybrid population means indicate that inter-population hybrids exhibit heterosis in some crosses but reduced fitness in others.
Figure 1.
Mean (± 1 Jackknife SEM) number of offspring produced by 40 sires when crossed to each of three dam populations (each data point: 200 crosses)
The heritabilities for offspring numbers and the associated standard errors on the intra-population and inter-population backgrounds are given in Table 2 and raw among sire variances are given in Table 3. Within each population, sires differed from one another in offspring numbers on the intra-population and inter-population backgrounds (Table 2). In all six cases, the phenotypic variation was greater in the inter-population crosses than in the intra-population crosses, despite all progeny being reared under identical environmental conditions. The genetic component of the phenotypic variation, the among-sire variation, was greater in five of the six cases when sires were crossed to inter-population females than it was when they were crossed to intra-populaiton females (Table 3, column 3). This increase in among-sire variance is consistent with some fixation between populations of genes with epistatic interactions which increases the additive variance by increasing the additive effects of genes (Goodnight, 2005). We estimated the genetic correlation between backgrounds from the sire half-sib mean offspring numbers (Table 4, genetic correlation plus or minus one jack-knifed standard error). In all cases, the genetic correlations were significantly less than +1.0. However, despite significant among-sire variations for four of the nine cases, none could be bounded away from zero. These observations are consistent with gene interactions affecting offspring numbers and differences among populations in interacting genes but provide no evidence that locally divergent natural selection has contributed to the observed genetic differentiation.
Table 2.
Narrow sense heritability (h2) of offspring numbers estimated on sympatric and allopatric backgrounds.
| Dam |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bhopal | cSM | Dar es salaam | ||||||||||
| Sire | h2 | SE | CI 95% | h2 | SE | CI 95% | h2 | SE | CI 95% | |||
| Bhopal | 0.593 | ± | 0.38 | -0.167, 1.352 | 0.850 | ± | 0.32 | 0.229, 1.471 | 0.292 | ± | 0.24 | -0.180, 0.764 |
| cSM | 0.546 | ± | 0.33 | -0.094, 1.186 | 0.575 | ± | 0.26 | 0.060, 1.090 | 1.219 | ± | 0.34 | 0.553, 1.885 |
| Dar es salaam | 0.887 | ± | 0.39 | 0.115, 1.658 | 0.867 | ± | 0.48 | -0.054, 1.789 | 0.513 | ± | 0.42 | -0.315, 1.341 |
Values in bold face are significantly greater than 0
Table 4.
Genetic correlations between intra- and inter-population performance estimated from half-sib family means.
| Sire | Dam | rgintra inter | SEM | 95% CI | |
|---|---|---|---|---|---|
| Bhopal | cSM | 0.307 | ± | 0.16 | -0.012, 0.627 |
| Dar es salaam | -0.094 | ± | 0.16 | -0.412, 0.225 | |
| cSM | Bhopal | 0.239 | ± | 0.20 | -0.152, 0.629 |
| Dar es salaam | 0.202 | ± | 0.19 | -0.164, 0.569 | |
| Dar es salaam | Bhopal | 0.094 | ± | 0.16 | -0.221, 0.409 |
| cSM | 0.074 | ± | 0.17 | -0.249, 0.398 |
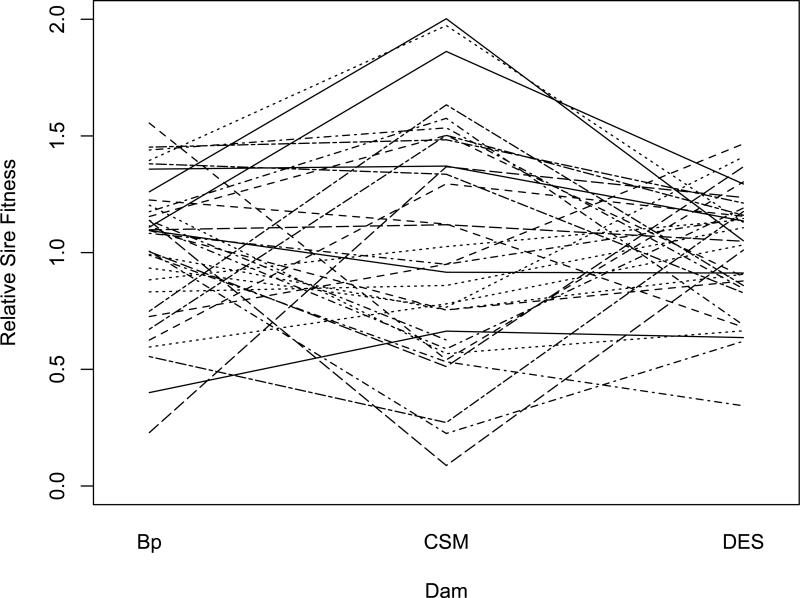
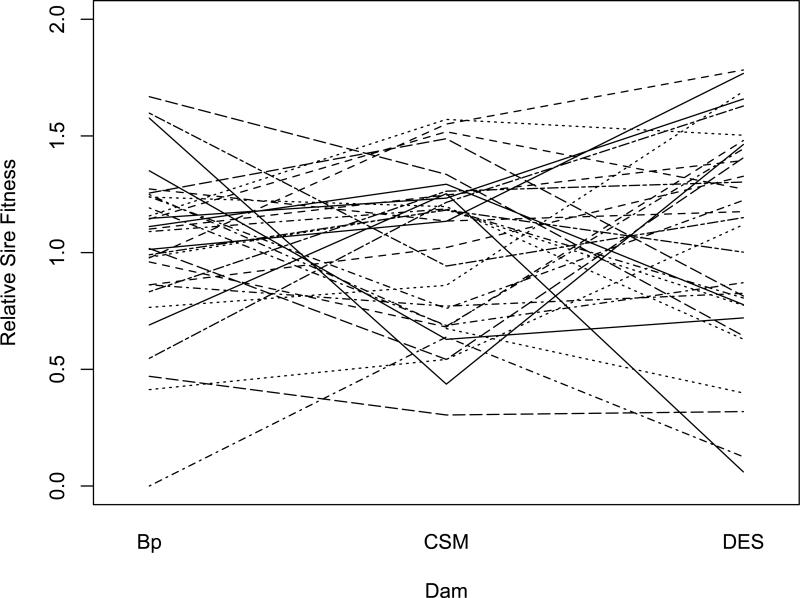
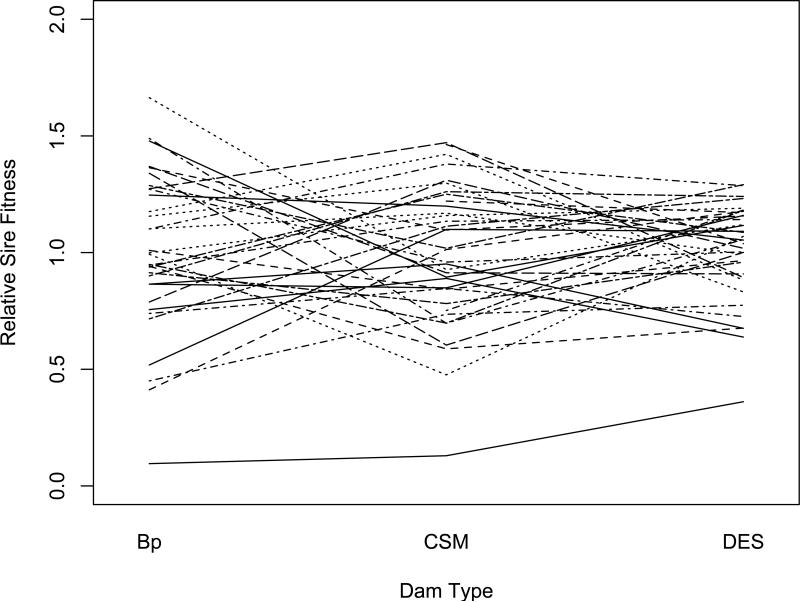
In Figures 2, 3, and 4, we show the effects of population background on the relative fitness based on sire mean offspring numbers when Bhopal, cSM, and Dar es Salaam sires are crossed to dams of their own and the other two allopatric populations. Like the illustration of ‘crossing-type’ genotype-by-environment interaction (Wade, 1990), the crossing of lines in Figures 2-4 is indicative of epistasis for fitness. In every case, the rank order among sires for offspring numbers changes with changes in the population background of the dams to which the sires were crossed. This change in the relative rankings of sire relative fitness from one background to another is the reason that the genetic correlations across genetic backgrounds are significantly less than one in Table 4.
Figure 2.
Norm of reaction for relative fitness of Bhopal sires across three genetic backgrounds (Dam populations). Each line represents as single sire and each inflection point is the mean relative fitness of 3 to 5 dams.
Figure 3.
Norm of reaction for relative fitness of c-SM sires across three genetic backgrounds (Dam populations). Each line represents as single sire and each inflection point is the mean of 3 to 5 dams.
Figure 4.
Norm of reaction for relative fitness of Dar es salaam sires across three genetic backgrounds (Dam populations). Each line represents as single sire and each inflection point is the mean of 3 to 5 dams.
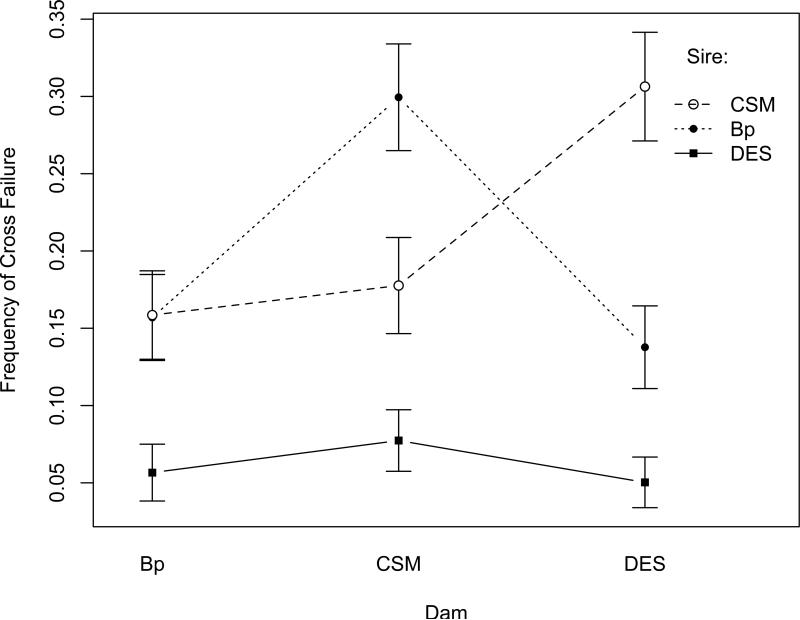
Cross Success and Failure
In each population pairing, we observed failed crosses that produced no adult offspring. As with adult offspring numbers, the propensity for crosses to fail to produce any adult offspring was significantly influenced by sire population (P < 0.001), dam population (P < 0.025), and sire-bydam interactions (P < 0.001) (Table 5). In Figure 5, we show the mean frequency of failed crosses for each of the nine population pairs. Males of the c-SM laboratory strain had twice as many failed crosses with Dar es Salaam females (0.306 ± .035) as with Bhopal females (0.158 ± .029) or cSM females (0.178 ± .031). Dar es Salaam males exhibited the lowest mean frequency of failed crosses (0.062 ± .011) and were relatively uniform in their success with females from all populations. Bhopal males had twice as many failed crosses with c-SM females (0.299 ± .034), than with either Dar es Salaam or their sympatric Bhopal dams (0.157 ± 0.028 and 0.138 ± 0.027, respectively). Reciprocal crosses often differed markedly in success rate. The most extreme difference occurred in the Dar es Salaam by c-SM cross: failed crosses were observed in 7% (.077 ± .021) of Dar es Salaam male by c-SM female crosses but were four-fold more common (30.6%; 0.306 ± .035) in the reciprocal crosses. Significant heritable genetic variation was found among c-SM sires for compatibility with Dar es Salaam dams, while the crosses with the second highest incidence of incompatibility (Bhopal sires × c-SM dams) were associated with nearly significant heritabilities (Table 6).
Table 5.
Results of an analysis of variance of cross failure as a function of sire population, dam population and their interaction.
| Source | d.f. | MS | F | P-value | Prand |
|---|---|---|---|---|---|
| Sire population | 2 | 3.7 | 29.4203 | < 0.001 | 0.0001 |
| Dam population | 2 | 0.534 | 4.2445 | 0.015 | 0.0176 |
| Sire population × Dam population | 4 | 0.974 | 7.7484 | < 0.001 | 0.0001 |
| Residuals | 1515 | 0.126 |
Prand refers to probabilities generated from 10 000 randomizations (see text)
Figure 5.
Incidence of cross failure among 40 sires when crossed to each of three dam populations (± 1 Jackknife SEM)
Table 6.
Inter- and intra-population narrow sense heritabilities (h2) related to incidence of cross failure
| Dam |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bhopal | cSM | Dar es salaam | ||||||||||
| Sire | h2 | SEM | CI 95% | h2 | SEM | CI 95% | h2 | SEM | CI 95% | |||
| Bhopal | 0.382 | ± | 0.22 | -0.052, 0.816 | 0.281 | ± | 0.15 | -0.017, 0.579 | 0.157 | ± | 0.15 | -0.137, 0.452 |
| cSM | 0.305 | ± | 0.23 | -0.159, 0.768 | 0.000 | ± | 0.00 | 0.00 | 0.571 | ± | 0.17 | 0.239, 0.903 |
| Dar es salaam | 0.147 | ± | 0.09 | -0.033, 0.327 | 0.000 | ± | 0.00 | 0.00 | 0.000 | ± | 0.00 | 0.00 |
Values in bold face are significantly greater than 0
Within each population, we observed significant variation among sires in the frequency of successful crosses and much of the variation in male success was associated with the population from which the dams were chosen. For example, for Bhopal sires, on average the fewest failed crosses occurred with intra-population, Bhopal females, while the failure rate was higher in crosses to females from the other two populations. However, some Bhopal males showed the opposite pattern, namely, a higher rate of cross failure with intra-population rather than inter-population females. Overall, males with the highest fitness within Bhopal have the lowest fitness in inter-population crosses; this is especially evident in crosses to c-SM dams. If selection within the Bhopal population were to continue on the basis of the male fitness differences with intra-population females (Figure 1), the degree of reproductive incompatibility (measured by cross failure) between Bhopal and each of the other two populations would increase over time as a correlated response. The significantly elevated frequency of failed crosses indicates that isolation in at least one direction is developing between the c-SM and Dar es Salaam populations and between the c-SM and Bhopal populations.
Discussion
As in earlier studies, our results show that the frequency of successful crosses and the mean number of offspring in hybrid crosses is significantly reduced for some population pairs. In addition, our results show that there is heritable variation segregating within populations affecting the reproductive success of inter-population hybridizations. We found significant sire by dam-population variation for both hybrid offspring numbers and crossing success (Tables 1 and 5; Figure 1). In every population, some sires had lower half-sib fitness when crossed to allopatric females than they did when crossed to sympatric females and vice versa. Because the paternal half-sib means changed rank order between the sympatric and allopatric backgrounds, the genetic correlations across backgrounds are significantly less than one (Table 4, Figures 2-4). We did not detect any significantly negative genetic covariances for breeding values estimated on the intra-population and inter-population backgrounds. This is expected from the pleiotropic consequence of adaptive genetic changes occurring within a population. This could be owing to a lack of statistical power with only forty sires per cross. However, if genes responsible for the hybrid incompatibilities are neutral in the parent or ancestral populations, then the expected genetic covariance would be zero, rather than one with only additive gene action or negative one with epistasis and divergent selection. We should also emphasize that the estimated genetic covariances represent an average across all contributing genes. Thus, although divergently selected genes contributing to Dobzhansky-Muller incompatibilities should exhibit a negative genetic covariance between intra-population and inter-population backgrounds, for other segregating genes, including additively acting genes, we would expect a positive genetic covariance. A preponderance of the latter would result in an average genetic covariance near to or exceeding zero despite the presence of some Dobzhansky-Muller incompatibilities sufficient to affect the hybrid means (see discussion in Johnson and Wade (1996)). The greater the divergence between genetic architectures, the more likely we are to observe a negative genetic covariance across backgrounds. Given the modest level of genetic divergence and the ability of all population pairs (though not all sire-dam pairs) to produce some viable, fertile hybrid offspring, it might be unreasonable to expect to find a negative genetic covariance in fitness at this early stage in the process of speciation.
Lastly, we also found that the heritability of adult offspring numbers varied with background (Tables 2, 3, and 6). All three findings are evidence that gene interactions are playing a role in the genetic differentiation among these Tribolium populations and are affecting hybrid fitness.
Our findings illustrate the importance of the evolution of divergent genetic architectures among geographically isolated populations and how the subsequent change in allelic effects is important in the initial stages of speciation. Geographically isolated populations experience genetic changes independent of one another, allowing the development of differing genetic architectures (co-adapted gene complexes). Upon secondary contact, the fitnesses of genes evolving in a sympatric background are tested on an independently derived allopatric background. Given different genetic architectures, alleles established in one population with positive or neutral effects on fitness may now have effects that range from mildly deleterious to lethal in an allopatric background. Our results in combination with the earlier findings of Demuth and Wade (2007 a and b) indicate that segregating genes as well as genes fixed between semi-allopatric populations are contributing to hybrid fitness reduction in T. castaneum.
In the classic Dohzhansky-Muller model, reproductive isolation between allopatric daughter populations descended from a common ancestor increases as an indirect effect of the process of new genes being introduced by mutation and subsequently becoming fixed by natural selection (Charlesworth et al., 1987). This process of adaptive fixation of novel mutants is necessarily a slow process because the waiting time for a mutation with a positive effect on fitness at a specific locus is approximately the inverse of the adaptive mutation rate. In addition, the probability of fixation of a new mutant ‘good gene’ is only 2s (Kimura, 1962), so that advantageous genes must be introduced many times by mutation before they are fixed (Orr, 2010). Adaptive fixations are believed to be sufficiently rare that it is routine in theoretical treatments (e.g., Gillespie 2004; Orr 2010) to assume that they happen sequentially, one at a time. Since only a portion of adaptive gene substitutions in different allopatric populations result in a decrease in fitness when the genes are brought together in hybrids and several such incompatible gene pairs are believed necessary for the evolution of complete reproductive isolation, the process of speciation by mutation, selection and fixation in allopatry will necessarily be very slow.
If the necessary genetic variation were segregating in an ancestral population, that variation could be partitioned between allopatric daughter populations by the combined action of random genetic drift and natural selection. This partitioning of standing genetic variation can occur at a faster rate than the introduction of de novo adaptive variation by mutation which must subsequently be fixed by selection. This would allow DBM incompatibilities to accumulate between daughter populations faster than the lower limit imposed by the waiting time for multiple adaptive mutations. Additive × additive epistasis specifically, a type of interaction with the necessary property that some allele combinations at multiple loci have higher fitness than others, could serve as the basis for DBM incompatibilities (Demuth & Wade, 2005). Only additive by additive epistasis changes the average additive effect of alleles at one locus when the other locus is fixed and allows changes in the sign of allelic effects at one locus when alternative alleles at the other locus are fixed (see Goodnight 1995, 2000 for a derivation or Figure 1 in Wade 2002 for a graphical illustration of the change in the additive effect of alleles at one locus resulting from fixing one of two interacting loci on the types of epistasis.) In addition, several theoretical investigations have shown that, with additive × additive epistasis, stabilizing selection on a quantitative trait can maintain substantial genetic variation, but not additive genetic variance, in a population at equilibrium, even in the absence of mutations (Crow & Kimura, 1970; Gimelfarb, 1989; Gavrilets & de Jong, 1993). (Although dominance × dominance epistasis could also maintain variation at equilibrium, fixing alleles at one or the other of two such interacting loci does not subsequently increase the additive genetic variance at the remaining segregating locus [Goodnight 1995, 2000; Figure 1 in Wade 2002].) It is this type of epistasis that causes the sire-by-dam-population interaction which we observed in these population crosses. This finding supports the inference that allelic variation segregating at interacting loci within each of two allopatric populations can contribute to DMB incompatibilities in the same way as interacting alleles fixed between them.
Acknowledgments
We thank N. Johnson for his helpful comments throughout this work as well as B. Furomoto and A. Siniard for help with animal rearing. We acknowledge NSF-IGERT Grant DGE-0504627 for supporting DWD and the support of NIH grant 5R01GM65414-4 to MJW.
References
- Banos G, Shook GE. Genotype by environment interaction and genetic correlations among parities for somatic cell count and milk yield. J Dairy Sci. 1990;73:2563–2573. doi: 10.3168/jds.S0022-0302(90)78942-4. [DOI] [PubMed] [Google Scholar]
- Carson HL. Genetics of Speciation at Diploid Level. American Naturalist. 1975;109:83–92. [Google Scholar]
- Charlesworth B, Coyne JA, Barton NH. The Relative Rates of Evolution of Sex Chromosomes and Autosomes. The American Naturalist. 1987;130:113–146. [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sinauer Associates; Sunderland, Mass.: 2004. [Google Scholar]
- Crow JF, Kimura M. An introduction to population genetics theory. Harper & Row; New York: 1970. [Google Scholar]
- Damon RA, Crown RM, Mccraine SE, Singletary CB, Harvey WR. Genetic Analysis of Crossbreeding Beef Cattle. Journal of Animal Science. 1961;20:849–&. [Google Scholar]
- Demuth JP, Wade MJ. On the theoretical and empirical framework for studying genetic interactions within and among species. American Naturalist. 2005;165:524–536. doi: 10.1086/429276. [DOI] [PubMed] [Google Scholar]
- Demuth JP, Wade MJ. Population differentiation in the beetle Tribolium castaneum. I. Genetic architecture. Evolution. 2007;61:494–509. doi: 10.1111/j.1558-5646.2007.00048.x. [DOI] [PubMed] [Google Scholar]
- Drury DW, Siniard AL, Wade MJ. Genetic Differentiation among Wild Populations of Tribolium castaneum Estimated Using Microsatellite Markers. Journal of Heredity. 2009;100:732–741. doi: 10.1093/jhered/esp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzo MA. Covariances among sire by breed group of dam interaction effects in multibreed sire evaluation procedures. Journal of Animal Science. 1990;68:4079–4099. doi: 10.2527/1990.68124079x. [DOI] [PubMed] [Google Scholar]
- Falconer DS. The Problem of Environment and Selection. The American Naturalist. 1952;86:293. [Google Scholar]
- Gavrilets S, de Jong G. Pleiotropic models of polygenic variation, stabilizing selection, and epistasis. Genetics. 1993;134:609–625. doi: 10.1093/genetics/134.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JH. Population Genetics : A Concise Guide. 2nd edn. The Johns Hopkins University Press; Baltimore: 2004. [Google Scholar]
- Gimelfarb A. Genotypic variation for a quantitative character maintained under stabilizing selection without mutations: epistasis. Genetics. 1989;123:217–227. doi: 10.1093/genetics/123.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodnight CJ. Epistasis and the Increase in Additive Genetic Variance - Implications for Phase-1 of Wrights Shifting-Balance Process. Evolution. 1995;49:502–511. doi: 10.1111/j.1558-5646.1995.tb02282.x. [DOI] [PubMed] [Google Scholar]
- Goodnight CJ. Quantitative trait loci and gene interaction: the quantitative genetics of metapopulations. Heredity. 2000;84:587–598. doi: 10.1046/j.1365-2540.2000.00698.x. [DOI] [PubMed] [Google Scholar]
- Goodnight CJ, Wade MJ. The ongoing synthesis: A reply to Coyne, Barton, and Turelli. Evolution. 2000;54:317–324. doi: 10.1111/j.0014-3820.2000.tb00034.x. [DOI] [PubMed] [Google Scholar]
- Goodnight CJ. Lamkey K, editor. Gene interaction and selection. Long Term Selection: A Celebration Of 100 Years Of Selection For Oil And Protein In Maize. 2004.
- Goodnight CJ. Multilevel selection: the evolution of cooperation in non-kin groups. Population Ecology. 2005;47:3–12. [Google Scholar]
- Ibanez-Escriche N, Fernando RL, Toosi A, Dekkers JC. Genomic selection of purebreds for crossbred performance. Genetics Selection Evolution. 2009;41:12. doi: 10.1186/1297-9686-41-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson NA, Wade MJ. Genetic covariances within and between species: Indirect selection for hybrid inviability. Journal of Evolutionary Biology. 1996;9:205–214. [Google Scholar]
- Kimura M. On the probability of fixation of mutant genes in a population. Genetics. 1962;47:713–719. doi: 10.1093/genetics/47.6.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp SJ, Bridges WC, Jr, Yang MH. Nonparametric Confidence Interval Estimators for Heritability and Expected Selection Response. Genetics. 1989;121:891–898. doi: 10.1093/genetics/121.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manly BFJ. Randomization and Monte Carlo methods in biology. 1st edn. Chapman and Hall, London; New York: 1991. [Google Scholar]
- Mayr E. Animal species and evolution. Belknap Press of Harvard University Press; Cambridge: 1963. [Google Scholar]
- Mccauley DE, Wade MJ. The Populational Effects of Inbreeding in Tribolium. Heredity. 1981;46:59–67. [Google Scholar]
- Orr HA. The Population-Genetics of Speciation - the Evolution of Hybrid Incompatibilities. Genetics. 1995;139:1805–1813. doi: 10.1093/genetics/139.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. The population genetics of beneficial mutations. Philos Trans R Soc Lond B Biol Sci. 2010;365:1195–1201. doi: 10.1098/rstb.2009.0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer ME, Feldman MW. Dynamics of Hybrid Incompatibility in Gene Networks in a Constant Environment. Evolution. 2009;63:418–431. doi: 10.1111/j.1558-5646.2008.00577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poivey JP, Cheng YS, Rouvier R, Tai C, Wang CT, Liu HL. Genetic parameters of reproductive traits in Brown Tsaiya ducks artificially inseminated with semen from Muscovy drakes. Poult Sci. 2001;80:703–709. doi: 10.1093/ps/80.6.703. [DOI] [PubMed] [Google Scholar]
- Robinson T, Johnson NA, Wade MJ. Postcopulatory, Prezygotic Isolation - Intraspecific and Interspecific Sperm Precedence in Tribolium Spp, Flour Beetles. Heredity. 1994;73:155–159. doi: 10.1038/hdy.1994.114. [DOI] [PubMed] [Google Scholar]
- Roff DA. The evolution of threshold traits in animals. Quarterly Review of Biology. 1996;71:3–35. [Google Scholar]
- Roff DA. Evolutionary quantitative genetics. Chapman & Hall; New York: 1997. [Google Scholar]
- Roff DA. Introduction to computer-intensive methods of data analysis in biology. Cambridge University Press; Cambridge: 2006. [Google Scholar]
- Roger M, Jilek AF, Burns WC, Crockett JR. Sire Effects for Specific Combining Ability in Purebred and Crossbred Cattle. Journal of Animal Science. 1975;40:230–234. [Google Scholar]
- Simons AM, Roff DA. The Effect of Environmental Variability on the Heritabilities of Traits of a Field Cricket. Evolution. 1994;48:1637–1649. doi: 10.1111/j.1558-5646.1994.tb02201.x. [DOI] [PubMed] [Google Scholar]
- Sokoloff A. The biology of Tribolium: with special emphasis on genetic aspects. Clarendon Press; Oxford: 1972. [Google Scholar]
- Templeton AR. The Theory of Speciation Via the Founder Principle. Genetics. 1980;94:1011–1038. doi: 10.1093/genetics/94.4.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade CM, James JW. Genetic Interaction between Sire and Population of Mates in Drosophila-Melanogaster. Genetics Selection Evolution. 1994;26:253–261. [Google Scholar]
- Wade MJ. Group selections among laboratory populations of Tribolium. Proc Natl Acad Sci U S A. 1976;73:4604–4607. doi: 10.1073/pnas.73.12.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade MJ. Experimental-Study of Group Selection. Evolution. 1977;31:134–153. doi: 10.1111/j.1558-5646.1977.tb00991.x. [DOI] [PubMed] [Google Scholar]
- Wade MJ, Mccauley DE. Group Selection - the Interaction of Local Deme Size and Migration in the Differentiation of Small Populations. Evolution. 1984;38:1047–1058. doi: 10.1111/j.1558-5646.1984.tb00375.x. [DOI] [PubMed] [Google Scholar]
- Wade MJ. Genotype-Environment Interaction for Climate and Competition in a Natural-Population of Flour Beetles, Tribolium-Castaneum. Evolution. 1990;44:2004–2011. doi: 10.1111/j.1558-5646.1990.tb04306.x. [DOI] [PubMed] [Google Scholar]
- Wade MJ. Genetic Variance for Rate of Population Increase in Natural-Populations of Flour Beetles, Tribolium Spp. Evolution. 1991;45:1574–1584. doi: 10.1111/j.1558-5646.1991.tb02664.x. [DOI] [PubMed] [Google Scholar]
- Wade MJ, Goodnight CJ. Wright Shifting Balance Theory - an Experimental-Study. Science. 1991;253:1015–1018. doi: 10.1126/science.1887214. [DOI] [PubMed] [Google Scholar]
- Wade MJ, Johnson NA. Reproductive Isolation between 2 Species of Flour Beetles, Tribolium-Castaneum and T-Freemani - Variation within and among Geographical Populations of T-Castaneum. Heredity. 1994;72:155–162. doi: 10.1038/hdy.1994.22. [DOI] [PubMed] [Google Scholar]
- Wade MJ, Johnson NA, Wardle G. Analysis of Autosomal Polygenic Variation for the Expression of Haldanes Rule in Flour Beetles. Genetics. 1994;138:791–799. doi: 10.1093/genetics/138.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade MJ. Epistasis, complex traits, and mapping genes. Genetica. 2001;112:59–69. [PubMed] [Google Scholar]
- Wright S. Evolution and the Genetics of Populations: Genetics and Biometric Foundations v. 3 (Experimental Results and Evolutionary Deductions) University of Chicago Press; Chicago: 1977. [Google Scholar]