Abstract
The basic helix–loop–helix transcription factor TAL1 (or SCL) is a critical regulator of hematopoietic and vascular development and is misexpressed in the majority of patients with T-cell acute lymphoblastic leukemia. We found previously that TAL1 could interact with transcriptional co-activator and co-repressor complexes possessing histone acetyltransferase and deacetylase activities, respectively. Here, we report that TAL1 is subject to acetylation in vivo and can be acetylated by p300 and the p300/CBP-associated factor P/CAF in vitro. P/CAF-mediated acetylation, which mapped to a lysine-rich motif in the loop region, increased TAL1 binding to DNA while selectively inhibiting its interaction with the transcriptional co-repressor mSin3A. Furthermore, P/CAF protein, TAL1–P/CAF interaction and TAL1 acetylation increased significantly in murine erythroleukemia cells induced to differentiate in culture, while enforced expression of an acetylation-defective P/CAF mutant inhibited endogenous TAL1 acetylation, TAL1 DNA-binding activity, TAL1-directed transcription and terminal differentiation of these cells. These results reveal a novel mechanism by which TAL1 activity is regulated and implicate acetylation of this transcription factor in promotion of erythroid differentiation.
Keywords: co-regulator interactions/DNA-binding activity/erythroid differentiation/protein acetylation/TAL1
Introduction
Acetylation of core histones has long been associated with transcriptionally active chromatin (reviewed in Wolffe and Pruss, 1996; Grunstein, 1997; Struhl, 1998). A critical advance in an understanding of this observation came with the discovery that a number of transcriptional co-activators, including GCN5 (Brownell et al., 1996), TAFII250 (Mizzen et al., 1996), p300 and the related CREB-binding protein (CBP) (Bannister and Kouzarides, 1996; Ogryzko et al., 1996), p300/CBP-associated factor (P/CAF) (Yang et al., 1996), ACTR (Chen et al., 1997) and steroid receptor co-activator-1 (SRC-1) (Spencer et al., 1997), possess a histone acetyltransferase (HAT) activity required for their function. The recognition that a mammalian histone deacetylase (HDAC) was the homolog of the yeast co-repressor RPD3 (Taunton et al., 1996) has led, in turn, to the concept that transcriptional activation and repression result from the opposing actions, respectively, of HATs and HDACs (reviewed in Glass and Rosenfeld, 2000). Histone acetylation is believed to destabilize local nucleosomal structure, thereby allowing transcription factors and the basal transcriptional machinery access to DNA (Hebbes et al., 1988). HATs may, in addition, have effects that extend beyond chromatin opening (Li et al., 1999).
A number of nuclear proteins beside histones are subject to reversible acetylation. These include transcription factors (Gu and Roeder, 1997; Boyes et al., 1998; Sakaguchi et al., 1998; Zhang and Bieker, 1998; Hung et al., 1999; Liu et al., 1999; Sartorelli et al., 1999; Martínez-Balbás et al., 2000), transcriptional co-regulators (Chen et al., 1999), retroviral gene products (Kiernan et al., 1999; Ott et al., 1999) and other chromatin components (Munshi et al., 1998; Herrera et al., 1999; Bergel et al., 2000). Analogously to phosphorylation, this post-translational modification appears to modulate diverse protein functions involving DNA recognition, protein–protein interaction, protein stability and transcriptional potency (reviewed in Kouzarides, 2000). With few exceptions (Sakaguchi et al., 1998; Hung et al., 1999; Liu et al., 1999; Sartorelli et al., 1999), however, the biological significance of acetylation of these non-histone substrates has not been determined.
The TAL1 (or SCL) gene encodes a critical regulator of normal and leukemic hematopoiesis. A member of the basic helix–loop–helix (bHLH) family of transcription factors, TAL1 is frequently misexpressed in patients with T-cell acute lymphoblastic leukemia (T-ALL) as a result of chromosomal rearrangement (Begley et al., 1989; Finger et al., 1989; Chen et al., 1990) and has been found to act as an oncoprotein in transgenic models (Condorelli et al., 1996; Kelliher et al., 1996). Gene targeting and overexpression studies have shown that TAL1 also has an essential role in hematopoietic and vascular development, promotes erythroid and megakaryocytic differentiation and inhibits myeloid differentiation (Aplan et al., 1992; Condorelli et al., 1995; Robb et al., 1995; Shivdasani et al., 1995; Elwood et al., 1998). Similarly to other tissue-restricted bHLH proteins, TAL1 binds a specific DNA sequence motif, the E-box, as a heterodimer with one of a group of more widely expressed bHLH proteins, including E12, E47, E2-2/ITF2 and HEB/HTF4 (reviewed in Massari and Murre, 2000).
Reflecting an ability to associate with transcriptional co-activators and co-repressors, TAL1 is capable of both stimulating and repressing transcription. In particular, its transcriptional potency is enhanced by p300/CBP (Huang et al., 1999) and the LIM domain proteins LMO1 and LMO2 (Ono et al., 1997, 1998) and decreased by a complex of the co-repressor mSin3A and HDAC1 (Huang and Brandt, 2000). Given its interaction with a protein possessing acetyltransferase activity (Huang et al., 1999), we investigated whether TAL1 was subject to acetylation. We report that TAL1 is acetylated differentially by the co-activators P/CAF and p300, that the effect of P/CAF-stimulated acetylation is to enhance TAL1 binding to DNA while destabilizing its interaction with mSin3A, and that P/CAF acetyltransferase activity contributes importantly to erythroleukemia cell differentiation. These studies reveal a novel mechanism by which TAL1 function is regulated and implicate transcription factor acetylation in a specific cellular differentiation program.
Results
P/CAF associates with TAL1 in vitro and in vivo
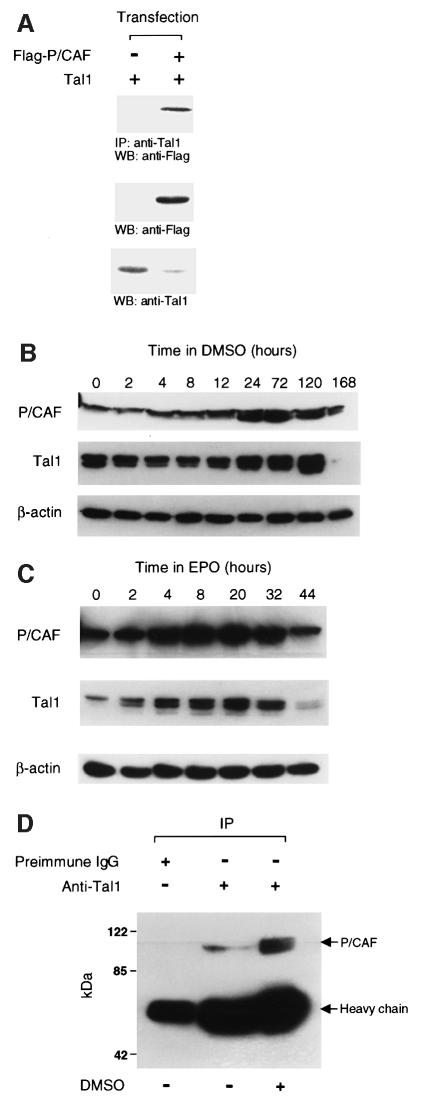
Prompted by its ability to associate with p300 (Huang et al., 1999), we investigated whether TAL1 would interact similarly with the p300/CBP-associated factor P/CAF, first identified through its competition with E1A for binding to p300/CBP (Yang et al., 1996). To determine whether P/CAF could co-immunoprecipitate with TAL1 in vivo, HeLa cells were transfected with a TAL1 expression vector, with and without a vector encoding Flag-tagged P/CAF. Cellular extracts were then immunoprecipitated with antibody to TAL1 and the resulting immunoprecipitates subjected to western blot analysis with a Flag epitope-specific antibody. In accord with results of immunolocalization studies (not shown), P/CAF immunoprecipitated with TAL1 only in extracts containing both proteins (Figure 1A).
Fig. 1. P/CAF co-immunoprecipitates with TAL1 in cellular extracts. (A) Co-immunoprecipitation of P/CAF with TAL1 in transfected HeLa cells. HeLa cells were transfected with expression vectors for TAL1 or Flag-tagged P/CAF, cellular extracts immunoprecipitated with TAL1 antibody, and immunoprecipitates subjected to western blot (WB) analysis with a Flag epitope-specific antibody. P/CAF protein was identified only in extracts from TAL1- and P/CAF-co-transfected cells (top). Direct western blot analysis confirmed that both P/CAF (middle) and TAL1 (bottom) proteins were expressed in transfected cells. (B) Increase in P/CAF and TAL1 expression in differentiating MEL cells. Lysates of MEL cells incubated with 1.8% DMSO for the indicated number of hours were fractionated by SDS–PAGE and transferred to a PVDF membrane, which was incubated sequentially with antibodies to P/CAF (top), TAL1 (middle) and β-actin (bottom). Antibody binding was visualized by enhanced chemiluminescence. Multiple forms of TAL1 are evident, reflecting phosphorylation, alternative translational initiation (‘leaky’ ribosomal scanning), acetylation or any combination of these. (C) Increase in P/CAF and TAL1 expression in Epo-stimulated FVA cells. Lysates of FVA cells incubated with Epo for the indicated number of hours were subjected to immunoblot analysis as described above. (D) Co-immunoprecipitation of P/CAF with TAL1 in extracts from differentiating MEL cells. Cellular extracts from 2 × 106 MEL cells cultured for 5 days with (+) or without (–) 1.8% DMSO were incubated with TAL1 pre-immune IgG or Tal1 antibody and the resulting immunoprecipitates subjected to western blot analysis with P/CAF-specific antibody.
To substantiate that the P/CAF–TAL1 interaction could occur in cells that express these gene products physiologically, a similar co-immunoprecipitation analysis was carried out on cultured murine erythroid leukemia (MEL) cells. Preliminary studies in MEL cells induced to differentiate with the chemical compound dimethylsulfoxide (DMSO) (Tsiftsoglou and Wong, 1985) and Friend virus-induced proerythroblasts treated with erythropoietin (Epo) (Koury et al., 1984) showed that P/CAF was expressed by these cells and that steady-state concentrations of TAL1 and P/CAF increased in parallel in these well-characterized models of terminal erythroid differentiation (Figure 1B and C). Co-immunoprecipitation analysis then showed that P/CAF interacted with TAL1 in MEL cellular extracts, with P/CAF–TAL1 interaction strongly increasing with DMSO-stimulated differentiation of these cells (Figure 1D), and in extracts from Epo-treated FVA cells (not shown). The increased amounts of P/CAF that co-immunoprecipitated with TAL1 with induced differentiation of these cells could have resulted from the increase in concentration of TAL1 and P/CAF that accompanies this process (Figure 1B and C), an increase in the affinity of one for the other, or both. In any event, these data demonstrate that TAL1 and P/CAF, expressed either endogenously or following transfection, interact in vivo and that their association in erythroid cells is influenced by the extent of cellular differentiation.
To characterize the region of TAL1 involved in this interaction, glutathione S-transferase (GST) pull-down assays were carried out with a series of bacterially expressed GST–TAL1 fusion proteins and in vitro translated P/CAF (Figure 2A). Fusions containing, or even limited to, the bHLH domain were able to bind radiolabeled P/CAF, whereas no binding was noted to beads adsorbed with GST alone or fusion proteins containing the N-terminal 144 or C-terminal 88 amino acids of TAL1 (Figure 2A). As the different fusion proteins used in these studies were added in comparable amounts (Figure 2A, bottom), their ability to mediate P/CAF binding was attributable to the specific TAL1 protein sequence included rather than to their overall abundance.
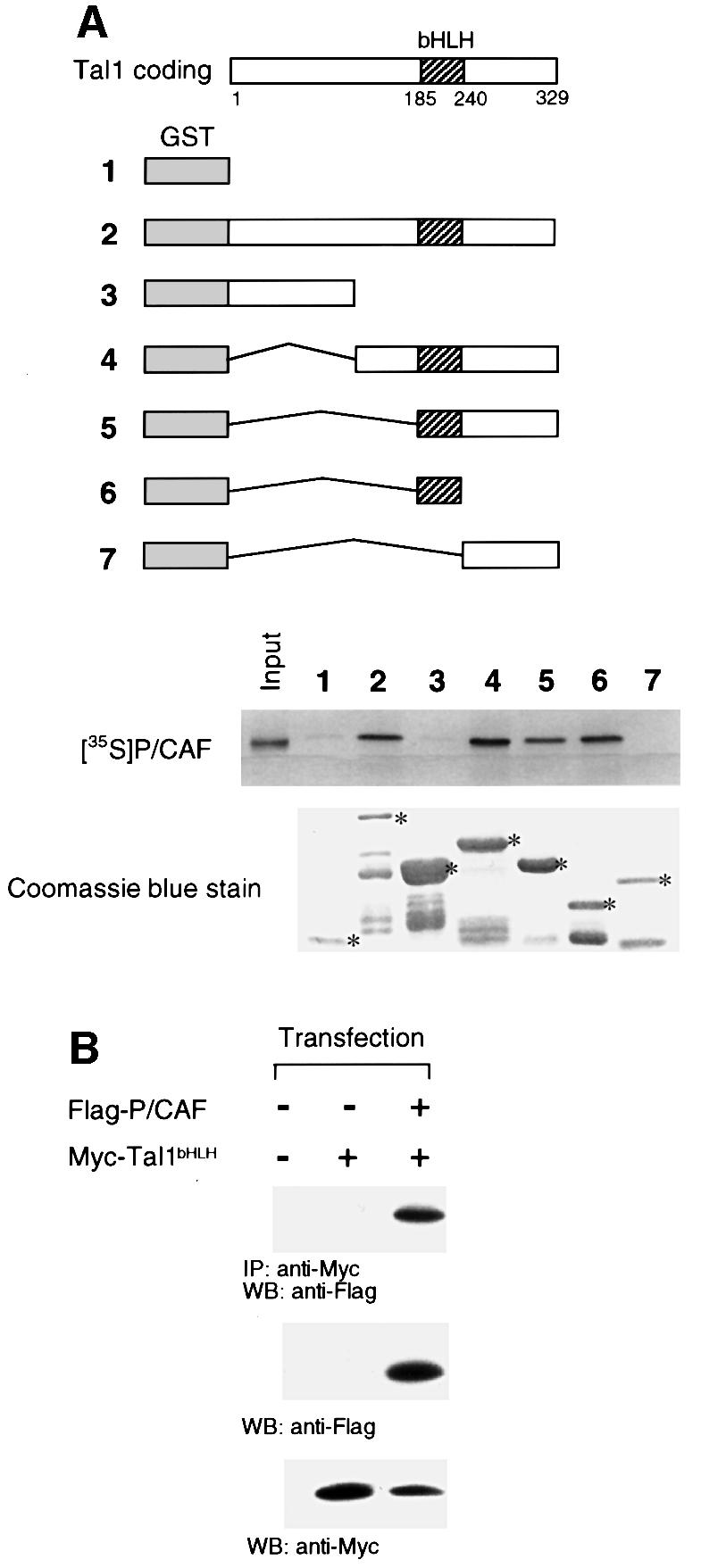
Fig. 2. The bHLH domain of TAL1 is necessary and sufficient for its interaction with P/CAF. (A) Schematic representation of GST–TAL1 fusion proteins used in GST pull-down assays. Constructs 2–7 contain amino acids 1–329, 1–144, 142–329, 185–329, 185–240 and 242–329, respectively, of the TAL1 coding sequence. 35S-labeled P/CAF was incubated with GST or GST–TAL1 proteins pre-adsorbed to glutathione–Sepharose beads. Specifically bound P/CAF was eluted from washed beads and visualized by fluorography following SDS–PAGE. Input represents 10% of the in vitro translated P/CAF protein used in the assay. The relative amounts of fusion proteins used (denoted by asterisks) are shown in the accompanying photograph of a Coomassie Blue-stained SDS–polyacrylamide gel. (B) HeLa cells were transfected with expression vectors for Myc-tagged TAL1 bHLH polypeptide (Myc-Tal1bHLH) or Flag-tagged P/CAF (Flag-P/CAF), cellular extracts immunoprecipitated (IP) with antibody to Myc epitope, and immunoprecipitates subjected to western blot (WB) analysis with antibody to the Flag epitope. P/CAF protein was identified only in extracts from TAL1 bHLH- and P/CAF-co-transfected cells (top). Direct western blot (WB) analysis confirmed that both P/CAF (middle) and TAL1 (bottom) were expressed in transfected cells.
To determine whether the TAL1 bHLH domain was also sufficient to mediate P/CAF interaction intracellularly, Flag-tagged P/CAF and Myc-tagged TAL1 bHLH expression vectors were co-transfected into HeLa cells and extracts of these cells subjected to co-immunoprecipitation analysis. Similarly to results obtained with full-length TAL1 (Figure 1A), P/CAF and this TAL1 bHLH polypeptide co-immunoprecipitated only in cells expressing both proteins (Figure 2B). Taken together, these data indicate that the bHLH domain, which mediates TAL1’s association with p300 (Huang et al., 1999), mSin3A (Huang and Brandt, 2000) and the LIM-only proteins (Wadman et al., 1994), is both necessary and sufficient for its interaction with P/CAF.
TAL1 is acetylated by P/CAF in vitro and in vivo
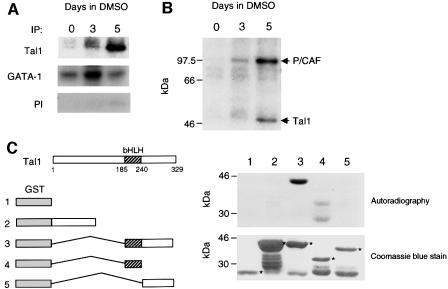
Given its ability to associate with two acetyltransferases, p300 (Huang et al., 1999) and P/CAF (Figure 1), we investigated whether TAL1 could also serve as their substrate. MEL cells incubated with or without DMSO were pulse-labeled with sodium [3H]acetate in the presence of the specific HDAC inhibitor trichostatin A (TSA) and then subjected to immunoprecipitation with TAL1 antibody. As a control, extracts were also immunoprecipitated with an antibody to GATA-1, which previously had been shown to undergo acetylation in erythroid cells (Boyes et al., 1998; Hung et al., 1999). Acetylated forms of both TAL1 and GATA-1 were immunoprecipitated in extracts of DMSO-induced MEL cells, with GATA-1 acetylation peaking at 3 days and then declining, while TAL1 acetylation increased progressively over 5 days of culture (Figure 3A). When adjusted for the change in TAL1 protein concentration, TAL1 acetylation increased a mean of 3.3- and 10.2-fold at 3 and 5 days, respectively, after addition of DMSO to culture. Significantly, a radiolabeled protein with the mobility predicted for P/CAF was also precipitated with TAL1 in extracts of differentiating MEL cells (Figure 3B). As P/CAF interacts with TAL1 (data herein) but not GATA-1 (Hung et al., 1999) and is known to undergo autoacetylation (Herrera et al., 1997; Puri et al., 1997; Sartorelli et al., 1999), it is likely that this protein represents P/CAF itself.
Fig. 3. TAL1 is acetylated by P/CAF in vitro and in vivo. (A) TAL1 acetylation in vivo. MEL cells treated with DMSO for the indicated numberof days were pulse-labeled with [3H]acetate in the presence of 50 nM TSA and lysed. Lysates were subjected to immunoprecipitation with TAL1 antibody (top), GATA-1 antibody (middle) or TAL1 pre-immune IgG (bottom). The resulting immunoprecipitates were fractionated by SDS–PAGE and analyzed for incorporation of [3H]acetate by autoradiography. (B) Replicate experiment of the above in which MEL cells were labeled with [3H]acetate and cellular extracts immunoprecipitated with TAL1 antibody. In addition to TAL1, a radiolabeled protein with the mobility expected for P/CAF was precipitated, consistent with P/CAF autoacetylation. (C) P/CAF acetylation of TAL1 in vitro. Purified GST (1) and GST fusion proteins containing amino acids 1–144 (2), 185–329 (3), 185–240 (4) and 242–329 (5) of the TAL1 coding sequence were incubated with [14C]acetyl-CoA and baculovirus-expressed P/CAF. Reaction products were fractionated by SDS–PAGE and the dried gel exposed to X-ray film. The relative amounts of fusion proteins used (denoted by asterisks) are shown in the accompanying photograph of a Coomassie Blue-stained SDS–polyacrylamide gel.
Finally, to determine whether TAL1 could be acetylated by P/CAF directly, purified GST–TAL1 fusion proteins were used as substrates in an in vitro acetylation assay with baculovirally expressed P/CAF (Gu and Roeder, 1997). Mirroring the results of the GST pull-down assay (Figure 2A), only TAL1 fusions containing its bHLH domain (amino acids 185–240) were acetylated (Figure 3C). As comparable amounts of the different fusion proteins were used in this assay, the changes in acetylation noted were attributable to the specific TAL1 sequence present in these proteins. Thus, TAL1 is subject to acetylation in differentiating erythroid cells in a region important for its interaction with P/CAF.
A conserved motif in the loop region of TAL1 is acetylated by P/CAF
To establish the functional relevance of P/CAF-stimulated acetylation of TAL1, site-directed mutagenesis was used to identify the specific residues in the protein acetylated by P/CAF. Although a specific molecular signature has not been determined for acetylation targets, they frequently reside in regions with closely spaced lysines (Gu and Roeder, 1997; Boyes et al., 1998; Chen et al., 1999). From its homology to the KXKK and KXXKK acetylation motifs identified in p53, GATA-1 and ACTR (Gu and Roeder, 1997; Boyes et al., 1998; Chen et al., 1999; Hung et al., 1999), a sequence in the loop region of TAL1, KKLSK (amino acids 221–225), was predicted to be subject to p300- and/or P/CAF-mediated acetylation. Accordingly, mutagenesis was used to substitute arginines for Lys221 and Lys222 (K221R and K222R), while Lys225, conserved in >95% of bHLH proteins (Atchley et al., 2000), was left intact. GST fusion proteins containing wild-type sequence (GST–TAL1) or these two arginine substitutions (GST–TAL1RR) were then expressed in bacteria, equivalent concentrations were incubated with [14C]acetyl-CoA and either full-length P/CAF or the p300 HAT domain (p300-HAT), and incorporation of [14C]acetate was determined by PAGE and autoradiography. While the efficiency with which they did so are not strictly comparable, both p300-HAT and P/CAF acetylated the GST fusion protein containing the wild-type TAL1 sequence. However, only P/CAF-stimulated acetylation was significantly attenuated by the K221R and K222R substitutions (Figure 4). Thus, these HATs acetylate TAL1 at distinct sites, with lysines 221 and/or 222 representing the principal, if not exclusive, site(s) of P/CAF acetylation.
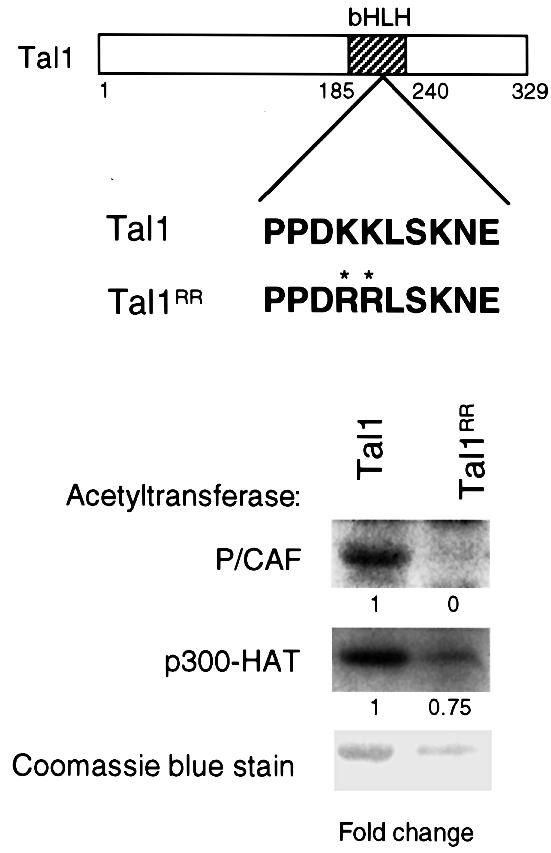
Fig. 4. TAL1 is differentially acetylated by P/CAF and p300. Purified GST fusion protein containing the complete TAL1 coding region (Tal1) and the corresponding fusion protein with arginine substitutions (K221R K222R) for the indicated lysine residues (denoted by asterisks) in the loop region of the bHLH domain (Tal1RR) were used as substrates in an in vitro acetylation assay. Baculovirally expressed P/CAF (top) or bacterially expressed p300-HAT (middle) was incubated with these fusion proteins and [14C]acetyl-CoA, and the reaction products were fractionated by SDS–PAGE and analyzed by autoradiography. The relative amount of acetylation (fold change) was adjusted for differences in fusion protein concentration as determined by Coomassie Blue staining of the gel (bottom).
P/CAF-mediated acetylation of TAL1 augments its DNA-binding activity in vitro and in vivo
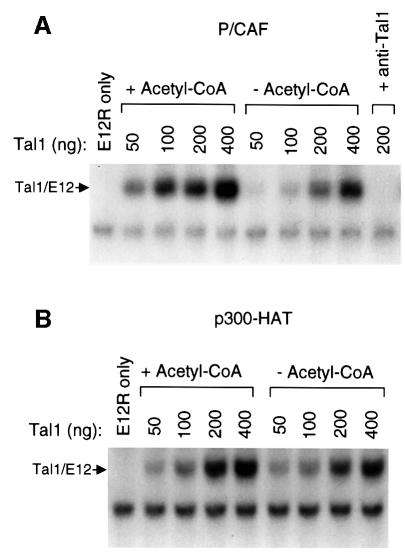
Given the stimulation of p53 (Gu and Roeder, 1997; Sakaguchi et al., 1998), MyoD (Sartorelli et al., 1999), E2F-1 (Martínez-Balbás et al., 2000) and, possibly, GATA-1 (Boyes et al., 1998) binding to DNA by acetylation, we investigated the effects on TAL1 DNA-binding activity of P/CAF- and p300-mediated acetylation. Purified GST–TAL1 and GST–TAL1RR proteins were incubated with each acetyltransferase in the presence and absence of acetyl-CoA, mixed with in vitro translated E12 protein, and then tested for their ability to bind to an E-box sequence preferred by TAL1–E2A heterodimers (Hsu et al., 1994). P/CAF acetylation of GST–TAL1 significantly increased its ability to bind with E12 to this site, which was particularly evident at lower concentrations of the TAL1 fusion (Figure 5A). Furthermore, the effect was specific to P/CAF, as p300-HAT did not increase TAL1/E12 binding to this E-box element (Figure 5B). Although the lysine to arginine substitutions made were expected to be neutral with regard to function, and another bHLH protein, the Drosophila proneural protein Amos (Goulding et al., 2000; Huang et al., 2000), contains arginines in both corresponding positions, the GST–TAL1RR mutant bound DNA with a significantly lower affinity than its wild-type counterpart (not shown), making an assessment of P/CAF’s effects on its DNA-binding activity problematic.
Fig. 5. TAL1 DNA-binding activity is augmented by P/CAF but notby p300 acetylation. (A) Purified GST–TAL1 protein in the indicated amounts and in vitro translated E12 protein (E12R) were incubated with baculovirally expressed P/CAF, with (+) or without (–) acetyl-CoA, for 1 h at 30°C. The reaction mixture was then incubated with a 32P-labeled double-stranded oligonucleotide corresponding to a preferred TAL1/E12-binding site for 30 min at room temperature, and DNA-binding activity was analyzed by EMSA. The presence of TAL1 in the binding complex marked Tal1/E12 was confirmed by the disappearance of this complex following addition of TAL1 antibody (+ anti-Tal1) to the binding reaction prior to electrophoresis. (B) Purified GST–TAL1 protein was incubated with the p300 HAT domain under acetylating and non-acetylating conditions as described in (A) and the effect of p300-stimulated acetylation on TAL1/E12 DNA binding determined by EMSA.
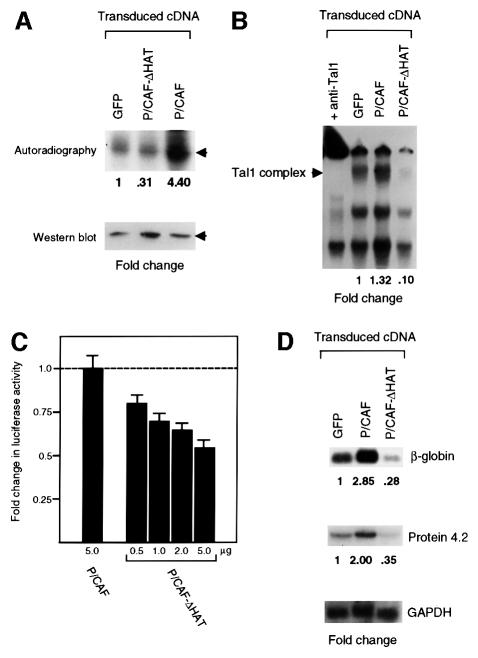
P/CAF interacts with its substrates in cells in the context of a large multiprotein complex (Ogryzko et al., 1998). To investigate the effect of P/CAF-mediated acetylation on TAL1 DNA binding in vivo, we infected MEL cells with a retroviral expression vector encoding full-length P/CAF or a dominant-negative mutant containing a deletion of its acetyltransferase domain (P/CAF-ΔHAT). Polyclonal populations of cells transduced with these cDNAs and vector controls were selected for expression of green fluorescent protein (GFP) by fluorescence-activated cell sorting (FACS). After confirming expression of the introduced proteins by immunoblot analysis (data not shown), transduced cells were treated with 1.8% DMSO for 5 days, pulse-labeled with [3H]acetate in the presence of TSA, and extracted for immunoprecipitation with TAL1 antibody. As shown in Figure 6A, enforced expression of wild-type P/CAF stimulated TAL1 acetylation ∼4-fold, while that of the P/CAF-ΔHAT mutant reduced TAL1 acetylation by over two-thirds (Figure 6A). Probably even underestimating the magnitude of these effects because they were obtained with polyclonal populations, these data establish that P/CAF also stimulates TAL1 acetylation in vivo. Having determined this, we investigated whether TAL1 DNA-binding activity would be stimulated accordingly. Following incubation with 50 nM TSA for 5 h, nuclear extracts were prepared from P/CAF- and P/CAF-ΔHAT-transduced cells and tested for their ability to interact with the preferred TAL1/E2A-binding site by electrophoretic mobility shift assay (EMSA). Compared with GFP vector controls, extracts of P/CAF-transduced cells exhibited somewhat increased TAL1 DNA-binding activity, while extracts from P/CAF-ΔHAT-transduced cells showed a marked reduction in this activity (Figure 6B). Thus, these results establish that P/CAF-stimulated acetylation enhances TAL1 DNA-binding activity in vivo.
Fig. 6. P/CAF acetyltransferase activity is important for TAL1 acetylation, DNA-binding activity, transcriptional activity and erythroleukemia cell differentiation. (A) The importance of P/CAF acetyltransferase activity for TAL1 acetylation in vivo. MEL cells transduced with retroviral vectors expressing wild-type P/CAF (P/CAF), a HAT-defective P/CAF mutant (P/CAF-ΔHAT) or MSCV-IRES-GFP parental vector (GFP) were selected for GFP expression by FACS, incubated with 1.8% DMSO for 5 days, and then pulse-labeled with [3H]acetate as described in Figure 3B. Cellular extracts were subjected to immunoprecipitation with TAL1 antibody and acetylation analyzed by autoradiography. The fold increase or decrease in acetylation was related to cells transduced with empty vector (GFP) and normalized for TAL1 protein expression (bottom panel). (B) The importance of P/CAF acetyltransferase activity for TAL1 DNA-binding activity in vivo. MEL cells were transduced with the retroviral vectors described above, selected for GFP expression by FACS and incubated with 50 nM TSA for 5 h. Nuclear extracts were incubated with a 32P-labeled, double-stranded oligonucleotide corresponding to a preferred TAL1/E2A-binding site for 30 min at room temperature, and DNA-binding activity was analyzed by EMSA. The presence of TAL1 in the binding complex marked Tal1 complex was confirmed by the shift in the mobility of this complex following addition of TAL1 antibody (+ anti-Tal1) to the binding reaction prior to electrophoresis. (C) MEL cells induced to differentiate by incubation with 1.5% DMSO for 3 days were transfected with a firefly luciferase reporter linked to four copies of the GAL4 DNA-binding site and minimum TK promoter (1.0 µg), an expression vector for a GAL4–full-length TAL1 fusion protein (1.0 µg), an expression vector for either full-length P/CAF (1.0–5.0 µg) or a mutant lacking the HAT domain (0.5–5.0 µg) and an expression vector for Renilla luciferase (0.1 µg). Transfected cells and controls transfected with the reporter construct alone were returned to culture for 24 h, treated with 100 nM TSA for an additional 24 h, and lysed for measurement of luciferase activity. The luciferase activity induced by GAL4–TAL1, as corrected for reporter activity elicited by GAL41–147 and normalized for variation in transfection efficiency, was assigned a value of 1, and the fold induction or repression of reporter activity by the co-transfected P/CAF constructs was determined. Plotted is the mean fold change in luciferase activity ± SE from four independent experiments. (D) Importance of P/CAF acetyltransferase activity for DMSO-induced differentiation of MEL cells. MEL cells were transduced with the retroviral vectors described above, selected for GFP expression by FACS and incubated with 1.8% DMSO for 5 days. RNA from these cells was fractionated in a formaldehyde–agarose gel, transferred to a Nytran membrane and sequentially hybridized with radiolabeled probes for β-globin, Protein 4.2 and GAPDH. The fold change in expression of β-globin and Protein 4.2 mRNAs was determined relative to vector-transduced cells and normalized for GAPDH mRNA expression.
P/CAF regulates TAL1-directed transcriptional activity
Given TAL1’s increased association with and acetylation by P/CAF with MEL cell differentiation, we investigated whether TAL1-directed transcription would be affected by P/CAF in these cells. As no target genes have been established definitively for this transcription factor, we tested the effect of co-expressed P/CAF and P/CAF-ΔHAT on transcription stimulated by a GAL4–TAL1 fusion protein from a minimum thymidine kinase promoter linked to four GAL4-binding sites. Although little or no increase in reporter activity was elicited by wild-type P/CAF transfected into DMSO-induced MEL cells, probably because this acetyltransferase was already present in considerable abundance under these conditions, the HAT-defective mutant inhibited TAL1-directed transcription in a dose-dependent manner, with almost 50% inhibition of luciferase activity noted at the highest concentration of P/CAF plasmid used (Figure 6C). These results showing quantitatively greater inhibition with the mutant than stimulation by wild-type P/CAF protein thus parallel those obtained in acetylation (Figure 6A) and DNA binding (Figure 6B) assays.
P/CAF acetyltransferase activity contributes to MEL cell differentiation
The increase in DMSO-induced MEL cells in P/CAF expression (Figure 1B) and autoacetylation (Figure 3B), and in P/CAF-stimulated TAL1 acetylation (Figure 6A) and DNA-binding activity (Figure 6B) suggested that this acetyltransferase could be required for, or at least contribute to, erythroid differentiation. To investigate this, we characterized the expression of several markers of differentiation in P/CAF-, P/CAF-ΔHAT- and vector-transduced cells treated with 1.8% DMSO for 5 days. Paralleling their effects on TAL1 acetylation (Figure 6A) and DNA-binding activity (Figure 6B), overexpression of wild-type P/CAF increased the steady-state levels of β-globin and Protein 4.2 mRNA by 3- and 2-fold, respectively, relative to vector-transduced cells (Figure 6D). In contrast, the acetylation-defective P/CAF mutant significantly decreased both β-globin and Protein 4.2 mRNA levels (Figure 6D). While P/CAF’s effects on TAL1 function may not constitute its only action, these results show that P/CAF acetyltransferase activity contributes importantly to erythroleukemia cell differentiation.
P/CAF-stimulated acetylation of TAL1 selectively impairs its interaction with the transcriptional co-repressor mSin3A
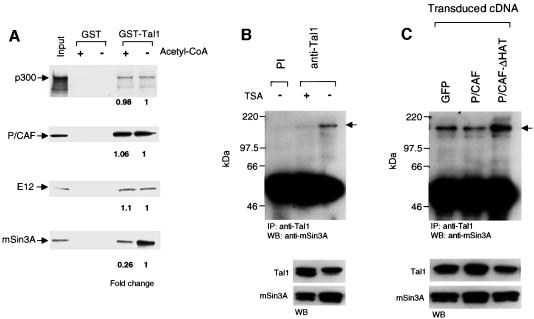
In addition to protein oligomerization and DNA binding, the bHLH domain of TAL1 mediates its interactions with a number of transcriptional co-regulators, including the LIM domain proteins LMO1 and LMO2 (Wadman et al., 1997), mSin3A (Huang and Brandt, 2000), p300 (Huang et al., 1999) and P/CAF (data herein). As acetylation influences protein–protein in addition to protein–DNA interactions, we investigated whether P/CAF-stimulated acetylation affected the ability of TAL1 to heterodimerize with E12 or to interact with any of these co-regulators, including P/CAF itself. Accordingly, a GST fusion protein encompassing the complete TAL1 coding region was incubated with P/CAF in the presence and absence of acetyl-CoA, and its interactions with p300, P/CAF, E12 and mSin3A were assessed semi-quantitatively in a GST pull-down assay. As shown in Figure 7A, P/CAF-stimulated acetylation did not alter the affinity of this GST–TAL1 fusion for E12, p300 or P/CAF, while it reduced binding of mSin3A by >4-fold.
Fig. 7. Acetylation of TAL1 by P/CAF selectively impairs its interaction with mSin3A. (A) Glutathione–Sepharose beads pre-adsorbed with GST or GST–TAL11–329 fusion protein treated with purified P/CAF in the presence or absence of acetyl-CoA were mixed with [35S]methionine-labeled p300, P/CAF, E12 or mSin3A for 1 h at room temperature. Specifically bound protein was eluted from washed beads and visualized by fluorography following SDS–PAGE. The fold difference in binding of the indicated proteins was determined from image analysis of exposed X-ray film. Input represents 10% of the radiolabeled protein used in the assay. (B) Reduced association of mSin3A with hyperacetylated TAL1 protein in vivo. Lysates of MEL cells that had been treated for 5 h with (+) or without (–) 50 nM TSA were incubated with antibody to TAL1 or TAL1 pre-immune immunoglobulin (PI) and the resulting immunoprecipitates (IP) subjected to western blot analysis with an mSin3A-specific antibody. mSin3A protein that co-immunoprecipitated with TAL1 was visualized by enhanced chemiluminescence and is marked by the arrowhead. The presence of both TAL1 and mSin3A protein in these cells was verified by direct western blot (WB) analysis of extracts. (C) The importance of P/CAF acetyltransferase activity for TAL1 interaction with mSin3A in vivo. Cellular lysates were prepared from MEL cells transduced with P/CAF, P/CAF-ΔHAT or parental retrovirus and treated with 50 nM TSA for 1 h. mSin3A protein associated with TAL1 was quantitated by co-immunoprecipitation analysis as described in (B). Comparable amounts of TAL1 and mSin3A protein were present in these cells as determined by western blot (WB) analysis of extracts.
To investigate whether acetylation of TAL1 similarly would reduce its association with mSin3A in vivo, MEL cells were cultured for 5 h with and without 50 nM TSA and extracted for co-immunoprecipitation analysis. Consistent with the results of in vitro studies, the amount of mSin3A that was immunoprecipitated with TAL1 was markedly lower in extracts from cells treated with TSA compared with vehicle (Figure 7B). To determine the extent to which TAL1 hyperacetylation in these cells was due to P/CAF-mediated modification, co-immunoprecipitation assays were also carried out in P/CAF-, P/CAF-ΔHAT- and vector-transduced MEL cells similarly treated with 50 nM TSA for 5 h. Relative to vector-transduced cells, overexpression of P/CAF significantly impaired the TAL1–mSin3A interaction, while enforced expression of P/CAF-ΔHAT significantly enhanced their association (Figure 7C). As the two P/CAF proteins interacted equally well with TAL1 in GST pull-down assays (data not shown), interference with the acetyltransferase activity of the endogenously expressed enzyme was probably the mechanism by which P/CAF-ΔHAT increased the TAL1– mSin3A interaction. Taken together with the results of the metabolic labeling studies (Figure 6A), these results demonstrate that P/CAF-mediated acetylation inhibits TAL1’s interaction with an mSin3A–HDAC1 complex present in these cells (Huang and Brandt, 2000).
Discussion
Transcribed chromatin is enriched in acetylated histones, and a considerable literature indicates that this modification, and the acetyltransferase activity of the enzymes that catalyze it, are critical for gene expression (reviewed in Kuo and Allis, 1998). In addition to histones, other nuclear proteins are subject to reversible acetylation, including the transcription factors p53 (Gu and Roeder, 1997; Sakaguchi et al., 1998; Liu et al., 1999), MyoD (Sartorelli et al., 1999), GATA-1 (Boyes et al., 1998; Hung et al., 1999), erythroid Krüppel-like factor (EKLF) (Zhang and Bieker, 1998), HNF-4 (Soutoglou et al., 2000), E2F-1 (Martínez-Balbás et al., 2000; Marzio et al., 2000), -2 (Marzio et al., 2000) and -3 (Marzio et al., 2000), the non-histone structural proteins HMG-14 (Bergel et al., 2000), HMG-17 (Herrera et al., 1999) and HMG-I(Y) (Munshi et al., 1998), the general transcription factors TFIIEβ and TFIIF (Imhof et al., 1997), and the retroviral protein Tat (Kiernan et al., 1999; Ott et al., 1999). As for histones, acetylation occurs on lysine residues, is reversible and affects the strength of protein–DNA or protein– protein interactions. With few exceptions (Sakaguchi et al., 1998; Hung et al., 1999; Liu et al., 1999; Sartorelli et al., 1999), however, the biological significance of acetylation of these non-histone substrates is unknown.
Our studies show that the hematopoietic transcription factor TAL1 is subject to acetylation in erythroid cells and can act as a substrate for two acetyltransferases, p300 and P/CAF, in vitro. P/CAF acetylation was mapped to TAL1’s bHLH domain and shown to augment its binding to DNA while inhibiting its interaction with the nuclear co-repressor mSin3A. Using DMSO-induced MEL cells as a model system, we demonstrate further that P/CAF-stimulated acetylation affects TAL1 actions and erythroleukemia cell differentiation more generally. These results provide insights into the regulation of TAL1 function and the role of non-histone protein acetylation in cellular growth and differentiation.
Both p300 and P/CAF proved capable of acetylating TAL1 in vitro, although at distinct sites and, at least with regard to DNA-binding activity, with different functional consequences. Histones H3 and H4 are acetylated similarly by both HATs (Schiltz et al., 1999), and a number of non-histone proteins (Imhof et al., 1997; Munshi et al., 1998; Sakaguchi et al., 1998; Kiernan et al., 1999; Liu et al., 1999; Martínez-Balbás et al., 2000) are substrates for two, or even three (Imhof et al., 1997), acetyltransferases. In contrast, histones H2A and H2B (Schiltz et al., 1999), and the hematopoietic transcription factors GATA-1 (Hung et al., 1999) and EKLF (Zhang and Bieker, 1998), appear to be substrates exclusively for p300/CBP. The lack of a consensus sequence for acetylation and results of studies with peptide competitors (Herrera et al., 1999) and synthetic inhibitors of HAT activity (Lau et al., 2000) suggest that site selection by P/CAF may be determined by more than contiguous amino acid sequence. Notwithstanding this, a subset of bHLH proteins, including TAL2 (Xia et al., 1991), NHLH1 and NHLH2 (Begley et al., 1992; Göbel et al., 1992), Mist1 (Lemercier et al., 1997, 1998), Mash2 (Johnson et al., 1990) and Math1 (Akazawa et al., 1995), demonstrate near identity to TAL1 in the distal loop region and could be predicted to be subject to P/CAF acetylation.
As for many of the proteins acetylated by p300/CBP or P/CAF (Boyes et al., 1998; Sakaguchi et al., 1998; Liu et al., 1999; Sartorelli et al., 1999; Martínez-Balbás et al., 2000; Marzio et al., 2000; Soutoglou et al., 2000), this post-translational modification increases TAL1 binding to DNA. X-ray crystallographic studies of the related bHLH protein MyoD (Ma et al., 1994) indicate that residues in the loop and proximal helix II contact the phosphodiester backbone of DNA directly, raising the possibility that acetylation of the corresponding region in TAL1 enhances such an interaction. The fact that multiple functions of TAL1 are affected, however, involving both protein– protein and protein–DNA interactions, is consistent with acetylation producing a more global change in protein conformation, as also suggested for GATA-1 (Boyes et al., 1998), MyoD (Sartorelli et al., 1999) and HNF-4 (Soutoglou et al., 2000).
An important finding of this work was that TAL1’s interaction with the co-repressor mSin3A (Huang and Brandt, 2000) could be significantly, and selectively, destabilized by P/CAF-mediated acetylation. Both co-activators and co-repressors have been shown to interact physically and functionally with TAL1 (Wadman et al., 1994; Huang et al., 1999; Huang and Brandt, 2000). In fact, TAL1 interacts reciprocally with mSin3A (Huang and Brandt, 2000) and with the co-activators p300 (Huang et al., 1999) and P/CAF (data herein) in the same cells, according to their stage of differentiation. While a decline in mSin3A concentration is likely to be responsible for this co-regulator switch in MEL cells (Huang and Brandt, 2000), it was not evident why TAL1 and mSin3A fail to interact in differentiating primary erythroid cells that continue to express both proteins (Huang and Brandt, 2000). These studies provide a potential explanation and define a novel mechanism for regulating transcription factor activity, involving acetylation-induced destabilization of transcription factor–co-repressor interaction. This contrasts with the reduced affinity of the transcriptional co-activator ACTR for the estrogen receptor as a result of its hormone-induced acetylation (Chen et al., 1999), and the enhanced affinity of HNF-4 for CBP as the result of its acetylation by CBP (Soutoglou et al., 2000). That TSA treatment alone destabilized the TAL1–mSin3A interaction, without the requirement for P/CAF overexpression, suggests that, as for histones (Yoshida et al., 1990), a dynamic equilibrium exists in cells between acetylation and deacetylation of non-histone substrates.
The ability of P/CAF to inhibit the TAL1–mSin3A interaction may also have implications for P/CAF’s possible function as a tumor suppressor (reviewed in Schiltz and Nakatani, 2000) and TAL1’s actions as an oncogene. In addition to counteracting E1A-mediated deregulation of cell cycle progression (Yang et al., 1996) and enhancing p53 function in response to DNA damage (Gu and Roeder, 1997; Scolnick et al., 1997; Sakaguchi et al., 1998; Liu et al., 1999), P/CAF-mediated TAL1 acetylation would serve to release a co-repressor complex that was found to be constitutively associated with this transcription factor in a T-ALL cell line (Huang and Brandt, 2000) and that can act as an oncogenic cofactor for a number of other leukemic oncoproteins (David et al., 1998; Dhordain et al., 1998; Gelmetti et al., 1998; Grignani et al., 1998; He et al., 1998; Lin et al., 1998; Lutterbach et al., 1998; Wang et al., 1998). It remains to be determined, however, if a deficiency in P/CAF acetylation is responsible for this increased interaction between TAL1 and mSin3A in leukemic cells and, in turn, whether overexpression of P/CAF would effect dissociation of mSin3A and phenotypic reversion of TAL1-expressing leukemic cells.
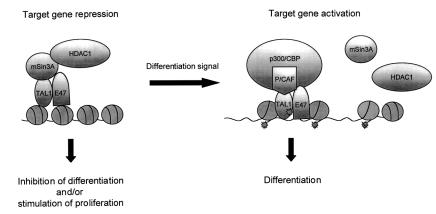
Beyond establishing the effects of acetylation on TAL1 function in vivo, the P/CAF gain- and loss-of-function experiments we carried out suggest a role for its acetyltransferase activity in erythroid differentiation. The involvement of HAT activity in this process was suggested by the rapid increase in histone acetylation observed in DMSO-treated MEL cells (Leiter et al., 1984) and Epo-stimulated proerythroblasts (Takaku et al., 1969) and, more directly, by the inhibition of MEL cell differentiation by the adenoviral protein E1A (Blobel et al., 1998), an inhibitor of multiple HATs (Arany et al., 1995; Bannister and Kouzarides, 1995; Lundblad et al., 1995; Reid et al., 1998) and other chromatin-modifying enzymes (Miller et al., 1996). As DNA binding appears to be required for TAL1 promotion of MEL cell differentiation (Aplan et al., 1992; our unpublished results) while mSin3A and HDAC1 act to oppose this action (Huang and Brandt, 2000), P/CAF’s acetylation of TAL1 could potentially account for its stimulation of erythroleukemia cell differentiation independently of its histone-acetylating and any acetylation-independent actions (Jiang et al., 1999) (Figure 8). Finally, the analogous involvement in myogenesis of P/CAF acetylation of MyoD (Sartorelli et al., 1999) suggests that transcription factor acetylation may be a general mechanism for regulating cellular differentiation.
Fig. 8. Model of TAL1–co-regulator interactions in erythroleukemia cell differentiation. TAL1 interacts with the nuclear co-repressor mSin3A and associated histone deacetylase HDAC1 in undifferentiated MEL cells. This co-repressor complex mediates TAL1-dependent transcriptional repression and restricts the ability of these cells to respond to differentiation inducers. Addition of DMSO results in formation of TAL1–P/CAF and TAL1–p300/CBP complexes and acetylation of both TAL1 and histones. P/CAF-mediated TAL1 acetylation, in addition, decreases TAL1 interaction with mSin3A-containing complexes. In combination, these effect the conversion of TAL1 from transcriptional repressor to activator and augment cellular differentiation. Although the figure shows both P/CAF and p300/CBP present in a single complex in differentiated cells, this has not been formally established.
Materials and methods
Cell culture and transfection
MEL (F4-12B2 line) and HeLa cells were cultured as previously described (Huang and Brandt, 2000). Purified Friend virus-induced proerythroblasts (Koury et al., 1984) were provided by Dr Mark Koury (Vanderbilt University, Nashville, TN) and cultured for the indicated times with 2 U/ml recombinant human Epo. HeLa cells were transfected with liposome–DNA complexes (LipofectAmine; Life Technologies, Gaithersburg, MD), washed with phosphate-buffered saline (PBS) following an additional 24–30 h of culture, and collected by low-speed centrifugation. A 5 µg aliquot of pcDNA1-Tal1, pcDNA-Myc-Tal1bHLH, pCX-P/CAF (Yang et al., 1996) or empty vector was transfected into HeLa cells for studies of TAL1–P/CAF interaction. The total mass of DNA transfected was normalized by addition of the appropriate vector plasmid.
A GAL4-thymidine kinase (GAL4-TK)-luciferase reporter was introduced using the DMRIE-C liposomal reagent (Life Technologies, Gaithersburg, MD) into cultures of MEL cells previously incubated with 1.5% DMSO for 3 days; 100 nM TSA was added 24 h later, and extracts were prepared 40–48 h after transfection. A Renilla luciferase expression vector (Dual-Luciferase Reporter Assay System; Promega, Madison, WI) was co-transfected to adjust for variation in transfection efficiency, and luciferase assays were performed essentially as described by De Wet et al. (1987). The amount of plasmid DNA used in each transfection is detailed in the figure legends. The total mass of DNA transfected was adjusted to 7 µg by addition, where necessary, of plasmid pCMV4, and each reporter assay was repeated three or more times.
Recombinant plasmids and protein expression
Plasmid pcDNA1-Tal1 was described previously (Kallianpur et al., 1994). Plasmid pCI-p300 was provided by Dr David Livingston (Dana Farber Cancer Center, Boston, MA), and p-Flag-p300 (1195–1810) was obtained from Dr Robert Roeder (The Rockefeller University, New York, NY). Plasmids pCX-P/CAF and pCX-P/CAF-ΔHAT containing cDNAs for full-length P/CAF and a deletion mutant lacking amino acids 497–526 (P/CAF-ΔHAT), respectively, were provided by Dr Yoshihiro Nakatani (National Institute of Child Health and Human Development, Bethesda, MD). P/CAF and P/CAF-ΔHAT cDNAs were cloned into the murine stem cell virus-based bicistronic retroviral vector MSCV-IRES-GFP (Persons et al., 1997) provided by Dr Arthur Nienhuis (St Jude Children’s Research Hospital, Memphis, TN). Plasmid pcDNA-Myc-Tal1bHLH was constructed by subcloning a PCR-generated cDNA encompassing the TAL1 bHLH domain into the pCS+NLS-MT vector to acquire a human Myc epitope tag and then transferring the resulting insert into vector pcDNA3.1 (Invitrogen, San Diego, CA). A full-length Tal1 cDNA was subcloned in-frame with the DNA-binding domain of GAL4 (GAL41–147) in vector pSG424 (Sadowski and Ptashne, 1989) for expression of a GAL4–TAL1 fusion protein (Huang et al., 1999; Huang and Brandt, 2000). The GAL4-TK-luciferase reporter construct was provided by Dr Scott Hiebert and has been described previously (Fenrick et al., 1999).
Plasmids expressing GST–TAL1 fusion proteins were generated previously (Huang and Brandt, 2000). Mutation of the TAL1 coding sequence was accomplished with the Quick Change Mutagenesis System (Stratagene, La Jolla, CA) and verified by nucleotide sequencing. Flag epitope-tagged p300-HAT (1195–1810) protein was expressed in a BL21 bacterial host at room temperature and purified on an M2 agarose affinity column (Sigma-Aldrich, St Louis, MO) as described previously (Gu and Roeder, 1997). A recombinant baculoviral vector expressing Flag-tagged full-length P/CAF was provided by Dr Yoshihiro Nakatani (Yang et al., 1996). Plaque-purified virus was used to transfect Sf9 insect cells, and Flag-tagged protein was purified on an M2 agarose affinity column according to the manufacturer’s instructions.
In vitro translation and GST pull-down assay
Translation-grade [35S]methionine was purchased from Amersham Pharmacia Biotech (Piscataway, NJ). 35S-labeled P/CAF, P/CAF-ΔHAT, p300, mSin3A and E12 proteins were synthesized using a coupled in vitro transcription–translation system in reticulocyte lysates (Promega). GST pull-down assays were performed as described (Huang et al., 1999).
Retroviral transduction
The amphotropic retroviral packaging cell line ΦNX-Ampho was provided by Dr Garry Nolan (Stanford University, Stanford, CA) and cultured as described previously (Huang and Brandt, 2000). Infectious virus was produced by liposome-mediated transfection of these cells and frozen in aliquots at –70°C. Retroviral infection of MEL cells was carried out exactly as described previously (Huang and Brandt, 2000). GFP-expressing transductants were isolated by FACS and expanded in culture.
Protein acetylation assays
Protein acetyltransferase assays were performed as described (Gu and Roeder, 1997) with slight modification: 4–5 µg of the indicated GST fusion protein and 200 ng of acetyltransferase protein were incubated for 1 h at 30°C in a 30 µl reaction volume containing 50 mM Tris–HCl pH 8.0, 10% glycerol, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 mM sodium butyrate and 1 µl [14C]acetyl-CoA (55 mCi/mmol; Amersham Pharmacia Biotech). Reaction products were resolved by SDS–PAGE and visualized following autoradiography of dried gels.
Metabolic labeling and immunoprecipitation analysis
In vivo acetylation experiments were performed as described (Huang et al., 1999) with slight modification. Briefly, 2 × 107 MEL cells were incubated with or without 1.8% DMSO for the indicated number of days, collected by low-speed centrifugation and incubated for 90 min at 37°C in 2 ml of tissue culture medium containing 1 mCi/ml sodium [3H]acetate (15 Ci/mmol; ICN Pharmaceuticals, Costa Mesa, CA) and 50 nM TSA. Cells were lysed in buffer containing 350 mM NaCl, 50 mM Tris–HCl pH 7.5, 0.5% Igepal, 1 mM EDTA, 0.5 mM DTT, 10 mM sodium butyrate and protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN). The resulting lysates were incubated with TAL1 pre-immune immunoglobulin, affinity-purified rabbit TAL1 antibody (Kallianpur et al., 1994) or GATA-1 monoclonal antibody (N6, Santa Cruz Biotechnology, San Diego, CA). Immunoprecipitates were collected with Sepharose beads, washed 4–5 times in buffer containing 150 mM NaCl, 50 mM Tris–HCl (pH 7.5), 0.5% Igepal, 1 mM EDTA, 0.5 mM DTT, 10 mM sodium butyrate and protease inhibitor cocktail, and resolved by SDS–PAGE. Labeled proteins were visualized following autoradiography of dried gels.
Immunoprecipitation and immunoblot analysis
Rabbit antibody to P/CAF (Yang et al., 1996) was generously provided by Dr Yoshihiro Nakatani. Monoclonal antibodies to the Flag epitope (M2), human c-Myc epitope (9E10) and β-actin (AC-74) were purchased from Sigma-Aldrich. Monoclonal antibodies to p300 (RW128) and mSin3A (K-20) were purchased from Upstate Biotechnology (Lake Placid, NY) and Santa Cruz Biotechnology, respectively. Immunoprecipitation and immunoblot analyses were carried out as described previously (Huang and Brandt, 2000).
Electrophoretic mobility shift assay
EMSA was carried out as described (Huang et al., 1999) with some modification. A double-stranded oligonucleotide corresponding to the preferred TAL1/E2A-binding site (Hsu et al., 1994), 5′-ACCTGAACAGATGGTCGGCT-3′, with the sequence of the E-box underlined, was end-labeled with 32P and incubated with the indicated amounts of GST–TAL1 fusion protein or with 15 µg nuclear extract in 20% glycerol, 50 mM HEPES pH 7.8, 200 mM KCl, 20 mM MgCl2, 1.2 mM EDTA, 5.6 mM DTT and 1 µg/reaction of poly(dI–dC) in a total volume of 30 µl. Protein–DNA complexes were resolved on 5% non-denaturing polyacrylamide gels in Tris-glycine buffer at 4°C. Dried gels were exposed to film and analyzed by autoradiography.
Northern blot analysis
Northern blot analysis was performed as previously described (Huang and Brandt, 2000). Following capillary transfer, a 1.2 kb murine β-globin genomic fragment, a 1.4 kb murine glyceraldehyde phosphate dehydrogenase (GAPDH) cDNA and a 2.2 kb murine Protein 4.2 cDNA were radiolabeled by random primer extension (Life Technologies) and hybridized as described.
Autoradiographic analysis
Band densities on X-ray film were quantitated using the NIH Image software package (version 1.5).
Acknowledgments
Acknowledgements
We are indebted to Yoshihiro Nakatani for P/CAF antibody and plasmids; Robert Roeder, David Livingston, Arthur Nienhuis, Long-Sheng Chang, Mark Koury, Maurice Bondurant and Garry Nolan for providing reagents; Suzanne Szak for advice on protein acetyltransferase assays; Roland Stein for support of Y.Q.; Raymond Mernaugh for assistance with baculoviral expression; and Scott Hiebert for reviewing the manuscript. This work was supported in part by National Institutes of Health grant R01 HL49118 (S.J.B.), a Merit Review Award from the Department of Veterans Affairs (S.J.B.) and an American Society of Hematology Fellow Scholar Award (S.H.). Photomicroscopy was carried out in the Cell Imaging Resource, and baculovirus culture was performed in the Molecular Recognition Resource of the Vanderbilt-Ingram Cancer Center supported by National Institutes of Health grant CA68485.
References
- Akazawa C., Ishibashi,M., Shimizu,C., Nakanishi,S. and Kageyama,R. (1995) A mammalian helix–loop–helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J. Biol. Chem., 270, 8730–8738. [DOI] [PubMed] [Google Scholar]
- Aplan P.D., Nakahara,K., Orkin,S.H. and Kirsch,I.R. (1992) The SCL gene product: a positive regulator of erythroid differentiation. EMBO J., 11, 4073–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arany Z., Newsome,D., Oldread,E., Livingston,D.M. and Eckner,R. (1995) A family of transcriptional adaptor proteins targeted by the E1A protein. Nature, 374, 81–84. [DOI] [PubMed] [Google Scholar]
- Atchley W.R., Wollenberg,K.R., Fitch,W.M., Terhalle,W. and Dress,A.W. (2000) Correlations among amino acid sites in bHLH protein domains: an information theoretic analysis. Mol. Biol. Evol., 17, 164–178. [DOI] [PubMed] [Google Scholar]
- Bannister A.J. and Kouzarides,T. (1995) CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J., 14, 4758–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A.J. and Kouzarides,T. (1996) The CBP co-activator is a histone acetyltransferase. Nature, 384, 641–643. [DOI] [PubMed] [Google Scholar]
- Begley C.G. et al. (1989) Chromosomal translocation in a human leukemic stem cell line disrupts the T-cell antigen receptor δ-chain diversity region and results in a previously unreported fusion transcript. Proc. Natl Acad. Sci. USA, 86, 2031–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley C.G., Lipkowitz,S., Gobel,V., Mahon,K.A., Bertness,V., Green,A.R., Gough,N.M. and Kirsch,I.R. (1992) Molecular characterization of NSCL, a gene encoding a helix–loop–helix protein expressed in the developing nervous system. Proc. Natl Acad. Sci. USA, 89, 38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergel M., Herrera,J.E., Thatcher,B.J., Prymakowska-Bosak,M., Vassilev,A., Nakatani,Y., Martin,B. and Bustin,M. (2000) Acetylation of novel sites in the nucleosomal binding domain of chromosomal protein HMG-14 by p300 alters its interaction with nucleosomes. J. Biol. Chem., 275, 11514–11520. [DOI] [PubMed] [Google Scholar]
- Blobel G.A., Nakajima,T., Eckner,R., Montminy,M. and Orkin,S.H. (1998) CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc. Natl Acad. Sci. USA, 95, 2061–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes J., Byfield,P., Nakatani,Y. and Ogryzko,V. (1998) Regulation of activity of the transcription factor GATA-1 by acetylation. Nature, 396, 594–598. [DOI] [PubMed] [Google Scholar]
- Brownell J.E., Zhou,J., Ranalli,T., Kobayashi,R., Edmondson,D.G., Roth,S.Y. and Allis,C.D. (1996) Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell, 84, 843–851. [DOI] [PubMed] [Google Scholar]
- Chen H., Lin,R.J., Schiltz,R.L., Chakravarti,D., Nash,A., Nagy,L., Privalsky,M.L., Nakatani,Y. and Evans,R.M. (1997) Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell, 90, 569–580. [DOI] [PubMed] [Google Scholar]
- Chen H., Lin,R.J., Xie,W., Wilpitz,D. and Evans,R.M. (1999) Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell, 98, 675–686. [DOI] [PubMed] [Google Scholar]
- Chen Q. et al. (1990) The tal gene undergoes chromosome translocation in T cell leukemia and potentially encodes a helix–loop–helix protein. EMBO J., 9, 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condorelli G., Vitelli,L., Valtieri,M., Marta,I., Montesoro,E., Lulli,V., Baer,R. and Peschle,C. (1995) Coordinate expression and developmental role of Id2 protein and TAL1/E2A heterodimer in erythroid progenitor differentiation. Blood, 86, 164–175. [PubMed] [Google Scholar]
- Condorelli G.L., Facchiano,F., Valtieri,M., Proietti,E., Vitelli,L., Lulli,V., Huebner,K., Peschle,C. and Croce,C.M. (1996) T-cell-directed TAL-1 expression induces T-cell malignancies in transgenic mice. Cancer Res., 56, 5113–5119. [PubMed] [Google Scholar]
- David G., Alland,L., Hong,S.H., Wong,C.W., DePinho,R.A. and Dejean,A. (1998) Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene, 16, 2549–2556. [DOI] [PubMed] [Google Scholar]
- De Wet J.R., Wood,K.V., DeLuca,M., Helinski,D.R. and Subramani,S. (1987) Firefly luciferase gene: structure and expression in mammalian cells. Mol. Cell. Biol., 7, 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhordain P., Lin,R.J., Quief,S., Lantoine,D., Kerckaert,J.P., Evans,R.M. and Albagli,O. (1998) The LAZ3 (BCL-6) oncoprotein recruits a SMRT/mSIN3A/histone deacetylase containing complex to mediate transcriptional repression. Nucleic Acids Res., 26, 4645–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwood N.J., Zogos,H., Pereira,D.S., Dick,J.E. and Begley,C.G. (1998) Enhanced megakaryocyte and erythroid development from normal human CD34(+) cells: consequence of enforced expression of SCL. Blood, 91, 3756–3765. [PubMed] [Google Scholar]
- Fenrick R., Amann,J.M., Lutterbach,B., Wang,L., Westendorf,J.J., Downing,J.R. and Hiebert,S.W. (1999) Both TEL and AML-1 contribute repression domains to the t(12;21) fusion protein. Mol. Cell. Biol., 19, 6566–6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger L.R., Kagan,J., Christopher,G., Kurtzberg,J., Hershfield,M.S., Nowell,P.C. and Croce,C.M. (1989) Involvement of the TCL5 gene on human chromosome 1 in T-cell leukemia and melanoma. Proc. Natl Acad. Sci. USA, 86, 5039–5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelmetti V., Zhang,J., Fanelli,M. and Minucci,S. (1998) Aberrant recruitment of the nuclear receptor corepressor–histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol. Cell. Biol., 18, 7185–7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass C.K. and Rosenfeld,M.G. (2000) The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev., 14, 121–141. [PubMed] [Google Scholar]
- Göbel V., Lipkowitz,S., Kozak,C.A. and Kirsch,I.R. (1992) NSCL-2: a basic domain helix–loop–helix gene expressed in early neurogenesis. Cell Growth Differ., 3, 143–148. [PubMed] [Google Scholar]
- Goulding S.E., zur Lage,P. and Jarman,P. (2000) amos, a proneural gene for Drosophila olfactory sense organs that is regulated by lozenge. Neuron, 25, 69–78. [DOI] [PubMed] [Google Scholar]
- Grignani F. et al. (1998) Fusion proteins of the retinoic acid receptor-α recruit histone deacetylase in promyelocytic leukaemia. Nature, 391, 815–818. [DOI] [PubMed] [Google Scholar]
- Grunstein M. (1997) Histone acetylation in chromatin structure and transcription. Nature, 389, 349–352. [DOI] [PubMed] [Google Scholar]
- Gu W. and Roeder,R.G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell, 90, 595–606. [DOI] [PubMed] [Google Scholar]
- He L.Z., Guidez,F., Tribioli,C., Peruzzi,D., Ruthardt,M., Zelent,A. and Pandolfi,P.P. (1998) Distinct interactions of PML-RARα and PLZF-RARα with co-repressors determine differential responses to RA in APL. Nature Genet., 18, 126–135. [DOI] [PubMed] [Google Scholar]
- Hebbes T.R., Thorne,A.W. and Crane-Robinson,C. (1988) A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J., 7, 1395–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera J.E., Bergel,M., Yang,X.-J., Nakatani,Y. and Bustin,M. (1997) The histone acetyltransferase activity of human GCN5 and PCAF is stabilized by coenzymes. J. Biol. Chem., 272, 27253–27258. [DOI] [PubMed] [Google Scholar]
- Herrera J.E., Sakaguchi,K., Bergel,M., Trieschmann,L., Nakatani,Y. and Bustin,M. (1999) Specific acetylation of chromosomal protein HMG-17 by PCAF alters its interaction with nucleosomes. Mol. Cell. Biol., 19, 3466–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H.L., Huang,L., Tsan,J.T., Funk,W., Wright,W.E., Hu,J.S., Kingston,R.E. and Baer,R. (1994) Preferred sequences for DNA recognition by the TAL1 helix–loop–helix proteins. Mol. Cell. Biol., 14, 1256–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M.-L., Hsu,C.-H. and Chien,C.-T. (2000) The proneural gene amos promotes multiple dendritic neuron formation in the Drosophila peripheral nervous system. Neuron, 25, 57–67. [DOI] [PubMed] [Google Scholar]
- Huang S. and Brandt,S.J. (2000) mSin3A regulates murine erythroleukemia cell differentiation through association with the TAL1 (or SCL) transcription factor. Mol. Cell. Biol., 20, 2248–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Qiu,Y., Stein,R.W. and Brandt,S.J. (1999) p300 functions as a transcriptional coactivator for the TAL1/SCL oncoprotein. Oncogene, 18, 4958–4967. [DOI] [PubMed] [Google Scholar]
- Hung H.-L., Lau,J., Kim,A.Y., Weiss,M.J. and Blobel,G.A. (1999) CREB-binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol. Cell. Biol., 19, 3496–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhof A., Yang,X.-J., Ogryzko,V.V., Nakatani,Y., Wolffe,A.P. and Ge,H. (1997) Acetylation of general transcription factors by histone acetyltransferases. Curr. Biol., 7, 689–692. [DOI] [PubMed] [Google Scholar]
- Jiang H., Lu,H., Schiltz,R.L., Pise-Masison,C.A., Ogryzko,V.V., Nakatani,Y. and Brady,J.N. (1999) PCAF interacts with Tax and stimulates Tax transactivation in a histone acetyltransferase-independent manner. Mol. Cell. Biol., 19, 8136–8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.E., Birren,S.J. and Anderson,D.J. (1990) Two rat homologues of Drosophila achaete-scute specifically expressed in neuronal precursors. Nature, 346, 858–861. [DOI] [PubMed] [Google Scholar]
- Kallianpur A.R., Jordan,J.E. and Brandt,S.J. (1994) The SCL/TAL-1 gene is expressed in progenitors of both the hematopoietic and vascular systems during embryogenesis. Blood, 83, 1200–1208. [PubMed] [Google Scholar]
- Kelliher M.A., Seldin,D.C. and Leder,P. (1996) Tal-1 induces T cell acute lymphoblastic leukemia accelerated by casein kinase IIα. EMBO J., 15, 5160–5166. [PMC free article] [PubMed] [Google Scholar]
- Kiernan R.E. et al. (1999) HIV-1 tat transcriptional activity is regulated by acetylation. EMBO J., 18, 6106–6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koury M.J., Sawyer,S.T. and Bondurant,M.C. (1984) Splenic erythroblasts in anemia-inducing Friend disease: a source of cells for studies of erythropoietin-mediated differentiation. J. Cell. Physiol., 121, 526–532. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. (2000) Acetylation: a regulatory modification to rival phosphorylation? EMBO J., 19, 1176–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M.H. and Allis,C.D. (1998) Roles of histone acetyltransferases and deacetylases in gene regulation. BioEssays, 20, 615–626. [DOI] [PubMed] [Google Scholar]
- Lau O.D. et al. (2000) HATs off: selective synthetic inhibitors of the histone acetyltransferases p300 and PCAF. Mol. Cell, 5, 589–595. [DOI] [PubMed] [Google Scholar]
- Leiter J.M.E., Helliger,W. and Puschendorf,B. (1984) Increase in histone acetylation and transitions in histone variants during Friend cell differentiation. Exp. Cell Res., 155, 222–231. [DOI] [PubMed] [Google Scholar]
- Lemercier C., To,R.Q., Swanson,B.J., Lyons,G.E. and Konieczny,S.F. (1997) Mist1: a novel basic helix–loop–helix transcription factor exhibits a developmentally regulated expression pattern. Dev. Biol., 182, 101–113. [DOI] [PubMed] [Google Scholar]
- Lemercier C., To,R.Q., Carrasco,R.A. and Konieczny,S.F. (1998) The basic helix–loop–helix transcription factor Mist1 functions as a transcriptional repressor of myoD. EMBO J., 17, 1412–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Imhof,A., Collingwood,T.N., Urnov,F.D. and Wolffe,A.P. (1999) p300 stimulates transcription instigated by ligand-bound thyroid hormone receptor at a step subsequent to chromatin disruption. EMBO J., 18, 5634–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R.J., Nagy,L., Inoue,S., Shao,W., Miller,W.H.,Jr and Evans,R.M. (1998) Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature, 391, 811–814. [DOI] [PubMed] [Google Scholar]
- Liu L., Scolnick,D.M., Trievel,R.C., Zhang,H.B., Marmorstein,R., Halazonetis,T.D. and Berger,S.L. (1999) p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol., 19, 1202–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad J.R., Kwok,R.P.S., Laurence,M.E., Harter,M.L. and Goodman,R.H. (1995) Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature, 374, 85–88. [DOI] [PubMed] [Google Scholar]
- Lutterbach B. et al. (1998) ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol. Cell. Biol., 18, 7176–7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P.C., Rould,M.A., Weintraub,H. and Pabo,C.O. (1994) Crystal structure of MyoD bHLH domain–DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell, 77, 451–459. [DOI] [PubMed] [Google Scholar]
- Martínez-Balbás M.A., Bauer,U.M., Nielsen,S.J., Brehm,A. and Kouzarides,T. (2000) Regulation of E2F1 activity by acetylation. EMBO J., 19, 662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzio G., Wagener,C., Gutierrez,M.I., Cartwright,P., Helin,K. and Giacca,M. (2000) E2F family members are differentially regulated by reversible acetylation. J. Biol. Chem., 275, 10887–10892. [DOI] [PubMed] [Google Scholar]
- Massari M.E. and Murre,C. (2000) Helix–loop–helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol., 20, 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.E., Cairns,B.R., Levinson,R.S., Yamamoto,K.R., Engel,D.A. and Smith,M.M. (1996) Adenovirus E1A specifically blocks SWI/SNF-dependent transcriptional activation. Mol. Cell. Biol., 16, 5737–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizzen C.A. et al. (1996) The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell, 87, 1261–1270. [DOI] [PubMed] [Google Scholar]
- Munshi N., Merika,M., Yie,J., Senger,K., Chen,G. and Thanos,D. (1998) Acetylation of HMG I(Y) by CBP turns off IFNβ expression by disrupting the enhanceosome. Mol. Cell, 2, 457–467. [DOI] [PubMed] [Google Scholar]
- Ogryzko V.V., Schiltz,R.L., Russanova,V., Howard,B.H. and Nakatani,Y. (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell, 87, 953–959. [DOI] [PubMed] [Google Scholar]
- Ogryzko V.V., Kotani,T., Zhang,X., Schiltz,R.L., Howard,B.H., Qin,J. and Nakatani,Y. (1998) Histone-like TAFs within the PCAF histone acetylase complex. Cell, 94, 35–44. [DOI] [PubMed] [Google Scholar]
- Ono Y., Fukuhara,N. and Yoshie,O. (1997) Transcriptional activity of TAL1 in T cell acute lymphoblastic leukemia (T-ALL) requires RBTN1 or -2 and induces TALLA1, a highly specific tumor marker of T-ALL. J. Biol. Chem., 272, 4576–4581. [DOI] [PubMed] [Google Scholar]
- Ono Y., Fukuhara,N. and Yoshie,O. (1998) TAL1 and LIM-only proteins synergistically induce retinaldehyde dehydrogenase 2 expression in T-cell acute lymphoblastic leukemia by acting as cofactors for GATA3. Mol. Cell. Biol., 18, 6939–6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M., Schnölzer,M., Garnica,J., Fischle,W., Emiliani,S., Rackwitz, H.R. and Verdin,E. (1999) Acetylation of the HIV-1 Tat protein by p300 is important for its transcriptional activity. Curr. Biol., 9, 1489–1492. [DOI] [PubMed] [Google Scholar]
- Persons D.A., Allay,J.A., Allay,E.R., Smeyne,R.J., Ashmun,R.A., Sorrentino,B.P. and Nienhuis,A.W. (1997) Retroviral-mediated transfer of the green fluorescent protein gene into murine hematopoietic cells facilitates scoring and selection of transduced progenitors in vitro and identification of genetically modified cells in vivo. Blood, 90, 1777–1786. [PubMed] [Google Scholar]
- Puri P.L. et al. (1997) Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell, 1, 35–45. [DOI] [PubMed] [Google Scholar]
- Reid J.L., Bannister,A.J., Zegerman,P., Martínez-Balbás,M.A. and Kouzarides,T. (1998) E1A directly binds and regulates the P/CAF acetyltransferase. EMBO J., 17, 4469–4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb L., Lyons,I., Li,R., Hartley,L., Köntgen,F., Harvey,R.P., Metcalf,D. and Begley,C.G. (1995) Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. Proc. Natl Acad. Sci. USA, 92, 7075–7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I. and Ptashne,M. (1989) A vector for expressing GAL4(1–147) fusions in mammalian cells. Nucleic Acids Res., 17, 7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi K., Herrera,J.E., Saito,S., Miki,T., Bustin,M., Vassilev,A., Anderson,C.W. and Appella,E. (1998) DNA damage activates p53 through a phosphorylation–acetylation cascade. Genes Dev., 12, 2831–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli V., Puri,P.L., Hamamori,Y., Ogryzko,V., Chung,G., Nakatani,Y., Wang,J.Y.J. and Kedes,L. (1999) Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol. Cell, 4, 725–734. [DOI] [PubMed] [Google Scholar]
- Schiltz R.L. and Nakatani,Y. (2000) The PCAF acetylase complex as a potential tumor suppressor. Biochim. Biophys. Acta, 1470, M37–M53. [DOI] [PubMed] [Google Scholar]
- Schiltz R.L., Mizzen,C.A., Vassilev,A., Cook,R.G., Allis,C.D. and Nakatani,Y. (1999) Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem., 274, 1189–1192. [DOI] [PubMed] [Google Scholar]
- Scolnick D.M., Chehab,N.H., Stavridi,E.S., Lien,M.C., Caruso,L., Moran,E., Berger,S.L. and Halazonetis,T.D. (1997) CREB-binding protein and p300/CBP-associated factor are transcriptional coactivators of the p53 tumor suppressor protein. Cancer Res., 57, 3693–3696. [PubMed] [Google Scholar]
- Shivdasani R.A., Mayer,E.L. and Orkin,S.H. (1995) Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature, 373, 432–434. [DOI] [PubMed] [Google Scholar]
- Soutoglou E., Katrakili,N. and Talianidis,I. (2000) Acetylation regulates transcription factor activity at multiple levels. Mol. Cell, 5, 745–751. [DOI] [PubMed] [Google Scholar]
- Spencer T.E. et al. (1997) Steroid receptor coactivator-1 is a histone acetyltransferase. Nature, 389, 194–198. [DOI] [PubMed] [Google Scholar]
- Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev., 12, 599–606. [DOI] [PubMed] [Google Scholar]
- Takaku F., Nakao,K., Ono,T. and Terayama,H. (1969) Changes in histone acetylation and RNA synthesis in the spleen of polycythemic mouse after erythropoietin injection. Biochim. Biophys. Acta, 195, 396–400. [DOI] [PubMed] [Google Scholar]
- Taunton J., Hassig,C.A. and Schreiber,S.L. (1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science, 272, 408–411. [DOI] [PubMed] [Google Scholar]
- Tsiftsoglou A.S. and Wong,W. (1985) Molecular and cellular mechanisms of leukemic hemopoietic cell differentiation: an analysis of the Friend system. Anticancer Res., 5, 81–99. [PubMed] [Google Scholar]
- Wadman I., Li,J., Bash,R.O., Forster,A., Osada,H., Rabbitts,T.H. and Baer,R. (1994) Specific in vivo association between the bHLH and LIM proteins implicated in human T cell leukemia. EMBO J., 13, 4831–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman I.A., Osada,H., Grutz,G.G., Agulnick,A.D., Westphal,H., Forster,A. and Rabbitts,T.H. (1997) The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J., 16, 3145–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Hoshino,T., Redner,R.L., Kajigaya,S. and Liu,J.M. (1998) ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-Cor/mSin3/HDAC1 complex. Proc. Natl Acad. Sci. USA, 95, 10860–10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A.P. and Pruss,D. (1996) Targeting chromatin disruption: transcription regulators that acetylate histones. Cell, 84, 817–819. [DOI] [PubMed] [Google Scholar]
- Xia Y., Brown,L., Yang,C.Y., Tsan,J.T., Siciliano,M.J., Espinosa,R.,III, Le Beau,M.M. and Baer,R.J. (1991) TAL2, a helix–loop–helix gene activated by the (7;9)(q34;q32) translocation in human T-cell leukemia. Proc. Natl Acad. Sci. USA, 88, 11416–11420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.J., Ogryzko,V.V., Nishikawa,J., Howard,B.H. and Nakatani,Y. (1996) A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature, 382, 319–324. [DOI] [PubMed] [Google Scholar]
- Yoshida M., Kijima,M., Akita,M. and Beppu,T. (1990) Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem., 265, 17174–17179. [PubMed] [Google Scholar]
- Zhang W. and Bieker,J.J. (1998) Acetylation and modulation of erythroid Krüppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc. Natl Acad. Sci. USA, 95, 9855–9860. [DOI] [PMC free article] [PubMed] [Google Scholar]