Abstract
Bnip3 (Bcl-2/adenovirus E1B 19-kDa-interacting protein 3) is a mitochondrial BH3-only protein that contributes to cell death through activation of the mitochondrial pathway of apoptosis. Bnip3 is also known to induce autophagy, but the functional role of autophagy is unclear. In this study, we investigated the relationship between mitochondrial dysfunction and upregulation of autophagy in response to Bnip3 in cells lacking Bax and Bak. We found that Bnip3 induced mitochondrial autophagy in the absence of mitochondrial membrane permeabilization and Bax/Bak. Also, co-immunoprecipitation experiments showed that Bnip3 interacted with the autophagy protein LC3 (microtubule-associated protein light chain 3). Although Bax-/Bak-deficient cells were resistant to Bnip3-mediated cell death, inhibition of mitochondrial autophagy induced necrotic cell death. When investigating why these mitochondria had to be removed by autophagy, we discovered that Bnip3 reduced both nuclear- and mitochondria-encoded proteins involved in oxidative phosphorylation. Interestingly, Bnip3 had no effect on other mitochondrial proteins, such as Tom20 and MnSOD, or actin and tubulin in the cytosol. Bnip3 did not seem to reduce transcription or translation of these proteins. However, we found that Bnip3 caused an increase in mitochondrial protease activity, suggesting that Bnip3 might promote degradation of proteins in the mitochondria. Thus, Bnip3-mediated impairment of mitochondrial respiration induces mitochondrial turnover by activating mitochondrial autophagy.
Keywords: Bnip3, Bcl-2, autophagy, apoptosis, mitochondria, oxidative phosphorylation
Autophagy is a highly conserved process that is involved in the degradation of long-lived proteins and removing damaged organelles.1 In this process, a double-membrane structure called the autophagosome sequesters cytoplasmic components such as ubiquitinated protein aggregates or organelles including mitochondria, peroxisomes, and endoplasmic reticulum. The autophagosome then fuses with a lysosome, resulting in the destruction of its content by the acid hydrolases provided by the lysosome. Although autophagy is generally considered nonspecific, there appears to be selection for specific organelles, such as the mitochondria, under certain conditions.2 Autophagy occurs constitutively in most cells, but is rapidly upregulated when there is a change in the cellular environment, such as a reduction in nutrients or oxygen. However, the functional role of this enhancement is not clear and increased autophagy has been reported to be both protective and detrimental. For instance, autophagy is an important survival mechanism during nutrient deprivation when the cell needs to recycle fatty acids and amino acids from lipids and proteins for survival.1 Autophagy can also promote survival by removing protein aggregates and damaged mitochondria that may be harmful to the cell.2 In contrast, autophagy has been implicated as a death mechanism in response to cardiac pressure overload3 and during reperfusion after ischemia.4
The Bcl-2 family of proteins has a key role in regulating the intrinsic (mitochondrial) apoptotic pathway,5 and display either anti- or pro-apoptotic function. Anti-apoptotic members, such as Bcl-2 and Bcl-XL, protect mitochondrial integrity, whereas pro-apoptotic proteins promote the release of apoptogenic factors from the mitochondria.5 The pro-apoptotic proteins can be divided into the BH3-only proteins including Bid, Bad, and Bim, and their effectors Bax and Bak. Studies using cells derived from knockout mice lacking both Bax and Bak have demonstrated that Bax and Bak are essential for the initiation of apoptosis through the mitochondria.6 The Bcl-2 proteins have also been shown to regulate autophagy. Bcl-2 and Bcl-XL can inhibit autophagy by sequestering Beclin-1,7 whereas BH3-only proteins can induce autophagy by competitively disrupting the interaction between Beclin-1 and Bcl-2/Bcl-XL.8
Bcl-2/adenovirus E1B 19-kDa-interacting protein 3 (Bnip3) is an atypical pro-apoptotic BH3-only protein and has a key role in the pathogenesis of many diseases, such as heart failure9 and cancer.10 Bnip3 is primarily localized to the mitochondria and it is well documented that Bnip3 induces loss of mitochondrial membrane potential (ΔΨm) and cell death.9, 11, 12 In addition, several studies have reported that Bnip3 is a potent inducer of autophagy in many different cell types.13, 14, 15, 16, 17 However, the functional role of Bnip3-mediated autophagy is not clear. Kanzawa et al.16 reported that Bnip3 induced autophagic cell death in malignant glioma cells. Bnip3 was also reported to contribute to autophagic cell death during hypoxia.15 In contrast, Bellot et al.14 found that Bnip3-induced autophagy during hypoxia was a survival mechanism that promoted tumor progression. We also found that autophagy protected against Bnip3-mediated cell death in HL-1 cells.13 As Bnip3 may represent a potential therapeutic target in both heart disease and cancer, it is important to understand how Bnip3 regulates mitochondrial function and autophagy. In this study, we investigated the functional role of Bnip3-induced mitochondrial autophagy in apoptosis resistant cells lacking Bax and Bak.
Results
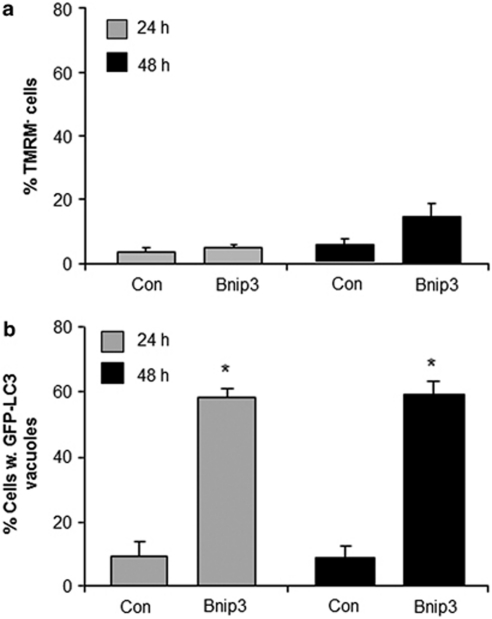
As Bnip3 is well known to induce mitochondrial dysfunction,9, 11, 12 we investigated the relationship between loss of ΔΨm and upregulation of autophagy in response to Bnip3 in HL-1 cells. We found that overexpression of Bnip3 in HL-1 myocytes led to the loss of ΔΨm as measured by loss of tetramethylrhodamine methyl ester (TMRM) uptake (Figure 1a). The microtubule-associated protein light chain 3 (LC3) is recruited to autophagic vesicles upon the initiation of autophagy, which can be detected as punctate accumulations of GFP-LC3.18 Using GFP-LC3 to monitor formation of autophagosomes, we confirmed that Bnip3 is a potent inducer of autophagy in HL-1 cells (Figure 1b). Interestingly, overexpression of the anti-apoptotic protein Bcl-2 preserved ΔΨm (Figure 1a), but did not prevent upregulation of autophagy in response to Bnip3 (Figure 1b). We found that the cellular localization of the autophagosomes changed over time in HL-1 cells. At 24 h, the autophagosomes were distributed throughout the cytosol, but by 36 h they started to aggregate in the perinuclear region (Figure 1c). We observed a similar aggregation of autophagosomes in cardiac myocytes,17 but it is currently unknown why the autophagosomes accumulates in the perinuclear region.
Figure 1.
Overexpression of Bcl-2 protects against Bnip3-mediated loss of mitochondrial membrane potential but does not prevent induction of autophagy in HL-1 myocytes. HL-1 cells were cotransfected with pcDNA3.1 or Bnip3, and vector or Bcl-2 plus GFP-LC3 for 24 h. (a) The mitochondrial membrane potential was measured by TMRM fluorescence and (b) autophagy was determined by analyzing staining patterns of GFP-LC3. Cells were examined at × 60 magnification, and classified as cells with diffuse GFP-LC3 fluorescence, or as cells with numerous GFP-LC3 puncta (>30 dots/cell) representing autophagosomes. (c) Representative images of HL-1 cells. Data are mean ± S.D. (n=3, *,**P<0.05 compared with con and con+Bcl-2)
Studies using cells derived from knockout mice lacking both Bax and Bak have demonstrated that Bax and Bak are essential for the initiation of cell death through the mitochondrial pathway.6 In our previous work, we found that Bax/Bak are essential downstream effectors of Bnip3-mediated mitochondrial permeabilization and cell death.11, 19 Therefore, we investigated whether Bnip3 could induce autophagy in mouse embryonic fibroblasts (MEFs) derived from Bax/Bak double knockout (DKO) mice. Although ΔΨm was preserved in cells overexpressing Bnip3 (Figure 2a), autophagy was still significantly upregulated (Figure 2b). Upregulation of autophagy by Bnip3 in Bax/Bak DKO MEFs was also confirmed by acridine orange staining which showed increased presence of acidic vesicular organelles, a characteristic of autophagy (Supplementary Figure S1). The Bnip3 homolog Nix (also called Bnip3L) also mediates cell death via activation of Bax/Bak,20 but induces autophagy independent of Bax/Bak in erythrocytes.21 In this study, we confirmed that Nix induced significant autophagy in Bax/Bak DKO MEFs (Supplementary Figure S2). This suggests that autophagy is induced in response to Bnip3 and Nix whether or not the intrinsic (mitochondrial) cell death pathway is activated.
Figure 2.
Bax/Bak DKO MEFs are resistant to Bnip3-mediated loss of mitochondrial membrane potential but not to Bnip3-induced autophagy. Bax/Bak DKO MEFs were cotransfected with vector or Bnip3 plus GFP-LC3 and (a) the ΔΨm was measured by TMRM fluorescence and (b) autophagy was determined by analyzing GFP-LC3 fluorescence. Data are mean±S.D. (n=3, *P<0.05 compared with control)
The increased levels of autophagosomes in the Bax/Bak MEFs overexpressing Bnip3 could reflect either increased autophagosome formation due to enhanced autophagic activity, or an accumulation of autophagosomes because of an impairment in the degradation pathway. Therefore, we investigated the effect of Bnip3 on autophagic flux in Bax/Bak DKO MEFs. The proton ATPase inhibitor Bafilomycin A1 prevents lysosomal acidification, which inhibits lysosomal degradation and results in accumulation of autophagosomes.22 Lysotracker red (LTR) accumulates in acidic compartments in the cell and we confirmed that the presence of Bafilomycin A1 abolished labeling of lysosomes with LTR (Figure 3a). Moreover, the presence of Bafilomycin A1 significantly increased the percentage of cells exhibiting GFP-LC3-positive autophagosomes in cells overexpressing Bnip3 (Figure 3b) and increased the conversion of endogenous LC3-I to LC3-II as assessed by western blotting (Figure 3c). The LC3-II abundance was increased in cells overexpressing Bnip3, and the LC3-II levels were increased even further when degradation was inhibited by Bafilomycin A1, consistent with the flux measurements made by fluorescence microscopy. This suggests that Bnip3 increases autophagic activity in the cells.
Figure 3.
Bnip3 increases autophagic activity in cells. Bax/Bak DKO MEFs were infected with adenoviruses encoding β-gal or Bnip3 plus GFP-LC3 for 24 h and treated with 100 nM Bafilomycin A1 for 1 h. (a) Representative images of live cells stained with 50 nM lysotracker red. (b) Quantitation of autophagy in the presence and absence of Bafilomycin A1 (n=3). (c) Western blot analysis and quantitation of LC-II/LC3-I levels (n=4, *P<0.05 compared with β-gal, **P<0.05 compared with Bnip3). Data are mean±S.D.
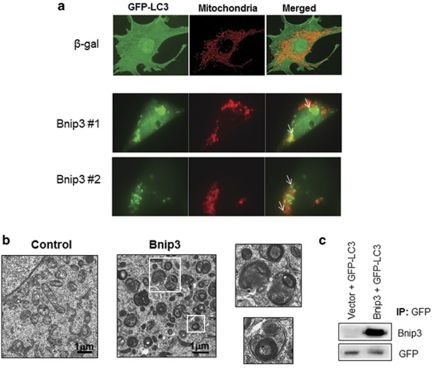
The autophagy–lysosomal pathway is important in removing damaged organelles, such as mitochondria. We have previously found that Bnip3 induces mitochondrial autophagy in HL-1 cells and cardiac myocytes.13, 17 Co-localization experiments confirmed that GFP-LC3-positive autophagosomes co-localized with mitochondria (see arrows) in Bax/Bak DKO MEFs overexpressing Bnip3 (Figure 4a) suggesting that mitochondrial autophagy occurs even in the absence of Bax/Bak. Electron microscopy (EM) confirmed the presence of numerous autophagic vacuoles containing mitochondria in Bax/Bak DKO MEFs overexpressing Bnip3 (Figure 4b). Nix was recently reported to interact directly with the autophagy proteins LC3 and GABARAP.23 To investigate whether Bnip3 also interacts with LC3, we performed co-immunoprecipitation experiments of Bnip3 and LC3. We found that Bnip3 co-immunoprecipitated with GFP-LC3 when overexpressed in Bax/Bak DKO MEFs (Figure 4c). This suggests that both Nix and Bnip3 might be functioning as receptors for autophagosomes.
Figure 4.
Bnip3 induces mitochondrial autophagy in Bax/Bak DKO MEFs. Cells were infected with β-gal or Bnip3 plus and GFP-LC3. (a) GFP-LC3 labeled autophagosomes colocalize (marked by arrows) with COX IV positive mitochondria in cells overexpressing Bnip3. Images show representative cells overexpressing Bnip3 from two separate experiments. (b) Electron micrographs reveal extensive mitochondrial autophagy in cells overexpressing Bnip3. (c) Co-immunoprecipitation of GFP-LC3 and Bnip3
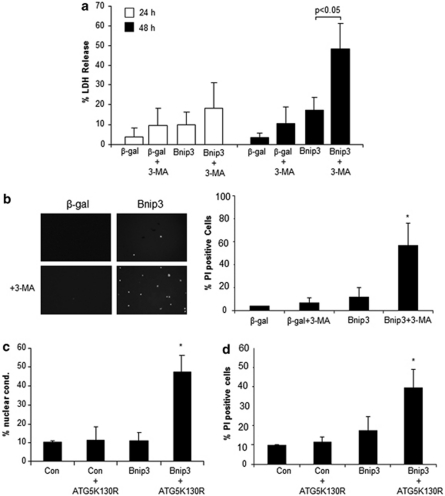
The functional role of autophagy is not clear and has been reported to contribute to as well as protect against cell death. To investigate the functional significance of mitochondrial autophagy in Bax/Bak DKO MEFs, we tested the effect of two different inhibitors of autophagy on cell viability. Although Bax/Bak DKO MEFs were resistant to Bnip3-mediated cell death, the presence of the autophagy inhibitor 3-methyladenine (3-MA) resulted in significant cell death in cell overexpressing Bnip3 (Figure 5a and b). Similarly, overexpression of ATG5K130R, a dominant negative of ATG5 previously shown to suppress vacuole formation,24 also led to activation of cell death (Figure 5c and d). This suggests that Bnip3-mediated mitochondrial autophagy is an important protective response and preventing removal of these mitochondria by autophagy leads to necrotic cell death.
Figure 5.
Bnip3 induces cell death when autophagy is inhibited in Bax/Bak DKO MEFs. Cells were infected with an adenovirus encoding β-gal or Bnip3 in the presence of 10 mM 3-MA. Cell death was assessed at 24 h and 48 h by measuring LDH release (a) or PI uptake (b). Cells were cotransfected with pcDNA3.1 or Bnip3, and GFP or GFP-ATG5K130R and cell death was measured after 48 h by analyzing nuclear condensation (c) or PI uptake (d). Data are mean±S.D. (n=3, *P<0.05 compared with Bnip3 alone)
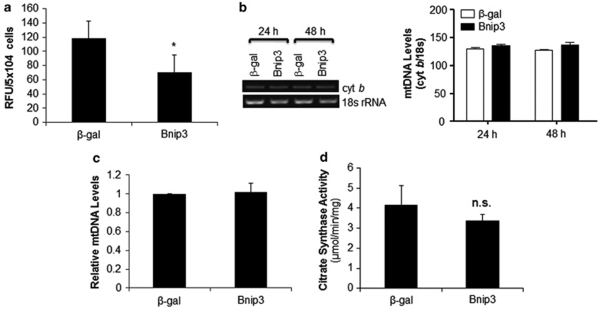
Bnip3 is unable to induce permeabilization of the mitochondrial membrane in the absence Bax/Bak.11, 19 This suggests that Bnip3 has impaired these mitochondria by some other unknown mechanism which requires their removal by autophagy. As the synthesis of ATP in eukaryotic cells occurs mainly through oxidative phosphorylation in mitochondria, we investigated whether Bnip3 had an effect on ATP levels in Bax/Bak DKO MEFs. Although cells were still viable, we found that the cellular levels of ATP were significantly reduced at 48 h after infecting cells with an adenovirus encoding Bnip3 compared with cells that were infected with β-galactosidase (β-gal; Figure 6a). Moreover, 5′-AMP-activated protein kinase (AMPK) is activated in response to a reduction in cellular ATP levels and has been reported to have an important role in the induction of autophagy during ischemia in cardiac myocytes.4 Although Bnip3 reduced cellular ATP levels, there was only a small (but not significant) increase in AMPK activation in cells overexpressing Bnip3 and inhibiting AMPK with compound C had no effect on cell viability (Supplementary Figure S3). To investigate whether the reduced ATP level was due to excess removal of mitochondria by autophagy, we assessed the number of mitochondria in the cells by measuring mitochondrial DNA (mtDNA) content. Surprisingly, we found that the mtDNA copy number was not significantly different in these cells (Figure 6b and c). In addition, there was no significant difference in citrate synthase activity, a key enzyme in the Krebs cycle and a commonly used marker for mitochondrial matrix proteins, between β-gal- and Bnip3-overexpressing cells (Figure 6d). This suggests that the reduced ATP level was not due to fewer mitochondria but a diminished capacity to produce ATP. Uncoupling proteins (UCPs) are inner mitochondrial membrane proton carriers that uncouple ATP synthesis. However, there were no differences in UCP2 and UCP3 protein levels in Bax/Bak DKO MEFs overexpressing Bnip3 (Supplementary Figure S4).
Figure 6.
Bnip3 reduces cellular ATP levels without decreasing the number of mitochondria. (a) At 48 h after infection, cellular ATP levels were measured in Bax/Bak DKO MEFs overexpressing β-gal or Bnip3 (n=6). (b) Mitochondrial DNA content in Bax/Bak DKO MEFs. Shown is a representative gel of PCR for mitochondrial cytochrome b (cyt b) and nuclear 18 s rRNA. Densitometry of cyt b normalized to 18S rRNA (n=3). (c) Real-time quantitative PCR of mtDNA levels at 24 h after infection (n=3). (d) Citrate synthase activity in cells after 48 h of β-gal or Bnip3 overexpression (n=4). Data are mean±S.D. (*P<0.05 compared with β-gal)
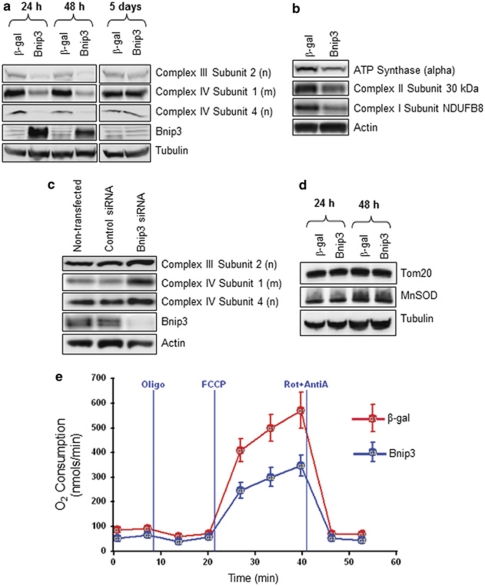
The oxidative phosphorylation system in the inner mitochondrial membrane is responsible for generating ATP and consists of five major membrane protein complexes, the mitochondrial complexes I–V. Thus, we investigated whether Bnip3 had affected the proteins involved in oxidative phosphorylation. Western blot analysis revealed that both nuclear- and mitochondria-encoded subunits in complex III and IV were reduced in Bax/Bak DKO MEFs overexpressing Bnip3 at 24 h and 48 h after infection (Figure 7a). However, the protein levels of all three subunits were restored to normal 5 days after infection when Bnip3 was no longer expressed (Figure 7a). Bnip3 also reduced the ATP synthase α-subunit (complex V) and subunits in complex I and II (Figure 7b). In addition, silencing of endogenous Bnip3 under normal conditions resulted in an increase in the mitochondrial subunits (Figure 7c). Interestingly, levels of other mitochondrial proteins, such as Tom20 and MnSOD, which are not involved in oxidative phosphorylation as well as cytosolic actin and tubulin were unaffected by elevated Bnip3 (Figure 7d). This suggests that the reduction in protein levels by Bnip3 were limited to proteins involved in oxidative phosphorylation. Also, inhibiting mitochondrial autophagy with 3-MA did not restore complex IV subunit 4 levels (Supplementary Figure S5), confirming that the reduced level of this subunit was not due to excessive mitochondrial autophagy. Consistent with our observations of lower levels of components of complexes I-V, maximal (uncoupler stimulated) rates of respiration in intact Bax/Bak DKO MEFs overexpressing Bnip3 were significantly lower (P<0.01) by an average of 59%±2 than in β-gal-overexpressing cells (Figure 7e). Moreover, Bellot et al.14 recently reported that both Bnip3 and Nix are involved in hypoxia-mediated autophagy in MEFs, and we found that 48 h of hypoxia caused upregulation of both Bnip3 and Nix. We also found that hypoxia caused a reduction in both nuclear and mitochondrial encoded subunits in complexes III and IV (Supplementary Figure 6S) and that silencing of Bnip3 or Nix using small interfering RNA (siRNA) reduced the hypoxia-mediated decrease in the protein levels. However, silencing of Bnip3 and/or Nix did not fully restore protein levels to levels observed during normoxia, suggesting that other proteins involved in protein degradation and/or stabilization are activated during hypoxia.
Figure 7.
Bnip3 reduces levels of both nuclear- and mitochondria-encoded proteins involved in mitochondrial respiration. Bax/Bak DKO MEFs were infected with adenoviruses encoding β-gal or Bnip3 for the indicated times and protein levels were analyzed by western blot analysis. (a) Increased levels of Bnip3 reduced the expression of both nuclear- (n) and mitochondrial (m)-encoded subunits in complex III and IV. The protein levels were restored to normal when Bnip3 levels were reduced 5 days after infection. (b) Bnip3 reduced the levels of subunits in complex V (ATP synthase), II, and I. (c) Downregulation of endogenous Bnip3 using siRNA for 72 h results in increased levels of both nuclear (n) and mitochondrial (m) encoded subunits in complex III and IV. (d) Mitochondrial proteins Tom20 and MnSOD were unchanged in response to Bnip3 in Bax/Bak DKO MEFs. (e) O2 consumption in Bax/Bak DKO MEFs was assessed in a Seahorse (North Billerika, MA, USA) XF24 extracellular analyzer. The graph is a representative experiment showing rates of endogenous, state 4 (oligo), maximal uncoupler-stimulated (FCCP), and non-mitochondrial (Rot/Anti-A) respiration
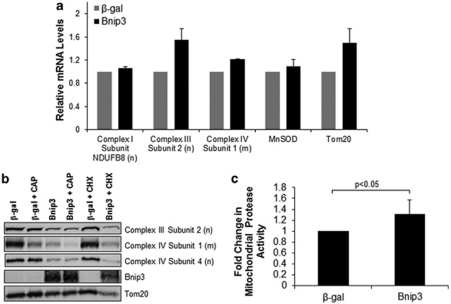
Next, we investigated potential mechanisms by which Bnip3 might cause a reduction in the components of the respiratory complexes. First, we examined the effects of Bnip3 on transcription in Bax/Bak DKO MEFs. Real-time quantitative PCR revealed that Bnip3 overexpression did not reduce transcription of nuclear and mitochondrial genes involved in oxidative phosphorylation (Figure 8a). Next, we investigated the effect of Bnip3 on protein translation. Treatment of cells with chloramphenicol (CAP), an inhibitor of mitochondrial protein synthesis, reduced the mitochondrial-encoded subunit in β-gal-infected cells as expected, but had no effect on the nuclear encoded subunits. Moreover, treatment of cells with cyclohexamide (CHX), an inhibitor of translation of nuclear encoded proteins, did not reduce levels of complex III and IV subunits in β-gal-infected cells (Figure 8b). As the inhibition of mitochondrial and nuclear translation failed to mimic the effect of Bnip3, it suggests that Bnip3 did not reduce the levels of these proteins by inhibiting translation. To investigate whether the reduced protein levels were due to increased degradation, we treated cells with MG-132 to inhibit proteasomal degradation. However, inhibition of the proteasome with MG-132 treatment had no effect on the protein levels in Bnip3-overexpressing cells (Supplementary Figure S7), suggesting that these proteins were not subjected to increased proteasomal degradation. The mitochondrial matrix contains proteases that are involved in degrading proteins in the matrix and inner membrane.25 Interestingly, we found that there was a significant increase in mitochondrial proteolytic activity in Bax/Bak DKO MEFs overexpressing Bnip3 (Figure 8c), suggesting that the reduction of components of complexes I–V might be due to increased degradation in the mitochondria.
Figure 8.
Bnip3 overexpression activates mitochondrial proteases in Bax/Bak DKO MEFs. (a) Real-time PCR for mRNA levels at 24 h after infection (n=3). (b) Representative western blot of cells treated with or without 50 μg/ml chloramphenicol (CAP) or 40 μg/ml cyclohexamide (CHX) for 24 h. (c) Mitochondrial protease activity assay (n=4). Values are expressed as mean±S.D.
Discussion
This study reports several new and important findings. First, Bnip3-mediated mitochondrial autophagy is not dependent on mitochondrial membrane permeabilization and activation of the mitochondrial cell death pathway. Second, Bnip3 reduces levels of essential proteins involved in mitochondrial respiration, which leads to reduced mitochondrial oxidative phosphorylation and cellular ATP levels. Third, removal of these dysfunctional mitochondria by autophagy is an essential survival response and inhibition of autophagy triggers necrotic cell death. Thus, our data suggest that maintaining a healthy population of mitochondria is essential for the survival of these cells and impairment of mitochondrial function by Bnip3 stimulates mitochondrial turnover.
The question of whether autophagy is a cell death or survival mechanism appears to be context dependent. We previously found that autophagy protects against Bnip3-mediated cell death in HL-1 cells.13 In this study, we provide further evidence that mitochondrial autophagy is a protective response against Bnip3 in Bax/Bak DKO MEFs, which are resistant to Bnip3-mediated cell death. As genetic deletion of Bax/Bak prevents activation of the mitochondrial apoptotic pathway, it suggests that necrotic cell death is activated when the defective mitochondria cannot be removed in these cells. Similarly, Huang et al.26 found that autophagy has a pro-survival role in H2O2-treated Bax/Bak DKO MEFs, in whom suppression of autophagy by knockdown of essential autophagy genes enhanced H2O2-induced cell death. In contrast, Shimizu et al.27 found that etoposide treatment induced a non-apoptotic cell death in Bax/Bak DKO MEFs that was dependent on autophagy. These studies demonstrate that autophagy can be both protective and detrimental in the same cell type depending on the stimulus.
Bnip3 and its homolog Nix/Bnip3L share several features with the BH3-only proteins of the Bcl-2 family, such as sequence homology in the BH3 domain,28, 29 residence in the mitochondrial membrane,30 and the ability to interact with Bcl-2 and Bcl-XL.28, 31 However, studies have revealed that Bnip3 and Nix do not function similar to typical BH3-only proteins, such as tBid. For instance, the transmembrane domains, but not the BH3 domains, of Bnip3 and Nix, have a major role in the induction of cell death.30 Bnip3 and Nix are weak inducers of cell death that induce opening of the mPTP,9, 12, 28 and they are strong activators of mitochondrial autophagy.13, 17, 21 We have also found that Bnip3 induces mPTP-independent permeabilization of the inner mitochondrial membrane.19 In addition, the BH3-only protein PUMA was found to induce autophagy, which was dependent on the presence of Bax/Bak.32 Although Bnip3-mediated cell death is dependent on Bax/Bak,11 we found that Bnip3-mediated autophagy is independent of Bax/Bak. Similarly, Nix was recently reported to be essential for mitochondrial autophagy during reticulocyte maturation, and this process was independent of Bax/Bak.21 In this study, we confirmed that Nix induces autophagy in the absence of Bax/Bak. Clearly, these two proteins are atypical BH3-only proteins and their function may not be limited to a pro-death role. On the basis of our study and others, it is clear that Nix and Bnip3 are also important regulators of mitochondrial turnover. Recently, Nix was reported to interact directly with the autophagy proteins LC3 and GABARAP, and to recruit GABARAP to depolarized mitochondria.23 In this study, we demonstrate that Bnip3 can also interact with LC3. This suggests Nix and Bnip3 might function as receptors for autophagosomes and that binding of Nix or Bnip3 to LC3 on the autophagosome tethers the mitochondrion to the autophagosome. Although our data suggest that mitochondria in the Bax/Bak DKO MEFs overexpressing Bnip3 retain their ΔΨm, the impairment of the electron transport chain and ATP synthesis will likely reduce the ΔΨm. Interestingly, a reduction in ΔΨm has been reported to target the mitochondria for degradation by the autophagosome.33 Thus, this could be a potential mechanism by which Bnip3 targets mitochondria for degradation.
It is interesting that Bnip3 reduced the levels of both nuclear- and mitochondrial-encoded proteins involved in oxidative phosphorylation, but had no effect on other mitochondrial proteins such as Tom20 and MnSOD in Bax/Bak DKO MEFs. The oxidative phosphorylation system is composed of five major membrane protein complexes. Biogenesis and assembly of these complexes is a complex process involving the coordination of two different genomes. The majority of the subunits for the different complexes are encoded by nuclear genes, but 13 subunits are encoded by the mitochondrial genome. Interestingly, we found that Bnip3 did not disrupt transcription or translation of nuclear and mitochondrial encoded subunits. Mitochondria contain endogenous proteases, such as the AAA proteases and the Lon protease, that are responsible for the degradation of internal mitochondrial proteins, including proteins in the electron transport chain.25, 34 The proteolysis products are transported to the cytosol by a specific peptide transporter, wherein they are degraded by the proteasome.35 Our data suggest that Bnip3 overexpression increases protease activity in the mitochondria. Thus, we hypothesize that this might lead to increased degradation of proteins in the mitochondria. However, further studies are required to determine exactly how Bnip3 activates mitochondrial proteases and whether it leads to specific degradation of proteins involved in oxidative phosphorylation.
It is clear that Bnip3 can perturb mitochondria by multiple mechanisms. First, Bnip3 can induce mitochondrial membrane permeabilization via Bax/Bak11 or via opening of the mPTP,9, 12 which leads to release of mitochondrial pro-death proteins and activation of cell death. In this study, we have discovered a novel function of Bnip3, wherein Bnip3 affects mitochondrial function by specifically reducing the levels of proteins involved in oxidative phosphorylation. Bnip3 contributes to the loss of cells in myocardial ischemia/reperfusion-injury11, 13 and post-infarct remodeling.36 Bnip3 also has a role in many cancers, wherein alteration in Bnip3 expression and localization contributes to the deregulation of cell death.10 Thus, our findings emphasize the importance of gaining an increased understanding of how Bnip3 regulates mitochondrial function and autophagy, as Bnip3 may represent a potential therapeutic target in both heart disease and cancer.
Materials and Methods
Cell culture
Bax/Bak DKO MEFs were generously provided by Dr. Craig B. Thompson and maintained in DMEM supplemented with 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin. Atrial-derived HL-1 myocytes were obtained from Dr. WC Claycomb37 and cultured in Claycomb medium (JHR Biosciences, Lenexa, KS, USA) supplemented with 10% FBS, 2 mM -glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 0.25 μg/ml amphotericin B, and 100 μM norepinephrine (Sigma, St Louis, MO, USA).
Transient transfections and adenoviral infections
MEFs were transiently transfected using Lipofectamine2000 (Invitrogen, Carlsbad, CA, USA) and HL-1 cells were transfected with Effectene (Qiagen, Valencia, CA, USA) according to the manufacturers' instructions. The transfection efficiency MEFs and HL-1 cells were ∼40%. For adenoviral infections, the MEFs were incubated with an adenovirus encoding β-gal or Bnip3 for 2 h in DMEM+2% heat inactivated FBS. After infection, the cells were rinsed and then incubated in regular cell culture media.
siRNA and hypoxia
A mixture of four ON-TARGETplus SMARTpool siRNAs (Dharmacon) specific to Bnip3, Bnip3L/Nix or non-targeting control was used for silencing experiments. The siRNA sequences are described in Supplementary Table S1. Bax/Bak DKO MEFs were transfected with 100 nM of each siRNA pool using DharmaFECT 4 (Dharmacon, Lafayette, CO, USA) and downregulation of Bnip3 and Bnip3L was verified by western blotting at 72 h after transfection. For hypoxia experiments, Bax/Bak DKO MEFs were transfected with the siRNA for 24 h before deprived of oxygen in hypoxic pouches (GasPakTM EZ; BD Biosciences, San Diego, CA USA) at 37°C. Cell lysates for western blotting experiments were prepared at after 48 h of hypoxia.
Assessment of autophagy, ΔΨm, propidium iodide (PI) uptake, and nuclear condensation
To assess autophagy cells overexpressing LC3-GFP were examined at × 60 magnification and classified as: (a) cells with diffuse LC3-GFP fluorescence, or as (b) cells with numerous LC3-GFP puncta (>30 dots/cell), representing autophagosomes. For analysis of ΔΨm, 24 h after transfection live cells were loaded with 20 nM TMRM, which accumulates within the matrix of respiring mitochondria for 20 min at 37°C. Only GFP-positive cells were scored for TMRM fluorescence. To measure PI uptake or nuclear condensation, live cells were stained with 30 μg/ml Hoechst 33342 for 5 min at room temperature and then examined by fluorescence microscopy. For transfected cells, only GFP-positive cells were scored and at least 150 cells were scored from two replicate dishes in three independent experiments. Cells were observed through a Nikon TE300 fluorescence microscope (Nikon, Melville, NY, USA) equipped with a × 10 lens (0.3 NA, Nikon), a × 40 Plan Fluor and a × 60 Plan Apo objective (1.4 and 1.3 NA oil immersion lenses; Nikon), a Z-motor (ProScanII, Prior Scientific, Rockland, MA, USA), and a cooled CCD camera (Orca-ER, Hamamatsu, Bridgewater, NJ, USA). Alternatively, cells were observed a Carl Zeiss AxioObserver Z1 (Carl Zeiss, Göttingen, Germany) fitted with a motorized Z-stage and an apotome for optical sectioning. For high-resolution microscopy, Z-stacks were acquired at × 63 magnification with 0.6 μm increments in ApoTome mode using a high-resolution AxioCam MRm digital camera, a × 63 Plan-Apochromat (Carl Zeiss) (oil immersion) objective and Zeiss AxioVision 4.8 software (Carl Zeiss).
Transmission EM
Bax/Bak DKO MEFs were fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer, after fixed in 1% osmium tetroxide, and then treated with 0.5% tannic acid, 1% sodium sulfate, cleared in 2-hydroxypropyl methacrylate and embedded in LX112 (Ladd Research, Williston, VT, USA). Sections were mounted on copper slot grids coated with parlodion and stained with uranyl acetate and lead citrate for examination on a Philips CM100 EM (FEI, Hillsbrough, OR, USA).
Lactate dehydrogenase (LDH) release
Bax/Bak DKO MEFs in growth medium without phenol red were infected with β-gal or Bnip3. Leakage of LDH from dead or dying cells was assessed at 24 and 48 h after infection by measuring LDH release into the medium using a cytotoxicity detection kit according to the manufacturer's protocol (Roche Applied Bioscience, Indianapolis, IN, USA). The released LDH is expressed as a percentage of total LDH in the well.
Immunoprecipitation
Bax/Bak DKO MEFs cultured in 60 mm dishes were co-transfected with vector or Bnip3 plus GFP-LC3 using Lipofectamine 2000. After 24 h, cells were lysed in buffer containing 50 mM Tris-HCl (pH 7.4) 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, and complete protease inhibitor cocktail (Roche Applied Bioscience) and the concentrations were adjusted to 1 mg/ml. Cell lysates were precleared with protein G PLUS agarose (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 1 h, and then incubated with anti-GFP o/n to immunoprecipitate GFP-LC3. Immune complexes were captured with protein G PLUS agarose beads, centrifuged, washed four times in PBS, and solubilized in 2 × SDS sample buffer. Proteins were analyzed by western blotting with anti-Bnip3 to determine how much of Bnip3 co-immunoprecipitated with GFP-LC3, or anti-GFP to verify that equal amounts of GFP-LC3 was immunoprecipitated.
Western blotting
Cells were lysed in buffer containing 50 mM Tris-HCl (pH 7.4) 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, and complete protease inhibitor cocktail (Roche Applied Bioscience), and cleared by centrifugation at 20 000 × g for 20 min.11 Proteins in the supernatants were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with an antibody against Bnip3 (1 : 1000, Sigma), LC3 (1 : 1000, Cell Signaling Technology, Danvers, MA, USA), complex III subunit 2 (1 : 1000, Invitrogen), complex IV subunit 1 (1 : 1000, Invitrogen), complex IV subunit 4 (1 : 1000, Invitrogen), Mitoprofile Total OXPHOS Rodent WB Antibody Cocktail (1 : 200, MitoSciences, Eugene, OR, USA), UCP-2 (1 : 200, Santa Cruz Biotechnology), UCP-3 (1 : 200, Santa Cruz Biotechnology), P-AMPK/AMPK (1 : 1000, Cell Signaling Technology), GAPDH (1 : 1000, Cell Signaling Technology), tubulin (1 : 1000, Sigma), or actin (1 : 1000, Sigma).
ATP Measurements
Bax/Bak DKO MEFs overexpressing β-gal or Bnip3 were counted at 48 h after infection and lysates from 5 × 104 cells were used per well (96-well plate). Cellular ATP levels were quantified using a luciferase reaction-based assay kit (CellTiter-Glo Reagent, Promega Biosciences, San Luis Obispo, CA, USA).
Citrate synthase assay
Citrate synthase activity was measured using a Citrate Synthase Activity kit (Sigma). Bax/Bak DKO MEFs were homogenized in the CelLytic MT Cell Lysis Reagent. The supernatant was transferred to the kit assay buffer supplemented with acetyl CoA (0.3 mM), and 5,5′-dithiobis-(2-nitrobenzoic acid; 0.1 mM). Oxaloacetate (0.1 mM) was added to start the reaction and changes in absorbance at 420 nm at 25°C were measured using a SpectraMax plate reader (Molecular Devises, Sunnyvale, CA, USA). The activity is expressed as μmol/min/mg protein.
Oxygen consumption
An XF24 extracellular flux analyzer (Seahorse Biosciences, North Billerica, MA, USA) was used to measure the rates of O2 consumption by monolayers of cells. Bax/Bak DKO MEFs infected with β-gal or Bnip3 were plated at 6 × 104 cells/well the day before the experimentation. Respiration was measured at 48 h after infection in unbuffered, serum-free DMEM (Sigma) supplemented with 10 mM glucose, 10 mM pyruvate, and 2 mM glutaMAX (Invitrogen; pH 7.4). Rates of endogenous respiration were initially measured followed by state 4 (resting) respiration by the addition of 1 μM oligomycin (Sigma). To measure maximal respiration, a titrated concentration of the uncoupler carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP, Sigma) was added, followed by measurement of non-mitochondrial O2 consumption in the presence of 200 nM rotenone and 4 μM antimycin A (Sigma). O2 consumption was measured in four to six replicate wells using cells from three separate experiments. An updated algorithm that accounts for oxygen diffusion through the plastic plate was used for data analysis.38
Analysis of mtDNA
Genomic DNA was extracted from MEFs using GenElute Mammalian Genomic DNA Kit (Sigma). Following the extraction, traditional PCR amplification was performed on 1 μg genomic DNA using PCR Master Mix (Fermentas, Glen Burnie, MD, USA). 18 s rRNA (aaacggctaccacatccaag and cctccaatggatcctcgtta) was used as a control for nuclear DNA and Cyt b (ttctgaggtgccacagttatt and gaaggaaaggtattagggctaaa) for mtDNA quantification. For real-time PCR experiments, primers were designed using Primer Express Software (Applied Biosystems, Carlsbad, CA, USA) and are described in Supplementary Table S1. In all, 40 ng of DNA were used as template in real-time PCR reaction and performed using an ABI PRISM 7500 (Applied Biosystems).
Quantification of RNA by quantitative real-time PCR
Total RNA was extracted from Bax/Bak DKO MEFs cells using TRIzol Reagent (Invitrogen) according to the manufacturer's instructions. First strand cDNA synthesis from total RNA was performed using High Capacity RNA-to-cDNA Master Mix (Applied Biosystems). Primers and probes for real-time PCR were designed by Applied Biosystems and are described in Supplementary Table S1. Real-time PCR was performed using ABI PRISM 7500 and relative quantification of RNA amount was accomplished by using the comparative Ct method (2−ΔΔCt). Relative amounts of mRNA for genes of interest were normalized to housekeeping gene GAPDH and expressed relative to the calibrator using the arithmetic formula:
where ΔCt = Ct (gene of interest) − Ct (GAPDH).
Inhibition of mitochondrial translation
Cells were infected with adenoviruses encoding β-gal or Bnip3 for 24 h and then treated with vehicle (DMSO), 50 μg/ml chloramphenicol or 40 μg/ml CHX to inhibit translation of nuclear and mitochondrial encoded proteins. These concentrations have previously been reported to inhibit protein translation in fibroblasts.39, 40 The inhibition was verified by western blotting for nuclear and mitochondrial encoded proteins. After 24 h of treatment with inhibitors, cell lysates were prepared for western blot analysis.
Mitochondrial protease assay
Protease activity was measured in isolated mitochondria using the Pierce Fluorescent Protease Assay Kit (Thermo Scientific, Pittsburg, PA, USA). Mitochondria were isolated from Bax/Bak MEFs overexpressing β-gal or Bnip3 at 48 h after infection. Cells were homogenized in a glass douncer in buffer containing 10 mM Tris-HCl (pH 7.4), 0.25 M sucrose, and 1 mM EDTA. The homogenates were centrifuged at 300 × g for 5 min to pellet unbroken cells. The supernatant was further spun at 6000 × g for 15 min to pellet mitochondria. The mitochondria were resuspended in assay buffer (50 mM Tris-Hcl, pH 7.9, 10 mM MgCl2, 1 mM DTT and 0.05% Triton X-100). Cleavage of FTC-casein was measured in a fluorescent plate reader (Molecular Devices) equipped with fluorescein excitation/emission filters (485/538 nm). TPCK trypsin was used as a positive control.
Statistical analysis
All values are expressed as means±S.D. Student's t-test was used to evaluate significance between two experimental conditions and P<0.05 was considered to be statistically significant.
Acknowledgments
We are grateful for the Bax/Bak DKO MEFs from Dr. Thompson (University of Pennsylvania, PA, USA) and the HL-1 cells from Dr. Claycomb (LSU Health Sciences Center, LA, USA). We appreciate the GFP-LC3 cDNA from Dr. T Yoshimori (National Institute of Genetics, Japan) and the help of Dr. Malcolm Wood (The Scripps Research Institute, La Jolla) with the electron microscopy experiments. This research was supported by NIH HL087023, NIH HL101217 and a Scientist Development Award from the American Heart Association. MNQ is supported by a Post-Baccalaureate Research Supplement from NHLBI.
Glossary
- Bnip3
Bcl-2/adenovirus E1B 19-kDa-interacting protein 3
- β-gal
β-galactosidase
- DKO
double knockout
- MEF
mouse embryonic fibroblast
- ΔΨm
mitochondrial membrane potential
- LTR
lysotracker red
- LC3
microtubule-associated protein light chain 3
- 3-MA
3-methyladenine
- mtDNA
mitochondrial DNA
- AMPK
5′-AMP-activated protein kinase
- UCPs
uncoupling proteins
- PI
propidium iodide
- LDH
lactate dehydrogenase
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by L Scorrano
Supplementary Material
References
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Tannous P, Johnstone JL, Kong Y, Shelton JM, Richardson JA, et al. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007;117:1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- Gustafsson AB, Gottlieb RA. Bcl-2 family members and apoptosis, taken to heart. Am J Physiol Cell Physiol. 2006;292:C45–C51. doi: 10.1152/ajpcell.00229.2006. [DOI] [PubMed] [Google Scholar]
- Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. Embo J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regula KM, Ens K, Kirshenbaum LA. Inducible expression of BNIP3 provokes mitochondrial defects and hypoxia-mediated cell death of ventricular myocytes. Circ Res. 2002;91:226–231. doi: 10.1161/01.res.0000029232.42227.16. [DOI] [PubMed] [Google Scholar]
- Burton TR, Gibson SB. The role of Bcl-2 family member BNIP3 in cell death and disease: NIPping at the heels of cell death. Cell Death Differ. 2009;16:515–523. doi: 10.1038/cdd.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubli DA, Ycaza JE, Gustafsson AB. Bnip3 mediates mitochondrial dysfunction and cell death through Bax and Bak. Biochem J. 2007;405:407–415. doi: 10.1042/BJ20070319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande VC, Cizeau J, Dubik D, Alimonti J, Brown T, Israels S, et al. BNIP3 and genetic control of necrosis-like cell death through the mitochondrial permeability transition pore. Mol Cell Biol. 2000;20:5454–5468. doi: 10.1128/mcb.20.15.5454-5468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamacher-Brady A, Brady NR, Logue SE, Sayen MR, Jinno M, Kirshenbaum LA, et al. Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ. 2007;14:146–157. doi: 10.1038/sj.cdd.4401936. [DOI] [PubMed] [Google Scholar]
- Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29:2570–2581. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad MB, Chen Y, Henson ES, Cizeau J, McMillan-Ward E, Israels SJ, et al. Hypoxia induces autophagic cell death in apoptosis-competent cells through a mechanism involving BNIP3. Autophagy. 2008;4:195–204. doi: 10.4161/auto.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzawa T, Zhang L, Xiao L, Germano IM, Kondo Y, Kondo S. Arsenic trioxide induces autophagic cell death in malignant glioma cells by upregulation of mitochondrial cell death protein BNIP3. Oncogene. 2005;24:980–991. doi: 10.1038/sj.onc.1208095. [DOI] [PubMed] [Google Scholar]
- Quinsay MN, Thomas RL, Lee Y, Gustafsson AB. Bnip3-mediated mitochondrial autophagy is independent of the mitochondrial permeability transition pore. Autophagy. 2010;6:17–24. doi: 10.4161/auto.6.7.13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinsay MN, Lee Y, Rikka S, Sayen MR, Molkentin JD, Gottlieb RA, et al. Bnip3 mediates permeabilization of mitochondria and release of cytochrome c via a novel mechanism. J Mol Cell Cardiol. 2009;48:1146–1156. doi: 10.1016/j.yjmcc.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Lewis W, Diwan A, Cheng EH, Matkovich SJ, Dorn GW., II Dual autonomous mitochondrial cell death pathways are activated by Nix/BNip3L and induce cardiomyopathy. Proc Natl Acad Sci USA. 2010;107:9035–9042. doi: 10.1073/pnas.0914013107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci USA. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem. 1991;266:17707–17712. [PubMed] [Google Scholar]
- Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo JO, Jang MH, Kwon YK, Lee HJ, Jun JI, Woo HN, et al. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem. 2005;280:20722–20729. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- Koppen M, Langer T. Protein degradation within mitochondria: versatile activities of AAA proteases and other peptidases. Crit Rev Biochem Mol Biol. 2007;42:221–242. doi: 10.1080/10409230701380452. [DOI] [PubMed] [Google Scholar]
- Huang Q, Wu YT, Tan HL, Ong CN, Shen HM. A novel function of poly(ADP-ribose) polymerase-1 in modulation of autophagy and necrosis under oxidative stress. Cell Death Differ. 2009;16:264–277. doi: 10.1038/cdd.2008.151. [DOI] [PubMed] [Google Scholar]
- Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- Imazu T, Shimizu S, Tagami S, Matsushima M, Nakamura Y, Miki T, et al. Bcl-2/E1B 19 kDa-interacting protein 3-like protein (Bnip3L) interacts with bcl-2/Bcl-xL and induces apoptosis by altering mitochondrial membrane permeability. Oncogene. 1999;18:4523–4529. doi: 10.1038/sj.onc.1202722. [DOI] [PubMed] [Google Scholar]
- Yasuda M, Theodorakis P, Subramanian T, Chinnadurai G. Adenovirus E1B-19K/BCL-2 interacting protein BNIP3 contains a BH3 domain and a mitochondrial targeting sequence. J Biol Chem. 1998;273:12415–12421. doi: 10.1074/jbc.273.20.12415. [DOI] [PubMed] [Google Scholar]
- Chen G, Cizeau J, Vande VC, Park JH, Bozek G, Bolton J, et al. Nix and Nip3 form a subfamily of pro-apoptotic mitochondrial proteins. J Biol Chem. 1999;274:7–10. doi: 10.1074/jbc.274.1.7. [DOI] [PubMed] [Google Scholar]
- Ray R, Chen G, Vande VC, Cizeau J, Park JH, Reed JC, et al. BNIP3 heterodimerizes with Bcl-2/Bcl-X(L) and induces cell death independent of a Bcl-2 homology 3 (BH3) domain at both mitochondrial and nonmitochondrial sites. J Biol Chem. 2000;275:1439–1448. doi: 10.1074/jbc.275.2.1439. [DOI] [PubMed] [Google Scholar]
- Yee KS, Wilkinson S, James J, Ryan KM, Vousden KH. PUMA- and Bax-induced autophagy contributes to apoptosis. Cell Death Differ. 2009;16:1135–1145. doi: 10.1038/cdd.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. Embo J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin S, Nolden M, Muller S, Hardt O, Arnold I, Langer T. Characterization of peptides released from mitochondria: evidence for constant proteolysis and peptide efflux. J Biol Chem. 2005;280:2691–2699. doi: 10.1074/jbc.M410609200. [DOI] [PubMed] [Google Scholar]
- Young L, Leonhard K, Tatsuta T, Trowsdale J, Langer T. Role of the ABC transporter Mdl1 in peptide export from mitochondria. Science. 2001;291:2135–2138. doi: 10.1126/science.1056957. [DOI] [PubMed] [Google Scholar]
- Diwan A, Krenz M, Syed FM, Wansapura J, Ren X, Koesters AG, et al. Inhibition of ischemic cardiomyocyte apoptosis through targeted ablation of Bnip3 restrains postinfarction remodeling in mice. J Clin Invest. 2007;117:2825–2833. doi: 10.1172/JCI32490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb WC, Lanson NA, Jr, Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, et al. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci USA. 1998;95:2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerencser AA, Neilson A, Choi SW, Edman U, Yadava N, Oh RJ, et al. Quantitative microplate-based respirometry with correction for oxygen diffusion. Anal Chem. 2009;81:6868–6878. doi: 10.1021/ac900881z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, You S, Farris J, Foster LK, Foster DN. Post-transcriptional inactivation of p53 in immortalized murine embryo fibroblast cells. Oncogene. 2001;20:3306–3310. doi: 10.1038/sj.onc.1204423. [DOI] [PubMed] [Google Scholar]
- Li CH, Cheng YW, Liao PL, Yang YT, Kang JJ. Chloramphenicol causes mitochondrial stress, decreases ATP biosynthesis, induces matrix metalloproteinase-13 expression, and solid-tumor cell invasion. Toxicol Sci. 2010;116:140–150. doi: 10.1093/toxsci/kfq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.