Abstract
The evolutionary history of the entire Escherichia coli chromosome was traced by examining the distribution of the ∼4300 open reading frames (ORFs) from E.coli MG1655 among strains of known genealogical relationships. Using this framework to deduce the incidence of gene transfer and gene loss, a total of 67 events—37 additions and 30 deletions—were required to account for the distribution of all genes now present in the MG1655 chromosome. Nearly 90% of the ORFs were common to all strains examined, but, given the variation in gene content and chromosome size, strains can contain well over a megabase of unique DNA, conferring traits that distinguish them from other members of the species. Moreover, strains vary widely in their frequencies of deletions, which probably accounts for the variation in genome size within the species.
Keywords: bacterial chromosomes/deletions/Escherichia coli/genome evolution/lateral gene transfer
Introduction
The analysis of total gene content, as recovered from complete genomic sequences, has begun to elucidate the manner in which bacterial genomes change over an evolutionary timescale. Initial comparisons of sequenced genomes have demonstrated that relatively few genes are shared by all bacterial species (Mushegian and Koonin, 1996; Watanabe et al., 1997; de Rosa and Labedan, 1998; Huynen and Bork, 1998; Snel et al., 1999), indicating that bacterial genomes are very dynamic and subject to repeated events of gene acquisition and loss (Doolittle, 1999; Jain et al., 1999). However, the set of organisms sequenced to date cannot fully reveal the rate and pattern of genetic events that shape their genomes. In most comparisons, the organisms are too distantly related and have sustained so many changes that their evolutionary histories have been erased. Even in cases where complete sequence information is available for two closely related or conspecific strains, e.g. Helicobacter pylori (Alm and Trust, 1999) or Chlamydia spp. (Read et al., 2000), it is usually not possible to establish: (i) the rate and order of events that led to the present day chromosome compositions; (ii) whether the differences in gene content are due to the acquisition of horizontally transferred genes or to the loss of ancestral sequences; or even (iii) whether shared genes are ancestral to the species as a whole or were later acquired by a more recent common ancestor.
An alternative approach that does not rely upon information from other sequenced organisms has been developed to establish the ancestry and ages of genes within a bacterial genome (Lawrence and Ochman, 1997). This method assesses the history of a gene by examining its degree of departure from sequence characteristics prevalent in the genome as a whole and, hence, is independent of both the number and the phylogenetic distribution of other completely sequenced organisms. Application of these procedures has revealed that bacteria vary widely in the amount of horizontally acquired sequences currently present in their genomes (Ochman et al., 2000). For example, in Escherichia coli strain MG1655 (Blattner et al., 1997), it was estimated that nearly 18% of its 4290 open reading frames (ORFs) were introduced in at least 234 transfer events and that the majority of the acquired genes appeared in this lineage relatively recently (Lawrence and Ochman, 1998). Coupled with the fact that natural isolates of E.coli may differ by nearly a megabase in genome size (Richmond et al., 1999; Tao et al., 1999), these findings suggest that gene content is highly variable within this species, with divergent strains having gained and deleted very different arrays of genes.
To investigate the dynamics of genome content over an evolutionarily relevant timescale, we applied a whole genome approach to trace the distribution of every gene from the sequenced E.coli MG1655 chromosome among natural strains of varying degrees of relatedness, known ancestries and representing the range of chromosome sizes in the species at large. By comparing lineages of varying degrees of relatedness, it is possible to reconstruct individual events of gene transfer and loss occurring over the entire E.coli genome, to estimate the amount of unique DNA within each strain and to determine the relative ages of every gene on the MG1655 chromosome. Ages of genes inferred from their phylogenetic distributions and derived from sequence characteristics (Lawrence and Ochman, 1997, 1998)—and hence the evolutionarily effective rates of gene transfer and loss calculated by both approaches—were in many cases found to be similar. Repeated events of gene acquisition and the concomitant loss of sequences have created a situation in which divergent lineages of E.coli possess, along with the basal set of genes, a unique complement of genes (and their encoded traits) that distinguishes them from other members of the species. Tracing the distribution of gene transfer events showed that certain strains are prone to deletions, which might account for the wide variation in genome size within the species.
Results and discussion
Strains of E.coli are highly variable in gene content and have experienced numerous episodes of gene acquisition and loss. The examination of E.coli strains of known phylogenetic relationships allows us to trace each event contributing to this variation and to recognize three classes of ORFs: (i) those absent from the ancestor of all E.coli but acquired by an ancestor of MG1655 after diverging from a particular lineage; (ii) those ancestral to all E.coli but lost by another lineage; and (iii) those ancestral to and present in all lineages of E.coli.
Genomic novelty and strain diversity
Gene content, like genome size, is thought to be closely related to phylogeny (Bergthorsson and Ochman, 1995, 1998) such that the evolutionary distance, as resolved by nucleotide divergence in homologous sequences, is also reflected in the total constellation of genes shared between strains and species. The relationship between phylogeny and gene content has also been observed for divergent bacterial taxa (Fitz-Gibbon and House, 1999; Snel et al., 1999), but there is no reason why such an association should exist within species where it might be erased by either very high or very low rates of gene transfer and loss.
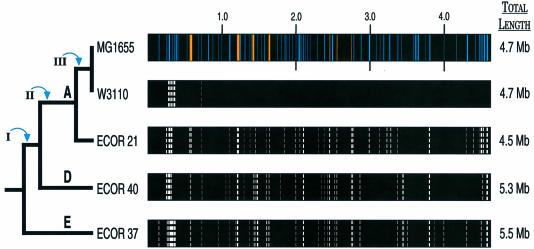
The gene contents of the two closely related and recently derived laboratory isolates, MG1655 and W3110, are very similar, and there is also some degree of correspondence between genetic distance and gene content among the more distantly related strains of E.coli. ECOR 21 and ECOR 40 have roughly the same number of ORFs in common with MG1655, although ECOR 21 is much more closely related to MG1655 based on sequence divergence and has much less unique DNA (Table I). It appears that additions and deletions within E.coli are sufficiently frequent for only very close relatives to display an excess of shared genes, whereas each divergent lineage of E.coli possesses several hundred unique genes.
Table I. Amounts of unique and conserved DNA among strains of E.coli.
| Strain | Number of ORFs missing (of the 4290 in MG1655) | Amount of unique DNA (relative to MG1655)a |
|---|---|---|
| W3110 | 82 | 65 kb |
| ECOR 21 | 318 | 77 kb |
| ECOR 37 | 392 | 1183 kb |
| ECOR 40 | 324 | 925 kb |
aCalculated from amounts of missing DNA as determined by filter hybridizations and total chromosome lengths.
The strains analyzed in this study differ by as much as 800 kb in chromosome length, and, given the variation in gene content, we can estimate the amount of unique DNA harbored by each relative to the E.coli MG1655 genome (Table I). For example, W3110 is of the same chromosome size as MG1655 but lacks ∼80 of the ORFs (65 kb) present in MG1655, implying that it harbors approximately the same amount of novel sequences. On the other hand, ECOR 37, in addition to having a larger genome, lacks >350 kb of coding sequences of MG1655, resulting in a difference of well over a megabase of unique DNA. The coding capacity of the unique DNA in ECOR 37 cannot be determined from these studies; however, this strain, which was originally isolated from a marmoset, is similar in chromosome size and is phylogenetically closely related to the pathogenic E.coli O157:H7, whose complete genomic sequence is almost fully resolved (Burland et al., 1998; Perna et al., 1998; Plunkett et al., 1999). Hence, future hybridization studies will establish which of the genomic regions unique to O157:H7 are also in ECOR 37, and the extent to which related pathogenic and non-pathogenic E.coli strains differ in their gene contents.
Of the 4290 ORFs present in MG1655, a maximum of 3782 are common to all strains of E.coli. This is certainly an overestimate of the minimal number of genes shared (and required) by E.coli because our analysis did not include non-contiguous (i.e. individual) ORFs whose distributions vary between strains. Consequently, the amounts of novel DNA calculated for each strain are underestimates. The number of addition or deletion events involving single genes could be very high, perhaps equal to, or exceeding the total number of events affecting more than one gene (Lawrence and Ochman, 1998). But because these regions are small—E.coli genes average 1 kb in length—they would be of relatively little consequence on the total amount of unique DNA per strain.
Another factor that enters into calculations of the amounts of unique DNA per strain is the presence of transposable or other repetitive elements whose copy numbers vary between strains. Naturally, such genome-wide hybridization studies can only reveal the presence or absence, and not the number of copies, of each type of repetitive element. However, the copy numbers of several classes of insertion sequences (IS1–IS5 and IS30) have already been enumerated for these strains (Sawyer et al., 1987) and range from seven chromosomally encoded copies in ECOR 37 to 35 copies in MG1655. Although IS elements account for very little of the variation in total lengths of chromosomes, this difference in the numbers of IS elements harbored by each strain reflects an additional 30 kb of novel DNA in ECOR 37 relative to MG1655.
Acquisition and deletion events in the E.coli genome
By tracking the presence or absence of each of the MG1655 ORFs among E.coli strains of known genealogical relationships, we can account for the present distribution of all 4290 genes by inferring a total of 67 events: 37 additions and 30 deletions (Table II). Each event was characterized by contiguous ORFs missing in one or several strains, and when missing from more than one strain, the variable regions generally spanned the identical set of ORFs. However, the 5–8 min region of the chromosome has an extremely complex history, involving overlapping addition and deletion events that could not be fully reconstructed by this approach. For this region, which represents <3% of the total chromosome, neighboring deletions (separated by fewer than five ORFs) with the same phylogenetic distribution were counted as a single event.
Table II. Distribution of insertion and deletion events.
| No. of Events | Percentage GCb | No. of ORFs (range)c | |
|---|---|---|---|
| Insertions intoa | |||
| branch I | 11 | 48.3 | 70 (2–24) |
| branch II | 10 | 49.6 | 70 (3–23) |
| branch III | 14 | 45.4 | 193 (4–36) |
| MG1655 | 2 | 58.4 | 35 (3–22) |
| Deletions from | |||
| W3110 | 3 | 54.2 | 57 (10–24) |
| ECOR 21 | 15 | 49.0 | 92 (2–17) |
| ECOR 37 | 4 | 48.0 | 34 (2–23) |
| ECOR 40 | 8 | 47.7 | 30 (2–6) |
aBranch positions (I, II and III) correspond to those shown in Figure 1.
bCalculated as the unweighted average of all events.
cCalculated as the total number of ORFs contained in events.
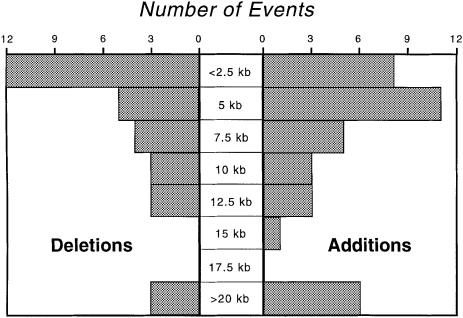
This analysis monitors addition events occurring along the lineage leading to MG1655 (i.e. branches I, II and III in Figure 1), as well as those deletions (involving MG1655 genes) occurring in any of the other lineages, therefore we anticipate an ∼2-fold excess of deletion events. Also, given that the E.coli chromosome has not been ever expanding over an evolutionary timescale (Lawrence and Ochman, 1998), and hence in the long term additions would be offset by an equal loss of DNA, the relative number of deletion events should be augmented further because deletions tend to be shorter than insertions (6.4 versus 8.3 kb; Figure 2). Thus, we expect an excess of deletions events; however, there was no significant difference in the numbers of insertions and deletions (χ2 = 0.4; P >0.5).
Fig. 1. Distribution of deletions among strains of E.coli. Chromosomes are represented linearly and drawn to equal length to show the locations of missing ORFs relative to their map positions in MG1655. Actual chromosome sizes, as determined by pulsed field gel electrophoresis (Bergthorsson and Ochman, 1998), are shown to the right. White dashed lines portray chromosomal positions where the corresponding MG1655 ORFs are lacking in a laboratory (W3110) or natural (ECOR) isolate of E.coli. The relative thickness of these lines denotes the amount of missing DNA. Blue bands show the positions and sizes of regions in the MG1655 chromosome deduced to be horizontally acquired, based on sequence features, and the orange bands represent the positions and sizes of known prophage in the MG1655 chromosome (Lawrence and Ochman, 1998). Upper case letters designate major phylogenetic subgroups within E.coli. Roman numerals indicate ancestral lineages of E.coli that acquired DNA and are referred to in the text and in Table II.
Fig. 2. Size distribution of addition and deletion events in strains of E.coli.
A notable feature of this analysis is that the numbers of deletion events are highly variable among natural strains of E.coli; the frequency of deletions is greatest in the lineage with the smallest chromosome (Table II). These differences in deletion frequencies are independent of the rates of sequence evolution: the strain enduring the most deletions is not on an unusually long branch of the phylogenetic tree. Given the dynamic nature of the E.coli chromosome, these results suggest that rates of deletion and gene loss govern the within-species variation in genome size. Because these studies only recognize changes in gene content relative to that of MG1655, it is possible that all strains have the same deletion rate but that many of the deletions occurring in more distantly related lineages (ECOR 37 and ECOR 40) involve unique sequences and go undetected. However, the slight association between phylogenetic distance and gene content, as well as the observation that each strain has experienced several independent deletions of MG1655 ORFs, argue that there is little predisposition towards the deletion of unique regions.
Both insertion and deletion events include regions of atypical composition; however, a higher proportion of the acquisition events have GC contents that deviate from those typical of E.coli genes (57% of the insertion events and 40% of the deletion events have base compositions either <48% or >55% GC). This is not surprising given that sequences are often acquired from organisms of very distinct base compositions, whereas deletion events include those acquired sequences that do not benefit the host as well as some ancestral sequences.
Characteristics of certain variable regions appear to make them prone to deletion. At least two of the acquired regions were subsequently deleted by a strain, and in other cases, the identical segment was lost independently by two lineages. Several of the insertion/deletion events involve genetic elements, such as rhs, insertion sequences or phage, that are known to be exchanged among strains or species. There are several prophage in MG1655 (and W3110) that were not present or intact in any of the natural isolates, implying their relatively recent acquisition by the lineage adopted for laboratory use. Aside from these prophage, ∼10% of the other acquisition/deletion events are situated next to tRNA loci, which are common integration sites of phage and other foreign sequences (Cheetham and Katz, 1995; Ochman et al., 2000). In addition, both of the MG1655-specific additions of ORFs (Table II) were adjacent to, and probably mediated by IS elements.
Gene distribution and function
These microarrays include all 4290 ORFs present in the E.coli MG1655 chromosome, therefore we can identify the specific genes that have a sporadic distribution among strains. Aside from prophage (which, as noted above, are present in the laboratory strains but missing from the ECOR strains; orange stripes in Figure 1), most of the larger regions of variable distribution have no assigned function or phenotype. However, many of the ‘named’ genes, whose distributions were found to be affected by acquisition or deletion events, have already been shown to be variable within E.coli. For example, the hsd genes, encoding a type I restriction and modification system, were known to be lacking in some of the ECOR strains (Barcus et al., 1995). Similarly, other variable regions such as the rfa and rfb operons, which specify the O-antigen (Klena et al., 1993; Lai et al., 1998), and the aga operon, conferring the ability to grow on N-acetyl-galactosamine (Charbit and Autret, 1998; Brinkkotter et al., 2000), are known to be subject to horizontal transfer or deletion in E.coli strains and other species. Other sequences that were lacking in one or more strains of E.coli include genes from the fec (iron transport; Angerer and Braun, 1998), chp (cell proliferation; Masuda et al., 1993), hip (peptidoglycan synthesis inhibition; Black et al., 1991), mcr (methylation restriction; Raleigh and Wilson, 1986) and rel (cytotoxin; Gotfredsen and Gerdes, 1998) operons.
Evolutionary timing of acquisition and deletion events
By tracing the phylogenetic distribution of each event of gene transfer or loss, we can establish the relative time of its appearance and, hence, the duration of ORFs within the species at large. The ages of genes within the E.coli MG1655 genome have also been estimated analytically by calculating the degree to which the base composition of a coding region conforms to that typical for an E.coli gene (Lawrence and Ochman, 1998). These algorithms assume that variation in GC contents across bacterial species is largely due to mutational biases (Sueoka, 1988, 1992), and that genes acquired from an organism of distinct base composition will ‘ameliorate’ to resemble the features of the resident genome (Lawrence and Ochman, 1998). Given the substitution rates estimated for E.coli and the mutational bias of this species, it is possible to predict the amount of time required after transfer for a gene to fully resemble native DNA and to estimate the amount of time that a horizontally transferred gene has been ameliorating (i.e. residing) in the genome, thus providing the age of acquired sequences.
In examining the correspondence between the dynamics of horizontally transferred regions as assessed by ‘amelioration’ algorithms and those resolved through the use of filter hybridizations (which shows the phylogenetic distribution of MG1655 ORFs among E.coli strains), there are three classes of results.
(i) Genes of sporadic phylogenetic distribution but not identified as being horizontally acquired in MG1655 based on sequence features. This category includes MG1655 genes that were deleted from individual strains or lineages, but also includes acquired regions whose sequence characteristics are sufficiently similar to those of ancestral genes such that they were not identified as being horizontally acquired.
At several sites of the chromosome there are ORFs that were originally viewed as ancestral to E.coli but whose phylogenetic distributions are best explained by lateral transfer. Perhaps not surprisingly, many of these genes have a base composition similar to that of the E.coli chromosome, which might account for the inability to infer their ancestry based solely on sequence characteristics. For example, a 20 kb region that maps to 31 min on the MG1655 chromosome, and that lies between an acknowledged horizontally transferred gene and an IS element, is missing from both ECOR 37 and ECOR 40 (Figure 1). The 17 genes within this region have an average base composition of 52.2% G+C, which is similar to that of the entire E.coli chromosome; hence, these genes were not identified previously as horizontally transferred sequences despite being situated between acquired sequences. Considering all such regions adds at least 80 genes to the number of acquired regions in the MG1655 chromosome and suggests that at least 20% of the current chromosome was not present in the ancestral E.coli.
(ii) Genes identified as being horizontally acquired in MG1655 based on sequence features and having a sporadic phylogenetic distribution among E.coli strains. Based on levels of sequence divergence, the common ancestor of all present-day strains of E.coli is estimated to have occurred between 25 and 40 million years ago (assuming a divergence time of 100–150 million years for the split between E.coli and Salmonella enterica) (Ochman and Wilson, 1987; Ochman et al., 1999). Therefore, regions acquired prior to this time are expected to be present in multiple strains whereas younger regions would be restricted to sets of closely related lineages.
How well do the ages of horizontally acquired regions, as estimated by amelioration algorithms, correspond to their patterns of occurrence among strains? There are 20 regions of the MG1655 chromosome for which both a single acquisition event can explain the observed phylogenetic distribution and a date of acquisition has been assigned (Lawrence and Ochman, 1998). In most cases, there is striking concordance between the estimated age of a gene and its representation among strains.
Based on filter hybridizations, five regions were introduced into an ancestral E.coli lineage along branch I (i.e. present in all strains except ECOR 37; Figure 1), and four of these five regions were estimated by amelioration algorithms to have been acquired between 3 and 50 million years ago. In contrast, five of the six regions introduced along branch II (i.e. present in all strains except ECOR 37 and ECOR 40; Figure 1), and six of the seven regions introduced at branch III (i.e. absent from ECOR 21, ECOR 37 and ECOR 40; Figure 1) were estimated to have been acquired much later—between 0 and 3 million years ago (the two remaining events involve regions confined to MG1655, one of which is estimated to be <3 million and the other >50 million years old). Thus, although the ages assigned to these regions span a broad period, genes displaying a narrower phylogenetic range typically have sequence characteristics denoting a more recent ancestry within E.coli.
For the other cases, where regions identified as being horizontally acquired in MG1655 have a scattered distribution among E.coli strains, the relationship is less straightforward. These include acquired regions that have been deleted from individual strains, as well as those whose histories involve more than one insertion and/or deletion event. Because phage can reside in many hosts and are often chimeric, there are several uncertainties in estimating their duration in a genome from the sequence of their encoded genes. However, none of the MG1655 prophage are present or intact in any of the ECOR strains, suggesting that these phage are rather recent additions to the laboratory lineage that includes strains MG1655 and W3110.
(iii) Genes identified as being horizontally acquired in MG1655 based on sequence features and distributed among all E.coli strains. This category includes: (i) regions that were acquired before the diversification of all present-day strains; (ii) recently acquired regions that have been exchanged among lineages; and (iii) regions that, based on sequence characteristics, were erroneously identified as being acquired.
Among the 14 regions (containing more than one ORF) estimated previously to have been acquired >50 million years ago (Lawrence and Ochman, 1998), 10 were distributed among all strains examined and three were subject to later deletion by an individual strain. Cumulatively, this bolsters the view that these regions were acquired by an ancestral strain prior to the diversification of E.coli, and it is anticipated that phylogenies based on genes within these acquired regions would be identical to those based on ancestral genes.
Aside from those genes acquired prior to the diversification of E.coli strains, it is difficult to account for the distribution of many of the ‘younger’ segments thought to be horizontally acquired but present in all E.coli strains. As evident from the number of these regions associated with IS elements and the capacity for natural isolates to exchange genes (Guttman and Dykhuizen, 1994; Wang et al., 1997; McGraw et al., 1999; Milkman et al., 1999), some recently acquired regions are likely to have been disseminated among strains by recombination. Sequences acquired by one member of a species and transferred horizontally to conspecifics would display the sequence characteristics of a newly acquired gene despite its widespread distribution within a species.
Alternatively, some of the MG1655 genes could have been previously misclassified as being acquired, based on sequence features, when they are actually ancestral to E.coli. Additional information about the distribution of these genes in enteric species closely related to E.coli, such as Salmonella or Klebsiella, has assisted in reconstructing the evolutionary history of these regions. However, because acquired genes retain the sequence characteristics of the donor genome for a very long time (Sueoka, 1988; Lawrence and Ochman, 1997) and due to their mobility may be subject to successive rounds of transfer and/or recombination, the phylogenetic distribution of a gene may not always provide a full clue to its ancestry.
The analysis of sequence features to explore the magnitude of lateral gene transfer has the advantage of being a ‘genome-independent’ approach, i.e. it does not require information from or comparison to any additional genomes to establish the ancestry of a sequence. However, this method generally underestimates the total amount of foreign DNA in a genome because transfer events from genetically similar organisms are only rarely recognized. Moreover, wide variation in the base compositions or codon usage patterns of ancestral genes, as apparent in some bacterial genomes, will confound the identification of horizontally acquired DNA. In contrast, the analysis of gene content via filter hybridizations has, for the first time, provided information on frequencies and features of both addition and deletion events occurring over the evolutionary history of E.coli. Repeated events of gene acquisition and the concomitant loss of sequences have created a situation in which divergent lineages of E.coli possess, along with a basal set, a unique complement of genes (and their encoded traits) that distinguishes them from other members of the species. And, although there is not complete concordance between estimates of the number and ages of acquired genes as determined by sequence features and those obtained by directly assessing gene content, both analyses reveal fluidity in the composition of the E.coli chromosome.
Materials and methods
Total genomic DNAs from five strains of E.coli were used to probe Panorama E.coli Gene Arrays (Sigma-Genosys Biotechnologies, Woodland, TX), which are nylon membranes spotted in duplicate with each the 4290 ORFs present in the completely sequenced E.coli strain MG1655 (Blattner et al., 1997; Richmond et al., 1999; Tao et al., 1999). The five strains consist of two closely related laboratory isolates (W3110 and MG1655) and three strains from the ECOR collection (ECOR 21, ECOR 37 and ECOR 40) (Ochman and Selander, 1984), which were selected to span the major phylogenetic lineages and the range of genome sizes within natural populations of E.coli (Bergthorsson and Ochman, 1995, 1998). Phylogenetic relationships among these strains have been established by multilocus enzyme electrophoresis at 38 polymorphic loci (Herzer et al., 1990) and the nucleotide sequences of several genes (Lecointre et al., 1998). Although some genes yield a slightly different branching order for some of the ECOR strains, the majority of studies have produced the topology shown in Figure 1.
DNA probes were labeled with [33P]dATP by the random hexamer labeling method according to the manufacturer’s directions (Boehringer Mannheim), and unincorporated nucleotides removed with G50 Sephadex spin columns. Prior to hybridization, Gene Array filters were rinsed in 2× SSPE, followed by a 1 h incubation at 65°C in 5 ml of hybridization solution (5× SSPE, 2% SDS, 1× Denhardt’s, 100 mg/ml sheared salmon sperm DNA). After the addition of probe denatured in 3 ml of hybridization solution, filters were incubated for 12–18 h at 65°C with rotation. Following hybridization, blots were washed three times in 0.5× SSPE, 0.2% SDS for 5 min at 25°C and three times for 20 min at 65°C. Washed filters were air dried and exposed overnight to a phosphoimager screen prior to scanning on a PhosphorImager 445SI (Molecular Dynamics) at a pixel density of 175 µM. Prior to rehybridization, the Gene Array filters were stripped of probe by incubation in 50% formamide, 0.5× SSC, 0.1% SDS for 30 min at 65°C, and rinsed in 0.1× SSC, 0.1% SDS for 30 min at 65°C. Complete removal of radioactivity was confirmed by scanning after overnight exposure to a phosphoimager screen.
The scanned Gene Arrays were analyzed with NIH Image software and subsequently scored by eye. Only those spots without any trace of a hybridization signal were recorded as lacking the corresponding ORF, and there were no cases where the duplicate spots gave contradictory results. Because of high variability in the intensity of hybridization signals among ORFs, false negatives—i.e. spots displaying no hybridization signal when the corresponding gene is present—were identified by using MG1655 as probe. We then aligned ORFs with their chromosomal locations to determine the absolute size and number of regions absent from a particular strain.
To control against the inclusion of any additional false negatives, only chromosomal regions lacking at least two adjoining ORFs were recorded as missing from a strain (because the ORFs spotted on these filters are arranged without regard to their map positions, the probability that two contiguous ORFs would be scored as absent due to chance anomalies in their hybridization signals is extremely low). Hence, this approach detects all but the very smallest (i.e. single gene) regions that are variable in their distributions among strains.
Each of these variable regions (containing two or more genes) was classified as either an addition or a deletion event by determining the most parsimonious explanation that accounts for its distribution among strains. Because preliminary results indicated that most insertions and deletions could be mapped onto the phylogenetic tree without invoking homoplasy, parsimony is likely to yield the most accurate reconstruction of evolutionary events. In cases where two or more equally parsimonious scenarios could be applied, we relied upon supplemental information (such as presence in an outside reference species, or proximity to a translocatable element) to determine the ancestry of a region.
For example, genes uniquely missing from ECOR 37 could have originated either by the strain-specific deletion of ancestral sequences or by an acquisition event after this lineage diverged (i.e. along branch I in Figure 1). Such addition and deletion events were distinguished by searching via BLAST for corresponding sequences in the Salmonella sequence databases (http://genome.wustl.edu/gsc/bacterial/salmonella.html) and calculating the synonymous divergence (Ks) between the corresponding E.coli and Salmonella sequences to establish homology. In previous comparisons (Lawrence and Ochman, 1997), Ks values range from 0.4 to 1.2 between E.coli and Salmonella orthologs, with the variation largely due to the degree of codon usage bias. Therefore, for a gene to be considered ancestral to E.coli, we considered whether the degree of divergence at synonymous sites was in line with that of other ancestral genes displaying similar levels of codon bias. In cases where no corresponding genes were recovered from the Salmonella databases, we examined the base composition and codon usage patterns of a gene to gain information about its ancestry. Use of these methods allowed us to distinguish the polarity of additional insertion and deletion events.
References
- Alm R.A. and Trust,T.J. (1999) Analysis of the genetic diversity of Helicobacter pylori: the tale of two genomes. J. Mol. Med., 77, 834–846. [DOI] [PubMed] [Google Scholar]
- Angerer A. and Braun,V. (1998) Iron regulates transcription of the Escherichia coli ferric citrate transport genes directly and through the transcription initiation proteins. Arch. Microbiol., 169, 483–490. [DOI] [PubMed] [Google Scholar]
- Barcus V.A., Titheradge,A.J. and Murray,N.E. (1995) The diversity of alleles at the hsd locus in natural populations of Escherichia coli. Genetics, 140, 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson U. and Ochman,H. (1995) Heterogeneity of genome sizes among natural isolates of Escherichia coli. J. Bacteriol., 177, 5784–5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson U. and Ochman,H. (1998) Chromosomal changes during experimental evolution in laboratory populations of Escherichia coli. Mol. Biol. Evol., 15, 6–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D.S., Kelly,A.J., Mardis,M.J. and Moyed,H.S. (1991) Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol., 173, 5732–5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkkotter A., Kloss,H., Alpert,C. and Lengeler,J.W. (2000) Pathways for the utilization of N-acetyl-galactosamine and galactosamine in Escherichia coli.Mol. Microbiol., 37, 125–135. [DOI] [PubMed] [Google Scholar]
- Blattner F.R. et al.(1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- Burland V., Shao,Y., Perna,N.T., Plunkett,G., Sofia,H.J. and Blattner,F.R. (1998) The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res., 26, 4196–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbit A. and Autret,N. (1998) Horizontal transfer of chromosomal DNA between the marine bacterium Vibrio furnissii and Escherichia coli revealed by sequence analysis. Microb. Comp. Genomics, 3, 119–132. [DOI] [PubMed] [Google Scholar]
- Cheetham B.F. and Katz,M.E. (1995) A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol. Microbiol., 18, 201–208. [DOI] [PubMed] [Google Scholar]
- de Rosa R. and Labedan,B. (1998) The evolutionary relationships between the two bacteria Escherichia coli and Haemophilus influenzae and their putative last common ancestor. Mol. Biol. Evol., 15, 17–27. [DOI] [PubMed] [Google Scholar]
- Doolittle W.F. (1999) Phylogenetic classification and the universal tree. Science, 284, 2124–2129. [DOI] [PubMed] [Google Scholar]
- Fitz-Gibbon S.T. and House,C.H. (1999) Whole genome-based phylogenetic analysis of free-living microorganisms. Nucleic Acids Res., 27, 4218–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotfredsen M. and Gerdes,K. (1998) The Escherichia coli relBE genes belong to a new toxin–antitoxin gene family. Mol. Microbiol., 29, 1065–1076. [DOI] [PubMed] [Google Scholar]
- Guttman D.S. and Dykhuizen,D.E. (1994) Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science, 266, 1380–1383. [DOI] [PubMed] [Google Scholar]
- Herzer P.J., Inouye,S., Inouye,M. and Whittam,T.S. (1990) Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol., 172, 6175–6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen M.A. and Bork,P. (1998) Measuring genome evolution. Proc. Natl Acad. Sci. USA, 95, 5849–5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R., Rivera,M.C. and Lake,J.A. (1999) Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl Acad. Sci. USA, 96, 3801–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klena J.D., Pradel,E. and Schnaitman,C.A. (1993) The rfaS gene, which is involved in production of a rough form of lipopolysaccharide core in Escherichia coli K-12, is not present in the rfa cluster of Salmonella typhimurium LT2. J. Bacteriol., 175, 1524–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai V., Wang,L. and Reeves,P.R. (1998) Escherichia coli clone Sonnei (Shigella sonnei) had a chromosomal O-antigen gene cluster prior to gaining its current plasmid-borne O-antigen genes. J. Bacteriol., 180, 2983–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecointre G., Rachdi,L., Darlu,P. and Denamur,E. (1998) Escherichia coli molecular phylogeny using the incongruence length difference test. Mol. Biol. Evol., 15, 1685–1695. [DOI] [PubMed] [Google Scholar]
- Lawrence J.G. and Ochman,H. (1997) Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol., 44, 383–397. [DOI] [PubMed] [Google Scholar]
- Lawrence J.G. and Ochman,H. (1998) Molecular archaeology of the Escherichia coli genome. Proc. Natl Acad. Sci. USA, 95, 9413–9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda Y., Miyakawa,K., Nishimura,Y. and Ohtsubo,E. (1993) chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J. Bacteriol., 175, 6850–6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw E.A., Li,J., Selander,R.K. and Whittam,T.S. (1999) Molecular evolution and mosaic structure of α, β and γ intimins of pathogenic Escherichia coli. Mol. Biol. Evol., 16, 12–22. [DOI] [PubMed] [Google Scholar]
- Milkman R., Raleigh,E.A., McKane,M., Cryderman,D., Bilodeau,P. and McWeeny,K. (1999) Molecular evolution of the Escherichia coli chromosome. V. Recombination patterns among strains of diverse origin. Genetics, 153, 539–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushegian A.R. and Koonin,E.V. (1996) A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl Acad. Sci. USA, 93, 10268–10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H. and Selander,R.K. (1984) Standard reference strains of Escherichia coli from natural populations. J. Bacteriol., 157, 690–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H. and Wilson,A.C. (1987) Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J. Mol. Evol., 26, 74–86. [DOI] [PubMed] [Google Scholar]
- Ochman H., Elwyn,S. and Moran,N.A. (1999) Calibrating bacterial evolution. Proc. Natl Acad. Sci. USA, 96, 12638–12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Lawrence,J.G. and Groisman,E.A. (2000) Lateral gene transfer and the nature of bacterial innovation. Nature, 405, 299–305. [DOI] [PubMed] [Google Scholar]
- Perna N.T., Mayhew,G.F., Posfai,G., Elliott,S., Donnenberg,M.S., Kaper,J.B. and Blattner,F.R. (1998) Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun., 66, 3810–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plunkett G. III, Rose,D.J., Durfee,T.J. and Blattner,F.R. (1999) Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol., 181, 1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh E.A. and Wilson,G. (1986) Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc. Natl Acad. Sci. USA, 83, 9070–9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read T.D. et al. (2000) Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res., 28, 1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond C.S., Glasner,J.D., Mau,R., Jin,H. and Blattner,F.R. (1999) Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res., 27, 3821–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S.A., Dykhuizen,D.E., DuBose,R.F., Green,L., Mutangadura- Mhlanga,T., Wolczyk,D.F. and Hartl,D.L. (1987) Distribution and abundance of insertion sequences among natural isolates of Escherichia coli. Genetics, 115, 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel B., Bork,P. and Huynen,M.A. (1999) Genome phylogeny based on gene content. Nature Genet., 21, 108–110. [DOI] [PubMed] [Google Scholar]
- Sueoka N. (1988) Directional mutation pressure and neutral molecular evolution. Proc. Natl Acad. Sci. USA, 85, 2653–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N. (1992) Directional mutation pressure, selective constraints and genetic equilibria. J. Mol. Evol., 34, 95–114. [DOI] [PubMed] [Google Scholar]
- Tao H., Bausch,C., Richmond,C., Blattner,F.R. and Conway,T. (1999) Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol., 181, 6425–6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.S., Whittam,T.S. and Selander,R.K. (1997) Evolutionary genetics of the isocitrate dehydrogenase gene (icd) in Escherichia coli and Salmonella enterica. J. Bacteriol., 179, 6551–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H., Mori,H., Itoh,T. and Gojobori,T. (1997) Genome plasticity as a paradigm of eubacteria evolution. J. Mol. Evol., 44, S57–S64. [DOI] [PubMed] [Google Scholar]