Abstract
Ribonuclease (RNase) P is the universal ribozyme responsible for 5′-end tRNA processing. We report the crystal structure of the Thermotoga maritima RNase P holoenzyme in complex with tRNAPhe. The 154 kDa complex consists of a large catalytic RNA (P RNA), a small protein cofactor, and mature tRNA. The structure shows that RNA-RNA recognition occurs through shape complementarity, specific intermolecular contacts, and base pairing interactions. Soaks with a pre-tRNA 5′ leader sequence with and without metal help identify the 5′ substrate path and potential catalytic metal ions. The protein binds on top of a universally conserved structural module in P RNA and interacts with the leader, but not with mature tRNA. The active site is composed of phosphate backbone moieties, a universally conserved uridine nucleobase, and at least two catalytically important metal ions. The active site structure and conserved RNase P/tRNA contacts suggest a universal mechanism of catalysis by RNase P.
Ribonuclease P (RNase P) is a ribonucleoprotein complex responsible for processing many different RNA molecules in the cell (for recent reviews, see 1–3). It is found in almost all organisms and is composed of one essential RNA subunit and one or more protein subunits. The RNA component is responsible for catalysis and can process RNA in vitro in the absence of protein, albeit with reduced efficiency4. The discovery that the RNA component is the catalytic moiety4 helped cement the notion that RNA can be directly involved in catalysis. RNase P is considered a remnant of an ancient RNA-based world and an example of an RNA-based catalyst with many features in common with protein-based catalysts.
RNase P recognizes its substrate in trans and is a multiple turnover enzyme. The preferred substrate is pre-tRNA and recognition involves features distant from the cleavage site, such as the TΨC loop of the tRNA acceptor stem5. RNA cleavage requires divalent metals4,6,7, yet the chemical mechanism and the location of the active site remain largely undefined as well as the exact role of the protein components. In the case of bacterial RNase P, the single essential protein improves the reaction rate by 2 to 3 orders of magnitude8,9, helps to stabilize the active P RNA fold8,10, binds the 5′ leader region of the pre-tRNA substrate11,12, and assists in product release13.
Structural studies of the RNA component reveal a two domain (S- and C-domains) molecule formed by single and coaxial stems linked together by a variety of tertiary interactions14–17, including five conserved regions I to V (CR-I to CR-V) of P RNA that are common to all organisms18. These conserved regions cluster into two areas, one involved in substrate recognition and the other forming the active site scaffold19.
Here we present the crystal structure of Thermotoga maritima RNase P holoenzyme in complex with mature tRNAPhe, and also the structure of the complex in the presence of a post-cleavage tRNA leader. The two structures help answer key questions about the mechanism of this crucial ribozyme with implications for a broader understanding of the general mechanisms of RNA-RNA based recognition and catalysis.
Structure determination
The components of the complex were purified separately and assembled by mixing and heating prior to crystallization (see full methods). The pre-tRNA was processed into mature tRNA and hence the structure represents a ribozyme/product complex. To promote crystal formation, two interaction modules20 were introduced, which did not affect catalytic activity (Fig. S1 and Table STI). The crystals diffract anisotropically to 3.8 Å and ~4.0 Å. An initial 6 Å map was obtained from phases from a Ta6Br12 derivative; these phases helped locate heavy atoms in other derivative data sets. MIRAS phases produced an excellent map to 4.1 Å where all three components were visible (Figs. 1, S2–3, and Tables STII-IV). Density was particularly clear for the RNA molecules, while density was only clear for the protein backbone and hence the high resolution model of the T. maritima protein21 was positioned without significant rebuilding. The P RNA was built into the map using the structures of T. maritima17 and B. stearothermophilus14 P RNA as guides, while T. maritima tRNAPhe utilized yeast tRNAPhe as a guide22. The structure was refined using anisotropic data to 3.8 Å resolution. Crystals with a tRNA leader present were obtained by soaking a short, oligonucleotide with and without samarium chloride and this structure was refined to 4.2 Å resolution.
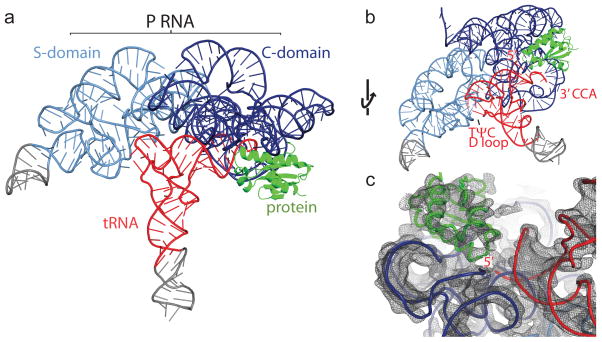
Figure 1. Crystal structure of the T. maritima RNase P holoenzyme in complex with tRNA.
a, Structure of bacterial RNase P, composed of a large RNA subunit (338 nucleotides, ~110 kDa) and a small protein component (117 amino acids, ~14.3 kDa), in complex with tRNA (76 nucleotides, ~26 kDa). The RNA component serves as the primary biocatalyst in the reaction and contains two domains, termed the catalytic (C, blue) and specificity (S, light blue) domains. The RNase P protein (green) binds the 5′ leader region of the pre-tRNA substrate and assists in product release. Transfer RNA (tRNAPhe) (red) makes multiple interactions with the P RNA (see Fig. 2 and S1 for details). Regions in grey denote additional RNA nucleotides required for crystallization. b, Alternate view of the RNase P/tRNA complex, identifying the tRNA recognition regions: the 5′ end where catalysis occurs, the 3′ CCA end, and the highly conserved TΨC and D loop regions. c, View of the 4.1 Å experimental electron density map centered on the 5′ end of tRNA. The map is represented as a dark grey mesh, contoured at 1.4 rmsd.
Overall structure
In the complex, the tRNA sits with the acceptor stem against RNase P, making several tRNA-P RNA intermolecular contacts (Figs. 1, S1). The TΨC and D loops of the tRNA contact the S-domain, while the acceptor stem extends from the S-domain into the C-domain crossing the main P1/P4/P5 coaxial stem (Figs. 2, S4). The 3′ CCA end of the tRNA enters a tunnel formed by P6/P15/P16/P17 and base pairs with nucleotides in the L15 region (Figs. 2, S4, S5), an interaction recognized previously23. The 5′ end of the tRNA indicates the location of the active site, which is close to the region where P4, P5, and CR-IV intersect. The protein component is also adjacent to the 5′ end of the tRNA, but does not contact it. The protein contacts include the CR-IV and CR-V regions, the P15 stem, and the P2/P3 helix interface (Figs. 3, S1, S6). The pre-tRNA leader makes extensive contacts with the protein, but few with the PRNA.
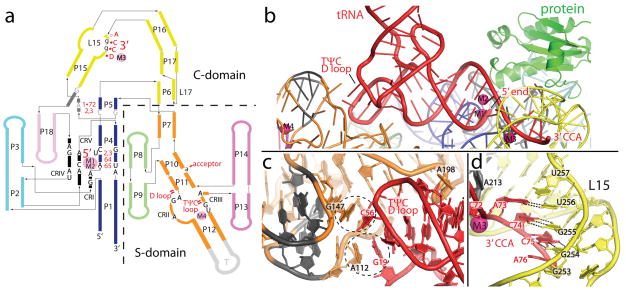
Figure 2. tRNA recognition by RNase P is mediated by RNA-RNA interactions.
a, Schematic of the P RNA secondary structure mapping the tRNA-P RNA contacts observed in the crystal structure. The tRNA nucleotides (1•72, 2, 3, 64, and 65) and regions (5′, 3′, TΨC loop, D loop, and acceptor) involved in direct interactions are shown in red. Intermolecular base pairs form between the 3′ end of tRNA (DCCA) and loop 15 (L15), where D is the discriminator nucleotide that serves as an identity element in tRNA biogenesis. P RNA nucleotides that are universally conserved (black, uppercase), conserved among all bacteria (grey, uppercase), or highly conserved in bacteria (black, lowercase) are identified. Metal ions are shown as filled pink circles, and denote the location of the active site (M1, M2), and other structurally important regions (M3, M4). Single and double dashes in red represent minor groove and base stacking interactions, respectively. All identified tRNA/P RNA contacts are within 4 Å. The crystallized T. maritima P RNA consists of eighteen paired helices (P), five universally conserved regions (CRI to CR-V) (black), two junctions containing conserved nucleotides in bacteria (dark grey), several loop (L) regions, and an engineered tetraloop region (T, light grey). The coaxial P1/P4/P5 stem is shown in blue, P2/P3 stems in cyan, P6/P15/P16 and L15/L17 in yellow, P7 and P10/P11/P12 in orange, P8/P9 in light green, and P13/P14 in pink (see Fig. S1 for additional details). b, Recognition of tRNA by the P RNA of RNase P. The acceptor stem of tRNA (red) docks onto the P RNA (colored as in a) making a series of interactions, including base stacking in the TΨC/D loops of tRNA and the S-domain, an A-minor interaction, and base pairing, ribose zippers, and stacking interactions between the 5′ and 3′ ends of tRNA and the C-domain. The protein (green) makes no direct contacts with mature tRNA. Critical metals ions (M1–M4) identified are shown as magenta spheres. c, tRNA recognition by the S-domain. Two universally conserved P RNA regions (CR-II and III, dark grey) facilitate base stacking interactions with unstacked bases in the structurally conserved TΨC and D loops of tRNA. Dashed circles highlight this stacking interaction between P RNA residues A112, G147 and tRNA residues G19, C56. A conserved P RNA adenosine (A198) stacks into the minor groove of the acceptor tRNA stem. d, Recognition of the tRNA 3′ CCA by the C-domain. Intermolecular base pairs form between the 3′ tRNA (ACC) and the L15 (GGU) loop of P RNA. This interaction is stabilized by a structural metal (M3, magenta sphere) and a L15 ribose zipper conformation.
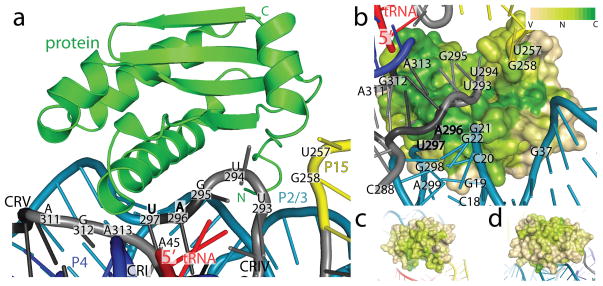
Figure 3. Protein-RNA contacts within the RNase P holoenzyme.
a, The protein sits on the P RNA surface formed by conserved regions I, IV, and V. The protein (green, shown as ribbons) additionally contacts the L15/P15 junction and the P2/3 helices (P RNA as colored in Fig. 2). Labeled P RNA nucleotides make protein contacts (within 4 Å) and include: A45 in CR-I, U257 and G258 in the L15/P15 junction, U293, U294, G295, A296, U297 in CR-IV, and A311, G312, and A313 in CR-V. Bold nucleotides are universally conserved. b, Surface representation of the protein colored by sequence conservation (Variable: tan, Neutral: light green, Conserved: green). A highly conserved patch in the protein extends from the vicinity of the 5′ end of the tRNA, and interacts with P RNA conserved regions IV (U293–U297) and V (A311–A313). Other P RNA nucleotides that make protein contacts include: the P2 helix (C18-G22, G298-A299), the P3 helix (G37), and the L15/P15 junction (U257-G258). Four hundred and ninety bacterial RNase P proteins were included in the analysis of the sequence conservation using the ConSurf server48. Panels (c) and (d) show different orientations to emphasize that high sequence conservation is concentrated in the region of the protein that faces the conserved regions of the P RNA. Neutral or slightly conserved regions shown in these two orientations correspond to a patch that interacts with the leader.
The components of the RNase P holoenzyme are largely unchanged when bound to tRNA (Fig. S7). A comparison between T. maritima P RNA alone17 and in the complex reveals an overall similar fold (backbone normalized rmsd ~1.1Å) with a small change in the relative orientation of the two domains (Fig. S7). The only major change in the P RNA structure occurs in the vicinity of the P15–P17 stems (Figs. S8, S9). A few additional residues at the N-terminus were clear and follow a similar path to the B. subtilis protein24 (Fig. S10); no changes in the structure of the protein component were detected. The structures of yeast and T. maritima tRNAPhe show remarkable resemblance (backbone normalized rmsd for acceptor stem ~0.8 Å) (Fig. S11). Further, a comparison with previous models reveals an excellent agreement with the predicted secondary structure25 and a good agreement with the models of the complex19,26,27 (Fig. S12).
tRNA recognition
The observed RNA-RNA interactions involved in substrate recognition agree with previous biochemical studies5,23,28 and include: 1) stacking between bases in the tRNA TΨC and D loops and the P RNA S-domain, 2) an A-minor interaction at the acceptor stem, and 3) the formation of canonical base pairs at the 3′ end of tRNA (Figs. 2, S1). The first interaction identifies the TΨC loop as a key element in recognition. Both the tRNA D and TΨC loops have unstacked bases (G19 and C56) that interact with unstacked bases in the P RNA (A112 and G147), forming G19-A112 and C56-G147 stacks in the complex. The second major interaction involves a highly conserved unstacked adenosine (A198) in the P11 stem entering the minor groove of the tRNA acceptor stem. These interactions facilitate shape complementarity and help explain the central role of the S-domain in recognition. The third major interaction involves intermolecular base pairing between the tRNA 3′ CCAD motif and the L15 loop. This interaction is likely conserved in all bacterial and most archaeal RNase Ps, but not in organisms where CCA is added post-transcriptionally1. The fourth to last nucleotide, A73, forms a Watson-Crick (WC) base pair with nucleotide U256. C74 and C75 form WC base pairs with G255 and G254, while the terminal A76 forms a weak interaction with G253. To accommodate these intermolecular base pairs, the two strands of L15 fold into a ribose zipper. In addition, a structural metal ion (M3) (Figs. 2, S13) binds adjacent to this P RNA-tRNA region and likely corresponds to a metal ion identified biochemically7. In the complex, the 3′ end of the tRNA separates from the 5′ end and enters a wide opening formed by P6/P15/P16/P17 (Figs. 1, 2, S4, S5). This opening is ~20 Å in diameter, can easily accommodate a single stranded RNA molecule, and is created when the P6/L17 pseudoknot forms (Figs. S1, S5).
Protein/RNA interactions
The bacterial RNase P protein structure is highly conserved, but has little or no sequence or structural similarity with the protein components of archaea or eukarya29. In the complex, the protein is near the 5′ end of tRNA, but is too far (over 6 Å) to make direct contacts. The protein sits between the P15 and P3 stems (Figs. 3, S6), and also contacts the CR-IV and CR-V loop regions of P RNA. Comparison of bacterial sequences reveals that the protein has a large, contiguous area with high sequence conservation (Figs. 3, S6) including important residues identified previously11,30,31. The conserved area extends in an arch along the surface of the protein, starting from a point close to the 5′ end of the tRNA and faces the universally conserved modules.
To investigate the interactions with the leader, crystals were soaked with a short oligoribonucleotide in the presence and absence of Sm3+. Fourier difference maps to 4.2 Å show 5 phosphates of the leader along the conserved surface of the protein (Figs. 4, S6), but the position of the nucleobases was ambiguous. The structure shows that the leader contacts residues Phe17, Phe21, Lys51, Arg52, and Lys90 and likely interacts with Ser26, Gln28, Lys56, Arg89, in agreement with biochemical results6,9,12,30,31. The 3′ end of the leader is located adjacent to the 5′ end of tRNA and near two conserved residues (Arg52 and Lys56). A metal ion is present in between the leader 3′ and the 5′ end of mature tRNA (Figs. 4, 5, S14), but is too distant (> 4 Å) to directly ligate protein residues. Leader nucleotides −1 to −3 are poised to interact with nucleotides A213, U294, G295, and A314 of P RNA (Figs. 4, S1, S6). These results indicate that the major role of the protein component is to interact with the leader to align the pre-tRNA in the complex, as previously observed9,11,31.
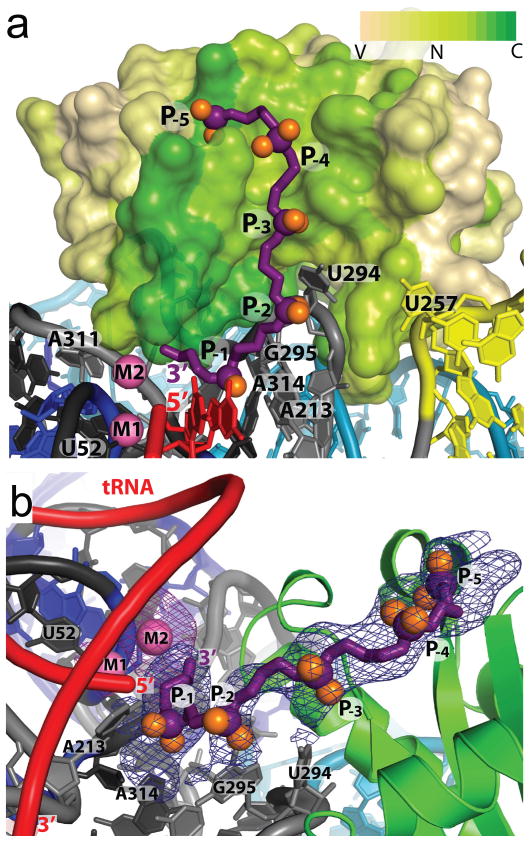
Figure 4. Pre-tRNA leader - protein interactions in the RNase P holoenzyme.
a, Surface representation of the protein colored by sequence conservation as in Fig. 3. The pre-tRNA 5′ leader (purple, with purple and orange spheres for the phosphorous and non-bridging oxygens, respectively) was modeled as a polyphosphate chain with five phosphates (P-1 to P-5). The leader follows a highly conserved patch in the protein extending from the 5′ end of the mature tRNA (red) and away from the P RNA. The addition of a 5′ leader with metal (Sm3+) reveals a second metal ion (M2). b, Alternative view of the pre-tRNA leader/protein interaction. Each phosphate position (P-1 though P-5) was visible in a 4.2 Å difference Fourier map (mFo-DFc) calculated from crystals where only the leader was soaked into the crystals (blue mesh, 3 rmsd contour levels). A second 4.2 Å difference Fourier map (mFo-DFc) calculated from crystals where the leader and Sm3+ metal were soaked into the crystals shows clearly the position of the second metal ion (magenta mesh, 3.5 rmsd contour level). P RNA residues poised to make contacts are labeled. Nucleotide U52 serves as a reference point in a and b and does not interact with the 5′ leader oligonucleotide.
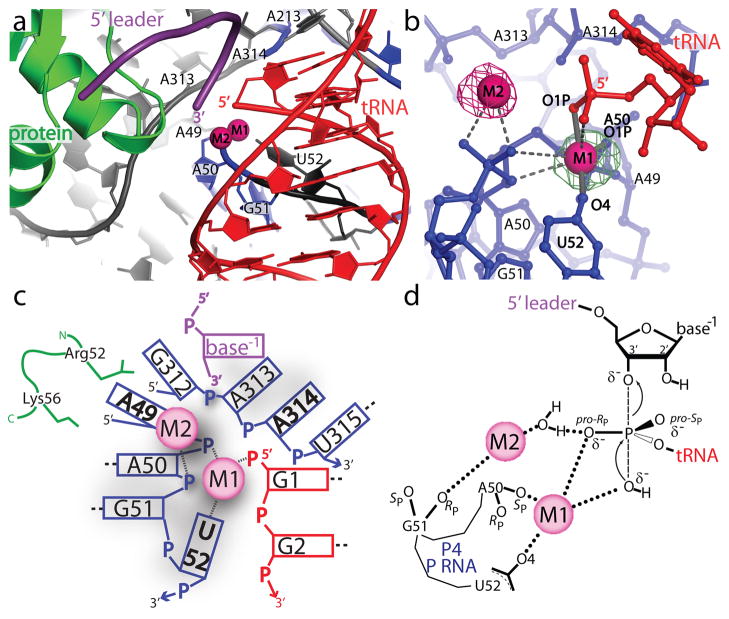
Figure 5. Structure of the RNase P active site environment.
a, The active site is inferred from the location of the mature 5′ end of tRNA. The diagram shows the position of the mature tRNA (red), the leader (purple), the protein component (green), and the P RNA (blue and grey). A group of conserved P RNA nucleotides (A49 - U52, A213, A313, and A314) form part of the active site. Two metal ions (magenta spheres) are found in the active site. b, The two active site metal ions (M1 and M2) are within 4 Å of the 5′ phosphate of tRNA and the M1–M2 metal-metal distance is ~4.8 Å. The M1 metal makes contacts (≤2.1 Å, solid grey bonds, labeled) with tRNA (G1 O1P) and P RNA (A50 O1P and U52 O4) oxygens. Other possible ligands within 3.5 Å of M1 or M2 are represented by dashed grey lines (Table STV). The figure shows two isomorphous difference Fourier (mFo-DFc) maps. The green mesh corresponds to a Eu3+ soak in the absence of leader and is contoured at the 9.5 rmsd level. The magenta mesh corresponds to a Sm3+ and 5′ leader soak and is contoured at the 5.5 rmsd level. The second metal is clearly visible only when the leader is present. c, Schematic diagram of the interactions around the active site. The diagram shows all residues within 8 Å of the 5′ phosphorus atom of tRNA. Short dashed lines represent metal ligand distances within 2.2 Å and longer dashed lines represent nucleotides which form canonical base pairs. Nucleotides in bold are universally conserved in P RNA. The P RNA, tRNA, 5′ leader, and protein side chains are shown in blue, red, purple and green, respectively. d, Proposed reaction mechanism for the endonucleoytic cleavage of pre-tRNA by RNase P based on the structure of the enzyme-product (E-P) complex and previous mechanistic studies49,50. The M1 metal distance to the 5′ phosphate ligands (Table STV) in the E-P complex is consistent with the proposed enzyme-substrate (E-S) transition state. In this proposed reaction scheme, M1 is ~180° from the apical O3′ position and activates a hydroxyl nucleophile for an in-line nucleophillic displacement, creating a new bond and displacing the 3′ scissile phosphate oxygen. As RNase P proceeds through an SN2 reaction pathway, the stereochemistry around the phosphorus atom undergoes a net inversion of configuration. If the pro-RP (O2P) oxygen coordinates metal in the E-S complex during catalysis, as previously observed49,50, this would subsequently allow for the pro-SP (O1P) oxygen to coordinate metal in the E-P complex, as observed in the crystal structure. Product release could be facilitated by a metal (M2) coordinated water, which would enable proton transfer to the 3′ scissile oxygen. The exact active site geometry and identity of other metal ligands in an E-S complex has yet to be established.
Active site
The location of the active site is inferred from the 5′ end of mature tRNA (Figs. 5, S15). The phosphate backbone of tRNA nucleotides (+1 to +3) sits on the major groove of the P4 stem (near A50, G51, and U52), and places the tRNA 5′ end next to the P4 phosphate backbone and nucleotides A313 and A314 (Figs. 5 and S15). The universally conserved U52 nucleotide is unstacked from the P4 stem and faces the tRNA 5′ end. In addition, the tRNA 1•72 base pair is stabilized by an adenosine stack with A213, a nucleotide conserved in all bacteria.
A metal ion (M1), putatively magnesium, is found trapped between the tRNA 5′ end, the A50 and G51 phosphates, and the O4 oxygen of the universal U52 nucleotide and was confirmed using crystals soaked with Sm3+ and Eu3+ (Figs. S14 and S15). Putative M1 metal contacts include the A50 non-bridging phosphoryl oxygen, the O4 oxygen of the U52 nucleobase, and the O1P oxygen at the 5′ end of tRNA. Other metal-ligand interactions may include: the backbone of A50, the phosphoryl oxygen of G51, and the 5′ end of tRNA (Table STV). Many of these oxygen ligands have been implicated in metal coordination and catalysis32–34. The M1 site may also coincide with a site (M6) observed in the structure of B. stearothermophilus P RNA35. The structure of the complex suggests that M1 participates in catalysis by directly binding P RNA and the 5′ phosphate of tRNA.
A second metal (M2) was located in experiments where the leader was soaked in the presence of Sm3+. The M2 metal is in close proximity to the phosphoryl oxygens of G51, the O3′ of the leader, and the 5′ end of tRNA (Table STV). The two metals observed in crystals soaked with the leader and Sm3+ are ~4.8 Å apart (Figs. 5, S15). The structures indicate that the active site includes at least two metal ions upon complex formation with pre-tRNA. Due to its location, the M2 metal ion could make additional contacts with both the tRNA and the P RNA during catalysis.
The structures of the active site of the complex and the apo-ribozyme structures are similar (Figs. S7, S16, and S17), including the presence of a metal ion next to the P4 helix35. With the exception of the U52 nucleobase (Fig. S16), no large changes are observed in the active site region. A fully occupied M2 site is observed only in the presence of leader, suggesting that a local metal-dependent conformation change may occur, as previously reported6. The structure also reveals that the tRNA 5′ and 3′ ends splay and separate to interact with the P RNA (Fig. S11), confirming the need for movement of the tRNA ends36,37. While accommodating the upstream RNA leader likely requires local protein and P RNA structural changes, the location of the active site is not significantly altered and is largely pre-assembled.
Mechanistic implications
RNase P can cleave a variety of substrates1,10,38, but pre-tRNA is the only one that is common among all organisms. To decipher its function, it is important to understand two different aspects of pre-tRNA processing by RNase P: substrate specificity and the chemical mechanism of cleavage.
tRNA recognition by RNase P involves the highly conserved tRNA TΨC and D loops and the CR-II and CR-III in the S-domain of P RNA. Thus, regions with high sequence and structure conservation are involved in specific tertiary interactions, suggesting a universal mode of recognition among all RNase P’s. The presence of unpaired nucleotides next to the cleavage site is also an important feature for pre-tRNA recognition, although it is unclear whether this is a universal feature of all natural substrates1. Finally, pre-tRNA is usually processed to form a 7 base pair long acceptor stem. An additional role of the interactions between CR-II and CR-III and tRNA may be to serve as a ‘ruler’ that ensures that the correct lengths are processed, although there is some flexibility as tRNA’s with acceptor stems 8 base pairs long can be processed39. The interaction with the 3′ CCA end is also a key recognition feature, but may not be necessarily an RNA-RNA interaction in higher organisms. The L15 loop of P RNA is not found in eukarya or some archea40 and its function may be replaced by additional protein(s), suggesting that 3′ CCA intermolecular base pairing is not a universal interaction.
The second important aspect of RNase P function is the chemical mechanism of cleavage. Hydrolysis of a phosphodiester bond generates the mature 5′ RNA product. While it is not possible to propose a complete mechanism from a structure at this resolution, the RNase P-tRNA structures, together with extensive biochemical information, help identify the major active site components. The structure suggests that at least two distinct metals play a direct role. It is possible to propose a transition state model (Fig. 5d) where the M1 metal directly positions the scissile phosphate oxygens of the substrate and enables a hydroxyl ion to perform an SN2-type nucleophilic substitution. In this scenario, the M2 metal ion stabilizes the transition state and mediates proton transfer to the 3′ scissile oxygen during product release, as previously proposed7. Other universally conserved nucleotides in the vicinity appear to play a structural role in forming the correct structure and are not directly involved in catalysis, consistent with proposals that sequence conservation is largely the result of strong structural constraints19. Hence, the RNaseP/tRNA complex reveals how the P RNA structure can serve as a scaffold to bind and orient metals and substrate properly. It appears that RNase P employs a two-metal ion catalytic mechanism, similar to other mechanisms proposed based on other large ribozyme structures41,42 and originally put forth as a general mechanism for many ribozymes43.
The structural studies of the holoenzyme/tRNA complex help to show that all RNase P ribozymes share a common, RNA-based mechanism of RNA cleavage and recognition that involves two universally conserved structural modules. Adaptation through the addition of protein increases RNase P functionality by accurately positioning the 5′ leader pre-tRNA substrate and by contacting conserved regions of the P RNA structure. The unique tertiary fold of the P RNA utilizes shape complementarity, specific RNA-RNA contacts, and intermolecular base pairing to recognize its substrate efficiently. Within this tertiary fold, the universally conserved regions are crucial to form the active site scaffold and to create regions involved in tRNA recognition. In addition, both P RNA and the pre-tRNA help to coordinate two catalytically important metal ions essential for the putative mechanism of pre-tRNA cleavage. The RNase P/tRNA complex offers a glimpse into the transition from an ancient, RNA-based world to the present, protein-catalyst dominated world and affirms that RNA molecules can display comparable versatility and complexity.
Methods summary
Crystallization
Preparation, purification, and folding of T. maritima RNase P and tRNAPhe have been described8,17,44. For crystallization, the components were mixed in a 1:1.1:1 (P RNA: pre-tRNA: protein) molar ratio to a concentration of 45 μM. The mixture was heated to 94 °C (2 min), cooled to 4 °C (2 min), and after the addition of MgCl2 to a final 10 mM concentration, further incubated at 50 °C (10 min) and 37 °C (40 min). Crystals were obtained by mixing 1 μl of complex with 1 μl of reservoir solution (1.8 M Li2SO4, 50 mM sodium cacodylate (pH 6.0)) and equilibrated by vapor diffusion at 30 °C. Crystals were cryo-protected using 15% xylitol plus reservoir solution.
Data collection and structure determination
Diffraction data were collected at 100 °K at the LS-CAT sector at the APS. Complete native and Ta6Br12, SmCl3, EuCl3, and iridium hexammine (Ir(NH3)6)3+ derivatives were collected. A weak Molecular Replacement45 solution using a trimmed model of the tRNA/P RNA complex19 located the Ta6Br12 cluster. MAD phases46 from the cluster extended to ~6 Å, with the map showing a clear envelope. These phases were used to locate the other heavy atoms that were used to calculate a 4.1 Å MIRAS map. To locate the pre-tRNA leader, crystals were soaked with a T. maritima 5′ tRNA 7 nucleotide leader sequence (final 0.2 mM concentration), with and without 14 mM SmCl3. Difference maps allowed the placement of five pre-tRNA nucleotides and the unambiguous identification of a second active site metal. The experimental electron density map was of excellent quality and allowed model building of nearly all RNA phosphate and nucleobase positions and accurate placing of the protein. Model building was guided by the known structures14,17,21. Final Rwork and Rfree are 24.9% and 27.0%, respectively, with rmsd of 0.007Å and 1.24° for bonds and angles. Figures were made with Pymol47.
Full Methods
Preparation of the T. maritima RNase P holoenzyme - tRNAPhe ternary complex
RNA transcriptions were performed in vitro using purified His6-tagged T7 RNA polymerase using standard protocols51. Sequences from the T. maritima RNase P RNA and tRNAPhe genes were inserted into a pUC19 vector at Fok I and Bsm AI restriction sites, respectively, allowing for run-off transcription of the DNA plasmid after digestion with the appropriate restriction enzyme (NEB). Constructions of modified RNA molecules with either mutations or additions were performed using the QuickChange mutagenesis kit (Strategene). RNA samples were purified by 6% denaturing polyacrylamide gel electrophoresis (PAGE), identified by UV absorbance, recovered by diffusion into 50 mM potassium acetate (pH 7) and 0.2 M potassium chloride, and precipitated with ethanol. tRNA was further purified by anion exchange [MonoQ (5/50 GL)] and gel filtration [HiPrep 26/60, Sephacryl S-200] chromatography (GE Health Sciences). Overexpression and purification of the RNase P protein from T. maritima was performed as described previously44.
To form the RNase P holoenzyme-tRNA complex, unfolded P RNA, unfolded tRNA, and P protein molecules were mixed at a 1:1.1:1 molar ratio in 66 mM HEPES, 33 mM Tris (pH 7.4), 0.1 mM EDTA (1X THE), and 100 mM NH4OAc (Ref. 8). The ternary mix, at a final concentration of 45 μM, was incubated at 94 °C for 2 minutes and then cooled to 4 °C over 2 minutes. After addition of MgCl2 to a final 10 mM concentration, the reaction mixture was incubated at 50 °C for 10 minutes, followed by incubation at 37 °C for 40 minutes, and finally cooled to 4 °C over 30 seconds.
Rational design of an RNA tertiary module to build a crystal lattice
In order to promote formation of a crystal lattice, intermolecular interactions were facilitated by introducing a tertiary structure interaction module. Based on the T. maritima RNA sequence and a proposed model of the P RNA-tRNA complex19, constructs were designed where a tetraloop was inserted into the P12 loop (L12) of the P RNA and a tetraloop-receptor into the anticodon stem of tRNA (Fig. S1). These two RNA regions were chosen as they were deemed to be far from the active site or other regions involved in specific interactions. In addition, the P12 stem of P RNA has a highly variable helix length across all organisms, lacks sequence conservation, and is non-essential or absent in several organisms40. The P12 and the anticodon loop of tRNA are not known to form any functional contacts. The length of the anticodon and the P12 stems were systematically varied by single base pair insertions adjacent to the tetraloop and tetraloop receptor module, thus altering the position (~2.7 Å per base pair added) and orientation (~36o per base pair added) of the tetraloop receptor and the tetraloop. Forty two combinations of molecules were screened for crystallization conditions using a sparse matrix approach employing a set of crystallization conditions developed locally. A few combinations of RNA molecules produced crystals, with most of them diffracting poorly. The best crystals were obtained from a construct where the P12 and anti-codon stems were elongated by five and three base pairs respectively. Insertion of two G-U wobble pairs adjacent to the tetraloop-tetraloop receptor module further improved diffraction, and also created a binding site for an iridium hexammine cation.
Crystallization and Data Collection
Crystals were obtained by mixing 1 μL of complex with 1 μL of reservoir solution (1.8 M LiSO4, 50 mM sodium cacodylate (pH 6.0)) and equilibrated by vapor diffusion hanging or sitting drops at 30 °C. Gel analysis of washed crystals show that all three components were present (data not shown). Attempts to crystallize the complex in the absence of protein yielded no crystals. Crystals suitable for data collection grew in approximately three weeks and were cryo-cooled in liquid nitrogen immediately after transfer to a 15% xylitol plus reservoir solution. Crystals of the RNase P holoenzyme/tRNA ternary complex suitable for data collection grew to approximately ~80–300 μm per side/edge, and diffract anisotropically to 3.8 Å in the best direction and ~4.0 Å in other directions. Crystals belong to space group P3121 (a=b=169.3 Å, c=185 Å) and contain 1 molecule per asymmetric unit.
A series of derivatized crystals were also prepared by soaking in heavy metal compounds. Derivatives were prepared by soaking the crystals in mother liquor plus the derivative and incubating for 2–24 hours before transferring them to cryo-protectant with the derivative present and freezing them in liquid nitrogen. Successful derivatizations were obtained by soaking the crystals in the following compounds: 2 mM Ta6Br12, 15 mM samarium chloride (Sm3+), 15 mM europium chloride (Eu3+), and 15 mM iridium hexammine (Ir(NH3)6)3+). However, several of the compounds partially precipitated upon addition to the mother liquor solution and hence the final concentration is not known precisely. In addition, crystals with a leader present were obtained by soaking in a 0.2 mM 7-mer oligonucleotide (5′-A-7 A-6 G-5 G-4 C-3 G-2 U-1 -3′) (ThermoFisher) for 4 hours with and without 14 mM samarium chloride present. The sequence was chosen by selecting the most common nucleotide in the T. maritima tRNA leaders at each position.
All diffraction data were collected at 100 °K at the Life Science-Collaborative Access Team (LS-CAT) sector located at the Advance Photon Source (APS) (Argonne, IL) using Rayonix CCD detectors. As the crystals are very radiation sensitive, the data collection range was optimized using the program MOSFLM52 to collect the most complete native or anomalous data set using the minimal rotation range. Multi-wavelength anomalous dispersion (MAD) data were collected from a tantalum bromide cluster (Ta6Br12) derivative at three different wavelengths. Single or multiple wavelength anomalous dispersion data were also collected fromthe samarium chloride (Sm3+), europium chloride (Eu3+3+), and iridium hexammine (Ir(NH3 )6) derivatives. Data were processed with XDS53 and scaled with SCALA54. All other processing was done with programs from the CCP4 suite54, except when noted. Data collection statistics for native and derivative data sets are shown in Table STII.
In all cases, the diffraction limits of the data were anisotropic. The extent of the anisotropy was determined using the Anisotropy Server55 and the data were treated in three different ways: 1) without any anisotropy correction, 2) carving the data to the limits suggested by the anisotropy server (3σ cut-off level on amplitudes), and 3) applying an anisotropic correction to the data using the server. For the second case, the integrated data from XDS was carved to the limits suggested by the server and then merged and scaled with SCALA before final processing. In many instances, the phasing and refinement calculations were done separately with the complete and carved data sets and the results compared. Overall, the different ways of treating the data had little effect on the final results, even though the data collection statistics were better for the carved data set (see Table STII and STIII).
Structure determination and model refinement
Molecular Replacement (MR) studies with the program PHASER45 using a proposed partial model of the P RNA/tRNA complex19 gave a weak low resolution (25–8 Å) MR solution (Z-scores: 5.4 and 9.0 for the rotation and translation functions respectively). Phases calculated from the MR solution were used to locate the position of the 3 sites in the Ta6Br12 cluster data set. The program SHARP46 was used to calculate MAD phases using data from three different wavelengths and spherically averaged form factors for the cluster. The solvent-flattened MAD map was of excellent quality but the phases were only good to ~6 Å resolution. The positions of the Eu3+, Sm3+, and (Ir(NH3 )6)3+ heavy atoms were determined using the cluster phases. The parameters from the cluster and other derivatives could not be refined simultaneously and instead Multiple Isomorphous Replacement with Anomalous Scattering (MIRAS) phases to 4.1 Å resolution were calculated using data from the single-atom derivatives together with phase information to 6 Å from the cluster data. The SOLOMON56 solvent-flattened map was very clear (Fig. S2) and all three molecules were apparent in the map. The model for the P RNA/tRNA complex19 fit well in many areas, but the map showed regions where the model needed to be changed, regions that were missing in the model, like the P12 extension and the pseudoknot region, and the position of the protein. The models for the tRNA and P RNA were rebuilt completely using the high resolution model of yeast tRNAPhe (ref. 22), T. maritima P RNA17, and B. stearothermophilus P RNA14 as guides. All regions of the RNA molecules were visible in the map and regions that were missing in the original T. maritima P RNA model were built. Some minor corrections to the original model were needed, but overall the models for P RNA agree well. The protein density was clear for the backbone, but not for the side chains and hence the high resolution model of the T. maritima protein21 was placed on the experimental electron density map as a rigid body with minimal rebuilding.
Refinement was performed using Refmac557 and BUSTER58. Due to the resolution of the data, the models were restrained to enforce good hydrogen bonding distance between Watson-Crick (WC) base pairs, planarity between base pairs (both for WC and non-WC base pairs), and C3′-endo sugar puckering for recognizable secondary structure elements. In addition, during BUSTER refinement the protein was restrained by the high resolution structure of the protein21. Model building with Coot59 was interspersed with either Refmac5 or BUSTER refinement. During rebuilding, missing nucleotides were added as well as some missing residues at the N-terminus of the protein. Mg2+ ions were included at positions that had high density peaks in residual maps and also coincided with heavy atom sites. Other large peaks in the native data set that coincided with phosphate positions in the leader-soaked crystals were modelled as PO4 ions. No individual atomic or group temperature factors were refined, only an overall anisotropic temperature factor. The final stages of the refinement were done with the program BUSTER. The refinement was done both with a carved data set where data outside the anisotropic diffraction limits (3σ cut-off) were excluded and also with a complete data set (isotropic) to the highest resolution limit (Table STIII). No significant difference was noted in the two refinements and the refinement statistics and electron density maps calculated from either data set were also virtually identical. It appears that the anisotropic temperature factor correction in the refinement programs adequately modeled the modest anisotropy of the data.
The final model for the P RNA includes nucleotides 1 to 338. Only the phosphate backbone was modelled for nucleotides 39, 241, and 314–317. In addition, 9 nucleotides were inserted between nucleotides 130 and 136 to account for the extension added for crystallization (Fig. S1). The final model for the tRNA includes nucleotides 1–76, but only the phosphate backbone was modelled for nucleotides 16, 17, and 20. The crystallization module added 8 extra nucleotides incorporated at the end of the anticodon stem (Fig. S1). Nearly the entire anticodon stem and anticodon loop were altered to accommodate the tetraloop receptor and altered anitcodon loop. The protein model includes residues 6 to 117. The positions of all side chains were ambiguous in the map and were not rebuilt, but kept as much as possible as in the original 1.2 Å model (PDB ID: 1NZ0) during refinement. Side chains that collided with the RNA were rebuilt when needed. There are 4 Mg2+ and 2 PO4 ions in the model of the complex. The final model to 3.8 Å resolution has an overall Rwork of 24.9% and Rfree of 27.0% with a root mean square deviation (rmsd) from target values of 0.007 Å and 1.24° for bonds and angles respectively. The model in the presence of the leader includes an additional polyphosphate molecule with five phosphates and two Mg2+ ions coinciding with metals ions M1 and M2. A total of 5 Mg2+ ions were modeled into the complex that contains the 5′ polyphosphate leader backbone. The final model to 4.21 Å resolution has an overall Rwork of 25.8% and Rfree of 26.7% with an rmsd of 0.007 Å and 1.23° for bonds and angles respectively (see Table STIII and STIV).
Model superpositions were done with programs from the CCP4 suite54, lsqman60, and Coot59. Diagrams were made with PYMOL47. Coordinates and structure factors have been deposited in the PDB with accession numbers 3OK7 and 3OKB.
Activity assays of RNase P holoenzyme
Cleavage assays measuring kcat/KM under single turnover conditions were performed on the RNase P and pre-tRNA constructs that gave the best diffracting crystals. The pre-tRNA (with a single nucleotide leader (−1)) which yielded crystals and a control pre-tRNA (containing a T. maritima nine nucleotide leader (−9)) were radioactively labelled at their 5′ ends. Labelled substrates were purified over a 10% denaturing polyacrylamide gel and identified by 32P-phosphorimaging. The holoenzyme was folded and cleavage reactions were performed in identical conditions as the folding reaction (1X THE, 10 mM MgCl2, 0.1 M NH4OAc, 37 °C). The enzyme activity of both the modified RNase P which gave crystals and the T. maritima wild-type RNase P were tested. The reaction was initiated by mixing pre-folded RNase P holoenzyme (25, 50, and 100 nM) with pre-folded pre-tRNA substrate (< 4 nM), incubated for various times (t = 0, 0.25, 1, 4, and 16 minutes), and subsequently quenched by adding 9 M urea, 50 mM EDTA. All reaction mixtures were directly loaded onto a 15% denaturing polyacrylamide gel which separated the substrate from the product(s). To observe unambiguously the products of the leader (−1) pre-tRNA, thin layer chromatography (TLC) was also performed with polyethyleneimine (PEI)-cellulose coated plates, where the quenched reaction mixture was spotted and run in a 5% acetic acid/100 mM NH4Cl solution. The dried gels and the TLC plates were exposed to a phosphorimaging screen and the reaction profile was quantified by a phosphorimager (Fuji Medical) using ImageGauge software. A plot of the percentage of product over time gave the cleavage reaction rate for each concentration. Single turnover conditions assuming a first order reaction follow the equation: [P] = [P]∞ (1 − e−kobst), where [P] is the fraction of pre-tRNA cleaved, [P]∞ is the fraction of uncleaved pre-tRNA at the end of the reaction, and kobs is the observed reaction rate constant. By measuring kobs at different concentrations it is possible to obtain Kcat/KM assuming Michaelis-Menten kinetics.
Supplementary Material
Acknowledgments
We thank E. Sontheimer and O. Uhlenbeck for comments and suggestions, N. Pace for the gift of the T. maritima RNase P protein plasmid, Obiter Research (Champaign, IL), A. Davis, and M. E. Duban for advice and preparation of iridium hexammine, and A. Samelson for discussions and assistance. In addition, we are grateful for data collection assistance from S. Anderson, Z. Wawrzak, and staff at LS-CAT. Research was supported by the NIH. NJR is an NRSA postdoctoral fellow.
Footnotes
Supplementary Information accompanies the paper on www.nature.com/nature.
Competing Interests statement The authors declare that they have no competing financial interests.
Author Contributions A.M. directed the work. A.T.-L. and A.M. conceived the project. All authors performed and designed experiments. N.J.R. obtained crystallographic data. N.J.R. and A.M. analyzed the crystallographic data. N.J.R. and A.M. wrote the paper with contributions from all authors.
Author Information Coordinates for the RNase P holoenzyme – tRNA complex, with and without 5′ tRNA leader sequence, have been deposited into the RCSB Protein Data Bank (accession code 3OK7 and 3OKB, respectively).
References
- 1.Hartmann RK, Gossringer M, Spath B, Fischer S, Marchfelder A. The making of tRNAs and more - RNase P and tRNase Z. Prog Mol Biol Transl Sci. 2009;85:319–368. doi: 10.1016/S0079-6603(08)00808-8. [DOI] [PubMed] [Google Scholar]
- 2.Kazantsev AV, Pace NR. Bacterial RNase P: a new view of an ancient enzyme. Nat Rev Microbiol. 2006;4:729–740. doi: 10.1038/nrmicro1491. [DOI] [PubMed] [Google Scholar]
- 3.Liu F, Altman S. Ribonuclease P. Vol. 10. Springer Science+Business Media, LLC; 2010. [Google Scholar]
- 4.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 5.Pan T, Loria A, Zhong K. Probing of tertiary interactions in RNA: 2′-hydroxyl-base contacts between the RNase P RNA and pre-tRNA. Proc Natl Acad Sci USA. 1995;92:12510–12514. doi: 10.1073/pnas.92.26.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsieh J, et al. A divalent cation stabilizes the active conformation of the B. subtilis RNase P pre-tRNA complex: a role for an inner-sphere metal ion in RNase P. J Mol Biol. 2010 doi: 10.1016/j.jmb.2010.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirsebom LA. RNase P. In: Liu F, Altman S, editors. Protein Reviews. Vol. 10. Vol. 7. Springer Science+Business Media, LLC; 2010. pp. 113–134. [Google Scholar]
- 8.Buck AH, Dalby AB, Poole AW, Kazantsev AV, Pace NR. Protein activation of a ribozyme: the role of bacterial RNase P protein. EMBO J. 2005;24:3360–3368. doi: 10.1038/sj.emboj.7600805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurz JC, Niranjanakumari S, Fierke CA. Protein component of Bacillus subtilis RNase P specifically enhances the affinity for precursor-tRNAAsp. Biochemistry. 1998;37:2393–2400. doi: 10.1021/bi972530m. [DOI] [PubMed] [Google Scholar]
- 10.Peck-Miller KA, Altman S. Kinetics of the processing of the precursor to 4.5 S RNA, a naturally occurring substrate for RNase P from Escherichia coli. J Mol Biol. 1991;221:1–5. doi: 10.1016/0022-2836(91)80194-y. [DOI] [PubMed] [Google Scholar]
- 11.Koutmou KS, et al. Protein-precursor tRNA contact leads to sequence-specific recognition of 5′ leaders by bacterial ribonuclease P. J Mol Biol. 2010;396:195–208. doi: 10.1016/j.jmb.2009.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun L, Campbell FE, Zahler NH, Harris ME. Evidence that substrate-specific effects of C5 protein lead to uniformity in binding and catalysis by RNase P. EMBO J. 2006;25:3998–4007. doi: 10.1038/sj.emboj.7601290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reich C, Olsen GJ, Pace B, Pace NR. Role of the protein moiety of ribonuclease P, a ribonucleoprotein enzyme. Science. 1988;239:178–181. doi: 10.1126/science.3122322. [DOI] [PubMed] [Google Scholar]
- 14.Kazantsev AV, et al. Crystal structure of a bacterial ribonuclease P RNA. Proc Natl Acad Sci U S A. 2005;102:13392–13397. doi: 10.1073/pnas.0506662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krasilnikov AS, Xiao Y, Pan T, Mondragón A. Basis for structural diversity in homologous RNAs. Science. 2004;306:104–107. doi: 10.1126/science.1101489. [DOI] [PubMed] [Google Scholar]
- 16.Krasilnikov AS, Yang X, Pan T, Mondragóon A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421:760–764. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- 17.Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragón A. Crystal structure of the RNA component of bacterial ribonuclease P. Nature. 2005;437:584–587. doi: 10.1038/nature04074. [DOI] [PubMed] [Google Scholar]
- 18.Chen JL, Pace NR. Identification of the universally conserved core of ribonuclease P RNA. RNA. 1997;3:557–560. [PMC free article] [PubMed] [Google Scholar]
- 19.Torres-Larios A, Swinger KK, Pan T, Mondragón A. Structure of ribonuclease P--a universal ribozyme. Curr Opin Struct Biol. 2006;16:327–335. doi: 10.1016/j.sbi.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Ferre-d’Amare AR, Zhou K, Doudna JA. A general module for RNA crystallization. J Mol Biol. 1998;279:621–631. doi: 10.1006/jmbi.1998.1789. [DOI] [PubMed] [Google Scholar]
- 21.Kazantsev AV, et al. High-resolution structure of RNase P protein from Thermotoga maritima. Proc Natl Acad Sci U S A. 2003;100:7497–7502. doi: 10.1073/pnas.0932597100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi H, Moore PB. The crystal structure of yeast phenylalanine tRNA at 1.93 A resolution: a classic structure revisited. RNA. 2000;6:1091–1105. doi: 10.1017/s1355838200000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirsebom LA, Svard SG. Base pairing between Escherichia coli RNase P RNA and its substrate. EMBO J. 1994;13:4870–4876. doi: 10.1002/j.1460-2075.1994.tb06814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stams T, Niranjanakumari S, Fierke CA, Christianson DW. Ribonuclease P protein structure: evolutionary origins in the translational apparatus. Science. 1998;280:752–755. doi: 10.1126/science.280.5364.752. [DOI] [PubMed] [Google Scholar]
- 25.Massire C, Jaeger L, Westhof E. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J Mol Biol. 1998;279:773–793. doi: 10.1006/jmbi.1998.1797. [DOI] [PubMed] [Google Scholar]
- 26.Buck AH, Kazantsev AV, Dalby AB, Pace NR. Structural perspective on the activation of RNase P RNA by protein. Nat Struct Mol Biol. 2005 doi: 10.1038/nsmb1004. [DOI] [PubMed] [Google Scholar]
- 27.Tsai HY, Masquida B, Biswas R, Westhof E, Gopalan V. Molecular modeling of the three-dimensional structure of the bacterial RNase P holoenzyme. J Mol Biol. 2003;325:661–675. doi: 10.1016/s0022-2836(02)01267-6. [DOI] [PubMed] [Google Scholar]
- 28.LaGrandeur TE, Huttenhofer A, Noller HF, Pace NR. Phylogenetic comparative chemical footprint analysis of the interaction between ribonuclease P RNA and tRNA. EMBO J. 1994;13:3945–3952. doi: 10.1002/j.1460-2075.1994.tb06710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mondragón A. Ribonuclease P. In: Liu F, Altman S, editors. Protein Reviews. 10 . Ch 4. Springer Science+Business Media, LLC; 2010. pp. 63–78. [Google Scholar]
- 30.Biswas R, Ledman DW, Fox RO, Altman S, Gopalan V. Mapping RNA-protein interactions in ribonuclease P from Escherichia coli using disulfide-linked EDTA-Fe. J Mol Biol. 2000;296:19–31. doi: 10.1006/jmbi.1999.3443. [DOI] [PubMed] [Google Scholar]
- 31.Niranjanakumari S, Stams T, Crary SM, Christianson DW, Fierke CA. Protein component of the ribozyme ribonuclease P alters substrate recognition by directly contacting precursor tRNA. Proc Natl Acad Sci USA. 1998;95:15212–15217. doi: 10.1073/pnas.95.26.15212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christian EL, Kaye NM, Harris ME. Helix P4 is a divalent metal ion binding site in the conserved core of the ribonuclease P ribozyme. RNA. 2000;6:511–519. doi: 10.1017/s1355838200000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crary SM, Kurz JC, Fierke CA. Specific phosphorothioate substitutions probe the active site of Bacillus subtilis ribonuclease P. RNA. 2002;8:933–947. doi: 10.1017/s1355838202025025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christian EL, Smith KM, Perera N, Harris ME. The P4 metal binding site in RNase P RNA affects active site metal affinity through substrate positioning. RNA. 2006;12:1463–1467. doi: 10.1261/rna.158606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kazantsev AV, Krivenko AA, Pace NR. Mapping metal-binding sites in the catalytic domain of bacterial RNase P RNA. RNA. 2009;15:266–276. doi: 10.1261/rna.1331809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pomeranz Krummel DA, Kent O, MacMillan AM, Altman S. Evidence for helical unwinding of an RNA substrate by the RNA enzyme RNase P: use of an interstrand disulfide crosslink in substrate. J Mol Biol. 2000;295:1113–1118. doi: 10.1006/jmbi.1999.3424. [DOI] [PubMed] [Google Scholar]
- 37.Gaur RK, Hanne A, Conrad F, Kahle D, Krupp G. Differences in the interaction of Escherichia coli RNase P RNA with tRNAs containing a short or a long extra arm. RNA. 1996;2:674–681. [PMC free article] [PubMed] [Google Scholar]
- 38.Forster AC, Altman S. Similar cage-shaped structures for the RNA components of all ribonuclease P and ribonuclease MRP enzymes. Cell. 1990;62:407–409. doi: 10.1016/0092-8674(90)90003-w. [DOI] [PubMed] [Google Scholar]
- 39.Burkard U, Soll D. The unusually long amino acid acceptor stem of Escherichia coli selenocysteine tRNA results from abnormal cleavage by RNase P. Nucleic Acids Res. 1988;16:11617–11624. doi: 10.1093/nar/16.24.11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walker SC, Engelke DR. Ribonuclease P: the evolution of an ancient RNA enzyme. Crit Rev Biochem Mol Biol. 2006;41:77–102. doi: 10.1080/10409230600602634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stahley MR, Strobel SA. Structural evidence for a two-metal-ion mechanism of group I intron splicing. Science. 2005;309:1587–1590. doi: 10.1126/science.1114994. [DOI] [PubMed] [Google Scholar]
- 42.Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krivenko AA, Kazantsev AV, Adamidi C, Harrington DJ, Pace NR. Expression, purification, crystallization and preliminary diffraction analysis of RNase P protein from Thermotoga maritima. Acta Crystallogr D Biol Crystallogr. 2002;58:1234–1236. doi: 10.1107/s0907444902007965. [DOI] [PubMed] [Google Scholar]
- 45.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fortelle E, Bricogne G. Maximum-Likelihood Heavy-Atom Parameter Refinement for Multiple Isomorphous Replacement and Multiwavelength Anomalous Diffraction Methods. Meth Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 47.The PyMol Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 48.Landau M, et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y, Li X, Gegenheimer P. Ribonuclease P catalysis requires Mg2+ coordinated to the pro-RP oxygen of the scissile bond. Biochemistry. 1997;36:2425–2438. doi: 10.1021/bi9620464. [DOI] [PubMed] [Google Scholar]
- 50.Warnecke JM, Furste JP, Hardt WD, Erdmann VA, Hartmann RK. Ribonuclease P (RNase P) RNA is converted to a Cd(2+)-ribozyme by a single Rp-phosphorothioate modification in the precursor tRNA at the RNase P cleavage site. Proc Natl Acad Sci U S A. 1996;93:8924–8928. doi: 10.1073/pnas.93.17.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Research. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leslie AGW. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992;26:27–33. [Google Scholar]
- 53.Kabsch W. Automatic indexing of rotation diffraction patterns. J Appl Cryst. 1988;21:67–71. [Google Scholar]
- 54.Collaborative Computational Project 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 55.Strong M, et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abrahams JP, Leslie AGW. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. 1996;D52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 57.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 58.Blanc E, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 59.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 60.Kleywegt GJ. Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr D Biol Crystallogr. 1996;52:842–857. doi: 10.1107/S0907444995016477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.