Abstract
The five genes of the human growth hormone (hGH) cluster are expressed in either the pituitary or placenta. Activation of the cluster is dependent on a locus control region (LCR) comprising pituitary- specific (HSI,II, –15 kb), placenta-specific (HSIV, –30 kb) and shared (HSIII, –28 kb; HSV, –32 kb) DNase I hypersensitive sites. Gene activation in the pituitary is paralleled by acetylation of a 32 kb chromatin domain 5′ to the cluster centered at HSI,II. In the present study we observed that acetylation of this region in placental chromatin was discretely limited to shared HSIII and HSV. Transgenic studies revealed placenta-specific activation of linked genes by a determinant (P-element) located 2 kb 5′ to each of the four placentally expressed genes. A localized peak of histone acetylation was observed at these P-elements in placenta but not pituitary. These data support a model for bifunctional action of the hGH LCR in which separate positive determinants, HSI,II and the P-elements, activate their respective target genes by tissue-specific recruitment of distinctly regulated histone acetyl transferase activities.
Keywords: chromatin/gene expression/histone acetylation/human growth hormone/locus control region
Introduction
In higher eukaryotes, the majority of DNA is packaged into a compact and repressive chromatin structure. This permits only a small portion of the genome to be expressed in any given cell or tissue type. Dominant control elements termed locus control regions (LCRs) may contribute to tissue-specific gene expression via localized alterations in chromatin structures (W.C.Forrester et al., 1987; Felsenfeld, 1996; Felsenfeld et al., 1996). LCR determinants can be detected and mapped by their ability to establish DNase I hypersensitive sites (HS) in the chromatin of expressing cells. LCRs are operationally defined by their ability to confer tissue-specific, copy-number dependent, high-level expression on linked transgenes irrespective of their sites of integration in the host genome (Grosveld et al., 1987; Festenstein et al., 1996; Li et al., 1999). Although the mechanism(s) underlying LCR function remains unclear, extensive histone acetylation of the chromatin encompassing LCRs has been associated with their activity in systems where this has been studied (Hebbes et al., 1994; Elefant et al., 2000; Schubeler et al., 2000). Since histone acetylation promotes chromatin decondensation and increases accessibility to trans-factor binding (Vettese-Dadey et al., 1994; Brownell and Allis, 1996; Gregory et al., 1998; Luger and Richmond, 1998), this modification provides a mechanism linking LCR function to chromatin opening and subsequent transcriptional activation.
The hGH cluster contains five closely linked genes spanning 48 kb on chromosome 17q22–24. These genes share >94% sequence identity and are arranged in a uniform transcriptional orientation: 5′-hGH-N, hCS-L, hCS-A, hGH-V, hCS-B-3′. This organization and structure suggests that the hGH cluster arose through a series of relatively recent gene duplication events (Chen et al., 1989). Despite the high level of sequence identity among these genes and their respective promoter elements, they exhibit mutually exclusive patterns of expression. hGH-N is selectively expressed in the somatotrope and somatolactotrope cells of the anterior pituitary (Bennani-Baiti et al., 1998a), whereas expression of the remaining four genes (collectively referred to as hCS) is restricted to the syncytiotrophoblast layer of the mid-to-late gestation placental villi (Liebhaber et al., 1989; Walker et al., 1991; MacLeod et al., 1992). The hCS-A and hCS-B genes are expressed at very high levels by term gestation whereas hCS-L and hGH-V are expressed only at trace levels. The mechanism(s) underlying this tissue specificity and the expression levels of these genes remains unclear.
The expression of the hGH cluster in both pituitary and placenta is under the control of its LCR. When introduced into the mouse genome on their own, hGH-N or hCS-A, even with extensive segments of contiguous flanking sequences, are either not expressed or expressed at low and sporadic levels (Jones et al., 1995). The hGH LCR is marked by a set of tissue-specific HS: HSI,II (because of their very close linkage, HSI and HSII are dealt with as a single entity), HSIII and HSV in pituitary nuclear chromatin, and HSIII, HSIV and HSV in the chromatin of placental syncytiotrophoblast nuclei. Linkage of the full set of HS to an hGH-N transgene selectively drives expression in somatotropes in a robust copy-number manner (Jones et al., 1995; Bennani-Baiti et al., 1998a; Shewchuk et al., 1999). Similarly, hCS, when linked to the LCR, is expressed selectively in the mouse placenta in a consistent, copy-number-dependent fashion (Su et al., 2000). These studies support a bifunctional role for the hGH LCR and demonstrate that the transgenic mouse model is a reliable system to study its action.
Distinct regulatory elements within the hGH LCR appear to be involved in promoting hGH-N and hCS expression and specification. HSI,II, the pituitary-specific set of HS, is fully sufficient to confer high-level, somatotrope-specific, position-independent expression on a linked hGH-N transgene (Jones et al., 1995; Bennani-Baiti et al., 1998a; Shewchuk et al., 1999). Of note, this expression is much higher than that of the endogenous mGH and is not copy-number dependent (Jones et al., 1995). These data suggest that HSI,II is the major positive regulator of LCR action in the pituitary but must be linked to other LCR determinants to establish a fully insulated and autonomously controlled locus. The basis for placenta-specific expression of the remaining genes in the cluster is less well understood. Cell culture-based studies by others (Nachtigal et al., 1993) have suggested that the specificity of hCS expression in the placenta reflects repression of its expression in the pituitary. Cell transfection studies have mapped a repressive activity to a 263 bp conserved element (P-element) located 2 kb 5′ to each of the placentally expressed genes. This element is embedded in an extensive region of conserved sequence identity associated with each placental gene. The repressive action of the P-element demonstrated in these cell transfection studies has not been addressed in a developmentally dynamic setting, and the possibility that the placental genes in the hGH cluster are regulated by a positively acting determinant has not been thoroughly tested. In the present study, we sought to expand our understanding of hGH LCR function by investigating the mechanistic basis for placenta-specific hCS activation. The results suggest a model in which specific positive regulatory elements can target distinct patterns of chromatin modification at the hGH locus in the pituitary and placenta. Such distinct patterns of chromatin modification may reflect the ability of the hGH LCR to activate genes within its multigene cluster via two selective and mutually exclusive pathways.
Results
HSIII and HSV were selectively enriched for acetylated histones in placental chromatin
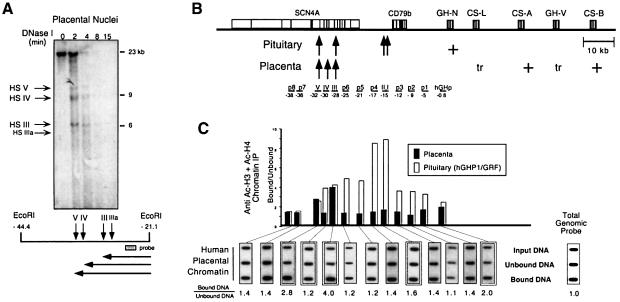
The genes of the hGH cluster are expressed at high levels in two distinct tissues; hCS-A and hCS-B in placental syncytiotrophoblasts and hGH-N in pituitary somatotropes. The remaining two genes, hGH-V and hCS-L, are also placenta specific, although they are expressed at very low levels. Expression of the genes of the hGH cluster in both tissues is LCR dependent (Jones et al., 1995; Su et al., 2000). Initial studies were designed to determine whether the acetylation pattern of the hGH LCR in the syncytiotrophoblasts paralleled that previously determined in the pituitary (Elefant et al., 2000). Syncytiotrophoblast nuclei were selectively released from the villi of a normal human term placenta (see Materials and methods) and subjected to DNase I mapping and chromatin immunoprecipitation (ChIP). The DNase I analysis of the LCR region confirmed the placenta-specific pattern of HS (HSIII, HSIV and HSV) (Figure 1A). The same nuclear preparation was then analyzed by ChIP; solubilized placental nucleosomes were immunoprecipitated with a mixture of antibodies recognizing acetyl-lysine residues on histones H3 and H4 (Figure 1C). Levels of acetylation at specific sites were determined by hybridization with a series of unique sequence probes. Hybridization signal intensities in each of the three ChIP fractions (input, unbound and bound) were normalized for DNA loading by rehybridizing the membrane with a total genomic DNA probe. The normalized ratios of histone acetylation enrichment (bound/unbound) were determined.
Fig. 1. HSIII and HSV but not HSIV are enriched for acetylated histones in placental chromatin. (A) Placental chromatin contains three prominent DNase I hypersensitive sites, HSIII, HSIV and HSV, located between 28 and 32 kb 5′ to the hGH multigene cluster. Nuclei selectively isolated from the hCS-expressing syncytiotrophoblasts of a normal human term placenta were digested with DNase I for increasing periods of time (indicated above respective lanes in minutes). DNA was isolated at each time point, digested to completion with EcoRI and analyzed by Southern blotting using the 32P-labeled probe (represented diagrammatically in the map below the autoradiograph). The identity of the HS that resulted in the generation of each of the sub-bands is indicated to the left of the autoradiograph and the size markers are shown to the right. The diagram below the Southern blot illustrates the position of each DNase I HS site (vertical arrows) relative to the EcoRI-defined 3′ terminus. The horizontal lines represent the lengths of each corresponding sub-band. The coordinates represent the number of kilobases 5′ to the transcription start site of hGH-N. Three major HS (HSIII, HSIV and HSV) and a minor HS (IIIa) were detected. (B) Diagram of the hGH cluster and its LCR. A schematic representation of the hGH multigene cluster and its associated LCR are shown. The positions of the closely linked B-lymphocyte-specific CD79b gene (Bennani-Baiti et al., 1998b) and the striated muscle-specific SCN4A gene (Bennani-Baiti et al., 1995) are also indicated. The shaded rectangles represent each of the genes (labeled) and the vertical lines within each rectangle indicate their respective exons. The presence of HS in placenta or pituitary are indicated (arrows). The expression patterns of each gene are indicated as strong (+) or trace (tr). The probes used to map chromatin acetylation levels across this region are labeled and underlined below the diagram, and their coordinates relative to the transcription start site of hGH-N are indicated. Three probes correspond to segments between hGH-N and HSI,II (p1, p2 and p3 located at –5, –9 and –12 kb, respectively); three to segments between HSI,II and HSIII (p4, p5 and p6 located at –17, –21 and –25 kb, respectively); and two to segments 5′ of the LCR (p7 and p8 located at –36 and –38 kb, respectively). (C) Acetylation of the hGH LCR in human placental chromatin is limited to segments encompassing HSIII and HSV. Chromatin from nuclei selectively released from placental syncytiotrophoblasts (the chromatin preparation analyzed in Figure 1A) was subjected to ChIP analysis. Soluble nuclear chromatin was immunoprecipitated with a mixture of antisera specific to acetylated histones H3 and H4. Equal amounts of DNA purified from starting chromatin (Input DNA), unacetylated chromatin (Unbound DNA) and acetylated antibody-bound chromatin (Bound DNA) were applied to nylon membranes via a slot-blot manifold. The membranes were then sequentially hybridized with the 32P-labeled probes underlined in (B). The autoradiographs generated using hybridization probes corresponding to each HS of the hGH LCR (including the pituitary-specific HSI,II) and by the hGH-N promoter are doubly boxed. Ratios of hybridization signal intensities in the bound and unbound chromatin fractions were normalized to the corresponding ratios obtained using a loading control probe (Total Genomic Probe) and are indicated below each autoradiograph. The normalized ratios are summarized in the histogram. Each bar in the histogram is centered below its respective probe. Black histogram bars represent placental chromatin. This figure shows the ChIP analysis from one representative experiment using chromatin isolated from a single placental preparation shown in (A). All chromatin immunoprecipitations and slot-blot analyses reported were repeated twice with consistent results. White histogram bars represent previously published data obtained from analyses of transgenic mouse lines carrying the entire hGH LCR and linked hGH gene cluster on a P1 transgene (hGH/P1) (Elefant et al., 2000). These data are included to facilitate direct comparison with the placental study.
The initial ChIP studies monitored levels of histone acetylation at specific sites 5′ to the hGH gene cluster. These sites included the hGH-N promoter, each of the hGH LCR HS, and regions between and surrounding the HS (Figure 1B). The positions of each of these probes could be exactly defined in relation to the hGH cluster and to the closely linked B-lymphocyte-specific CD79b (Bennani-Baiti et al., 1998b) and striated muscle-specific SCN4A genes (Bennani-Baiti et al., 1995) based on the sequence of this entire contiguous region (DDBJ/EMBL/GenBank accession No. AC005803). This analysis revealed a pattern of acetylation in placental chromatin that was clearly distinct from the extensive pattern of acetylation of the LCR previously determined in the pituitary. The pituitary ChIP data are summarized in Figure 1C as white bars for reference (data taken from Elefant et al., 2000). Acetylation in the placental chromatin was limited to HSIII and HSV (bound:unbound DNA ratios of 4.0 and 2.8, respectively). The acetylation levels at these two sites were essentially identical to those observed in the pituitary (compare black and white bars; Figure 1C). Surprisingly, acetylation at the placenta-specific HSIV was not significantly enriched over that of total genomic DNA (bound:unbound DNA <2.0; the background level was observed in non-expressing tissues; Elefant et al., 2000). Thus the patterns of hGH LCR chromatin acetylation in pituitary and placenta shared the modification of HSIII and HSV, but were otherwise distinct.
The P-element failed to repress expression of hGH-N in the transgenic mouse pituitary
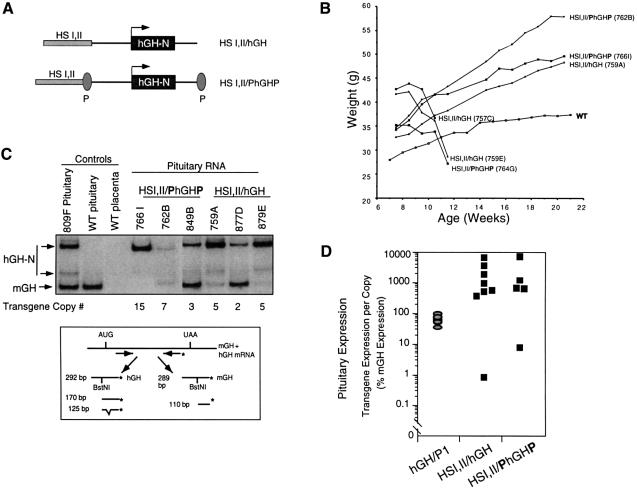
The lack of a placenta-specific site of histone acetylation in the 5′-flanking domain of the hGH gene cluster suggested that one or more element(s) relevant to activation of the hCS might exist elsewhere. A candidate for such a placental restriction determinant was the conserved P-element. The P-element is a 263 bp fragment positioned 2 kb 5′ to each of the placentally expressed genes in the hGH cluster (see Figure 4A) (Chen et al., 1989). A previous study, based upon cell transfection assays, concluded that the P-element controlled the placental restriction of the hCS genes by repressing their expression in the pituitary. In these studies, the repressive effect of the P-element did not appear to be hCS specific because it was equally effective in repressing a juxtaposed hGH-N promoter (Nachtigal et al., 1993). To determine whether the P-element could repress hGH-N expression in vivo, as it does in transfected pituitary cells, a transgene was constructed in which hGH-N linked to HSI,II was flanked by P-elements (HSI,I/PhGHP; Figure 2A). We have demonstrated previously that linking HSI,II to hGH-N (HSI,II/hGH transgene) results in consistent high-level expression in somatotrope cells of the anterior pituitary, causing a consistent phenotype of gigantism in host mice (Jones et al., 1995). A repressive effect of the P-element on transgene expression would be predicted to repress this phenotype of gigantism. Six transgenic mouse lines were generated carrying HSI,II/PhGHP and five lines were generated in a parallel set of injections carrying the control HSI,II/hGH. Transgene expression was initially monitored by growth curves of the male founders for each of the lines (Figure 2B). In comparison with the growth rate of wild-type littermates, each of the HSI,II/PhGHP lines demonstrated clear-cut gigantism. This phenotype was indistinguishable from that of mice carrying the corresponding transgene lacking the P-element. Three founders died secondary to complications of this gigantism (Brem et al., 1989) during the weighing period (Figure 2B). Quantitative analysis of pituitary hGH-N mRNA levels in three founders from each of the two transgenic groups (three HSI,II/PhGHP lines and three HSI,II/hGH lines) revealed equivalent high levels of transgene expression (Figure 2C and D). These data suggested that the P-element was unable to repress hGH-N expression in the transgenic mouse pituitary.
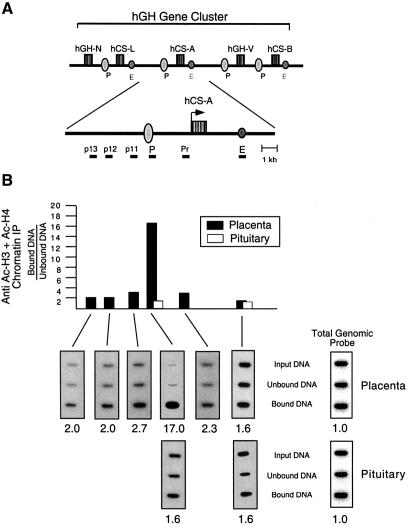
Fig. 4. The P-element displays a localized peak of hyperacetylation in placental chromatin. (A) Diagram of the 48 kb hGH multigene cluster with an enlargement of the hCS-A gene region. An expanded schematic of the hCS-A gene and flanking regions is provided to show the precise spacing of the probes used in this study. The conserved P-elements (P) are located 2 kb 5′ to each of the placentally expressed genes. A set of conserved enhancer elements (E) (Jiang and Eberhardt, 1997) are located 2 kb downstream from hCS-L, hCS-A and hCS-B. Due to the high level of sequence identity (92–98%) in the immediate flanking and intervening regions among these genes, we were unable to generate probes specific for each placental locus (see Materials and methods). Therefore, only p13 is specific for the hCS-A locus; each of the remaining probes hybridize to conserved regions adjacent to each of the placentally expressed genes (see Materials and methods). None of these probes, however, are represented elsewhere in the genome (confirmed by Southern blotting; data not shown). (B) Representative ChIP analyses of placental and pituitary chromatin at the hCS-A locus. Soluble nuclear chromatin prepared from each of the indicated sources was immunoprecipitated with anti-acetylated histone H3 and H4 antibodies, and analyzed as in Figure 2. All ratios were normalized to the ratio obtained using a probe for total genomic human DNA as a loading control (shown to the right). The normalized ratios detected in the bound versus unbound chromatin fractions are shown below each respective autoradiograph and are summarized in the histogram. The black histogram bars represent human placental chromatin. White histogram bars represent analysis of human pituitary chromatin at the P-element and enhancer regions.
Fig. 2. The P-element fails to repress expression of hGH-N in the transgenic pituitary. (A) HSI,II/hGH and HSI,II/PhGHP transgene constructs. The P-element, previously demonstrated to silence expression of linked genes in transfected pituitary-derived cell lines (Nachtigal et al., 1993), was inserted on either side of hGH-N in the context of the pituitary-specific HSI,II/hGH transgene to generate the HSI,II/PhGHP transgene. (B) Growth curves of mice containing the HSI,II/hGH or the derivative HSI,II/PhGHP transgenes. Body weights (ordinate) of each male founder transgenic for either transgene were monitored for 22 weeks (abscissa). Each growth curve is labeled with the respective transgene construct and identifying line number. The growth curve for a control, non-transgenic mouse is shown for comparison (WT). The three founders exhibiting the most extreme initial weights experienced rapid pre-morbid decreases in their body weights and died during the study period. (C) hGH-N mRNA levels in the pituitaries of HSI,II/hGH or HSI,II/PhGHP transgenic mice. A diagram depicting the assay is shown below the autoradiograph. hGH-N and mGH mRNAs were co-amplified from pituitary RNA samples isolated from each of the indicated lines. The 32P-end-labeled (asterisk) PCR product was digested with BstNI to differentiate between the co-amplified mGH and hGH mRNA species (see Materials and methods for details). The identity of each band is indicated by the corresponding labeled arrows to the left of the gel. Analyses of mGH and hGH mRNA content in the pituitaries of three HSI,II/hGH and three HSI,II/PhGHP transgenic mice are shown. Three of the lines (849B, 877D and 879E) were generated subsequent to the growth curve experiment to compensate for founder deaths. Controls included mRNA from the pituitary of the mouse line expressing the hGH-N transgene (line 809F; Jones et al., 1995), mRNA isolated from a wild-type (WT) mouse pituitary (showing only the 110 bp mGH cDNA band) and mRNA from wild-type mouse placenta (showing no hGH-N or mGH expression). (D) Equivalent expression of HSI,II/hGH and HSI,II/PhGHP transgenes in the mouse pituitary. Band intensities in (C) were quantified by PhosphorImager (Molecular Dynamics) analyses and the levels of hGH-N mRNA for each line were normalized to both mGH mRNA and transgene copy numbers (hGH-N mRNA/transgene copy/mGH RNA = % mGH expression). This value represents the level of expression from a single transgene copy as a percentage of the expression of a single endogenous mGH gene (ordinate; logarithmic scale). These results are compared to previously documented, copy-number-dependent hGH-N expression in the pituitaries of mice carrying the hGHP1 transgene encompassing the entire hGH multigene cluster and LCR (gray ovals) (Su et al., 2000). Each data point represents expression from a single transgenic line with a unique transgene insertion site.
The P-element stimulated linked transgene expression in the mouse placenta
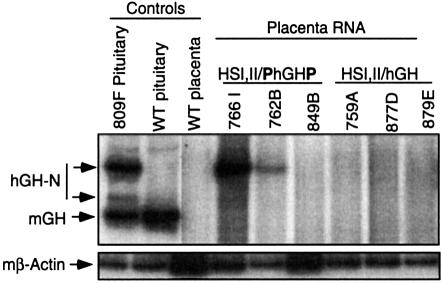
The lack of a repressive effect of the P-element on gene expression in the pituitary led us to consider that the placental restriction might reflect a positive effect in the placenta. Because the transgenic mouse accurately models placental as well as pituitary expression from the hGH gene cluster (Su et al., 2000), the possibility that the P-element serves a direct stimulatory role in placental expression could be tested in this system. The six transgenic founders surviving the morbidity of chronic hGH oversecretion (Figure 2B plus the female founders, not shown) were bred to generate transgenic embryos. Placental RNA was isolated on embryonic day (e) 18.5 from each of the embryos identified as transgenic. The hGH-N mRNA content in the placenta was analyzed by RT–PCR. Three HSI,II/hGH lines (759A, 877D and 879E) and three HSI,II/PhGHP lines (766I, 762B and 849B) were studied. hGH-N mRNA was detected in the placentas of two of the three HSI,II/PhGHP lines and in none of the three HSI,II/hGH lines (Figure 3). Expression of these transgenes in other tissues was limited to trace levels of ectopic expression in brain, heart or liver that varied from line to line, and was unrelated to the presence or absence of the P-element (data not shown). These data suggested that the P-element was driving the expression of hGH-N in the placenta and thus might be involved in the normal activation of the hCS genes.
Fig. 3. The P-element activates hGH-N transgene expression in the mouse placenta. mRNA was isolated from the placentas of transgenic embryos derived from each of the six surviving transgenic lines and was analyzed for the presence of hGH-N mRNA by RT–PCR as described in Figure 2C. hGH-N expression was detected in two of three HSI,IIPhGHP line placentas (766I and 762B) and in none of three HSI,IIhGH line placentas. RT–PCR for mouse β-actin RNA served as control for RNA quality.
The P-element was associated with a focused peak of histone acetylation in placental chromatin
Histone acetylation is associated with positive transcriptional activation in a number of studied systems and tends to be maximal at sites of the corresponding cis-acting regulatory element (see Discussion). HSI,II, the major somatotrope-specific positive regulatory element of the hGH LCR, is acetylated in primary human pituitary chromatin and in the pituitaries of transgenic mice carrying the hGH gene cluster linked to and contiguous with the LCR (Elefant et al., 2000). To determine whether acetylation was also involved in gene activation by the conserved P-element, histone modification was assessed at this site in human placental syncytiotrophoblast chromatin. Acetylation enrichment was also assessed at a region 2 kb 3′ to hCS-L, hCS-A and hCS-B genes. These regions each contain a conserved element shown to enhance hCS expression in transfected, cultured placental cell lines and, on the basis of these studies, have been referred to as the ‘hCS enhancers’ (Walker et al., 1990) (Figure 4A). ChIP analysis revealed a peak of histone acetylation at the P-elements that was 17-fold above total genomic control levels. In marked contrast, the 3′ hCS enhancer elements were not enriched for acetylation, and neither were the P-elements and enhancer elements in primary human pituitary chromatin (Figure 4B). Therefore, the chromatin encompassing the P-element was highly and specifically acetylated in the placenta.
Extension of P-element chromatin acetylation in placental cells was examined next. We had previously demonstrated that the high level of acetylation at HSI,II in pituitary chromatin extends at lower, but still substantial levels bidirectionally for ∼32 kb (see Figure 1C, gray bars). To determine whether acetylation within the hGH gene cluster in the placental chromatin was similarly distributed, regions flanking the P-element were probed in the ChIP assay. Probes located –1.95 and –2.4 kb upstream of the P-element (p12 and p13, respectively) revealed essentially no acetylation above background, and the two probes most closely bracketing the P-elements (p11 at –0.5 kb upstream and Pr at 1.5 kb downstream of the P-element) revealed only low levels of acetylation (Figure 4B). Thus, placental syncytiotrophoblast chromatin acetylation was highly localized to the P-element.
Discussion
Previous studies of the hGH multigene cluster have demonstrated that pituitary-restricted hGH-N expression as well as placenta-restricted hCS-L, hCS-A, hGH-V and hCS-B expression are dependent upon the hGH LCR. Gene expression in these two tissues is paralleled by the establishment of overlapping sets of tissue-specific DNase I HS in the LCR (Jones et al., 1995; Su et al., 2000). The findings of previous studies, when combined with the present data, suggest that the hGH LCR functions along distinct pathways of chromatin activation to effect mutually exclusive patterns of expression in pituitary and placenta. LCR-mediated hGH-N activation in the pituitary correlates with histone acetylation of a 32 kb chromatin domain encompassing the entire LCR. This pattern of chromatin modification, which is found in human pituitaries and is reliably reproduced in transgenic models (Elefant et al., 2000), demonstrates a distinct central peak at the pituitary-specific HSI,II (see Figure 1C, white bars). HSI,II has been demonstrated in transgenic studies to comprise the major positive regulatory element in the LCR for somatotrope expression of hGH-N (Jones et al., 1995; Bennani-Baiti et al., 1998a; Shewchuk et al., 1999). These data suggest that histone acetylation, targeted primarily to HSI,II, contributes to pituitary-specific changes in chromatin structure that direct and restrict hGH-N expression in this tissue.
The data in the present study demonstrated that the pattern of histone acetylation in placental chromatin was distinct from that in the pituitary. Overlap in the acetylation patterns at the hGH locus in pituitary and placental chromatin was limited to HSIII and HSV; these two sites form in both tissues and were modified to the same extent. The conservation of chromatin structure at these two sites (Figure 1C) implied that they mediate a function(s) shared in pituitary and placenta. The positioning of HSV at the 5′ border of the acetylated LCR domain in pituitary chromatin (Elefant et al., 2000) suggested that it may have an insulator role. The 5′ and 3′ borders of the chicken β-globin gene cluster are similarly bounded by LCR elements that are highly acetylated (Hebbes et al., 1994). These elements have been shown to exert local insulator functions and to contain cis-acting elements shared among insulators in unrelated gene systems (Chung et al., 1993; Pikaart et al., 1998; Prioleau et al., 1999; Saitoh et al., 2000). An insulator function for HSV, and possibly HSIII, in the hGH LCR is directly supported by previous studies demonstrating that a fragment containing HSIII and HSV can shield a weakly expressed hGH transgene from position effects in transgenic mice (Jones et al., 1995). Thus, the distal LCR region encompassing HSIII and HSV may serve as the boundary that marks the 5′ end of the active hGH locus chromatin domain in both the pituitary and placenta. Additional tests of this function are currently under way.
The initial DNase I mapping of the hGH LCR suggested that HSI,II would be of particular importance to pituitary expression and HSIV in placental expression. The subsequent studies, summarized above, have confirmed that HSI,II is the main positive regulatory element within the hGH LCR (Jones et al., 1995; Bennani-Baiti et al., 1998a; Shewchuk et al., 1999). HSIV, being unique to the placenta, might have a comparable function in that tissue. However, its role remains less well defined. Although it forms in placental chromatin (Figure 1A), the region encompassing HSIV is not acetylated (Figure 1C). The specific lack of HSIV acetylation in the placenta implies that any effect that it might be exerting on gene expression in this tissue is mediated by a mechanism independent of histone modification. These results suggested that one or more regulatory element(s) in addition to the 5′-flanking LCR might be involved in the selective expression of the subset of the hGH gene cluster in the placenta. The P-element may fill this role. Tissue culture-based studies had suggested that the P-elements, located 2 kb upstream of each placentally expressed gene in the hGH cluster, selectively repress the placentally expressed genes in the pituitary (Nachtigal et al., 1993). Our in vivo test of the P-element using transgenic mice failed to support this repressive model (Figure 2). Instead, the in vivo expression studies (Figure 3) and dramatic acetylation of the P-element in placental chromatin (Figure 4) support a model in which the placental activation of hCS reflects action by the P-element.
The mechanism of P-element function is not known. The P-elements upstream of hCS-L, hGH-V, hCS-A and hCS-B are each directly preceded by a 30- to 50- nucleotide-long polypurine tract (AnG) (Chen et al., 1989). Such tracts have been shown to form unusual helical structures that influence the binding of specific trans-acting factors and exclude nucleosomes (Gross and Garrard, 1988). Such a role for the P-element in chromatin modulation may explain its lack of stimulatory activity in transient transfection assays, because such assays, based on expression of loosely packaged episomes, may not accurately recapitulate chromatin-based mechanisms of gene activation (Jeong and Stein, 1994). Similarly, the lack of acetylation at the chromatin segment encompassing the hCS 3′ enhancer elements in primary syncytiotrophoblast nuclei was unexpected. These enhancers demonstrate a clear stimulatory action on hCS expression in cell transfection studies (Walker et al., 1990; Jiang and Eberhardt, 1995). These data run counter to the general correlation between enhancer action and histone acetylation (Sheridan et al., 1997; Krumm et al., 1998; Forrester et al., 1999; Forsberg et al., 1999; McMurry and Krangel, 2000). Because the hCS enhancer has only been defined in the context of cell transfection studies, a firm resolution of its effects in vivo and in the context of chromatin must await further analysis in appropriate systems.
The acetylation of the P-element was spatially localized, substantial, and restricted to placental chromatin (Figure 4). This suggested specific targeting of histone acetyl transferase (HAT)-containing co-activators to this site. Similarly, targeted and localized acetylation has been found at a limited number of studied genes and has been demonstrated to be tightly linked to transcriptional activation (Kuo et al., 1998; Chen et al., 1999; Parekh and Maniatis, 1999; Schubeler et al., 2000; Vignali et al., 2000). In support of a stimulatory role for the P-element in placental hCS activation, two of three HSI,II/PhGHP transgenic lines expressed hGH mRNA in the mouse placenta whereas no hCS expression was detected in any of the three HSI,II/hGH lines tested (Figure 3). The line-to-line variability in the levels of HSI,II/PhGHP transgene expression in the placenta (Figure 3; lines with high, low and no appreciable expression) suggested site-of-integration position effects. This position dependence was consistent with our previous data demonstrating a similar lack of copy-number dependence for the HSI,II/hGH transgene in the pituitary (Jones et al., 1995) (Figure 2D). In both cases, the lack of copy-number-dependent expression may reflect the absence of LCR component(s) such as HSV and HSIII that serve to insulate the transgene from surrounding influences at the insertion site (see above).
Of note, the present ChIP data were generated with a mix of anti-H3 and anti-H4 antibodies. Using antibodies specific for each of the histones in higher eukaryotic systems may (Schubeler et al., 2000) or may not (Parekh and Maniatis, 1999) reveal selectivity in the distribution of acetylation. In the present case, the observation of a dramatic peak of acetylation at the P-element and a lack of acetylation in the domain upstream of the hGH cluster (between the hGH gene and HSIII) clearly distinguish the placental from pituitary patterns of chromatin modification. This distinction would not be altered by more defined studies. The present data suggests that the stimulatory effect attributed to the P-element could be mediated by the recruitment to that site of a tissue-specific and highly restricted HAT activity. Distinctions between the acetylation of separate classes of histones, and among the various target lysines on each particular histone, may be of interest in future studies as the HAT complexes responsible for the modifications in the placenta and pituitary are identified.
The distinct differences in the patterns of histone acetylation at the hGH LCR in placenta and pituitary appear to reflect distinct mechanisms by which HAT activities are involved in the regulation of gene transcription over long distances. The mechanism mediated by HSI,II in the pituitary may reflect extended propagation of histone acetylation with resultant creation and/or maintenance of an activated chromatin domain (Hebbes et al., 1994; Madisen et al., 1998; Elefant et al., 2000; Schubeler et al., 2000). In contrast, the P-element appears to exert a more localized effect on chromatin structure (Kuo et al., 1998; Struhl, 1998; Parekh and Maniatis, 1999; Schubeler et al., 2000). The recent identification of two distinct HAT-containing complexes that mediate either restricted or extended modification of chromatin templates in vitro supports the two modes of chromatin modification observed in the expression of the GH locus (Vignali et al., 2000). Thus the distinct patterns of acetylation observed within the hGH locus in pituitary and placental chromatin suggest a model in which specific positive regulatory elements can in a tissue-specific manner target distinct HATs to a single LCR and multigene locus. It will be of considerable interest to identify the specific HATs involved in the extensive versus localized pattern of hyperacetylation in pituitary and placental chromatin, in order to test this model in vivo.
Materials and methods
Transgene construction
To create HSI,II/hGH, a 1.6 kb BglII restriction fragment containing HSI,II was isolated and inserted in the native orientation upstream of hGH-N (2.6 kb EcoRI fragment R2 of hGH-N; Chen et al., 1989) at the BamHI site of pBSIIKS-hGH-N (pBSIIKS; Stratagene). This hGH-N gene fragment contained 494 bp of 5′- and 530 bp of 3′-flanking sequences. To create HSI,II/PhGHP, EcoRI linkers were added to a Klenow-generated and blunt-ended 263 bp BamHI and HinfI restriction fragment encompassing the P-element sequence (Nachtigal et al., 1993). This end-modified P-element was inserted in its native orientation both 5′ and 3′ of hGH-N in the HSI,II/hGH plasmid.
Generation and analysis of transgenic mice
Transgene inserts released from vector sequences by BssHII digestion were resolved on agarose gels, isolated using a Qiaex II gel extraction kit, purified through an Elutip (Schleicher and Schuell) and diluted to 2 ng/µl in 10 mM Tris–HCl pH 7.6, 0.1 mM EDTA. This DNA solution was then micro-injected into fertilized C57BL/6J × SJL mouse embryos. Transgenic founders were detected by dot-blot analysis of tail DNA using a 1.37 kb SmaI fragment derived from hGH-N (coordinates –494 to +876 relative to the start of hGH-N transcription) as probe. The integrity of the transgenes and their copy numbers were determined by Southern blot analyses of BglII-digested tail DNA using the hGH genomic probe. Only founders carrying intact transgenes (as detected by a 4.8 kb hybridizing band for HSI,II/PhGHP lines and a 4.2 kb hybridizing band for HSI,II/hGH lines) were used for subsequent studies. Transgene copy number was established by comparing the signal intensity of the repeated transgene hybridization band with that of the hybridizing band corresponding to the single-copy junction fragment.
Detection of hGH transcripts in transgenic pituitary and placental RNA by RT–PCR
Pituitary and placental RNAs were prepared from tissues isolated from mice immediately following decapitation as described (Jones et al., 1995). hGH-N expression in the pituitary and placenta was detected by RT–PCR using individual organs as previously described (Jones et al., 1995; Su et al., 2000). Briefly, reverse transcription using AMV RT (Promega, Madison, WI) was performed using 0.2–0.5 µg of total RNA. Subsequent PCR was carried out using primers corresponding to perfectly conserved regions between mouse (m) GH and hGH (primers in Table I). Following 30 cycles of PCR, the 3′-end-labeled cDNA products were digested with BstNI to yield fragments specific to hGH-N (170 and 125 bp) and mGH (110 bp) mRNAs. To compare the levels of hGH mRNA relative to endogenous mGH mRNA in the pituitary, the intensities of the mGH- and hGH-specific bands were quantified by PhosphorImager (Molecular Dynamics, Inc., Sunnyvale, CA) analysis using ImageQuant software. The ratio was then divided by transgene copy number (see above) to establish the transgene expression per copy.
Table I. Oligonucleotide primers used in mRNA detection or as probes for ChIP analyses.
| Probe | Size | Primer set |
|---|---|---|
| mhGH | 290 bp | (5′-GCCTGCTCTGCCTGC-3′) (5′-GACTGGATGAGCAGCAG-3′) |
| p11 | 180 bp | (5′-GAATCCCAAGTCTAATGC-3′) (5′-CCTTTATAAGGGGTTACC-3′) |
| p12 | 194 bp | (5′-CCTGGACATATCATATGGC-3′) (5′-CAAGGAACTTCAGCCACAG-3′) |
| p13 | 175 bp | (5′-GAGCTCTGATTCCTAGCCC-3′) (5′-CATGGTTCTCACTGCCC-3′) |
Isolation of nuclei from placenta
Intact nuclei were selectively isolated from the syncytiotrophoblast cells of human term placentas as described previously (Jones et al., 1995) with the following minor modifications. Villous fragments were excised from placenta, resuspended in cold phosphate-buffered saline and finely minced. Fragments that passed through a 10-gauge screen were pelleted, resuspended in 150 mM NH4Cl containing 0.5 g of NH4HCO2 per 10 g of tissue, and incubated at 4°C. After 45 min, the majority of syncytiotrophoblast cells had undergone osmotic lysis due to their high levels of carbonic anhydrase. The preparation was pelleted at 1000 g to remove unlysed cells and residual NH4HCO2. The nuclear pellets were resuspended in 45 ml cold RB [0.1 M NaCl, 50 mM Tris–HCl pH 8.0, 3 mM MgCl2, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 10 mM sodium butyrate] and the preparation was passed through a cheesecloth to separate the syncytiotrophoblast nuclei from tissue debris. Preparations were examined under the microscope to confirm the presence of intact nuclei.
DNase I HS mapping
Nuclei were washed and resuspended in RB buffer and digested with 1.0 µg/ml of DNase I (Gibco-BRL, Grand Island, NY) at 37°C for increasing amounts of time. Reactions were terminated by the addition of Na2EDTA to 50 mM and the nuclei were lysed by overnight incubation at 37°C in 0.8 M NaCl, 50 mM EDTA, 0.5% SDS, 200 µg/ml of proteinase K. The samples were then phenol-chloroform extracted, ethanol precipitated, and digested with EcoRI prior to Southern blotting. DNA samples were resolved on 0.8% agarose gels, transferred to Zetabind nylon membranes (Cuno Inc., Meriden, CT), and pre-hybridized at 65°C overnight as previously described (Su et al., 2000). The membranes were subsequently incubated overnight at 65°C with hybridization solution containing 1–2 × 106 c.p.m./ml of random primer-labeled probe. Subsequent washes were at 60°C in 0.1% SDS and 0.1 × SSC, followed by autoradiography.
Immunoprecipitation of unfixed chromatin
Preparation of unfixed chromatin and the chromatin immunoprecipitation (ChIP) procedure were both carried out as previously described (Hebbes et al., 1994; O’Neill and Turner, 1995, 1996; Crane-Robinson et al., 1997) with certain modifications (Elefant et al., 2000). Briefly, 0.3 mg nuclei were digested with 25 U of micrococcal nuclease at 37°C for 6 min in 1 ml of digestion buffer containing 50 mM NaCl, 20 mM Tris–HCl pH 7.5, 3.0 mM MgCl2, 1.0 mM CaCl2, 10 mM sodium butyrate, 0.1 mM PMSF. The digestion was terminated by the addition of Na2EDTA to 0.5 mM and the salt-soluble chromatin was isolated as described (Hebbes et al., 1994). Soluble chromatin was concentrated using a Microcon centrifugal filter (Amicon Inc., Bedford, PA) and 250 µg of this chromatin (input) was incubated with 10 µl each of antisera specific for the acetylated forms of histones H3 or H4 (Upstate Biotech., Lake Placid, NY) in a total volume of 500 µl. Protein A–Sepharose (Amersham Pharmacia Biotech) precipitates were generated, washed, and DNA was purified from the pellets (bound) and supernatants (unbound) as previously described (O’Neill and Turner, 1996). DNA samples from the input, bound and unbound fractions were analyzed by electrophoresis on 1% agarose gels to determine the size distribution of the resulting oligonucleosomes. The majority of DNA ranged from 160 bp (mononucleosomes) to 1 kb; only trace DNA could be visualized above 2 kb.
Equal masses of DNA (1.0 µg) from input, unbound and antibody-bound DNAs were loaded onto Zetabind nylon membranes using a slot-blot manifold. The blots were incubated overnight at 65°C with hybridization solution containing 1–2 × 106 c.p.m./ml random primer-labeled probe. Subsequent washes were at 60°C in 0.1% SDS and 0.5 × SSC. Signals were quantified by PhosphorImager analysis. The ratios between bound and unbound DNA fractions were calculated for each probe used. All ratios were normalized for total DNA loading onto the slot blot by rehybridizing the membrane with 32P-labeled total genomic DNA (Elefant et al., 2000). All ratios were verified in a minimum of two separate experiments.
Hybridization probe preparation
The majority of hybridization probes were generated by PCR using AmpliTaq DNA polymerase (Perkin-Elmer). A P1 clone encompassing the hGH LCR and the first four genes of the hGH cluster (Su et al., 2000) served as the template. The primer sets used for generating probes p1–p8 and probes corresponding to HSI,II, HSIII, HSIV and HSV (Figure 1) were as previously described (Elefant et al., 2000). The sequences of the additional primer sets used for the present studies and the predicted sizes of the generated probes in bp are summarized in Table I. The P-element probe consisted of a subcloned 263 bp EcoRI fragment (a kind gift from Dr Peter Cattini, University of Manitoba, Canada) (Nachtigal et al., 1993). The hCS enhancer probe was a 240-bp amplified fragment of the repeated enhancer element sequence (Jiang and Eberhardt, 1997). Probe p13 is specific for the hCS-A upstream region; the p12, p11, P-element and E probes hybridize to conserved elements adjacent to each of the placental genes. The hGH promoter probe was a 500 bp BamHI fragment encompassing the hGH-N gene promoter region. This probe cross hybridized with the hCS and hGH-V promoters due to their high degree of sequence identity. A genomic DNA loading control probe was generated by random primer labeling of sonicated human total genomic DNA.
Acknowledgments
Acknowledgements
Transgenic mice were generated by the University of Pennsylvania School of Medicine Transgenic and Chimeric Mouse Core. This work was supported by NIH ROI-HD-25147 (N.E.C. and S.E.L.) and NIH NRSA F32-HD-08471 (F.E.).
References
- Bennani-Baiti I.M., Jones,B.K., Liebhaber,S.A. and Cooke,N.E. (1995) Physical linkage of the human growth hormone gene cluster and the skeletal muscle sodium channel α-subunit gene (SCN4A) on chromosome 17. Genomics, 29, 647–652. [DOI] [PubMed] [Google Scholar]
- Bennani-Baiti I.M., Asa,S.L., Song,D., Iratni,R., Liebhaber,S.A. and Cooke,N.E. (1998a) DNase I-hypersensitive sites I and II of the human growth hormone locus control region are a major developmental activator of somatotrope gene expression. Proc. Natl Acad. Sci. USA, 95, 10655–10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennani-Baiti I.M., Cooke,N.E. and Liebhaber,S.A. (1998b) Physical linkage of the human growth hormone gene cluster and the CD79b (Igβ/B29) gene. Genomics, 48, 258–264. [DOI] [PubMed] [Google Scholar]
- Brem G., Wanke,R., Wolf,E., Buchmuller,T., Muller,M., Brenig,B. and Hermanns,W. (1989) Multiple consequences of human growth hormone expression in transgenic mice. Mol. Biol. Med., 6, 531–547. [PubMed] [Google Scholar]
- Brownell J.E. and Allis,C.D. (1996) Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr. Opin. Genet. Dev., 6, 176–184. [DOI] [PubMed] [Google Scholar]
- Chen E.Y., Liao,Y.-C., Smith,D.H., Barrera-Saldaña,H.A., Gelinas,R.E. and Seeburg,P. (1989) The human growth hormone locus: nucleotide sequence, biology, and evolution. Genomics, 4, 479–497. [DOI] [PubMed] [Google Scholar]
- Chen H., Lin,R.J., Xie,W., Wilpitz,D. and Evans,R.M. (1999) Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell, 98, 675–686. [DOI] [PubMed] [Google Scholar]
- Chung J.H., Whiteley,M. and Felsenfeld,G. (1993) A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell, 74, 505–514. [DOI] [PubMed] [Google Scholar]
- Crane-Robinson C., Hebbes,T.R., Clayton,A.L. and Thorne,A.W. (1997) Chromosomal mapping of core histone acetylation by immunoselection. Methods, 12, 48–56. [DOI] [PubMed] [Google Scholar]
- Elefant F.E., Cooke,N.E. and Liebhaber,S.A. (2000) Targeted recruitment and spreading of histone acetyltransferase activity by a locus control region. J. Biol. Chem., 275, 13827–13834. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. (1996) Chromatin unfolds. Cell, 86, 13–19. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G., Boyes,J., Chung,J., Clark,D. and Studitsky,V. (1996) Chromatin structure and gene expression. Proc. Natl Acad. Sci. USA, 93, 9384–9388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festenstein R., Tolaini,M., Corbella,P., Mamalaki,C., Parrington,J., Fox,M., Milou,A., Jones,M. and Kioussis,D. (1996) Locus control region function and heterochromatin-induced position effect variegation. Science, 271, 1123–1125. [DOI] [PubMed] [Google Scholar]
- Forrester W.C., Takegawa,S., Papayannopoulou,T., Stamatoyannopoulos,G. and Groudine,M. (1987) Evidence for a locus activation region: the formation of developmentally stable hypersensitive sites in globin-expressing hybrids. Nucleic Acids Res., 15, 10159–10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester W.C., Fernandez,L.A. and Grosschedl,R. (1999) Nuclear matrix attachment regions antagonize methylation-dependent repression of long-range enhancer–promoter interactions. Genes Dev., 13, 3003–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg E.C., Johnson,K., Zaboikina,T.N., Mosser,E.A. and Bresnick,E.H. (1999) Requirement of an E1A-sensitive coactivator for long-range transactivation by the β-globin locus control region. J. Biol. Chem., 274, 26850–26859. [DOI] [PubMed] [Google Scholar]
- Gregory P.D., Schmid,A., Zavari,M., Lui,L., Berger,S.L. and Horz,W. (1998) Absence of Gcn5 HAT activity defines a novel state in the opening of chromatin at the PHO5 promoter in yeast. Mol. Cell, 1, 495–505. [DOI] [PubMed] [Google Scholar]
- Gross D.S. and Garrard,W.T. (1988) Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem., 57, 159–197. [DOI] [PubMed] [Google Scholar]
- Grosveld F., van Assendelft,G., Greaves,D.R. and Kollias,G. (1987) Position-independent, high-level expression of the human β-globin gene in transgenic mice. Cell, 51, 975–985. [DOI] [PubMed] [Google Scholar]
- Hebbes T.R., Clayton,A.L., Thorne,A.W. and Crane-Robinson,C. (1994) Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J., 13, 1823–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S.W. and Stein,A. (1994) Micrococcal nuclease digestion of nuclei reveals extended nucleosome ladders having anomalous DNA lengths for chromatin assembled on non-replicating plasmids in transfected cells. Nucleic Acids Res., 22, 370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S.-W. and Eberhardt,N.L. (1995) Involvement of a protein distinct from transcription enhancer factor-1 (TEF-1) in mediating human chorionic somatomammotropin gene enhancer function through the GT-IIC enhanson in choriocarcinoma and COS cells. J. Biol. Chem., 270, 13906–13915. [DOI] [PubMed] [Google Scholar]
- Jiang S.-W. and Eberhardt,N.L. (1997) The human chorionic somatomammotropin enhancers form a composite silencer in pituitary cells in vitro. Mol. Endocrinol., 11, 1233–1244. [DOI] [PubMed] [Google Scholar]
- Jones B.K., Monks,B.R., Liebhaber,S.A. and Cooke,N.E. (1995) The human growth hormone gene is regulated by a multicomponent locus control region. Mol. Cell. Biol., 15, 7010–7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm A., Madisen,L., Yang,X.J., Goodman,R., Nakatani,Y. and Groudine,M. (1998) Long-distance transcriptional enhancement by the histone acetyltransferase PCAF. Proc. Natl Acad. Sci. USA, 95, 13501–13506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M.-H., Zhou,J., Jambeck,P., Churchill,M.E.A. and Allis,C.D. (1998) Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev., 12, 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Harju,S. and Peterson,K.R. (1999) Locus control regions coming of age at a decade plus. Trends Genet., 15, 403–408. [DOI] [PubMed] [Google Scholar]
- Liebhaber S.A., Urbanek,M., Ray,J., Tuan,R.S. and Cooke,N.E. (1989) Characterization and histological localization of human growth hormone-variant gene expression in the placenta. J. Clin. Invest., 83, 1985–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K. and Richmond,T.J. (1998) The histone tails of the nucleosome. Curr. Opin. Genet. Dev., 8, 140–146. [DOI] [PubMed] [Google Scholar]
- MacLeod J.N., Lee,A.K., Liebhaber,S.A. and Cooke,N.E. (1992) Developmental control and alternative splicing of the placentally expressed transcripts from the human growth hormone gene cluster. J. Biol. Chem., 267, 14219–14226. [PubMed] [Google Scholar]
- Madisen L., Krumm,A., Hebbes,T.R. and Groudine,M. (1998) The immunoglobulin heavy chain locus control region increases histone acetylation along linked c-myc genes. Mol. Cell. Biol., 18, 6281–6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry M.T. and Krangel,M.S. (2000) A role for histone acetylation in the developmental regulation of V(D)J recombination. Science, 287, 495–498.10642553 [Google Scholar]
- Nachtigal M.W., Nickel,B.E. and Cattini,P.A. (1993) Pituitary-specific repression of placental members of the human growth hormone gene family. A possible mechanism for locus regulation. J. Biol. Chem., 268, 8473–8479. [PubMed] [Google Scholar]
- O’Neill L.P. and Turner,B.M. (1995) Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J., 14, 3946–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill L. and Turner,B.M. (1996) Immunoprecipitation of chromatin. Methods Enzymol., 274, 189–203. [DOI] [PubMed] [Google Scholar]
- Parekh B.S. and Maniatis,T. (1999) Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-β promoter. Mol. Cell, 3, 125–129. [DOI] [PubMed] [Google Scholar]
- Pikaart M.J., Recillas-Targa,F. and Felsenfeld,G. (1998) Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev., 12, 2852–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prioleau M.-N., Nony,P., Simpson,M. and Felsenfeld,G. (1999) An insulator element and condensed chromatin region separate the chicken β-globin locus from an independently regulated erythroid-specific folate receptor gene. EMBO J., 18, 4035–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh N., Bell,A.C., Recillas-Targa,F., West,A.G., Simpson,M., Pikaart,M. and Felsenfeld,G. (2000) Structural and functional conservation at the boundaries of the chicken β-globin domain. EMBO J., 19, 2315–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D., Francastel,C., Cimbora,D.M., Reik,A., Martin,D.I.K. and Groudine,M. (2000) Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human β-globin locus. Genes Dev., 14, 940–950. [PMC free article] [PubMed] [Google Scholar]
- Sheridan P.L., Mayall,T.P., Verdin,E. and Jones,K.A. (1997) Histone acetyltransferases regulate HIV-1 enhancer activity in vitro. Genes Dev., 11, 3327–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewchuk B.M., Asa,S.L., Cooke,N.E. and Liebhaber,S.A. (1999) Pit-1 binding sites in the somatotrope-specific HS I,II region of the hGH LCR are essential for in vivo hGH-N gene activation. J. Biol. Chem., 274, 35725–35733. [DOI] [PubMed] [Google Scholar]
- Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev., 12, 599–606. [DOI] [PubMed] [Google Scholar]
- Su Y., Liebhaber,S.A. and Cooke,N.E. (2000) The human growth hormone gene cluster locus control region supports position-independent pituitary- and placenta-specific expression in the transgenic mouse. J. Biol. Chem., 275, 7902–7909. [DOI] [PubMed] [Google Scholar]
- Vettese-Dadey M., Walter,P., Chen,H., Juan,L.-J. and Workman,J.L. (1994) Role of the histone amino termini in facilitated binding of a transcription factor, Gal4-AH to nucleosome cores. Mol. Cell. Biol., 14, 970–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignali M., Steger,D.J., Neely,K.E. and Workman,J.L. (2000) Distribution of acetylated histones resulting from Gal4-VP16 recruitment of SAGA and NuA4 complexes. EMBO J., 19, 2629–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker W.H., Fitzpatrick,S.K. and Saunders,G.F. (1990) Human placental lactogen transcriptional enhancer: tissue specificity and binding with specific proteins. J. Biol. Chem., 265, 12940–12948. [PubMed] [Google Scholar]
- Walker W.H., Fitzpatrick,S.L., Barrera-Saldaña,H.A., Resendez-Perez,D. and Saunders,G.F. (1991) The human placental lactogen genes: structure, function, evolution and transcriptional regulation. Endocrinol. Rev., 12, 316–328. [DOI] [PubMed] [Google Scholar]