Abstract
We have shown previously that the TFIIIC1/TFIIIC1′ fraction interacts specifically with the VA1 terminator regions to affect both termination and initiation/reinitiation of transcription by human RNA polymerase III. Here, we further purified the VA1 terminator-binding factor to apparent homogeneity and found, by peptide sequence analysis, that it belongs to the NF1 protein family. NF1 interacts specifically with the NF1-binding sites within the terminator regions of the VA1 gene and with two subunits (TFIIIC220 and TFIIIC110) of human TFIIIC2. Immunodepletion with anti-NF1 antibodies dramatically decreases transcription from the VA1 template in nuclear extract, and mutation at the NF1-binding site in the terminator region of the VA1 gene selectively affects multiple-round transcription (reinitiation of transcription) and termination. In addition, NF1 acts in conjunction with TFIIIC to promote accurate termination by RNA polymerase III on a C-tailed VA1 template.
Keywords: nuclear factor 1/RNA polymerase III/transcription
Introduction
RNA polymerase III transcribes genes for small structural RNAs such as 5S RNA, tRNA, adenovirus VA1 RNA and U6/7SK RNA (reviewed in Geiduschek and Kassavetis, 1992; White, 1994). The subclass II genes (tRNA and VA1 RNA) have a simple internal promoter containing A and B box elements that are recognized by TFIIIC, which in turn directs the sequential binding of TFIIIB and RNA polymerase III. The subclass I genes (5S RNA) have internal A box, I box and C box elements that are first recognized by TFIIIA, forming a DNA–protein complex that is then recognized sequentially by TFIIIC, TFIIIB and RNA polymerase III. Consistent with the conservation of these activation mechanisms from yeast to human (reviewed in Paule and White, 2000), components of the three subunit TFIIIB and the 17 subunit RNA polymerase III are highly conserved in structure and function (Wang and Roeder, 1997; M.Teichmann, Z.Wang, M.Ito, W.Meisner, K.H.Seifart and R.G.Roeder, manuscript submitted). Unlike yeast TFIIIC, which is composed of six polypeptides and binds both A and B box elements in tRNA templates (Deprez et al., 1999 and references therein), TFIIIC activity from HeLa cells can be separated into two fractions (TFIIIC1 and TFIIIC2), which are both required for transcription by RNA polymerase III (Yoshinaga et al., 1987; Wang and Roeder, 1996; Oettel et al., 1997). Highly purified human TFIIIC2 contains five polypeptides (220, 110, 102, 90 and 63 kDa) and binds the B box region (Yoshinaga et al., 1989; Kovelman and Roeder, 1992; Wang and Roeder, 1996). Although the composition and exact function of TFIIIC1 are still not clear, TFIIIC1 contains a terminator-binding activity and facilitates strong TFIIIC interactions over the entire promoter (Yoshinaga et al., 1987; Wang and Roeder, 1996; Oettel et al., 1997).
A cluster of four or more T residues surrounded by GC-rich sequences on the non-coding strand acts as a signal to terminate transcription by RNA polymerase III (Cozzarelli et al., 1983). However, termination and (re)initiation of RNA polymerase III do not appear to be isolated events. Thus, changes in the termination regions of the VA1 gene (Wang and Roeder, 1996), various tRNA genes (Allison and Hall, 1985; Young et al., 1991; Dieci and Sentenac, 1996) and Alu sequences (Chu et al., 1995) affect their overall transcription levels as well. An important question concerns the mechanism for the observed correlation between termination and (re)initiation of RNA polymerase III transcription. A model of facilitated recycling of RNA polymerase III on the transcribed gene was proposed by the Sentenac group and by our laboratory (Dieci and Sentenac, 1996; Wang and Roeder, 1996). This model explains both the correlation between efficient termination and efficient reinitiation, and the very high efficiency of multiple-round transcription of RNA polymerase III. In the case of human RNA polymerase III transcription, a critical role in the efficient termination and recycling of RNA polymerase III is played by holo TFIIIC. Holo TFIIIC contains TFIIIC and other factors that include PC4 and topoisomerase I, and strongly interacts not only with A and B box but also with the termination regions of tRNA genes (Wang and Roeder, 1998).
This study reports purification of the VA1 terminator-binding factor to homogeneity and identification of this factor as NF1 family proteins. It is further demonstrated that NF1 interacts directly with two TFIIIC2 subunits (TFIIIC220 and TFIIIC110), which are localized in B box and 3′ regions (Kovelman and Roeder, 1992; Bartholomew et al., 1993), and that it affects multiple-round transcription and accurate termination from the VA1 template.
Results
Holo TFIIIC interacts with downstream termination regions of the VA1 gene
We demonstrated previously that the TFIIIC1/TFIIIC1′ fraction interacts with the termination region of the VA1 template and that deletion of this region affects both the synergistic interaction between TFIIIC1 and TFIIIC2 on the DNA template and the level of transcription initiation from VA1 and tRNA templates (Wang and Roeder, 1996). Since a holo TFIIIC complex, immunopurified from the FLAG-tagged TFIIICα cell line at 100 mM KCl, contained both TFIIIC1 and TFIIIC2 activities (Wang and Roeder, 1998), we tested whether it could also interact with the termination region of the VA1 gene. The DNase I footprint analysis in Figure 1 shows that, in addition to showing strong interactions with A and B box regions, holo TFIIIC also interacts with the termination region of the VA1 gene (positions +175 to +153 on the lower strand and positions +154 to +170 on the upper strand) (lanes 3, 6, 9 and 10). This downstream interaction parallels that observed with the TFIIIC′ fraction (lane 8; Wang and Roeder, 1996). Also evident (lane 6) is a hypersensitive site (+88) similar to that generated by TFIIIC2 plus TFIIIC1 (Wang and Roeder, 1996).
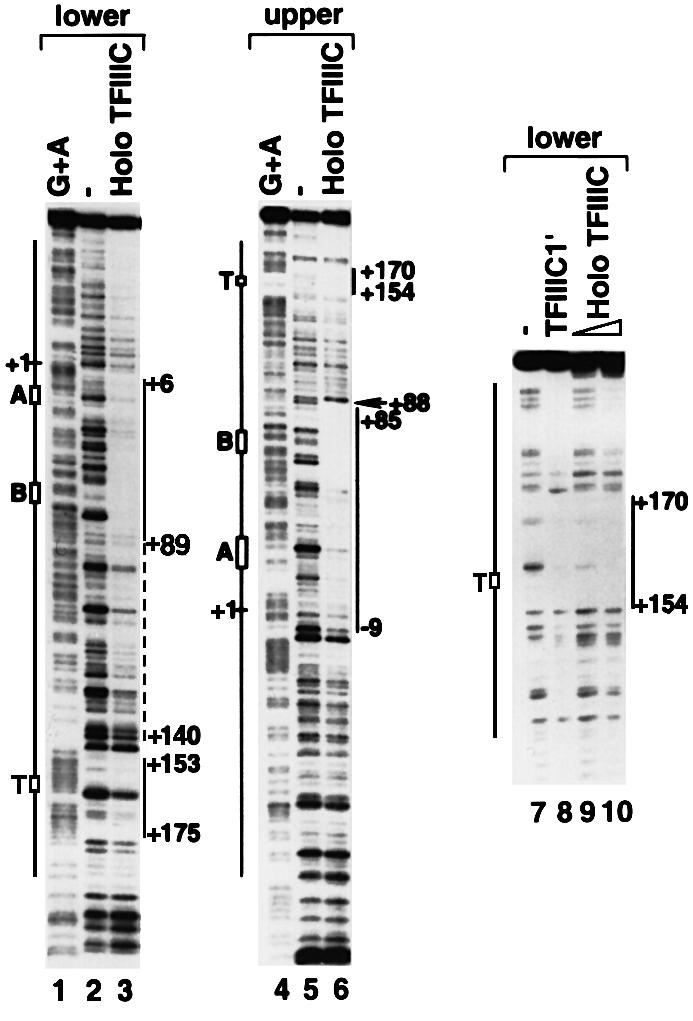
Fig. 1. Holo TFIIIC protects A box, B box and terminator regions of the VA1 gene. DNA fragments containing the full-length (–96 to +232) (lanes 1–6) or the terminator region (+101 to +232) (lanes 7–10) of the VA1 gene were labeled at the lower (lanes 1–3) or the upper (lanes 4–10) strand end with 32P. The labeled DNA fragments were incubated with mock immunoprecipitate (2 µl) (lanes 2, 5 and 7) or with 100 (2 µl) (lane 9) or 200 (4 µl) (lanes 3, 6 and 10) fmol of immunopurified holo TFIIIC, or 2 µl (0.2 µg protein) of the purified TFIIIC1′ fraction (lane 8). Lanes 1 and 4 are the Maxam–Gilbert G + A reactions. The protected regions are indicated on the left. The hypersensitive site (+88) is indicated by an arrow.
Since TFIIIC1′ and holo TFIIIC were purified by different methods, these results strongly support our previous conclusion that the downstream interactions are relevant to transcription by RNA polymerase III. Since a single TFIIIC-containing complex interacts with two sites (A/B boxes and termination region) that are separated by an ∼80 bp DNA sequence, this intervening DNA fragment may either form a loop or wrap around TFIIIC. A weak, but clear, footprint over this region (positions +89 to +140) supports the latter possibility (Figure 1, lane 3). TFIIIC immunopurified under high salt conditions (either 300 or 800 mM KCl) loses the terminator-binding activity (data not shown), indicating that the downstream interacting component(s) is not strongly bound to TFIIIC. This prompted speculation about the existence of other factor(s) in holo TFIIIC that interact directly with downstream regions of RNA polymerase III-transcribed genes.
Four groups of polypeptides co-purify with the VA1 terminator-binding activity
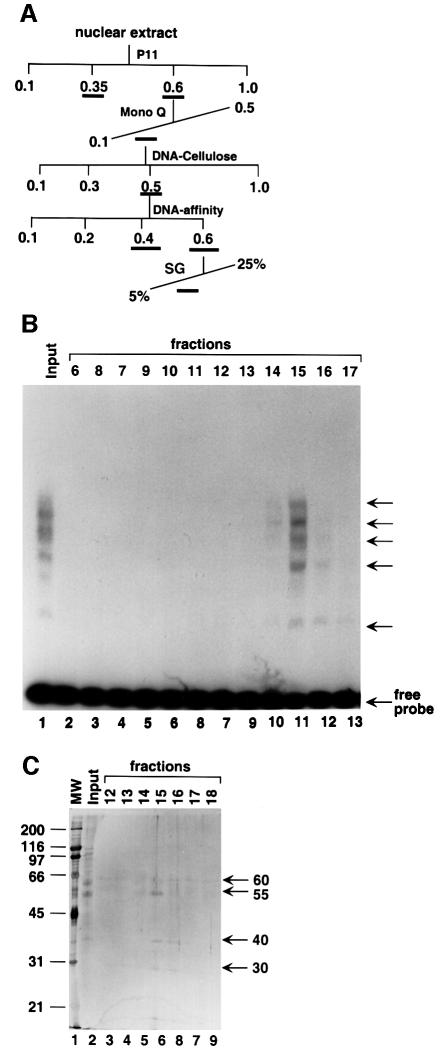
Given indications that the downstream-binding factor(s) may play an important role in transcription by RNA polymerase III, we established a procedure for its further purification (Figure 2A). Based on the footprint analysis, we adopted a gel shift assay with a 32P-labeled probe containing the protected VA1 terminator region to monitor activity through the purification steps. Multiple DNA–protein complexes were observed with a wild-type probe (Figure 2B, lane 1) and were competed by a wild type, but not by a mutant (TTTT to ACTG), VA1 terminator probe (data not shown). The first two chromatographic steps for purification of this binding activity are almost identical to those for TFIIIC1 fractionation, and the behavior of the binding activity on these two columns is also similar to that of TFIIIC1 (Wang and Roeder, 1996). The Mono Q fraction was purified on a double-stranded DNA–cellulose column to remove both non-binding and strong DNA-binding proteins, and the recovered terminator-binding activity was further purified through a DNA-affinity column that contained the first terminator region of the VA1 gene. The activity split evenly between the BC400 and BC600 fractions on the DNA-affinity column, and the BC600 fraction was subjected to a 5–25% sucrose gradient sedimentation analysis. The VA1 terminator-binding activity was centered at fraction number 15 (Figure 2B). SDS–PAGE and subsequent silver staining of the sucrose gradient fractions revealed four major protein groups, with molecular masses of ∼60, 55, 40 and 30 kDa, respectively, which co-migrated with the VA1 terminator-binding activity (Figure 2C). Each group contains several closely adjacent bands. Closer inspection revealed that there are also ‘ladders’ of thin, weak bands between the major band groups, especially between the 66 and 50 kDa groups.
Fig. 2. Purification of the VA1 terminator-binding factor. (A) Purification scheme. (B) Gel shift analysis of the sucrose gradient sedimentation fractions. One microliter of input and 5 µl of the gradient fractions were subjected to gel shift assay with a probe containing the VA1 terminator sequence. The multiple protein–DNA complexes are marked with arrows at the right. (C) SDS–PAGE analysis of the sucrose gradient sedimentation fractions. Input (2.5 µl) (lane 2) and 10 µl of the sucrose gradient fractions (lanes 3–9) were resolved on 10% SDS–PAGE and visualized by silver stain. The positions of molecular markers are indicated on the left and the four major groups of polypeptide bands co-eluted with the VA1 terminator-binding activity are marked with arrows on the right.
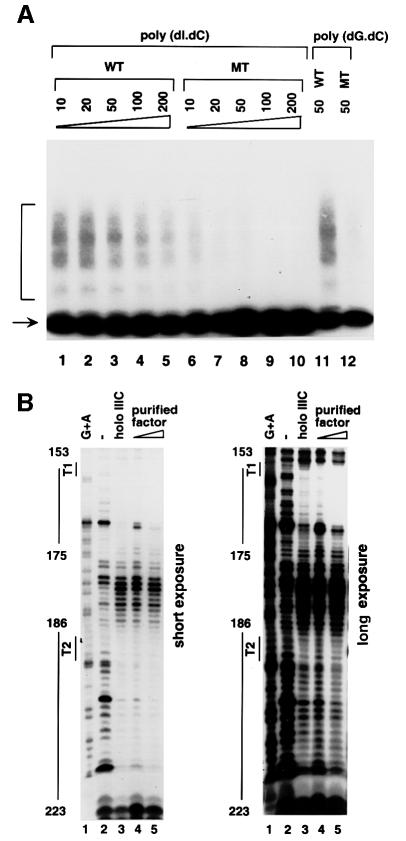
The DNA binding specificity of the purified VA1 terminator-binding factor was further determined by gel shift and footprint assays. As shown in the gel shift assay of Figure 3A, the purified factor bound avidly to the wild-type terminator probe (lane 3), but only very weakly to the mutant probe (TTTT to ACTG) (lane 8) under the standard assay conditions that employed 50 ng of poly(dI⋅dC) as non-specific DNA competitor. In addition, interaction of the purified VA1 terminator-binding factor with the wild-type VA1 terminator was more resistant to competition by poly(dI⋅dC) and poly(dG⋅dC) than that with the mutant VA1 terminator (lanes 1–5 versus 6–10, and lane 11 versus 12). These results indicate that the T-run sequence affects the DNA binding affinity of the purified factor.
Fig. 3. The purified factor specifically recognizes the VA1 terminator region. (A) Comparison of the purified factors binding to wild-type and mutant VA1 terminators. The factor purified through the oligo-affinity column was tested for its binding to either wild-type (lanes 1–5 and 11) or mutant (TTTT to ACTG) (lanes 6–10 and 12) VA1 terminator sequence by a gel shift assay in the presence of different concentrations of competitor DNA. The type of competitor and the concentrations(µg/ml) used are marked above the lanes. (B) The purified factor and holo TFIIIC generate identical footprint patterns over the terminator regions (+101 to +232) of the VA1 gene. Immunopurified holo TFIIIC (100 fmol) (lane 3) and 2 (lane 4) or 6 (lane 5) µl of the purified factor from sucrose gradient sedimentation were examined for their footprint on a fragment of the VA1 gene containing both the T1 and T2 terminator regions. The two panels show different exposures of the same gel to show clearly the protection over both terminator regions. The probe was labeled at the 5′ end of the lower strand. Lane 1 is a Maxam–Gilbert G + A reaction and lane 2 is without proteins. The protected sequences and the terminators (T1 and T2) are indicated on the left.
To compare the purified proteins with holo TFIIIC in relation to interactions with the terminator region of the VA1 gene, footprint assays were performed with templates containing both terminators (T1 and T2) of the VA1 gene (Figure 3B). Very significantly, the purified factor and holo TFIIIC generated identical footprint patterns over the termination region of the VA1 gene (lanes 4 and 5 versus 3). In the first terminator region (T1), both the purified factor and holo TFIIIC covered the region from 3 bases upstream to 16 bases downstream of the T-run. In the second terminator region (T2), both the purified factor and holo TFIIIC covered the region from 1 base upstream to 18 bases downstream of the T-run. These results indicate that the purified factor has a VA1 terminator-binding activity indistinguishable from that of holo TFIIIC.
The terminator-binding factor belongs to the NF1 protein family
The proteins from the BC600 fraction of the DNA-affinity column were separated by SDS–PAGE, transferred to a PVDF membrane and then subjected to microsequence analysis. Multiple peptide sequences were obtained for the four groups of proteins (60 kDa group: peptide 1, ED/FE/PM/QMDXD/GYE/LE; peptide 2, RPEYREDFVLS/TI/VTGK; peptide 3, PQCD/R/SNPGLCVQPHHIGV; 55 kDa group: peptide 1, AAQCGHPVLCVQPHHIGVAV; peptide 2, WASRLLAK; 40 kDa group: peptide 1, PQCSNP/AGLCVQPHHIGVSVK; 30 kDa group: peptide 1, IXPEFXEDFVLXI; peptide 2, TLACTGSL). Gene bank searching with these peptide sequences revealed that the four groups of proteins belong to the NF1 protein family.
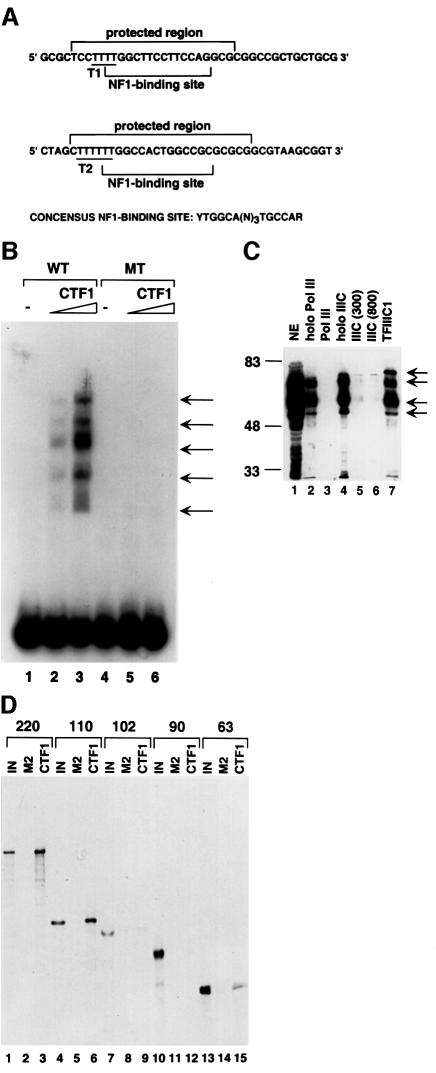
NF1 is a family of proteins with multiple functions (Qian et al., 1995 and references therein). They are derived from extensive alternative splicing of transcripts of four genes, NF1-A, -B, -C and -X, respectively. Individual NF1 proteins can dimerize with themselves, with splicing variants from the same NF1 gene, and with splicing variants from the other NF1 genes (Kruse and Sippel, 1994). The peptide sequences from the 55 kDa protein group are all derived from the NF1-C gene, while those from the 30 kDa group are from the NF1-X gene. The complexity of the various splicing variants from the four genes explains the protein patterns of the purified fraction on SDS–PAGE and the existence of multiple complexes in the gel shift assay. The various NF1 proteins have a similar DNA binding specificity, and the consensus DNA binding sequence for the family of proteins is 5′-YTGGCANNNTGCCAR-3′ (Borgmeyer et al., 1984). Inspection of the VA1 gene sequence shows that there is a probable, albeit imperfect, NF1-binding site in each terminator region immediately following the T-run (Figure 4A). This fits the footprint patterns of the purified factors and may explain the prior result that a TTTT to ACTG mutation in the VA1 terminator region reduces factor binding several fold.
Fig. 4. The VA1 terminator-binding factor belongs to the NF1 protein family and interacts directly with TFIIIC2 subunits. (A) DNA sequence of two terminator (T1 and T2) regions of the VA1 gene. The protected regions observed in the footprint assays and the consensus binding site for the NF1 protein family are indicated. (B) The interaction of CTF1 with the terminator region of the VA1 gene is dependent on theNF1 consensus sequence. The recombinant CTF1 shifted the VA1 terminator-containing probe in a dosage-dependent manner (lanes 2and 3) and the mutation of the NF1 consensus sequence in the probe diminished the shift (lanes 5 and 6). One (lanes 2 and 5) or 5 (lanes 3 and 6) ng of CTF1 expressed in bacteria were used in gel shift assay. (C) NF1 family proteins are present in holo RNA polymerase III, holo TFIIIC and TFIIIC1 fractions. HeLa cell nuclear extract (2.5 µl) (lane 1), holo RNA polymerase III (lane 2), core RNA polymerase III (lane 3), holo TFIIIC (lane 4), core TFIIIC purified under the salt concentrations of 300 (lane 5) or 800 (lane 6) mM KCl and TFIIIC1 fraction (lane 7) were resolved by 10% SDS–PAGE and subjected to immunoblot analysis with antibodies against the N-terminus of CTF1. Arrows indicate the polypeptides recognized by the antibodies and positions of the molecular size makers (kDa) are indicated on the left. (D) CTF1 interacts directly with TFIIIC220 and TFIIIC110. M2 agarose beads alone (lanes 2, 5, 8, 11 and 14) or M2 agarose beads containing 100 ng of the FLAG-tagged CTF1 expressed in Sf21 cells were subjected to individual pull-down assays with the 35S-labeled subunits of TFIIIC2 indicated at the top of lanes. Lanes 1, 4, 7, 10 and 13 are 20% of input.
The very large number of NF1 family proteins, their high degrees of sequence homology and functional similarity, and their complex interactions make it very difficult to pinpoint which NF1 proteins are involved in RNA polymerase III-mediated transcription. Our initial test employed CTF1 (Santoro et al., 1988), the largest NF1 variant from the human NF1-C gene, which was expressed in bacterial and Sf21 cells and purified via an affinity tag. Aside from availability of a full-length cDNA clone, the most important consideration was that CTF1 contains almost all of the exons present in the NF1-C gene. Similar to the purified fraction, the recombinant CTF1 bound strongly to the VA1 terminator region in the gel shift assay (Figure 4B, lanes 1–3). This interaction was abolished by mutation of the NF1 consensus sequence from TTGGCTTCCTTCCAG to TTccCTTCCTTCtAG (lanes 4–6). The bases that were mutated have been shown to play an important role in NF1 binding by methylation interference analysis and by binding assays with mutant probes (Zorbas et al., 1992). The multiple protein–DNA complexes observed in the present analysis probably reflect heavy degradation of the bacterially expressed recombinant CTF1.
For further analysis, rabbit polyclonal antibodies were raised against the N-terminal 1–233 amino acid residues of human CTF1, which has strong sequence similarities with the other NF1 proteins. Although it is not clear whether the polyclonal antibodies can recognize most NF1 proteins, an immunoblot analysis none the less revealed the presence of several distinct NF1 species in HeLa nuclear extract (Figure 4C, lane 1), holo RNA polymerase III (lane 2), holo TFIIIC (lane 4) and the TFIIIC1 fraction (lane 7); trace amounts of NF1 proteins were observed in TFIIIC preparations immunopurified under the higher salt conditions (lanes 5 and 6) but not in purified RNA polymerase III (lane 3). Although the purified fraction contained both 40 and 30 kDa groups of proteins (Figure 2C), the anti-CTF1 antibodies did not recognize them in HeLa nuclear extract (Figures 4C, lane 1 and 5B, lane 1). Assuming that the anti-CTF1 antibodies recognize CTF1 and other NF1 family proteins with similar affinities, quantitative western blot analysis revealed that the molar ratio of NF1 to TFIIIC2 subunits was ∼0.5 in the holo TFIIIC complex (data not shown).
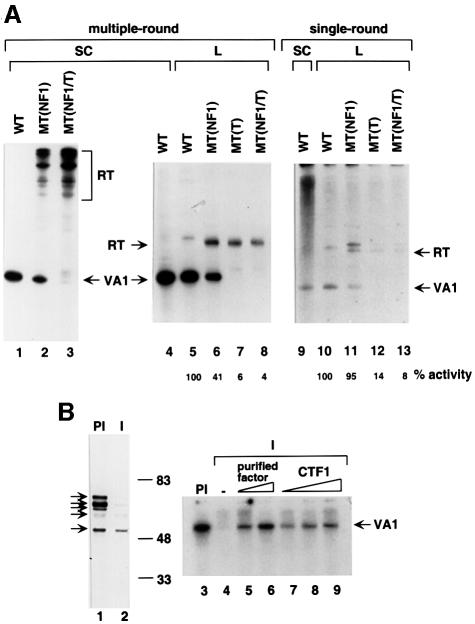
Fig. 5. NF1 protein is involved in multiple-round transcription from the VA1 template. (A) The NF1-binding site in the terminator region of the VA1 gene contributes to the multiple-round transcription by RNA polymerase III. In vitro transcription reactions were performed with HeLa nuclear extract using super-coiled (SC) (lanes 1–4 and 9) or linear (L) (lanes 5–8 and 10–13) templates under conditions for the multiple- (lanes 1–8) or single-round (lanes 9–13) transcription. VA1 templates with the wild-type terminator (lanes 1, 4, 5, 9 and 10), a mutated NF1-binding site (lanes 2, 6 and 11), a mutated terminator (lanes 7 and 12) or a mutated NF1-binding site and a mutated terminator (lanes 3, 8 and 13) are indicated as WT, MT(NF1), MT(T) and MT(NF1/T), respectively. The ratio of the signals obtained with the mutant templates (MT) relative to that obtained with the wild-type template (WT) is indicated at the bottom as percentage activity. The origin of the weak bands (lanes 5, 10 and 11) just above the RT products is unknown, but they may be generated from the vector sequence during transcription. (B) NF1 is required for transcription from the VA1 template. Western blot analysis was performed with anti-CTF1 antibodies to determine the nature and levels of NF1 proteins in the HeLa nuclear extract depleted with pre-immune (PI, lane 1) or immune (I, lane 2) anti-CTF1 antibodies. In vitro transcription reactions with the VA1 template were performed with HeLa nuclear extracts immunodepleted with either pre-immune (lane 3) or immune (lanes 4–9) antibodies against human CTF1. Reaction mixtures were complemented with either 2 (lane 5) or 6 µl (lane 6) of the purified factor or with 5 (lane 7), 10 (lane 8) or 20 ng (lane 9) of recombinant CTF1.
NF1 proteins interact directly with TFIIIC220 and TFIIIC110
Since NF1 proteins were co-immuopurified with TFIIIC from HeLa cell nuclear extract, we tested for direct interactions with TFIIIC2 subunits. For this purpose the FLAG-tagged human CTF1 was expressed in and purified from Sf21 cells and, after immobilization on M2 agarose beads, incubated with rabbit reticulocyte lysates containing in vitro 35S-labeled TFIIIC220, TFIIIC110, TFIIIC102, TFIIIC90 and TFIIIC63, respectively. An analysis of bound proteins (Figure 4D) shows that CTF1 interacted strongly with both TFIIIC220 (30% of input) and TFIIIC110 (34% of input), but only weakly with TFIIIC63 (4% of input) (lanes 3, 6 and 15) and not at all with TFIIIC102 (lane 9) or TFIIIC90 (lane 12). As a control, the five subunits of TFIIIC2 did not show any detectable interactions with unliganded M2 agarose beads (lanes 2, 5, 8, 11 and 14). Given that TFIIIC220 can be photo-crosslinked to the B box region (Kovelman and Roeder, 1992) and that the yeast homolog of human TFIIIC110 has been localized (by crosslinking) to the terminator region (Bartholomew et al., 1993), these interactions are consistent with the localization of the NF1-binding site in the VA1 gene and its function in transcription termination and reinitiation (see below).
RNA polymerase III involves NF1 protein in multiple-round transcription
In order to investigate further the mechanism of action of NF1 in RNA polymerase III-mediated transcription, mutations were introduced into the first terminator region of the VA1 gene and effects on transcription were analyzed in HeLa nuclear extract. As expected, almost no transcripts were terminated properly when both the T-run and the NF1-binding site were mutated in supercoiled templates (Figure 5A, lane 3). Instead, several long read-through transcripts appeared, probably due to transcription termination at downstream sites inside the vector. Mutation of just the NF1-binding site reduced the level of properly terminated transcripts to about one-half of the level from the wild-type template and generated several long read-through products (lane 2 versus 1). Thus, the NF1 site may contribute to the overall efficiency of transcription termination. Similar to what was observed with super-coiled templates, a linearized template mutated at the NF1-binding site generated a significantly reduced number of transcripts terminated at T1 (VA1) as well as read-through products (RT) (lane 6 versus 5). However, mutations either at the T-run or at both the T-run and the NF1-binding site completely abolished correct termination at T1 (VA1) on this template (lanes 7 and 8). Interestingly, the overall level of transcription (terminated plus read-through) from the template mutated at the NF1-binding site was only ∼41% of that from the wild-type template (lane 6 versus 5). This result suggests that the NF1-binding site affects initiation or reinitiation as well as termination. In a single-round transcription assay (lanes 9–13), the total transcription level (terminated plus read-through) obtained from the template mutated at the NF1-binding site was approximately the same (95%) as that obtained from the wild-type template (lane 11 versus 10). However, the levels observed with the templates mutated only at the T-run or at both the T-run and the NF1-binding site were much lower (14 and 8%, respectively) than the level observed with the wild-type template (lanes 12 and 13 versus 10). These results suggest that the NF1 consensus sequence within the terminator region of the VA1 gene affects mainly transcription reinitiation and termination whereas the T-run affects primary initiation, reinitiation and termination.
In a further study of NF1 function, HeLa nuclear extract was incubated with antigen-purified anti-CTF1 antibodies immobilized on protein A–Sepharose beads in order to deplete CTF1 and other NF1 proteins. Western blot analysis revealed that most of the NF1 proteins recognized by anti-CTF1 antibodies were removed by the depletion, the most notable exception being an ∼50 kDa polypeptide (Figure 5B, lane 2). As a control, HeLa nuclear extract that was mock depleted with IgG purified from the pre-immune serum showed a number of immunoreactive proteins (lane 1). As shown in Figure 5B (lanes 3–9), the anti-CTF1 depletion reduced the level of transcription from the VA1 gene ∼10- to 15-fold (lane 4 versus 3). Additions of the purified VA1 terminator-binding factor (lanes 5 and 6) and recombinant CTF1 (lanes 7–9) restored VA1 transcription, respectively, to 67% (lane 6 versus 3) and 25% (lane 9 versus 3) of the levels observed with the mock depleted extract (lane 3). These results imply that NF1 proteins indeed have a functional role in transcription of the VA1 gene.
NF1 promotes accurate termination by RNA polymerase III
A C-tailed template (Wang and Roeder, 1998) was used to investigate further the role of NF1 in termination of VA1 transcription. RNA polymerase can bind to the upstream tail of this template and initiate transcription by itself, generating similar amounts of three distinct transcripts, T1, T2 and RT (Figure 6A and B, lanes 1 and 6), which correspond to transcription termination at the first terminator, the second terminator and the end of the template, respectively. This result confirmed previous observations (Cozzarelli et al., 1983) indicating that RNA polymerase III has an intrinsic ability to recognize terminator sequence and to terminate transcription. However, the efficiency of recognition and termination is low. Addition of core TFIIIC or NF1 had no obvious effect (lanes 3, 8, 12 and 13). In contrast, addition of holo TFIIIC caused almost all transcripts to terminate at the first terminator, the primary site of termination in vivo, and reduced the overall transcription level ∼5-fold (lanes 2 and 7 versus 1 and 6). This indicated that a holo TFIIIC component(s) not present in core TFIIIC has an ability, in this assay, to help RNA polymerase III terminate transcription and to reduce the overall transcription as well. PC4/Topo I plus TFIIIC can mimic the effect of holo TFIIIC on the C-tailed VA1 template to some extent (Figure 6A, lane 9 versus 7; Wang and Roeder, 1998). Similarly, inclusion of NF1 with core TFIIIC greatly reduced the proportion of T2 and RT transcripts in the total population, and decreased the overall transcription level as well (lanes 4 and 5 versus 3). However, the combination of TFIIIC and NF1 did not result in termination by Escherichia coli RNA polymerase on the same template (data not shown), suggesting that the effect is RNA polymerase III specific and that NF1 does not function simply by binding to the termination region to serve as a road block. These results further suggest that NF1 is a genuine part of holo TFIIIC and performs its function in conjunction with core TFIIIC in transcription termination and reinitiation. In the same assay system, NF1 and PC4/Topo I did not show any synergy (lanes 10 and 11). Although the reason for the apparent redundancy of NF1 and PC4/Topo I in this assay needs further investigation, it is relevant that the molar ratios of NF1 and PC4 to TFIIIC are ∼1 and 100, respectively, in these assays (see Discussion).
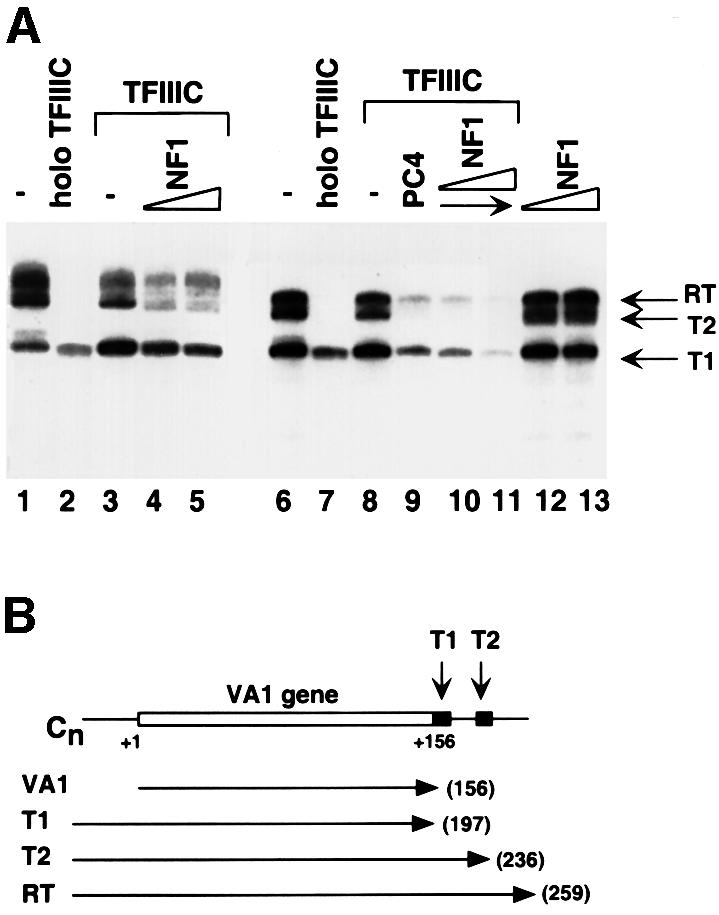
Fig. 6. NF1 protein enhances proper termination of transcription by RNA polymerase III in conjunction with TFIIIC. (A) Each reaction contained 20 fmol of C-tailed VA1 template and 25 fmol of immunopurified RNA polymerase III. The reaction mixtures were supplemented with no additional factors (lane 1), 200 fmol of holo TFIIIC (lanes 2 and 7), 200 fmol of core TFIIIC (lanes 3–5 and 8–11), 100 ng of recombinant PC4 (lanes 9–11), or 1 (lanes 4, 10 and 12) or 3 (lanes 5, 11 and 13) µl of the purified NF1. Transcripts terminated at the first terminator site (T1), the second terminator site (T2) or the end of the template (RT) are indicated on the right. (B) Diagram of the C-tailed VA1 template and derived transcripts. The positions of the two T stretches, T1 and T2, are indicated with arrows.
Discussion
A factor that interacts specifically with the terminator regions of the VA1 gene has been purified and identified as NF1 family proteins. The NF1-binding site within the terminator region was implicated in multiple-round transcription and termination by RNA polymerase III and, indicative of NF1 function, the depletion of NF1 proteins from HeLa nuclear extract dramatically decreased transcription from the VA1 template. NF1 affected accurate termination by RNA polymerase III in the presence of TFIIIC and was shown to interact directly with two TFIIIC2 subunits that are situated near the terminator regions. These results imply that termination and initiation/reinitiation events may be coupled through NF1 interactions with both the downstream sequence element and TFIIIC.
NF1 is a component of the TFIIIC-containing complex and interacts with the terminator region of the VA1 gene
The terminator-binding activity in the TFIIIC1 fraction was purified on the basis of its binding to the first terminator region of VA1 gene. In footprint assays, the purified factor showed interactions over the two terminator regions of the VA1 gene that were similar to those observed with TFIIIC1 and holo TFIIIC. Peptide sequence analysis revealed that the purified fraction contained a heterogeneous pool of NF1 family proteins, and immunoblot assays confirmed the presence of abundant NF1 proteins in holo TFIIIC, holo RNA polymerase III and the previously defined TFIIIC1 fraction. Although highly heterogeneous, NF1 proteins all bind to a similar consensus sequence [5′-YTGGCA(N)3TGCCAR-3′] through their N-terminal regions (Borgmeyer et al., 1984). An NF1-binding site is found in each of the two terminator regions of the VA1 gene, and the locations of these sites match the footprint patterns of holo TFIIIC, TFIIIC1 and the purified terminator-binding factor. These results thus indicate that the terminator-binding activity in holo TFIIIC and TFIIIC1 is indeed NF1.
The Seifart laboratory also reported a partially purified VA1 gene terminator-binding activity that stimulated transcription from class III genes with internal promoters, but not from class III genes with external promoters (Oettel et al., 1997, 1998). Based on its chromatographic and DNA-binding properties relative to the present results, this activity probably contains NF1 family proteins. Sturges et al. (1999) recently identified an enhancer element in the termination region of the Xenopus somatic 5S RNA gene and partially purified a corresponding binding activity designated EP3. Since our inspection of the enhancer sequence reveals an NF1-binding site in one of the two regions bound by the EP3 activity, NF1 family proteins may likewise account for at least part of the EP3 activity.
NF1 is involved in transcription by RNA polymerase III
Immunodepletion of NF1 proteins with polyclonal antibodies against the highly conserved N-terminal region of CTF1 severely (10- to 15-fold) diminished VA1 gene transcription in HeLa nuclear extract, and transcription was restored almost completely by native purified terminator-binding factor and partially by recombinant CTF1. Similarly, VA1 transcription in a heat-inactivated HeLa nuclear extract was stimulated strongly by the purified terminator-binding factor and by recombinant CTF1 (data not shown). However, in both assays the effects of recombinant CTF1 were always weaker than those of the pool of NF1 proteins in the natural purified factor, and full-length intact recombinant CTF1 (from Sf21 cells) was consistently less active than degraded preparations from bacteria (data not shown). A plausible explanation of this latter result is that with some degradation intact CTF1 preparations can mimic better the function of active natural NF1 variants.
Immunodepletion of nuclear extract with anti-CTF1 antibodies reduced transcription from 5S RNA, tRNA and U6 RNA genes only several fold, but, as with the more severely inhibited VA1 gene, transcription was restored almost completely by the native purified terminator-binding factor and partially by recombinant CTF1. Given that the anti-CTF1 antibodies failed to remove NF1 family proteins completely, leaving substantial amounts of the 30, 40 and 50 kDa groups, one possible explanation of the partial inhibition of transcription is that different variants of NF1 protein are preferred for transcription of different RNA polymerase III-transcribed genes. Moreover, although CTF1 by itself is a relatively less active NF1 variant, this does not exclude the possibility that hetero-dimerization with other NF variant(s) may form a functionally more active form. Thus, while the present data clearly implicate NF1 proteins in RNA polymerase III-mediated transcription, it remains to be determined which NF1 gene(s) and splicing variants are utilized for the different class III genes. The general idea of gene-specific functions (or preferences) for NF1 variants is supported by studies of genes transcribed by RNA polymerase II (Santoro et al., 1988; Altmann et al., 1994; Monaci et al., 1995; Nebl and Cato, 1995; Chaudhry et al., 1997, 1998).
NF1 promotes accurate termination by RNA polymerase III
Although a stretch of T bases signals termination of RNA polymerase III transcription, the surrounding sequences also affect the termination efficiency (Bogenhagen and Brown, 1981). Given the low efficiency of transcription termination by RNA polymerase III alone, the delocalization of TFIIIC2 subunits to downstream regions, and the ability of holo TFIIIC both to interact with downstream regions of some class III genes and to promote proper termination, the presence of a termination factor within holo TFIIIC is an attractive model. Our documentation of termination functions for NF1-binding sites and for NF1 family proteins that can recognize these sites and that are present in holo TFIIIC provides support for such a model with NF1 proteins as terminator factors. This result mimics the situation observed for proper termination by RNA polymerase I, which requires binding of a separate terminator protein to a specific site in the termination region of large rRNA genes (for review see Reeder and Lang, 1997).
NF1 proteins could stimulate transcription termination on the VA1 template either directly or by enhancing interactions of TFIIIC in the termination region. We have reported a similar termination-promoting function for PC4/Topo I, involving enhanced downstream interaction of TFIIIC, on the C-tailed VA1 template (Wang and Roeder, 1998). Although the basis for this apparent redundancy is not understood, the large molar excess (100-fold relative to TFIIIC) of PC4/Topo I necessary for this activity may somehow mimic the NF1 function observed here or, alternatively, may stimulate trace amounts of NF1 remaining in the TFIIIC preparations. The potentially greater relevance of NF1 is indicated not only by its presence in holo TFIIIC and by its function at near stoichiometric levels with purified core TFIIIC, but also by the stringent NF1 requirement for VA1 transcription in nuclear extracts. Additionally, since PC4/Topo I and NF1, either alone or together, were unable (with TFIIIC) to promote termination by RNA polymerase III as efficiently as holo TFIIIC, efficient termination at the major (T1) VA1 site may require still other factors present in holo TFIIIC. It is also noteworthy that NF1 (Nagata et al., 1983; Sock et al., 1991; Altmann et al., 1994; Nebl and Cato, 1995), like TTF1 (reviewed in Langst et al., 1998), has a number of functions other than that described for termination (Connelly and Manley, 1989a,b), consistent with interactions with a variety of other factors.
Mechanism for the involvement of NF1 family proteins in RNA polymerase III-mediated transcription
We have shown previously that the VA1 terminator sequence is required for stable TFIIIC–DNA complex formation, therefore affecting not only termination, but also initiation (single-round transcription) and reinitiation (multiple-round transcription) by human RNA polymerase III (Wang and Roeder, 1996). However, in the present study the NF1 protein affected reinitiation in a multiple-round transcription assay, but not initiation in a single-round transcription assay. These results fit a model of facilitated recycling of RNA polymerase III in which accurate and efficient transcription termination is tightly linked to efficient transcription reinitiation. Deletion of the terminator region of the U6 gene dramatically decreased transcription (unpublished observations), indicating that this model also applies to other class III genes. This mechanism may be conserved between human and yeast since it has also been reported that the high efficiency of transcription by yeast RNA polymerase III is due mainly to rapid RNA polymerase recycling on DNA–TFIIIC–TFIIIB complexes and dependent upon the intact terminator sequences (Dieci and Sentenac, 1996).
In most other class III genes tested, including human and mouse U6 RNA, human 5S RNA and human tRNAmet genes, well-conserved NF1-binding sites are not apparent in the immediate vicinities of the termination regions. One exception is the human 7SK RNA gene, which has an NF1-binding site immediately upstream of the T-run. Although NF1-binding sites do exist within these genes and/or their immediate neighboring regions, as shown by their strong binding to recombinant CTF1 in gel shift assays, it is unclear how NF1 proteins stimulate transcription in these cases. NF1 recruitment to the template and function at the terminator could be mediated or enhanced by interaction at the alternatively positioned NF1-binding sites. Alternatively, NF1 proteins could be recruited to the template, for subsequent function in termination, mainly through their documented association with TFIIIC and potentially through secondary interactions with other components of the general transcription machinery. This latter possibility, with less dependency on a consensus NF1-binding site at the termination region, may also explain the observation that mutation of the NF1-binding site in the VA1 gene decreased transcription ∼2.5-fold, whereas depletion of NF1 proteins from HeLa nuclear extract almost completely abolished transcription.
In a highly purified reconstituted system containing recombinant TFIIIB, immunopurified TFIIIC and RNA polymerase III, we failed to see an effect of recombinant CTF1 or purified terminator-binding factor on transcription from the VA1 template in either the presence or absence of recombinant PC4 (data not shown). This result suggests that negative factor(s) present in HeLa nuclear extract may be required in order to see the NF1 requirement for RNA polymerase III transcription. This situation mimics that described for the terminator protein for RNA polymerase I transcription, which only affects transcription on a chromatin template (Langst et al., 1998). Thus, it will be important to determine whether transcription from chromatin templates (or with other negative cofactors) elicits a more stringent NF1 requirement for VA1 in the purified reconstituted system or for tRNA/5S RNA gene transcription in nuclear extracts.
Materials and methods
Gel shift and DNase I footprint
Wild-type terminator probe was made by annealing the following two oligonucleotides: 5′-GATCCGTCAGACAACGGGGGAGCGCTCCTTTTGGCTTCTTCCAGGCGCGGCGGCT-3′ and 5′-CGGATCAGCCGCCGCGCCTGGAAGGAAGCCAAAAGGAGCGCTCCCCCGTTGTCTGA-3′. Mutant VA1 terminator probe was made by annealing the following two oligonucleotides: 5′-GATCCGTCAGACAACGGGGGAGCGCTCCACTGGGCTTCTTCCAGGCGCGGCGGCT-3′ and 5′-CGG ATCAGCCGCCGCGCCTGGAAGGAAGCCCAGTGGAGCGCTCCCCCGTTGTCTGA-3′. The underlined bases were mutated from their corresponding bases in the wild-type VA1 gene sequence. Wild-type NF1-binding probe was made by annealing the following two oligonucleotides: 5′-CTCCTTTTGGCTTCCTTCCAGGCGCGGCGG-3′ and 5′-CCGCCGCGCCTGGAAGGAAGCCAAAAGGAG-3′. Mutant NF1-binding probe was made by annealing the following two oligonucleotides: 5′-CTCCTTTTCCCTTCCTTCTAGGCGCGGCGG-3′ and 5′-CCGCCGCGCCTAGAAGGAAGGGAAAAGGAG-3′. The probes were end-labeled with 32P by Klenow enzyme. For gel shift assays, each 20 µl reaction contained 20 fmol of labeled probe, 20 mM HEPES–KOH pH 7.9, 70–80 mM KCl, 4% Ficoll 400, 1 mM spermidine, 1 µg of poly(dI⋅dC)–poly(dI⋅dC), 1 mM dithiothreitol, 0.03% NP-40 and 0.1 mg/ml bovine serum albumin. The binding reaction was initiated by the addition of protein, and the mixture was incubated for 20 min at 30°C. The reaction mixture was loaded directly onto a 4% (37.5:1 acrylamide:bisacrylamide) non-denaturing polyacrylamide gel with 0.5× TBE containing 0.03% NP-40 and run at 150 V for 2 h at room temperature.
The footprint probe was made by 32P-labeling the 5′ or 3′ end of a fragment of the pGVA1 construct, containing nucleotides –96 to +232 of the VA1 gene subcloned in the pGEM vector. The footprint assays were performed as described previously (Wang and Roeder, 1996).
In vitro transcription
To make plasmids pVA1-wt, pVA1-mNF1, pVA1-mT and pVA1-m(NF1/T), four PCRs were performed on the pVA1 template. All reactions used the same upstream primer: 5′-TAGACCGTGCAAAAGGAGAAGCCTG-3′. The downstream primers were: 5′-cgcctggaaggaagccaaaaggagcgCTCCCCCGTTGTC-3′ for making the template with wild-type first terminator region (pVA1-wt); 5′-cgcctAgaaggaagGGaaaaggagcgCTCCCCCGTTGTC-3′ for making the NF1 mutant template (pVA1-mNF1); 5′-cgcctggaaggaagccCAGTggagcgCTCCCCCGTTGTC-3′ for making the template with mutant first terminator (pVA1-mT); and 5′-cgcctAgaaggaagGGTGACggagcgCTCCCCCGTTGTC-3′ for making the NF1 and terminator double mutant template (pVA1-mNF1/T). The underlined bases were mutated in the two mutants. The PCR products were subcloned into pT7-Blue vector (Novagen). Transcription reactions were performed in the presence of 2 µg/ml α-amanitin (Kovelman and Roeder, 1990). The single-round transcription assay and the transcription reaction with the C-tailed VA1 template were performed as described (Kovelman and Roeder, 1990; Wang and Roeder, 1998).
Purification of the VA1 terminator-binding factor
Throughout the purification process, the VA terminator-binding activity was monitored by gel shift assay with a DNA probe containing the VA1 terminator region. All procedures were carried out at 4°C. One hundred milliliters of HeLa nuclear extract were applied to a 100 ml phosphocellulose (P11) column equilibrated with BC100. After washing with BC100, the column was eluted step-wise with BC350, BC600 and BC1000. The activity was eluted in the BC350 and BC600 fractions. After dialysis to 100 mM KCl, the BC600 fraction was applied to a Mono Q column (HR8/8, Pharmacia) equilibrated with BC100. The column was washed with BC100, and proteins were eluted with a linear gradient from BC100 to BC500. The activity was eluted at ∼200 mM KCl. Fractions with VA1 terminator-binding activity were pooled, dialyzed to BC100, and then applied to a 10 ml double-stranded DNA–cellulose (Fluka) column equilibrated with BC100. After washing with BC100, the column was sequentially eluted with BC300, BC500 and BC1000. The activity was concentrated in the BC500 fraction. The wild-type and mutant VA1 terminator affinity columns employed the same oligos used for gel shift assays. The single-stranded oligos were gel-purified, phosphorylated at the 5′ ends, annealed, ligated and coupled to Sepharose CL-2B (Pharmacia) resin as described (Kadonaga and Tjian, 1986). After dialysis to BC100, the fraction was mixed with 1 ml of DNA-affinity resin containing the sequence of the first terminator region of VA1 gene, and incubated for 1 h with gentle rotation. The resin was washed with BC100 and loaded into a disposable column. The column was then eluted step-wise with BC200, BC400, BC600 and BC1000. The activity was split almost evenly between the BC400 and BC600 fractions. Finally, a portion of the BC600 fraction, which was more pure than the BC400 fraction, was adjusted to BC500 with 25% glycerol and subjected to a 5–15% sucrose gradient sedimentation at 55 000 r.p.m. (SW60; Beckman) and 4°C for 24 h. Fractions (0.4 ml each) were collected drop-wise from the bottom of the centrifuge tube.
Recombinant CTF1 preparations, antibody production and immunodepletion
The full-length CTF1 cDNA (Santoro et al., 1988) was subcloned into the PET15d vector and expressed in bacterial cells. Recombinant protein was purified with NTA Sepharose, S Sepharose and double-stranded DNA–cellulose columns and consisted of a heterogeneous population as a result of significant (>50%) degradation. This preparation was used for all gel shift and transcription assays. Full-length CTF1 with a His-tag at the N-terminus and a FLAG-tag at the C-terminus was expressed in a baculovirus expression system in Sf21 cells. Full-length protein was obtained by sequential purification purified through NTA Sepharose and M2 agarose chromatography. Rabbit anti-CTF1 polyclonal antibodies were raised by using the N-terminal region (amino acids 1–233) as an antigen (HRP Inc.). The anti-CTF1 antibodies were purified through the antigen column and used to immunodeplete HeLa nuclear extract in the presence of 400 mM KCl and 0.1% NP-40 (Harlow and Lane, 1988).
Protein–protein interactions
One hundred microliters of M2 agarose beads (Sigma) were incubated with 10 ml of the whole cell extract made from Sf21 cells infected with the baculovirus containing Flag-tagged CTF1 for 3 h at 4°C. The beads were washed extensively with BC500–0.1% NP-40. Ten microliters of the beads (containing 100 ng CTF1) were incubated with 1–5 µl of TNT rabbit reticulocyte lysates (Promega) containing 35S-labeled TFIIIC2 subunits in BC150–0.1% NP-40 for 2 h at 4°C. After washing with BC150–0.1% NP-40, the beads were boiled with SDS sample buffer and subjected to SDS–PAGE.
Immunopurification of factors
Holo RNA polymerase III and RNA polymerase III were immunopurified from the BN51 cell line that expresses the FLAG-tagged 53 kDa subunit of human RNA polymerase III (Wang and Roeder, 1997). Holo TFIIIC was immunopurified from the Cα cell line that expresses the FLAG-tagged 220 kDa subunit of TFIIIC2 (Wang and Roeder, 1998). TFIIIC was immunopurified from the Cα cell line under high-salt conditions (300 and 800 mM KCl) and was similar to the purified TFIIIC obtained by sucrose gradient sedimentation of holo TFIIIC (Wang and Roeder, 1998) in both polypeptide composition and transcriptional activity. TFIIIC1 and TFIIIC1′ were purified as reported previously (Wang and Roeder, 1996).
Acknowledgments
Acknowledgements
The authors thank Martin Teichmann for discussion and suggestions. This work was supported by NIH grant CA 42567 to R.G.R.
References
- Allison D.S. and Hall,B.D. (1985) Effects of alterations in the 3′ flanking sequence on in vivo and in vitro expression of the yeast SUP4-o tRNATyr gene. EMBO J., 4, 2657–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann H., Wendler,W. and Winnacker,E.L. (1994) Transcriptional activation by CTF proteins is mediated by a bipartite low-proline domain. Proc. Natl Acad. Sci. USA, 91, 3901–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew B., Durkovich,D., Kassavetis,G.A. and Geiduschek,E.P. (1993) Orientation and topography of RNA polymerase III in transcription complexes. Mol. Cell. Biol., 13, 942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen D.F. and Brown,D.D. (1981) Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell, 24, 261–270. [DOI] [PubMed] [Google Scholar]
- Borgmeyer U., Nowock,J. and Sippel,A.E. (1984) The TGGCA-binding protein: a eukaryotic nuclear protein recognizing a symmetrical sequence on double-stranded linear DNA. Nucleic Acids Res., 12, 4295–4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry A.Z., Lyons,G.E. and Gronostajski,R.M. (1997) Expression patterns of the four nuclear factor I genes during mouse embryogenesis indicate a potential role in development. Dev. Dyn., 208, 313–325. [DOI] [PubMed] [Google Scholar]
- Chaudhry A.Z., Vitullo,A.D. and Gronostajski,R.M. (1998) Nuclear factor I (NFI) isoforms differentially activate simple versus complex NFI-responsive promoters. J. Biol. Chem., 273, 18538–18546. [DOI] [PubMed] [Google Scholar]
- Chu W.M., Liu,W.M. and Schmid,C.W. (1995) RNA polymerase III promoter and terminator elements affect Alu RNA expression. Nucleic Acids Res., 23, 1750–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly S. and Manley,J.L. (1989a) RNA polymerase II transcription termination is mediated specifically by protein binding to a CCAAT box sequence. Mol. Cell. Biol., 9, 5254–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly S. and Manley,J.L. (1989b) A CCAAT box sequence in the adenovirus major late promoter functions as part of an RNA polymerase II termination signal. Cell, 57, 561–571. [DOI] [PubMed] [Google Scholar]
- Cozzarelli N.R., Gerrard,S.P., Schlissel,M., Brown,D.D. and Bogenhagen,D.F. (1983) Purified RNA polymerase III accurately and efficiently terminates transcription of 5S RNA genes. Cell, 34, 829–835. [DOI] [PubMed] [Google Scholar]
- Deprez E., Arrebola,R., Conesa,C. and Sentenac,A. (1999) A subunit of yeast TFIIIC participates in the recruitment of TATA-binding protein. Mol. Cell. Biol., 19, 8042–8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G. and Sentenac,A. (1996) Facilitated recycling pathway for RNA polymerase III. Cell, 84, 245–252. [DOI] [PubMed] [Google Scholar]
- Geiduschek E.P. and Kassavetis,G.A. (1992) RNA polymerase III transcription complex. In Transcription Regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 247–279. [Google Scholar]
- Harlow E. and Lane,D. (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Kadonaga J.T. and Tjian,R. (1986) Affinity purification of sequence-specific DNA binding proteins. Proc. Natl Acad. Sci. USA, 83, 5889–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovelman R. and Roeder,R.G. (1990) Sarkosyl defines three intermediate steps in transcription initiation by RNA polymerase III: application to stimulation of transcription by E1A. Genes Dev., 4, 646–658. [DOI] [PubMed] [Google Scholar]
- Kovelman R. and Roeder,R.G. (1992) Purification and characterization of two forms of human transcription factor TFIIIC. J. Biol. Chem., 267, 24446–24456. [PubMed] [Google Scholar]
- Kruse U. and Sippel,A.E. (1994) Transcription factor nuclear factor I proteins form stable homo- and heterodimers. FEBS Lett., 348, 46–50. [DOI] [PubMed] [Google Scholar]
- Langst G., Becker,P.B. and Grummt,I. (1998) TTF-I determines the chromatin architecture of the active rDNA promoter. EMBO J., 17, 3135–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaci P., Nuzzo,M., Stampfli,S., Tollervey,D., De Simone,V. and Nicosia,A. (1995) A complex interplay of positive and negative elements is responsible for the different transcriptional activity of liver NF1 variants. Mol. Biol. Rep., 21, 147–158. [DOI] [PubMed] [Google Scholar]
- Nagata K., Guggenheimer,R.A. and Hurwitz,J. (1983) Specific binding of a cellular DNA replication protein to the origin of replication of adenovirus DNA. Proc. Natl Acad. Sci. USA, 80, 6177–6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebl G. and Cato,A.C. (1995) NFI/X proteins: a class of NFI family of transcription factors with positive and negative regulatory domains. Cell. Mol. Biol. Res., 41, 85–95. [PubMed] [Google Scholar]
- Oettel S., Hartel,F., Kober,I., Iben,S. and Seifart,K.H. (1997) Human transcription factors IIIC2, IIIC1 and a novel component IIICo fulfill different aspects of DNA binding to various pol III genes. Nucleic Acids Res., 25, 2440–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oettel S., Kober,I. and Seifart,K.H. (1998) The activity binding to the termination region of several pol III genes represents a separate entity and is distinct from a novel component enhancing U6 snRNA transcription. Nucleic Acids Res., 26, 4324–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paule M.R. and White,R.J. (2000) Transcription by RNA polymerases I and III. Nucleic Acids Res., 28, 1283–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian F., Kruse,U., Lichter,P. and Sippel,A.E. (1995) Chromosomal localization of the four genes (NFIA, B, C, and X) for the human transcription factor nuclear factor I by FISH. Genomics, 28, 66–73. [DOI] [PubMed] [Google Scholar]
- Reeder R.H. and Lang,W.H. (1997) Terminating transcription in eukaryotes: lessons learned from RNA polymerase I. Trends Biochem. Sci., 22, 473–477. [DOI] [PubMed] [Google Scholar]
- Santoro C., Mermod,N., Andrews,P.C. and Tjian,R. (1988) A family of human CCAAT-box-binding proteins active in transcription and DNA replication: cloning and expression of multiple cDNAs. Nature, 334, 218–224. [DOI] [PubMed] [Google Scholar]
- Sock E., Wegner,M. and Grummt,F. (1991) DNA replication of human polyomavirus JC is stimulated by NF-I in vivo. Virology, 182, 298–308. [DOI] [PubMed] [Google Scholar]
- Sturges M.R., Bartilson,M. and Peck,L.J. (1999) Enhancer of RNA polymerase III gene transcription. Nucleic Acids Res., 27, 690–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. and Roeder,R.G. (1996) TFIIIC1 acts through a downstream region to stabilize TFIIIC2 binding to RNA polymerase III promoters. Mol. Cell. Biol., 16, 6841–6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. and Roeder,R.G. (1997) Three human RNA polymerase III-specific subunits form a subcomplex and participate in specific transcription initiation. Genes Dev., 11, 1315–1326. [DOI] [PubMed] [Google Scholar]
- Wang Z. and Roeder,R.G. (1998) DNA topoisomerase I and PC4 can interact with human TFIIIC to promote both accurate termination and transcription reinitiation by RNA polymerase III. Mol. Cell, 1, 749–757. [DOI] [PubMed] [Google Scholar]
- White R.J. (1994) RNA Polymerase III Transcription. R.G. Landes Company, Austin, TX. [Google Scholar]
- Yoshinaga S.T., Boulanger,P.A. and Berk,A.J. (1987) Resolution of human transcription factor TFIIIC into two functional components. Proc. Natl Acad. Sci. USA, 84, 3585–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga S.T., L’Etoile,N.D. and Berk,A.J. (1989) Purification and characterization of transcription factor TFIIIC2. J. Biol. Chem., 264, 10726–10731. [PubMed] [Google Scholar]
- Young L.S., Rivier,D.H. and Sprague,K.U. (1991) Sequences far downstream from the classical tRNA promoter elements bind RNA polymerase III transcription factors. Mol. Cell. Biol., 11, 1382–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorbas H., Rein,T., Krause,A., Hoffmann,K. and Winnacker,E.L. (1992) Nuclear factor I (NF I) binds to an NF I-type site but not to the CCAAT site in the human α-globin gene promoter. J. Biol. Chem., 267, 8478–8484. [PubMed] [Google Scholar]