Abstract
Akata and Mutu cell lines are derived from Burkitt’s lymphoma (BL) and retain the in vivo phenotype of Epstein–Barr virus (EBV) expression that is characterized by expression of EBV-determined nuclear antigen 1 (EBNA1), EBV-encoded RNAs (EBERs) and transcripts from the BamHI A region (BARF0). We found that EBV-positive Akata and Mutu cell clones expressed higher levels of interleukin (IL)-10 than their EBV-negative subclones at the transcriptional level. Transfection of an individual EBV latent gene into EBV-negative Akata cells revealed that EBERs were responsible for IL-10 induction. Recombinant IL-10 enabled EBV-negative Akata cells to grow in low (0.1%) serum conditions. On the other hand, growth of EBV-positive Akata cells was blocked by treatment either with an anti-IL-10 antibody or antisense oligonucleotide against IL-10. EBV-positive BL biopsies consistently expressed IL-10, but EBV-negative BL biopsies did not. These results suggest that IL-10 induced by EBERs acts as an autocrine growth factor for BL. EBERs, EBER1 and EBER2, are non-polyadenylated RNAs and are 166 and 172 nucleotides long, respectively. The present findings indicate that RNA molecules could regulate cell growth.
Keywords: Burkitt’s lymphoma/EBV-encoded small RNA/Epstein–Barr virus/growth factor/IL-10
Introduction
Epstein–Barr virus (EBV), a human herpesvirus, is associated with >90% of Burkitt’s lymphoma (BL) in the African regions of endemicity and less frequently (15–20%) with the sporadic BL occurring worldwide (Epstein and Achong, 1979; Rickinson and Kieff, 1996). The most consistent finding in BL, whether EBV infected or not, is a chromosome translocation involving immunoglobulin (Ig) and c-myc genes (Taub et al., 1982). The translocation results in deregulation of c-myc expression and is regarded as the most important step in the genesis of BL (Klein, 1981). On the other hand, the role of EBV has not been studied for a long time. Tumor cells maintain the entire EBV genome mostly in a plasmid form and express a limited number of viral gene products (termed type I latency), including EBV-associated nuclear antigen 1 (EBNA1), EBV-encoded small RNAs (EBERs) and transcripts from the BamHI A region (BARF0) (Rickinson and Kieff, 1996). The lack of a suitable in vitro system that represents BL-type EBV expression has hampered study of the role of EBV in the genesis of BL. The Japanese BL-derived Akata cell line (Takada, 1984; Takada and Ono, 1989; Takada et al., 1991) allowed us to overcome this problem. The Akata line is unique in that it retains the in vivo phenotype of EBV expression even after long-term culture in vitro (Shimizu et al., 1994). By contrast, most BL cell lines convert EBV expression to that of EBV-immortalized lymphoblastoid cells, in which all the latent viral gene products, including six EBNAs and three latent membrane proteins (LMPs), are expressed (Rowe et al., 1987; Gregory et al., 1990). We isolated EBV-positive and -negative subclones from parental Akata cells by the limiting dilution method (Shimizu et al., 1994). Comparison of EBV-positive and -negative cell clones revealed that the presence of EBV in Akata cells was required for them to be more malignant and apoptosis resistant (Shimizu et al., 1994; Chodosh et al., 1998; Komano et al., 1998; Ruf et al., 1999), which emphasized the oncogenic role of EBV in the genesis of BL.
In this report, we demonstrate that EBERs support the growth of BL through activation of interleukin (IL)-10 expression. EBERs, EBER-1 and EBER-2, are non-polyadenylated RNAs transcribed by the RNA polymerase III system (Howe and Shu, 1989; Clemens, 1993). They are 166 and 172 nucleotides long, respectively, the most abundant EBV RNAs in latently infected cells. Most of the EBERs localize to the cell nucleus (Howe and Steitz, 1986), where they are complexed with cellular La protein, which is recognized by specific antisera from patients with systemic lupus erythematosus (Lerner et al., 1981). EBERs have extensive primary sequence similarity to adenovirus VA1 and VA2 (Rosa et al., 1981) and U6 cell small RNA (Howe and Shu, 1989), both of which also complex with La protein, although the significance of their interactions is not known.
Results
IL-10 expression in EBV-positive BL is dependent on the presence of EBV
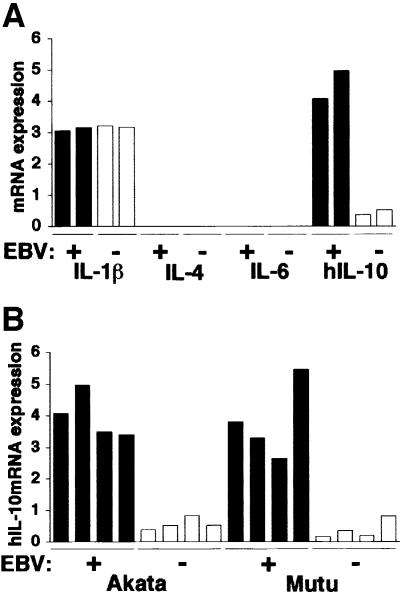
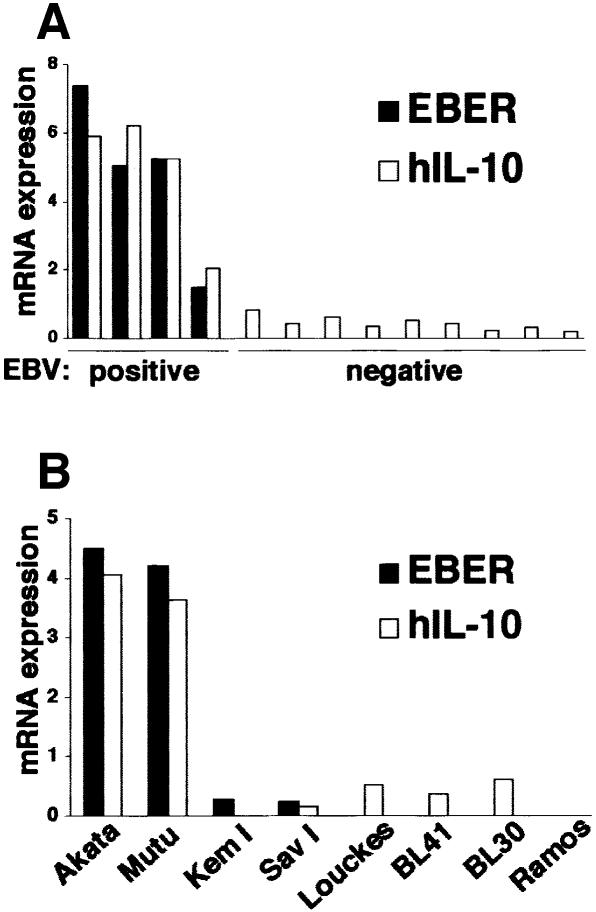
To assess the role of cytokines in the growth of BL cells, EBV-positive and -negative Akata cells were studied to examine the expression of various cytokines such as IL-1β, IL-4, IL-6 and IL-10, which are known to act as growth factors of B lymphocytes (Itoh and Hirohata, 1995; Kelso, 1998). Real-time quantitative RT–PCR analysis (Fink et al., 1998) revealed that expression of IL-10 was higher in EBV-positive cell clones than in EBV-negative clones (Figure 1A).
Fig. 1. Cytokine expression in BL-derived Akata and Mutu cell lines. Both cell lines were originally 100% EBV positive, and EBV-negative subclones were isolated by limiting dilution from the parental cultures. All analyses were performed by the real-time quantitative RT–PCR assay using a LightCycler™ (Fink et al., 1998). The results are expressed as the ratio to the value of GAPDH (K × cytokine copies/5 × 103 copies GAPDH; K = constant). (A) Cytokine mRNA expression in EBV-positive and -negative Akata cell clones (two clones each). (B) IL-10 mRNA expression in EBV-positive and -negative Akata and Mutu cell clones (four clones each).
Similar results were also obtained when EBV-positive and -negative Mutu cell clones were compared (Figure 1B). The Mutu cell line is derived from BL and retains BL-type EBV expression, as with Akata (Gregory et al., 1990). We have recently isolated EBV-negative subclones from the Mutu line by the limiting dilution method. Figure 2A and B shows that EBV-negative Mutu cell clones are truly negative for EBV by Southern blotting and PCR analyses. Western blot analysis indicated that EBV-positive Mutu cell clones were positive for EBNA1, but negative for other EBNAs and LMP1 (Figure 2C). RT–PCR analysis indicated that EBV-positive Mutu cell clones utilized the Q promoter for EBNA transcription, and were positive for BARF0, but negative for LMP2A and LMP2B (Figure 2D). These results indicated that EBV-positive Mutu cell clones retained type I latency. On the other hand, EBV-negative cell clones were completely negative for these messages, confirming the EBV negativity of these cell clones.
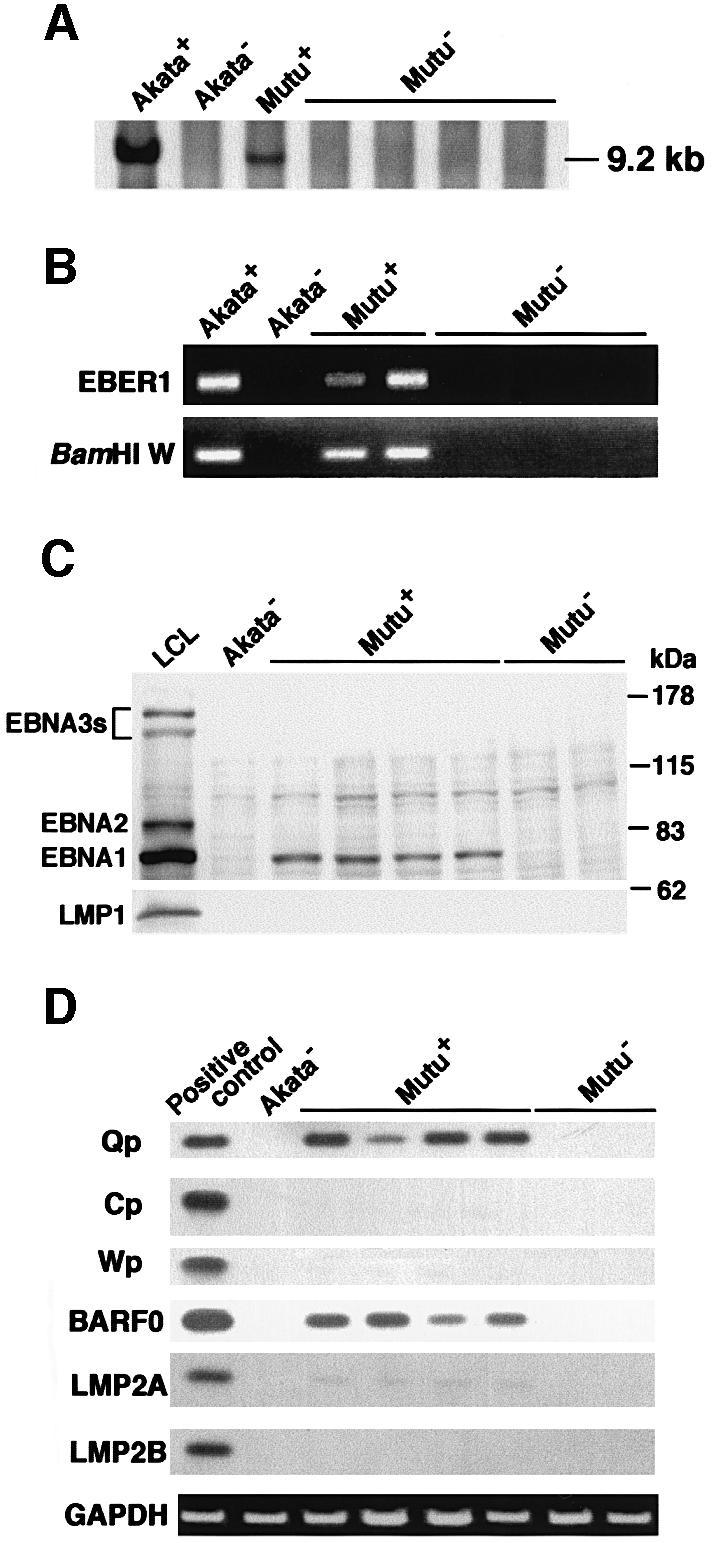
Fig. 2. Detection of EBV in EBV-positive and -negative Mutu cell clones. (A) Southern blot analysis of EBV genomes. DNA samples (5 µg each) were digested with BamHI, and the blot was probed with the BamHI C fragment of EBV DNA. (B) PCR analysis of EBV genomes. DNA samples (100 ng each) were amplified with primers for EBER1 and BamHI W regions. (C) Immunoblot analysis for detection of EBNAs and LMP1. The blots were probed with EBNA-positive human serum (upper blot) and an anti-LMP1 monoclonal antibody (lower blot). Protein samples extracted from 105 cells were loaded per slot. (D) RT–PCR analysis of EBNA promoter usage and EBV latent gene expression. Akata cells were used as a positive control for detection of Qp-initiated EBNA mRNA, and a lymphoblastoid cell line immortalized by Akata EBV (LCL) was used as a positive control for detection of Cp- or Wp-initiated EBNA mRNAs and BARF0, LMP2A and LMP2B mRNAs.
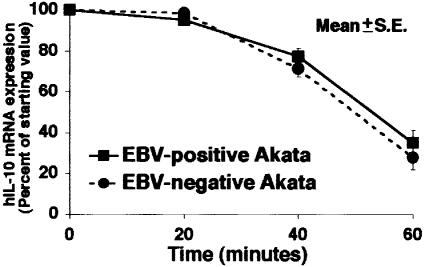
The half-life of IL-10 mRNA did not differ between EBV-positive and -negative Akata cells (Figure 3), thus indicating that increased expression of IL-10 mRNA in EBV-positive Akata cells was at the transcriptional level.
Fig. 3. IL-10 mRNA degradation in EBV-positive and -negative Akata cells. Actinomycin D (5 µg/ml) was added to the culture to block mRNA synthesis. At varying times thereafter, RNA was isolated and the amount of IL-10 mRNA remaining was determined. A representative result of several experiments is shown. In EBV-positive cells, t1/2 = 51.1 min; it was 49.3 min in EBV-negative cells. There was no significant difference between EBV-positive and -negative cells.
IL-10 induction was further confirmed by ELISA assay of culture supernatants of both Akata and Mutu cells (Figure 4A). After 3 days of culture of 106 cells, EBV-positive Akata and Mutu cells secreted 80–100 pg of IL-10. In contrast, the IL-10 levels of EBV-negative counterparts were ∼10 pg (Figure 4B). EBV encodes a viral homolog of the cellular IL-10 gene (vIL-10), which is induced in a viral lytic cycle. Figure 4C shows that both Akata and Mutu cells are negative for vIL-10.
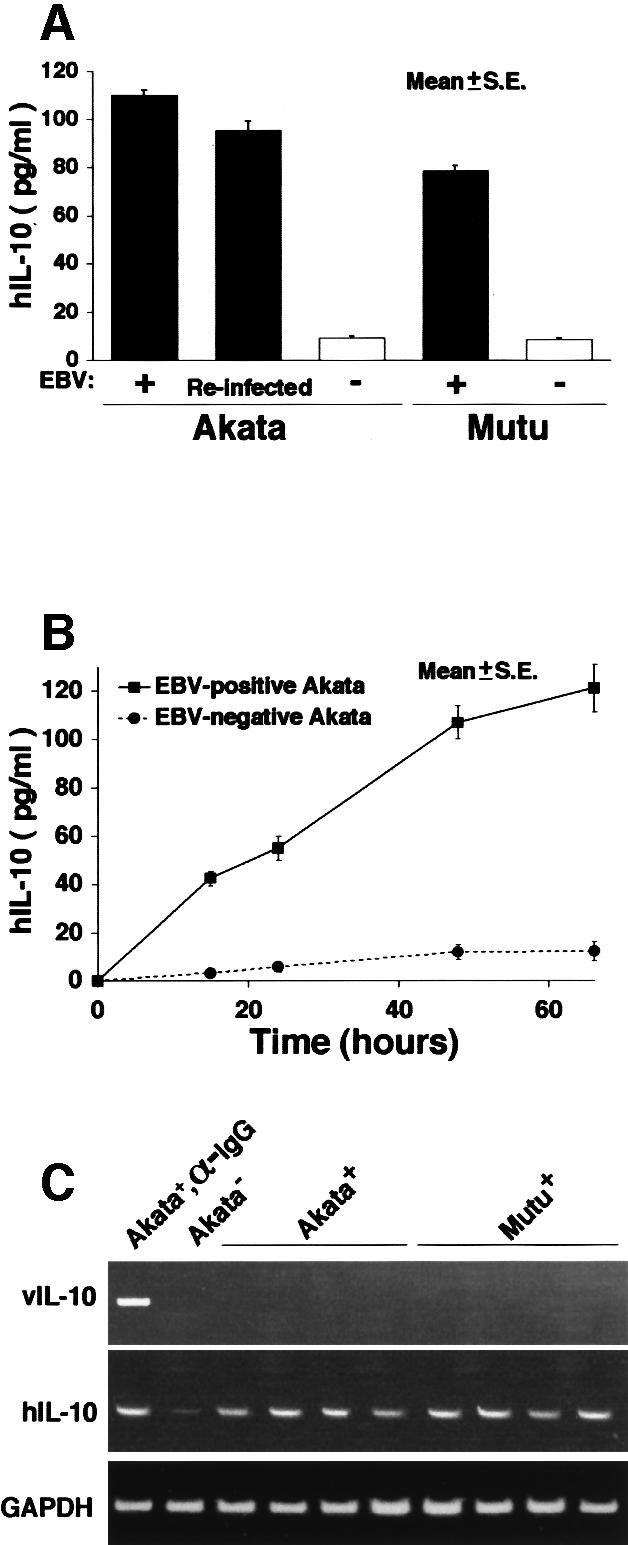
Fig. 4. (A) IL-10 production in EBV-positive and -negative Akata and Mutu cell clones. After 3 days of culture of 106 cells in 1 ml of RPMI with 10% FCS, the amount of IL-10 in the culture supernatant was measured with a high-sensitivity IL-10 human ELISA system (Amersham Pharmacia Biotech). (B) Time course of IL-10 production in EBV-positive and -negative Akata cells. (C) RT–PCR analysis of viral IL-10 expression in EBV-positive Akata and Mutu cell clones. EBV-positive Akata cells treated with anti-Ig antibody were used as a positive control for detection of viral IL-10.
EBERs are responsible for IL-10 induction inBL cells
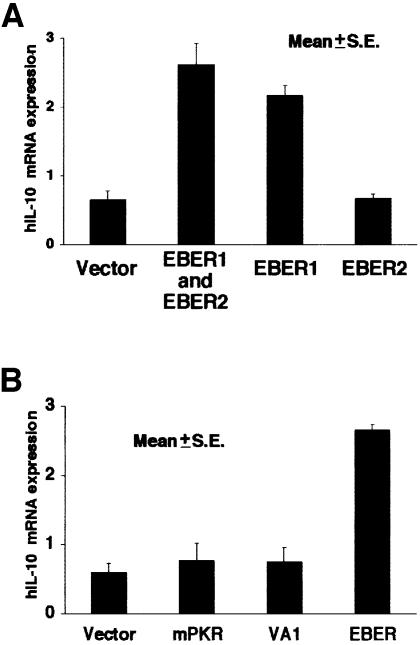
Next we examined which EBV gene among three latent genes (EBNA1, EBERs and BARF0) was responsible for IL-10 induction. The EBV-negative Akata cell clone was transfected with an individual EBV latent gene, and cell clones that stably expressed similar levels of EBV-positive Akata cells were selected and analyzed (Figure 5A and B). The results indicated that all cell clones transfected with the EBER gene expressed higher levels of IL-10 than those transfected with the other latent genes (Figure 5C). There was a good correlation between the level of EBER expression and that of IL-10 expression.
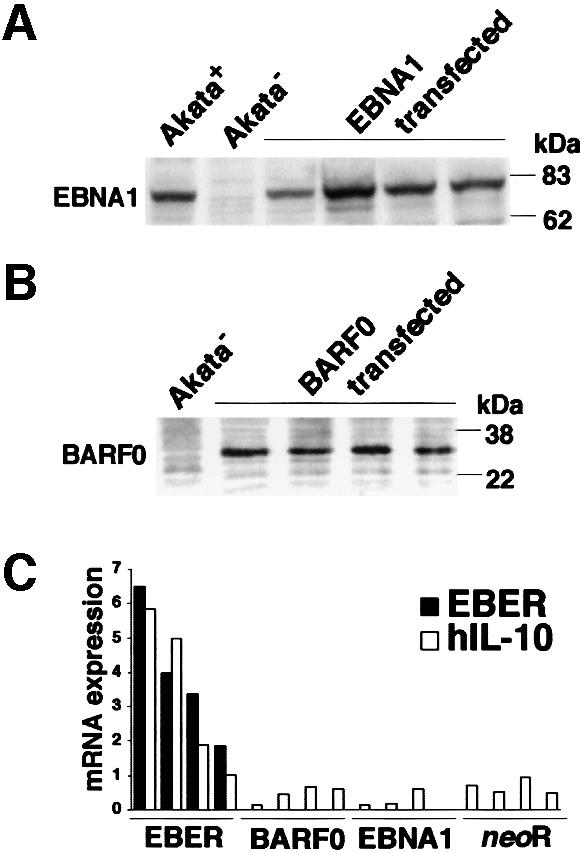
Fig. 5. IL-10 expression in EBV-negative Akata cell clones transfected with an individual EBV latent gene expressed in BL. An EBV-negative Akata cell clone was transfected with an individual EBV gene, and cell clones that stably expressed similar levels of EBV-positive Akata cells were selected and analyzed. (A) EBNA1 expression in EBV-negative Akata cell clones transfected with the EBNA1 gene. EBNA1 was detected by immunoblotting using EBNA1-positive human serum. (B) BARF0 expression in EBV-negative Akata cell clones transfected with the FLAG epitope-tagged BARF0 gene. BARF0 was detected by immunoblotting using an anti-FLAG M2 antibody (Upstate Technology Inc.). (C) IL-10 expression. Four clones each were examined for the expression of IL-10 and EBER by the real-time quantitative RT–PCR assay. The results are expressed as the ratio to the value of GAPDH (K × IL-10 copies/5 × 103 copies GAPDH; K × EBER copies/1 copy GAPDH; K = constant).
EBV reinfection of EBV-negative Akata cells induced IL-10 expression (Figure 6C). To further confirm that EBERs were responsible for IL-10 induction, we generated an EBV recombinant lacking EBER genes. EBER-knockout EBV-infected Akata cells revealed a pattern of EBV expression similar to that of wild-type EBV-infected Akata cells except for the absence of EBER expression (Figure 6A and B). Real-time quantitative RT–PCR analysis indicated that EBER-knockout EBV-infected Akata cells were negative for IL-10 expression (Figure 6C). These results clearly demonstrated that the EBERs were responsible for IL-10 induction.
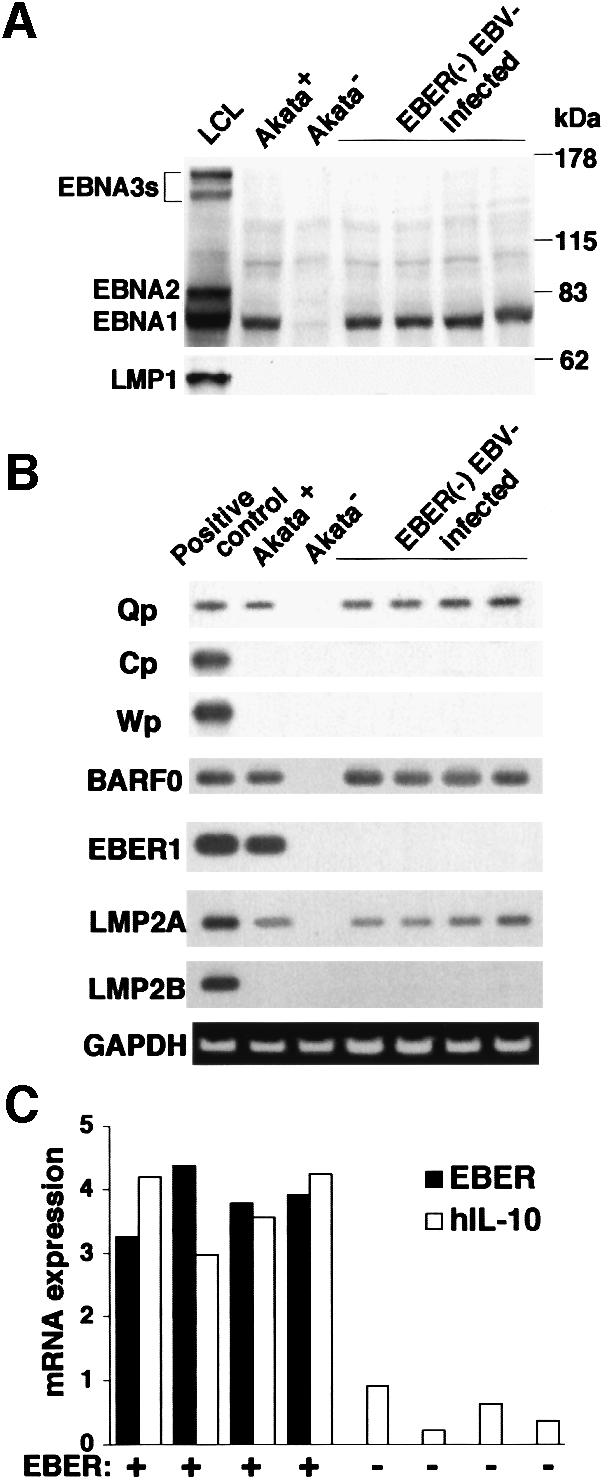
Fig. 6. IL-10 expression in EBER-knockout EBV-infected Akata cells. An EBV-negative Akata cell clone was reinfected with EBER-positive or -negative EBV, and 100% EBV-positive cell clones were isolated. (A) Immunoblot analysis for detection of EBNAs and LMP1. The blots were probed with EBNA-positive human serum (upper blot) and anti-LMP1 monoclonal antibody (lower blot). Protein samples extracted from 105 cells were loaded per slot. (B) RT–PCR analysis of EBNA promoter usage and EBV latent gene expression. Akata cells were used as a positive control for detection of Qp-initiated EBNA mRNA, and a lymphoblastoid cell line immortalized by Akata EBV (LCL) was used as a positive control for detection of Cp- or Wp-initiated EBNA mRNAs and BARF0, EBER1, LMP2A and LMP2B mRNAs. (C) IL-10 expression. Four clones each were examined for the expression of IL-10 and EBER by the real-time quantitative RT–PCR assay. The results are expressed as the ratio to the value of GAPDH (K × IL-10 copies/5 × 103 copies GAPDH; K × EBER copies/1 copy GAPDH; K = constant).
By transient assay, transfection of the EBER1 plasmid, but not the EBER2 plasmid, was found to induce IL-10 expression in EBV-negative Akata cells (Figure 7A). As with adenovirus-encoded VA1, EBERs were reported to bind an interferon-induced protein kinase (PKR; Clarke et al., 1991). Thus, based on the known function of VA1 (Schneider et al., 1985), EBERs were proposed to act in the cytoplasm to inhibit activation of PKR (Bhat and Thimmappaya, 1983), which would phosphorylate protein synthesis initiation factor eIF-2α and block translation, although purified EBERs had a lesser effect on eIF-2α kinase activity in in vitro assays (Clarke et al., 1991). Therefore, we examined the effect of VA1 on IL-10 expression in EBV-negative Akata cells. The results indicated that IL-10 expression was not influenced by VA1 expression (Figure 7B). The introduction of a mutant form of PKR into EBV-negative Akata cells, which could block the binding of EBER to PKR in a competitive manner, also had no effect on IL-10 expression (Figure 7B). These results suggested that PKR was not involved in IL-10 induction by EBERs.
Fig. 7. (A) IL-10 induction by transient expression of EBER1 or EBER2 in EBV-negative Akata cells. Cells (5 × 106) were transfected with the EBER plasmid (50 µg) by the electroporation method, harvested after 48 h of incubation, and subjected to the real-time quantitative RT–PCR assay. The results are expressed as the ratio to the value of GAPDH (K × IL-10 copies/5 × 103 copies GAPDH; K = constant). (B) Effect of adenovirus VA1 and a mutant form of PKR (mPKR) on expression of IL-10 in EBV-negative Akata cells. Cells (5 × 106) were transfected with the VA1 or mPKR plasmid (50 µg) by the electroporation method. Cells were harvested after 48 h of incubation and subjected to the real-time quantitative RT–PCR assay. The results are expressed as the ratio to the value of GAPDH (K × IL-10 copies/5 × 103 copies GAPDH; K = constant).
Growth of EBV-positive BL cells is dependenton IL-10
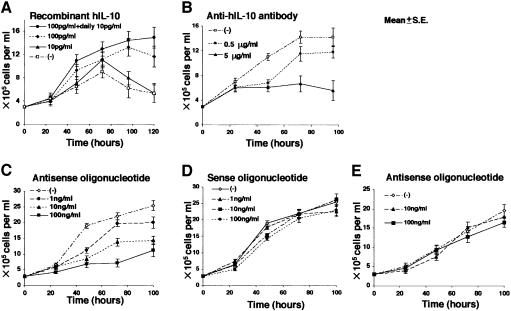
Next we examined whether the growth of EBV-positive Akata cells was dependent on IL-10. EBV-negative Akata cells could not grow under low (0.1%) serum conditions, while EBV-positive Akata cells could. Addition of 100 pg/ml recombinant IL-10 to the culture medium enabled EBV-negative Akata cells to grow just like EBV-positive Akata cells (Figure 8A). On the other hand, the growth of EBV-positive Akata cells was blocked by 5 µg/ml anti-IL-10 antibody (Figure 8B). Similar growth inhibition was also obtained using 100 ng/ml IL-10 antisense oligonucleotide, but not with IL-10 sense oligonucleotide (Figure 8C and D). The antisense oligonucleotide had no effect on the growth of myeloid leukemia-derived K562 cells that were negative for IL-10 expression (Figure 8E). These results indicated that IL-10 was an autocrine growth factor for EBV-positive Akata cells.
Fig. 8. (A) Effect of recombinant IL-10 on growth of EBV-negative Akata cells in low (0.1%) FCS conditions. Recombinant IL-10 (Endogen) was added to the culture medium in various concentrations. (B) Effect of anti-IL-10 antibody on growth of EBV-positive Akata cells. Purified rat anti-human IL-10 antibody (PharMingen), at various concentrations, was added to the culture medium containing 0.1% FCS. (C) Effect of antisense oligonucleotide against IL-10 on growth of EBV-positive Akata cells. Antisense oligonucleotide, at various concentrations, was added to the culture medium containing 10% FCS. (D) Effect of sense oligonucleotide against IL-10 on growth of EBV-positive Akata cells. Sense oligonucleotide, at various concentrations, was added to the culture medium containing 10% FCS. (E) Effect of antisense oligonucleotide against IL-10 on growth of K562 cells. Cell proliferation was determined by counting the viable cells.
IL-10 expression in BL biopsies and BL cell lines
We examined the IL-10 expression of BL biopsies from Japanese patients by the real-time RT–PCR assay (Figure 9A). All four EBV-positive BL biopsies were positive for IL-10. There was a good correlation between the level of EBER expression and of IL-10 expression. On the other hand, nine EBV-negative BL biopsies were negative for EBER expression and did not express detectable amounts of IL-10. These results, although only a small number of cases were analyzed, suggested that BL cells utilized IL-10 as an autocrine growth factor.
Fig. 9. IL-10 mRNA expression in BL biopsies (A), type I BL cell lines and EBV-negative BL cell lines (B). IL-10 mRNA was quantified by the real-time RT–PCR assay. The results are expressed as the ratio to the value of GAPDH (K × IL-10 copies/5 × 103 copies GAPDH; K × EBER copies/1 copy GAPDH; K = constant).
Two type I BL cell lines were available: KemI and SavI. Both cell lines were negative for LMP1, but were weakly positive for EBNA2, and thus a little different from real type I cells. Moreover, both cell lines had very weak EBER expression and were negative for IL-10 expression (Figure 9B). Four EBV-negative BL lines, Louckes, BL41, BL30 and Ramos, were negative for EBER expression and expressed very low levels of IL-10 (Figure 9B).
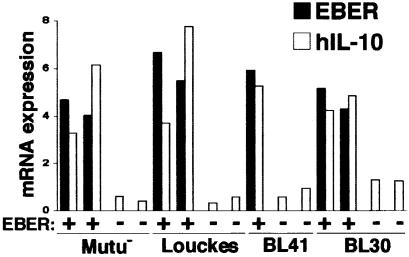
To further confirm that EBERs induced IL-10 expression, we transfected the EBER plasmid into four EBV-negative BL cell lines, and cell clones that stably expressed EBERs were selected and analyzed. The results indicated that in all four cell lines, EBERs induced IL-10 expression (Figure 10).
Fig. 10. IL-10 mRNA expression in EBER-transfected cell clones of EBV-negative BL cell lines. EBV-negative Mutu, Louckes, BL41 and BL30 cells were transfected with the EBER plasmid, and cell clones that stably expressed EBERs were selected and analyzed. Cell clones transfected with the neoR gene were used as controls. IL-10 mRNA was quantified by the real-time RT–PCR assay. The results are expressed as the ratio to the value of GAPDH (K × IL-10 copies/5 × 103 copies GAPDH; K × EBER copies/1 copy GAPDH; K = constant).
Tumorigenicity of IL-10-transfected EBV-negative Akata cells
In a previous study we demonstrated that EBERs confer tumorigenicity in EBV-negative Akata cells (Komano et al., 1999). Therefore, we tested whether IL-10 could confer tumorigenicity in EBV-negative Akata cells. An EBV-negative Akata cell clone was transfected with the IL-10 gene, and cell clones that stably expressed IL-10 were isolated and inoculated into SCID mice. As shown in Table I, IL-10-transfected Akata cell clones were not tumorigenic in SCID mice.
Table I. Tumorigenicity of hIL-10-transfected Akata cell clones in SCID mice.
| Cell clones | No. of mice with tumors |
|---|---|
| EBV-positive clones | 3/3 |
| EBV-negative clones | 0/3 |
| EBV-reinfected clones | 8/9 (2/3, 3/3, 3/3) |
| EBER-transfected clones | 7/15 (1/3, 2/3, 2/3, 1/3, 1/3) |
| neoR-transfected clones | 0/15 (0/3, 0/3, 0/3, 0/3, 0/3) |
| hIL-10-transfected clones | 0/15 (0/3, 0/3, 0/3, 0/3, 0/3) |
Five cell clones transfected with neoR or EBER or hIL-10, three EBV-reinfected clones derived from an EBV-negative Akata cell clone (1.5 × 107 cells each) were individually inoculated into 4-week-old male SCID mice (FOX CHASE C.B-17/ Icr-scid Jcl; Clea, Tokyo, Japan). Mice were killed 8 weeks after inoculation, and the developed tumors were measured. The tumors ranged from 0.8 to 4.5 cm.
Discussion
The present findings have revealed an ingenious strategy of EBV to survive in immune hosts. IL-10 allows it to escape the immune response by suppressing T-helper 1 (TH1) cytotoxic T lymphocytes (Fiorentino et al., 1989) whilst, on the other hand, the virus utilizes it as a growth factor for infected cells.
In a previous paper we reported that EBERs played an oncogenic role in Akata cells (Komano et al., 1999). Transfection of the EBER gene into EBV-negative Akata cells restored the capacity for growth in soft agar, tumorigenicity in SCID mice, resistance to apoptotic inducers and upregulated expression of bcl-2 oncoprotein, which were originally retained in parental EBV-positive Akata cells and lost in EBV-negative subclones. However, transfection of the IL-10 gene into EBV-negative Akata cells could not confer these capabilities. Therefore, IL-10 alone is not sufficient for mediating the oncogenicity of EBERs.
Since the half-life of IL-10 mRNA did not differ between EBV-positive and -negative Akata cells, increased expression of IL-10 mRNA in EBV-positive Akata cells was considered to occur at the transcriptional level. How, then, could EBERs (RNAs) activate transcription of the IL-10 gene? EBERs have been reported to bind some cellular proteins, La (Lerner et al., 1981), EAP/L22 (Toczyski and Steiz, 1991; Toczyski et al., 1994) and PKR (Clarke et al., 1991). Among them, the association of EBERs with PKR has been the most intensively studied (Bhat and Thimmappaya, 1983; Clarke et al., 1991; Clemens, 1993). In in vitro assays, they can bind to PKR, inhibit its activation, and block phosphorylation of protein synthesis initiation factor eIF2α, thus resulting in the blockage of protein synthesis. However, this activity has not been proved in in vivo assays. The present results suggested that PKR was not involved in IL-10 induction by EBERs, since adenovirus VA1 and a mutant form of PKR had no effect on IL-10 expression.
Several reports have indicated that IL-10 is induced by EBV infection. The majority of EBV-transformed lymphoblastoid cell lines and EBV-carrying cell lines derived from BL and AIDS-related lymphoma produce detectable amounts of IL-10 (Benjamin et al., 1992; Burdin et al., 1993). Furthermore, IL-10 expression was induced in EBV-negative BL cells by EBV conversion in vitro (Vieira et al., 1991). Most of these cells expressed LMP1, and Nakagomi et al. (1994) demonstrated that LMP1 was responsible for IL-10 induction by EBV infection. In the present study, we demonstrated that BL cells, which were negative for LMP1 expression, expressed detectable amounts of IL-10 and that EBERs were responsible for IL-10 induction. Since EBERs are expressed ubiquitously in latently EBV-infected cells, it is possible that EBERs contribute to the growth of not only BL, but also other EBV-associated malignancies like AIDS-associated lymphoma, by inducing IL-10 expression.
Among four type I BL cell lines examined in this study, the KemI and SavI cell lines expressed a very low level of EBERs and were negative for IL-10 expression. We do not know whether these two cell lines originally expressed a low level of EBERs or EBER expression decreased during cultivation. Since all four EBV-positive BL biopsies expressed a fair amount of EBERs, it is likely that EBER expression decreased during cultivation in the KemI and SavI cell lines. Transfection of the EBER gene induced IL-10 expression in all four EBV-negative BL lines examined. Together with the finding that EBV-positive BL biopsies consistently expressed the IL-10 gene, these results suggested that IL-10 induction by EBER is a general phenomenon beyond the Akata cell line.
In contrast to the consistent expression of the IL-10 gene in EBV-positive BL biopsies, EBV-negative BL biopsies were negative for IL-10 expression. The only common feature of EBV-positive and -negative BL is activation of the c-myc gene through reciprocal translocation between c-myc and Ig genes (Taub et al., 1982). Our results imply different growth mechanisms for EBV-positive and -negative BL, i.e. the growth of the former depends on IL-10 whereas that of the latter does not.
The present findings suggest a new therapeutic strategy. Anti-IL-10 antibody or antisense oligonucleotide against IL-10 could be used for the treatment of EBV-positive BL.
Materials and methods
Cell lines and culture
Akata (Takada et al., 1991), Mutu (Gregory et al., 1990), SavI and KemI are type I BL cell lines. We isolated EBV-negative Akata and Mutu cell clones from the parental cultures by the limiting dilution method. BL30 (Calender et al., 1987), BL41 (Calender et al., 1987), Louckes (van Santen et al., 1981) and Ramos (Klein et al., 1975) are EBV-negative BL cell lines. Cells were maintained in RPMI 1640 medium (Sigma) supplemented with 10% fetal calf serum (FCS; Gibco-BRL), penicillin (40 U/ml) and streptomycin (50 µg/ml) at 37°C in a 5% CO2 humidified atmosphere.
Plasmids, transfection and cell cloning
EBER1 and EBER2 open reading frames are located at 6628–6796 and at 6958–7129 bp, respectively, on the EcoRI K fragment of Akata EBV DNA, which corresponds to the EcoRI J fragment of B95-8 EBV DNA (Baer et al., 1984). Since the plasmid that contained a single copy of EBER could not induce levels of EBER expression in transfected cells equivalent to those in EBV-infected cells, we used the EBER plasmid that contained 10 tandem repeats of the EBER1 and EBER2 subfragment (6297–7325 bp) from the EcoRI K fragment of Akata EBV DNA and neomycin-resistant (neoR) gene driven by the simian virus 40 (SV40) promoter. The EBER1 plasmid and the EBER2 plasmid contained 10 tandem repeats of the EBER1 subfragment (6288–6797 bp) and EBER2 subfragment (6800–7317), respectively, from the EcoRI K fragment of Akata EBV DNA and neoR driven by the SV40 promoter (Komano et al., 1999). The pEBO plasmid carries the SV40 promoter-driven neoR, the EBNA1 gene and ori-P sequence of B95-8 origin. The BARF0 plasmid carries the SV40 promoter-driven neoR and the FLAG-tagged BARF0 gene derived from Akata EBV DNA. The VA1 expression plasmid was from Promega (pAdVAntage™). The plasmid for a mutant form (296Lys to Arg) of PKR (pmPKR) carries the SV40 promoter-driven FLAG-tagged mPKR. The plasmid for IL-10 carries the SV40 promoter-driven neoR and the human IL-10 gene.
Each of the plasmids was introduced into cells by the electroporation method. For transient analysis, cells were harvested 2 days after transfection. For isolation of stable transfectants, transfected cells were cultured for 2 days and transferred to 96-well, flat-bottom plates at 5000–10 000 cells per well in complete culture medium containing an appropriate concentration of G418 (Gibco-BRL) for selection (700 µg/ml for Akata and BL41 cells, 1 mg/ml for BL30 cells, 1.5 mg/ml for Mutu cells and 1.7 mg/ml for Louckes cells). Cultures were fed every 5 days by replacement of half of the medium until colonies emerged.
Generation of recombinant EBV and EBV infection
As described (Shimizu et al., 1996), the EBER-positive EBV recombinant carries neoR inserted into the EBV thymidine kinase gene, which is non-essential for infection and replication. An EBER-knockout EBV was generated by replacing the EBER coding region (6609–7270 bp) with neoR. Preparation of the virus solution was as described previously (Shimizu et al., 1996). For EBV infection, 5 × 106 cells were suspended in 2 ml of diluted EBV solution (1:2 to 10) for 60 min at room temperature with continuous gentle mixing. Then they were washed three times and cultured for 2 days. Selection and cloning procedures are described above.
Real-time quantitative RT–PCR
Total RNA was extracted from cells or tissues with Trizol reagent (Gibco-BRL), and then treated with DNase I (Gibco-BRL) according to the manufacturer’s instructions. For reverse transcription, 1 µg of total RNA was incubated at 37°C for 60 min in the presence of 20 mM Tris–HCl pH 8.3, 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 0.5 mM deoxynucleoside triphosphate (Takara), 100 pmol of random hexamer (Takara) and 200 U of Moloney murine leukemia virus reverse transcriptase (RTase; Gibco-BRL) in a total volume of 20 µl.
Quantitative PCR was performed in 20 µl glass capillary tubes using a LightCycler™ (Roche Molecular Biochemicals), which was equipped with a thermal cycler and real-time detector of fluorescence. Fifty nanograms of first-strand cDNA were amplified specifically by PCR at a final concentration of 1× PCR Reaction Buffer (Gibco-BRL), 0.01% bovine serum albumin (Takara), 3 mM MgCl2, 200 µM dNTP (Takara), 0.05 U/µl recombinant Taq polymerase (Gibco-BRL), 0.5 µM sense and antisense gene-specific primers and 0.2 µM up- and downstream adjacent hybridization probes in a total volume of 20 µl. For amplification of IL-1β, IL-4 and IL-6, a final concentration of 1:50 000 of SYBR green I (FMC Bioproducts) was added to the mixture instead of hybridization probes. Each PCR cycle consisted of denaturation for 0 s at 94°C, annealing for 5 s at 50–65°C, and extension for 6–20 s at 72°C. Sequences of the primers and probes are listed in Table II.
Table II. Sequences of primers/probes and PCR conditions.
| Transcripts | Type | Sequences (5′ to 3′) | Product size (bp) |
|---|---|---|---|
| IL-1β | 5′ primer | AATCCCCAGCCCTTTTGTTGAG | 214 |
| 3′ primer | ACTGCTACTTCTTGCCCCCTTTGA | ||
| IL-4 | 5′ primer | ACTGCTTCCCCCTCTGTTCTTCCTG | 354 |
| 3′ primer | ACGTACTCTGGTTGGCTTCCTTCACA | ||
| IL-6 | 5′ primer | AAAGAGGCACTGGCAGAAAACAAC | 421 |
| 3′ primer | TTAAAGCTGCGCAGAATGAGATGA | ||
| vIL-10 | 5′ primer | AGAGATGCCTTCAGTCGTGTT | 396 |
| 3′ primer | GGCTTTAATTGTCATGTATGCTTCTAT | ||
| BamHI W | 5′ primer | CGGTCGCCCAGTCCTACCAG | 125 |
| 3′ primer | CCTGGAGAGGTCAGGTTACT | ||
| Cp | 5′ primer | AGGCGCGGGATAGCGTGCGCTACCGGA | 339 |
| 3′ primer | TCCTCGTCCATGGTTATCAC | ||
| probe | AGACCTGGGAGCAGATTCAC | ||
| Qp | 5′ primer | CACTACAAGACCTACGCCTCTCCATCCATC | 297 |
| 3′ primer | TCTCCCCTAGGCCCTGAAGGTGAACCGCTT | ||
| probe | GCGACCGGTGCCTTCTTAGGAGCTGTCCGA | ||
| Wp | 5′ primer | TCAGAGCGCCAGGAGTCCACACAAAT | 235 |
| 3′ primer | TCTCCCCTAGGCCCTGAAGGTGAACCGCTT | ||
| probe | GCGACCGGTGCCTTCTTAGGAGCTGTCCGA | ||
| LMP2A | 5′ primer | ATGACTCATCTCAACACATA | 280 |
| 3′ primer | CATGTTAGGCAAATTGCAAA | ||
| probe | ATCCAGTATGCCTGCCTGTA | ||
| LMP2B | 5′ primer | CAGTGTAATCTGCACAAAGA | 325 |
| 3′ primer | CATAGTTAGGCAAATTGCAAA | ||
| probe | ATCCAGTATGCCTGCCTGTA | ||
| BARF0 | 5′ primer | AGAGACCAGGCTGCTAAACA | 232 |
| 3′ primer | AACCAGCTTTCCTTTCCCAG | ||
| probe | AAGACGTTGGAGGCACGCTG | ||
| hIL-10 | 5′ primer | TGCTCTGTTGCCTGGTCCTCCTGA | 154 |
| 3′ primer | GCTCCACGGCCTTGCTCTTGTTTT | ||
| 5′ probe | GGTTGCCAAGCCTTGTCTGAGATGATCCAGT | ||
| 3′ probe | ACCTGGAGGAGGTGATGCCCCAGCTT | ||
| EBER | 5′ primer | AGGACCTACGCTGCCCTAGA | 167 |
| 3′ primer | AAAACATGCGGACCACCAGC | ||
| 5′ probe | GGAGACGTGTGTGGCTGTAGCC | ||
| 3′ probe | CCCGTCCCGGGTACAAGTCCT | ||
| GAPDH | 5′ primer | GCCTCCTGCACCACCAACTG | 455 |
| 3′ primer | CGACGCCTGCTCACCACCTTCT | ||
| 5′ probe | CCTCCGGGAAACTGTGGCGTGATGGT | ||
| 3′ probe | GCGGGGCTCTCCAGAACATCATCCT |
The hybridization probes were designed according to guidelines recommended by Roche Molecular Biochemicals. In brief, the 3′-end of the 5′ probe was labeled by fluorescein, the 5′-end of the 3′ probe was labeled by LC Red 640 and the 3′-end of the probe was phosphorylated. Primers and probes were synthesized by Hokkaido System Science (Sapporo, Japan) and Nihon Gene Research Labs (Sendai, Japan), respectively.
For quantification of IL-10, EBER and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), the fluorescent intensity of LC Red 640, reflecting the amount of the PCR product, was read at the end of each annealing step. For measurement of IL-1β, IL-4 and IL-6, the fluorescent intensity of SYBR green I was read at the end of each extension step. After PCR reaction, background subtraction of the initial cycles was followed by determination of the optimal threshold level (5% of the full scale) of fluorescent intensity (Ft) using LightCycler™ software version 3 (Roche Molecular Biochemicals). Then, quantitative results for each sample were assessed by the threshold cycle number, which was determined from the crossing point between Ft and the plotted curve (Fink et al., 1998). Each RNA value was expressed as the ratio to the GAPDH value. Each PCR product was electrophoresed on 2% agarose gels to confirm specific DNA bands.
Southern blot analysis
Purified cellular DNA (5 µg) was digested with BamHI, size-fractionated by electrophoresis in a 0.8% agarose gel, and transferred to a nylon membrane (Amersham). To detect the EBV genome, the BamHI C fragment of Akata EBV was used as a probe. Probe labeling and detection of viral DNA signals were carried out using the ALKphos Direct Kit (Amersham) following the manufacturer’s instructions.
PCR
For PCR analysis of EBV genomes, purified cellular DNA (100 ng, corresponding to 1.6 × 104 cells) was subjected to PCR, with primer pairs specific for EBER and BamHI W regions of EBV DNA (Table I). The PCR products were size separated on 2% agarose gels and visualized by ethidium bromide staining.
Immunoblotting
Cells were lysed in SDS–PAGE loading buffer, sonicated and boiled for 5 min. A volume of lysate equal to 105 cells was separated in 10% polyacrylamide gels and transferred to a nitrocellulose membrane (Schleicher & Schuell). After blocking with 5% non-fat dry milk in Tris-buffered saline (TBS-M pH 7.6), the membrane was incubated for 2 h at room temperature with human serum diluted at 1:50 in TBS-M to detect EBNAs, washed three times with TBS-M containing 0.1% Tween 20 (TBS-TM), and then serially reacted for 30 min with biotinylated rabbit anti-human Ig G [diluted 1:500 in TBS-M (Dako)] and alkaline phosphatase-conjugated streptavidin [diluted 1:3000 in TBS-M (Amersham)] with washing between each reaction. Expression of EBNA2, LMP1 and FLAG-tagged proteins was examined using monoclonal antibodies PE2 (a gift of E.Kieff, Harvard Medical School, Boston, MA), CS1-4 (Dako) and M2 (Upstate Technology Inc.), respectively, and antibody reaction and washing were performed in TBS and TBS-T solutions, respectively. The second antibody reaction was carried out with horseradish peroxidase-conjugated sheep antibodies to human IgG [diluted 1:3000 in TBS-M (Amersham)]. After the second antibody reaction, the filters were washed five times with TBS-T, immersed in the enhanced chemiluminescence (ECL) solutions (Amersham) as specified by the manufacturer, and subjected to autoradiography.
RT–PCR
RT–PCR analysis was carried out to investigate the expression of EBV latent and lytic genes and the utilization of EBNA promoters (Qp, Cp and Wp) as described (Table I; Imai et al., 1998). Total cellular RNA was isolated by guanidium isothiocyanate–phenol extraction using TRIzol reagent (Gibco-BRL) according to the manufacturer’s protocol. Extracted RNA was heated for 10 min at 70°C, and rapidly cooled on ice. cDNA synthesis was performed for 60 min at 37°C with Molony murine leukemia virus RTase (Gibco-BRL) using 100 pmol of random hexamer (Takara) followed by 10 min of heating at 94°C to inactivate RTase. The cDNA samples were then subjected to 30 cycles of PCR in a thermal cycler. Each cycle consisted of denaturation for 30 s at 94°C, annealing for 30 s at 45–55°C and extension for 1 min at 72°C. The reaction mixture contained buffers and reagents as described, with 20 pmol of each primer and cDNA (equivalent to 5 × 104 cells/tube) in a volume of 50 µl. Five microliters of the PCR products were electrophoresed on a 2% agarose gel, blotted onto nylon membranes (Hybond N+, Amersham) and specific amplified DNA was detected by ECL 3′-oligolabeling and detection systems (Amersham). The quality of RNA was checked by parallel amplification of GAPDH mRNA.
IL-10 sense and antisense oligonucleotides
A 20-base phosphothionated sense oligonucleotide (5′-ATGCAC AGCTCAGCACTGCT-3′) and a 19-base phosphothionated antisense oligonucleotide (5′-AGCAGTGCTGAGCTGTGAT) (Masood et al., 1995) were synthesized and purified by Hokkaido System Science (Sapporo, Japan).
BL biopsies
BL biopsies were obtained from Japanese patients. The diagnosis of BL was made pathologically based on clinical and laboratory findings. All cases carried a chromosomal translocation between c-myc and Ig genes.
Acknowledgments
Acknowledgements
We thank R.F.Margolskee for pEBO, K.Shimotohno for pmPKR, J.Sample for EBV-positive Mutu, KemI and SavI cell lines, E.Kieff for BL30, BL41 and Louckes cell lines, and M.Imamura for helpful suggestions. We especially thank K.Adachi for her excellent technical assistance. This work has been supported by grants-in-aid from the Ministry of Education, Science, Sports and Culture, Japan, and from the Princess Takamatsu Research Fund.
References
- Baer R. et al. (1984) DNA sequence and expression of the B95-8 Epstein–Barr virus genome. Nature, 310, 207–211. [DOI] [PubMed] [Google Scholar]
- Benjamin D., Knobloch,T.J. and Dayton,M.A. (1992) Human B-cell interleukin-10: B-cell lines derived from patients with acquired immunodeficiency syndrome and Burkitt’s lymphoma constitutively secrete a large quantity of interleukin-10. Blood, 80, 1289–1298. [PubMed] [Google Scholar]
- Bhat R.A. and Thimmappaya,B. (1983) Two small RNAs encoded by Epstein–Barr virus can functionally substitute for the virus-associated RNAs in the lytic growth of adenovirus 5. Proc. Natl Acad. Sci. USA, 80, 4789–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdin N., Peronne,C., Banchereau,J. and Rousset,F. (1993) Epstein–Barr virus transformation induces B lymphocytes to produce human interleukin 10. J. Exp. Med., 177, 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calender A., Billaud,M., Aubry,J.P., Banchereau,J., Vuillaume,M. and Lenoir,G. (1987) Epstein–Barr virus (EBV) induces expression of B-cell activation markers on in vitro infection of EBV-negative B-lymphoma cells. Proc. Natl Acad. Sci. USA, 84, 8060–8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodosh J., Holder,V.P., Gan,Y., Belgaumi,A., Sample,J. and Sixbey,J. (1998) Eradication of latent Epstein–Barr virus by hydroxyurea alters the growth-transformed cell phenotype. J. Infect. Dis., 177, 1194–1201. [DOI] [PubMed] [Google Scholar]
- Clarke P.A., Schwemmle,M., Schickinger,J., Hilse,K. and Clemens,M.J. (1991) Binding of Epstein–Barr virus small RNA EBER-1 to the double-stranded RNA-activated protein kinase DAI. Nucleic Acids Res., 19, 243–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens M.J. (1993) The small RNAs of Epstein–Barr virus. Mol. Biol. Rep., 17, 81–92. [DOI] [PubMed] [Google Scholar]
- Epstein M.A. and Achong,B.G. (1979) The relationship of the virus to Burkitt’s lymphoma. In Epstein,M.A. and Achong,B.G. (eds), The Epstein–Barr Virus. Springer-Verlag, Berlin, Germany, pp. 321–337. [Google Scholar]
- Fink L., Seeger,W., Ermert,L., Hanze,J., Stahl,U., Grimminger,F., Kummer,W. and Bohle,R.M. (1998) Real-time quantitative RT–PCR after laser-assisted cell picking. Nature Med., 4, 1329–1333. [DOI] [PubMed] [Google Scholar]
- Fiorentino D.F., Bond,M.W. and Mosmann,T.R. (1989) Two types of mouse T helper cell. IV. Th2 clones secrete a factor that inhibits cytokine production by Th1 clones. J. Exp. Med., 170, 2081–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory C.D., Rowe,M. and Rickinson,A.B. (1990) Different Epstein–Barr virus-B cell interactions in phenotypically distinct clones of a Burkitt’s lymphoma cell line. J. Gen. Virol., 71, 1481–1495. [DOI] [PubMed] [Google Scholar]
- Howe J.G. and Shu,M.D. (1989) Epstein–Barr virus small RNA (EBER) genes: unique transcription units that combine RNA polymerase II and III promoter elements. Cell, 57, 825–834. [DOI] [PubMed] [Google Scholar]
- Howe J.G. and Steitz,J.A. (1986) Localization of Epstein–Barr virus-encoded small RNAs by in situ hybridization. Proc. Natl Acad. Sci. USA, 83, 9006–9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S., Nishikawa,J. and Takada,K. (1998) Cell-to-cell contact as an efficient mode of Epstein–Barr virus infection of diverse human epithelial cells. J. Virol., 72, 4371–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K. and Hirohata,S. (1995) The role of IL-10 in human B cell activation, proliferation, and differentiation. J. Immunol., 154, 4341–4350. [PubMed] [Google Scholar]
- Kelso A. (1998) Cytokines: principles and prospects. Immunol. Cell Biol., 76, 300–317. [DOI] [PubMed] [Google Scholar]
- Klein G. (1981) The role of gene dosage and genetic transpositions in carcinogenesis. Nature, 294, 313–318. [DOI] [PubMed] [Google Scholar]
- Klein G., Giovanella,B., Westman,A., Stehlin,J.S. and Mumford,D. (1975) An EBV-genome-negative cell line established from an American Burkitt lymphoma: receptor characteristics, EBV infectability and permanent conversion into EBV-positive sublines by in vitro infection. Intervirology, 5, 319–334. [DOI] [PubMed] [Google Scholar]
- Komano J., Sugiura,M. and Takada,K. (1998) Epstein–Barr virus contributes to the malignant phenotype and to apoptosis resistance in Burkitt’s lymphoma cell line Akata. J. Virol., 72, 9150–9156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komano J., Maruo,S., Kurozumi,K., Oda,T. and Takada,K. (1999) Oncogenic role of Epstein–Barr virus-encoded RNAs in Burkitt’s lymphoma cell line Akata. J. Virol., 73, 9827–9831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner M.R., Andrews,N.C., Miller,G. and Steitz,J.A. (1981) Two small RNAs encoded by Epstein–Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc. Natl Acad. Sci. USA, 78, 805–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masood R. et al. (1995) Interleukin-10 is an autocrine growth factor for acquired immunodeficiency syndrome-related B-cell lymphoma. Blood, 85, 3423–3430. [PubMed] [Google Scholar]
- Nakagomi H., Dolcetti,R., Bejarano,M.T., Pisa,P., Kiessling,R. and Masucci,M.G. (1994) The Epstein–Barr virus latent membrane protein-1 (LMP-1) induces interleukin-10 production in Burkitt lymphoma cells. Int. J. Cancer, 57, 240–244. [DOI] [PubMed] [Google Scholar]
- Rickinson A.B. and Kieff,E. (1996) Epstein–Barr virus. In Fields,B.N., Knipe,D.M. and Howley,P.M. (eds), Fields Virology, 3rd edn. Lippincott-Raven, Philadelphia, PA, pp. 2397–2446. [Google Scholar]
- Rosa M.D., Gottlieb,E., Lerner,M.R. and Steitz,J.A. (1981) Striking similarities are exhibited by two small Epstein–Barr virus-encoded ribonucleic acids and the adenovirus-associated ribonucleic acids VAI and VAII. Mol. Cell. Biol., 1, 785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe M., Rowe,D.T., Gregory,C.D., Young,L.S., Farrell,P.J., Rupani,H. and Rickinson,A.B. (1987) Differences in B cell growth phenotype reflect novel patterns of Epstein–Barr virus latent gene expression in Burkitt’s lymphoma cells. EMBO J., 6, 2743–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf I.K., Rhyne,P.W., Yang,H., Borza,C.M., Hutt-Fletcher,L.M., Cleveland,J.L. and Sample,J.T. (1999) Epstein–Barr virus regulates c-MYC, apoptosis, and tumorigenicity in Burkitt lymphoma. Mol. Cell. Biol., 19, 1651–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R.J., Safer,B., Munemitsu,S.M., Samuel,C.E. and Shenk,T. (1985) Adenovirus VAI RNA prevents phosphorylation of the eukaryotic initiation factor 2 α subunit subsequent to infection. Proc. Natl Acad. Sci. USA, 82, 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu N., Tanabe-Tochikura,A., Kuroiwa,Y. and Takada,K. (1994) Isolation of Epstein–Barr virus (EBV)-negative cell clones from the EBV-positive Burkitt’s lymphoma (BL) line Akata: malignant phenotypes of BL cells are dependent on EBV. J. Virol., 68, 6069–6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu N., Yoshiyama,H. and Takada,K. (1996) Clonal propagation of Epstein–Barr virus (EBV) recombinants in EBV-negative Akata cells. J. Virol., 70, 7260–7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada K. (1984) Cross-linking of cell surface immunoglobulins induces Epstein–Barr virus in Burkitt lymphoma lines. Int. J. Cancer, 33, 27–32. [DOI] [PubMed] [Google Scholar]
- Takada K. and Ono,Y. (1989) Synchronous and sequential activation of latently infected Epstein–Barr virus genomes. J. Virol., 63, 445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada K., Horinouchi,K., Ono,Y., Aya,T., Osato,T., Takahashi,M. and Hayasaka,S. (1991) An Epstein–Barr virus-producer line Akata: establishment of the cell line and analysis of viral DNA. Virus Genes, 5, 147–156. [DOI] [PubMed] [Google Scholar]
- Taub R., Kirsch,I., Morton,C., Lenoir,G., Swan,D., Tronick,S., Aaronson,S. and Leder,P. (1982) Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc. Natl Acad. Sci. USA, 79, 7837–7841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toczyski D.P. and Steitz,J.A. (1991) EAP, a highly conserved cellular protein associated with Epstein–Barr virus small RNAs (EBERs). EMBO J., 10, 459–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toczyski D.P., Matera,A.G., Ward,D.C. and Steitz,J.A. (1994) The Epstein–Barr virus (EBV) small RNA EBER1 binds and relocalizes ribosomal protein L22 in EBV-infected human B lymphocytes. Proc. Natl Acad. Sci. USA, 91, 3463–3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Santen V., Cheung,A. and Kieff,E. (1981) Epstein–Barr virus RNA VII: size and direction of transcription of virus-specified cytoplasmic RNAs in a transformed cell line. Proc. Natl Acad. Sci. USA, 78, 1930–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira P. et al. (1991) Isolation and expression of human cytokine synthesis inhibitory factor cDNA clones: homology to Epstein–Barr virus open reading frame BCRF1. Proc. Natl Acad. Sci. USA, 88, 1172–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]