Abstract
The cold shock domain (CSD) is an evolutionarily conserved nucleic acid binding domain that exhibits binding activity to RNA, ssDNA, and dsDNA. Mammalian CRHSP-24 contains CSD, but its structure-functional relationship has remained elusive. Here we report the crystal structure of human CRHSP-24 and characterization of the molecular trafficking of CRHSP-24 between stress granules and processing bodies in response to oxidative stress. The structure of CRHSP-24 determined by single-wavelength anomalous dispersion exhibits an α-helix and a compact β-barrel formed by five curved anti-parallel β strands. Ligand binding activity of the CSD is orchestrated by residues Ser41 to Leu43. Interestingly, a phosphomimetic S41D mutant abolishes the ssDNA binding in vitro and causes CRHSP-24 liberated from stress granules in vivo without apparent alternation of its localization to the processing bodies. This new class of phosphorylation-regulated interaction between the CSD and nucleic acids is unique in stress granule plasticity. Importantly, the association of CRHSP-24 with stress granules is blocked by PP4/PP2A inhibitor calyculin A as PP2A catalyzes the dephosphorylation of Ser41 of CRHSP-24. Therefore, we speculate that CRHSP-24 participates in oxidative stress response via a dynamic and temporal association between stress granules and processing bodies.
Keywords: Intracellular Processing, P-body, PP2C, Stress Granule, Trafficking, X-ray Crystallography, β-Barrel, Calyculin A, Phosphorylation
Introduction
Cold shock domains (CSD)4 are the most evolutionarily conserved of the nucleic acid binding domains, able to bind single-stranded DNA (ssDNA) and/or ssRNA with micromolar to nanomolar dissociation constants (KD) (1). This domain is named after the 70-amino acid prokaryotic cold-shock protein (CSP) up-regulated during cold stress. These CSPs have been shown to function as mRNA chaperones or transcriptional anti-terminators, which help to maintain transcription or translation of cold-induced genes both in vitro and in vivo at low temperature (2, 3). CSD is a key component of the eukaryotic Y-box proteins, which contain extra variable N and C termini. Among three Y-box proteins identified in vertebrates (YB-1, MSY2, and MSY4), YB-1 is the most widely characterized member of the family in both germ and somatic mammalian cells (4, 5). In the cytoplasm, YB-1 participates in the formation of message ribonucleoprotein particles and may act as a translational repressor (6, 7). YB-1 may shuttle between the cytoplasm and nucleus in response to physiological cues and stress-induced DNA damages (8, 9). Within the nucleus, YB-1 functions as a transcription factor and is able to activate transcription of a wide range of genes by recognizing Y-box elements (5′-CTGATTGG(C/T)(C/T)AA-3′) in their promoters (e.g. human MDR1) (10). Thus, YB-1 has been implicated as a multifunctional coordinator for the control of gene expression at both the transcriptional and the posttranscriptional levels (10). Binding experiments and crystal structures with bound oligonucleotides have shown that Bs-CspB from Bacillus subtilis adopts a five-stranded anti-parallel β-barrel with the oligonucleotide/oligosaccharide binding fold and has higher affinities for thymine (T)- or uracil (U)-rich sequences than the Y-box sequence (11–13). The solution structure of the CSD of the human YB-1 and the Y-box core sequence, 5′-ATTGG-3′, revealed that the flanking domains of CSD of intact YB-1 are required for strong interaction, although the conserved fold alone is sufficient to bind to ssDNA (14, 15).
Ca2+-regulated heat-stable protein of 24 kDa (CRHSP-24) was originally identified as a physiological substrate for calcineurin (16), and an interacting protein with the STYX/dead phosphatase in developing spermatids (17). CRHSP-24 exhibits a broad tissue distribution and localizes to the cytoplasm (16). CRHSP-24 possesses a CSD and shares ∼62% identity with its brain-specific paralog, PIPPin (18, 19), which binds to the 3′-untranslated region of histone H1° and H3.3 mRNAs to inhibit translation of these messages in vitro (20). Recently, it was shown that CRHSP-24 Ser52 is phosphorylated by protein kinase Bα and ribosomal S6 kinase in response to growth factors, whereas the Ser41 is a substrate of a DYRK isoform (21). Subsequently, four serines (Ser-30, -32, -41, and -52) were mapped in which Ser30 and Ser32 are dephosphorylated by calcineurin (22). However, the precise structure-functional relationship of CRHSP-24 has remained elusive.
Here we report the 2.8 Å crystal structure of the human CRHSP-24. Our data reveal that the conserved CSD region exhibits a five-stranded anti-parallel β-barrel with an oligonucleotide/oligosaccharide-binding fold. Ligand binding by the CSD is regulated by residues Ser41 to Leu43. Moreover, the phosphomimetic mutant S41D exhibits perturbed association between CRHSP-24 and ssDNA due to the negative charge on Ser41. Importantly, phosphorylation of Ser41 causes CRHSP-24 to be liberated from stress granules in vivo, suggesting that phosphorylation regulates CRHSP-24 conformation and spatial dynamics in cells. Taken together, our data suggest that CRHSP-24 participates in oxidative stress response via a dynamic and temporal association between stress granules and processing bodies.
EXPERIMENTAL PROCEDURES
Protein Expression, Preparation, and Purification
Full-length CRHSP-24 from Homo sapiens was cloned into a pGEX-6p-1 vector (GE Healthcare) and expressed in Escherichia coli BL21 (DE3) in rich (LB) medium as a fusion protein with an N-terminal GST tag. Expression and purification of GST-CRHSP-24 was carried out according to standard protocol.
Surface Plasmon Resonance (SPR) Analysis of CRHSP-24 Interaction with Nucleotides
The synthetic thymine-rich nucleotide fragment from Histone3.3 (NM_005324.3 1085 ∼ 1117) were purchased from Invitrogen. The interaction of CRHSP-24 with the nucleotide fragment was analyzed by SPR (23) using a BIACORE 3000 optical biosensor (Biacore AB, Uppsala, Sweden) according to the user's manual.
Crystallization
Conditions were identified by the hanging drop vapor diffusion method with Crystal Screen reagent kits I and II (Hampton Research). Crystals suitable for diffraction were obtained after 9 days under 0.1 m sodium acetate trihydrate, pH 4.9, 2.0 m sodium formate.
Data Collection and Model Check
Diffraction data from a single crystal of Se-CRHSP-24 were collected on an ADSC-Q270t CCD detector (Marresearch GmbH, Norderstedt Germany) at beamline BL17A of the High Energy Accelerator Research Organization (KEK, Tsukuba, Japan). Data from the native CRHSP-24 crystal were collected at wavelength 0.97945 Å at the Advanced Photon Source (Argonne National Laboratory). Model quality was checked with PROCHECK (24).
cDNA Construction and Cell Culture
The GST-CRHSP-24, GFP-CRHSP-24, and FLAG-CRHSP-24 plasmids were built using standard protocols. HeLa cells from the American Type Culture Collection were maintained as subconfluent monolayers in DMEM (Invitrogen) with 10% fetal bovine serum (Hyclone, Logan, UT) and 100 units/ml penicillin plus 100 μg/ml streptomycin (Invitrogen) at 37 °C with 8% CO2. Cells were transfected with plasmids or siRNA by Lipofectamine 2000 (Invitrogen) according to the manufacturer's manuals.
Antibodies and siRNA
Monoclonal antibodies to CRHSP-24 and G3BP were purchased from Santa Cruz Biotechnology and used according to user's manual. For siRNA studies, the 21-mer of siRNA duplexes against CRHSP-24 was purchased from Santa Cruz Biotechnology. The efficiency of siRNA-mediated protein suppression was judged by Western blotting analysis. After trial experiments using a series of concentration and time course assay, treatment of 100 nm for 48 h was finally selected as the most efficient conditions for repressing the target protein.
Immunofluorescence and Immunoprecipitation
Cells were transfected with GFP-tagged plasmids in a 24-well plate by Lipofectamine 2000 (Invitrogen) according to the manufacturer's manuals. Immunofluorescence microscopy and co-immunoprecipitation were performed as described previously (25).
RESULTS
Crystal Structure of CRHSP-24
CRHSP-24 is a major regulatory phosphoprotein in pancreatic acinar cells in response to secretagogues. To gain a better understanding of the structure-activity relationship, we sought to study the three-dimensional structure of CRHSP-24. To this end, full-length human CRHSP-24 (residues 1–147) was expressed in E. coli and purified by affinity and size-exclusion chromatography. After removal of the N-terminal GST tag, CRHSP-24 was first crystallized in space group P61, and diffraction data were collected to 2.8 Å resolution. Phases were obtained from a selenomethionine derivative by the single-wavelength anomalous dispersion method using the program PHENIX (26). The resulting experimental electron density maps were clear and readily traceable. The structure was refined to final Rwork/Rfree values of 22.8/27.2% using the program PHENIX.REFINE (26). The asymmetric unit of the crystal contains four protein molecules, each traced from residues 41–141 except for a disordered loop L45 (residues 114 and 117) connecting strands β4 and β5 in molecule A/C and residues 116–118 in molecule D. Data collection and refinement statistics are summarized in Table 1.
TABLE 1.
CRHSP-24 data collection and refinement statistics
SeMet, selenomethionine.
| Data collection | ||
| Crystal | Native CRHSP-24 | SeMet CRHSP-24 |
| Wavelength (Å) | 0.97945 | 0.97895 |
| Resolution (Å) | 50.00-2.80 | 50.00-3.30 |
| Last shell (Å) | 2.90-2.80 | 3.42-3.30 |
| Space group | P61 | P61 |
| Temperature (K) | 100 | 100 |
| Detector | AdscQ315 | AdscQ270t |
| Unit cell parameters | ||
| a (Å) | 79.9 | 84.6 |
| b (Å) | 79.9 | 84.6 |
| c (Å) | 182.0 | 186.5 |
| Total observations | 62,285 | 77,006 |
| Unique reflections | 16,210 | 11,010 |
| Refinement | ||
| Resolution (Å) | 45.60-2.80 | |
| Unique reflections (last shell) | 16,140 (1,620) | |
| Working set | 15,328 | |
| Free set (5.03%) | 812 | |
| I/σ(I) (last shell) | 14.759 (2.587) | |
| Data completeness (%) | 99.8 (100.0) | |
| Rmeansa (%) | 8.6 (35.9) | |
| Rwork/Rfreeb (%) | 22.8/27.2 | |
| Number of protein molecules | 4 | |
| Number of water molecules | 32 | |
| Mean B factor (Å2) | 59.96 | |
| Root mean square deviation | ||
| Bond lengths (Å) | 0.008 | |
| Bond angles (°) | 1.266 | |
| Planarity (Å) | 0.006 | |
| Ramachandran statistics (%) | ||
| Residues in allowed regions | 84.1 | |
| Residues in add. allowed regions | 15.2 | |
a Rmeans, redundancy independent R factor that correlates intensities from symmetry related reflections (43).
b Rwork and Rfree = Σ|Fobs − Fcalc|/Σ|Fobs|, where the working and free R factors are calculated using the working and free reflection sets, respectively. The free reflections were held aside throughout refinement.
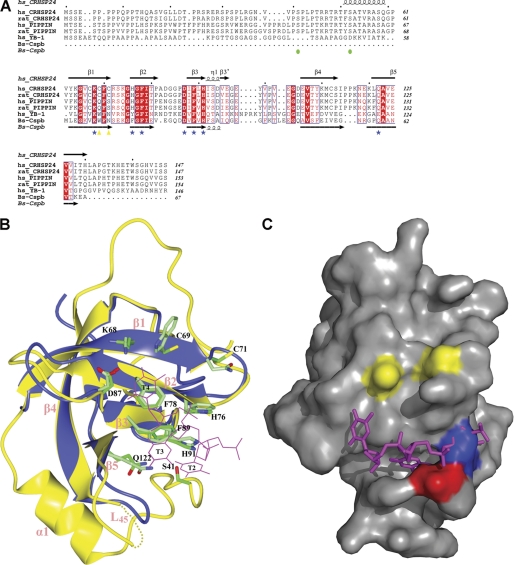
The CSD, the central element of CRHSP-24 (residues 62–126), features a compact β-barrel formed by five curved anti-parallel β strands linked by four loops and a short 310 helix at the C terminus of β3. The overall structure closely resembles that of human YB-1 and the fold of structural models of CSP (13, 15). The alignment with the structure of Bs-CspB (2ES2) yields root mean square deviation values of 1.30 Å for 65 α-carbon atoms (Fig. 1B). The sequence (residues 41–61) between CSD and the N-terminal proline-rich region, invisible in the crystal structure, shares a high degree of homology with PIPPin (Fig. 1A). The structure of CRHSP-24 consists of one α-helix, α1, which exists on the opposite side of the β-barrel from the ligand binding surface of CSD (Fig. 1B) and is involved in oligomerization (Fig. 2D).
FIGURE 1.
Structural analyses of features underlying cold shock domain proteins. A, from top to bottom, the sequences are: CRHSP-24 from H. sapiens (NP_001035941) and Rattus norvegicus (NP_690003), PIPPin from H. sapiens (NP_055275) and R. norvegicus (NP_663448), YB-1 from H. sapiens (NP_004550) and Bs-CspB (NP_388791). Secondary structure elements are indicated for CRHSP-24 and Bs-CspB; highly conserved residues among these proteins are indicated in red. Residues involved in DNA binding in the Bs-CspB·dT6 crystal structure are marked by blue stars. Amino acids that can make CRHSP-24 become aggregates are indicated by yellow triangles, and the sequences of CRHSP-24 that can be phosphorylated are marked by green circles. Sequences were aligned using T-coffee. B, shown is structural superposition of Bs-CspB bound to dT6 (blue) and CRHSP-24 (yellow). The dotted lines denote the region (residues 115–116) that is not resolved of L45. Secondary structural elements are indicated in pink. Residues involved in DNA binding in Bs-CspB·dT6 and equivalent residues in CRHSP-24 are shown in light and dark green, respectively. Potential binding between thymine that CRHSP-24 is marked by magenta and annotated by T2∼4. The figure was drawn using CCP4 mg. C, the structural surface of chain A of CRHSP-24 is shown. Three thymines (colored in magenta) are docked in the pocket of chain A. Ser41 is shown in red. Cys69 and Cys71 are marked in yellow. Leu43 is indicated in blue; it makes van der Waals contacts and occupies the similar site like T2 in B.
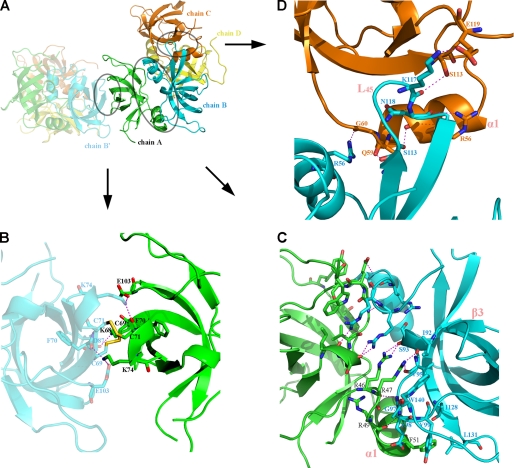
FIGURE 2.
Molecular structure of CRHSP-24 and its asymmetry. A, the asymmetry of CRHSP-24 is shown as a semitransparent color. The chains A, B, C, D are marked in green, cyan, orange, and yellow, respectively. The interface between chain A and B, chain B and C, and chain A and B′ (in the asymmetric unit) is circled. B, the interface between asymmetric units (chain A and B′). Five hydrogen bonds depicted as magenta dotted lines, whereas two disulfide bonds are shown in yellow. C, the interaction surface between chain A and chain B (the dimer interface) is shown. There is a non-crystallographic 2-fold axis between chains A and B. Hydrogen bonds are depicted as dotted lines, and residues involving in hydrophobic interactions and van der Waals contacts are shown in stick representation. D, shown is the contact surface between chain B and chain C; it is the major contact in tetramer formation. L45 of chain B is marked in pink. Residues that form hydrogen bonds are marked, and hydrogen bonds are shown as dotted lines.
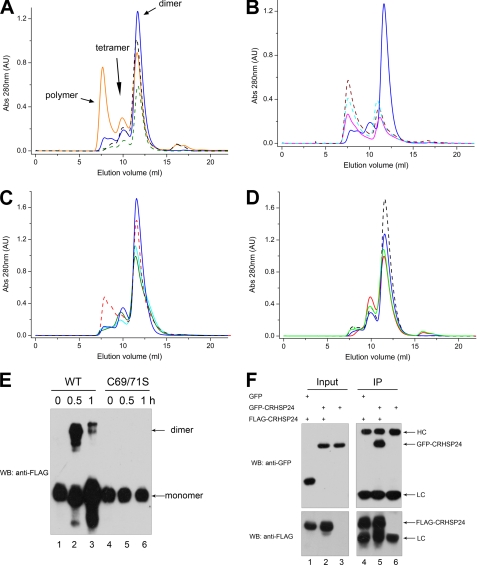
CRHSP-24 primarily forms dimers in solution, although about 15% exist as a tetramers based on gel filtration analysis (Fig. 3A). CRHSP-24 forms a tetramer within the asymmetric unit of the crystal structure, where four molecules are essentially identical to each other with a root mean square deviation of 0.4–0.6 Å between the Cα atoms of residues 41–141. The only significant differences between the four chains are observed in the connecting loop L45 (residues 114–118) between strands β4 and β5, which is wholly visible only in molecule B. Molecules A and B form identical dyad symmetrical dimers with contact areas of ∼1070 Å2, the same as molecules C and D. In the dimer interface the residues before the start of the α1 helix make contacts with the residues immediately after strand β3 in another molecule. Arg46, Arg47, Arg49, and Phe51 in one molecule form hydrogen bonds with Ile92, Ser93, Val95, Gly97, and Glu98 in another molecule. The hydrophobic side chain of Phe51 makes van der Waals contacts with Tyr99, Ile128, and Leu131. In addition, the side chains of Trp140, Arg46, and Arg47 in two molecules stack in parallel (Fig. 2C). Two dimers further interact to form a tetramer depending on the interaction of molecule B with molecule C and molecule D (contact area 589 and 380 Å2, respectively). The majority of residues participating in tetramer formation in molecule B are located on the connecting loop L45, whose contacts with molecules C or D make it ordered only in molecular B of the crystal structure (Fig. 2D). These residues involved in oligomerization are conserved in PIPPin, suggesting that PIPPin is also able to oligomerize in the same way as CRHSP-24.
FIGURE 3.
Molecular characterization of CRHSP-24 polymerization in vitro and in vivo. A, the blue solid line represents native CRHSP-24 in solution. There are three peaks representing polymers, dimers, and monomers. The orange line represents CRHSP-24 treated with 0.5 mm GSSG, whereas the dark green line represents the C69S/C71S double mutant without GSSG, and the black line represents the C69S/C71S double mutant in the presence of GSSG. AU, absorbance units. B, the peak native CRHSP-24 is shown by a blue solid line. The cyan line indicates the H91A mutant, the brown line represents the F89A mutant, and the magenta line indicates the F78A mutant. These mutants have apparently different peak profiles compared with the native CRHSP-24, suggesting that the mutations change the properties of CRHSP-24. C, the blue solid line represents native CRHSP-24, the red line shows the H76Q mutant, the cyan line shows the S41D mutant, and the light green line shows the S52D mutant. These mutants have similar peak profiles as the native CRHSP-24. D, C69S in the absence of GSSG, C71S in the absence of GSSG, and C69S in the presence of 0.5 mm GSSG for 2 h at room temperature and C71S in the presence of 0.5 mm GSSG for 2 h at room temperature are indicated by black, blue, red, and green lines, respectively. The polymeric form did not increase with 0.5 mm GSSG for 2 h at room temperature. E, overexpression of FLAG-CRHSP-24 and FLAG-C69S/C71S in HEK293 cells is shown. After 0.5 mm arsenite treatment for 30 min or 1 h, cell lysates were prepared and immunoprecipitated and fractionated on a non-reduced SDS-PAGE followed by anti-FLAG Western blotting (WB). Lane 1, FLAG-CRHSP-24 intraperitoneal; lane 2, FLAG-CRHSP-24 intraperitoneal after arsenite treatment for 0.5 h; lane 3, FLAG-CRHSP-24 intraperitoneal after arsenite treatment for 1 h; lane 4, FLAG-C69S/C71S intraperitoneal; lane 5, FLAG-C69S/C71S intraperitoneal after arsenite treatment for 0.5 h; lane 6, FLAG-C69S/C71S intraperitoneal after arsenite treatment for 1 h. Disulfide bond formation was found in the dimeric form of FLAG-CRHSP-24 wild type. F, FLAG-CRHSP-24 was transiently co-transfected with GFP-CRHSP-24 into HEK293 cells. After 24 h, the cells were harvested for immunoprecipitation (IP) with FLAG-M2 beads. The sample was resolved by SDS-PAGE and immunoblotted with anti-GFP antibody (upper) and anti-FLAG antibody (lower). HC, heavy chain; LC, light chain.
Surprisingly, crystal packing between two asymmetric units contain two intermolecular disulfide bonds formed by residues Cys69 and Cys71, which are located on strand β1 (Fig. 2B). The two residues are unique to CRHSP-24 and its CSD domain (Fig. 1A).
CRHSP-24 Oligomerizes under Oxidative Conditions Both in Vivo and in Vitro
As the crystal structure appears to be composed of hundreds of CRHSP-24 molecules linked by intermolecular disulfide bonds, we used the size exclusion column to determine the oligomeric state of CRHSP-24 in solution. Treatment of 0.5 mm GSSG for 2 h dramatically promotes polymer formation. As predicted, mutation of both Cys69 and Cys71 to Ser abolished the polymeric form regardless of GSSG addition and did not change the ratio of the dimer and tetramer forms (Fig. 3A). These results suggest that intermolecular disulfide linkages can be formed in oxidative solution and contribute to polymer formation, as seen in the crystal structure. We also constructed the single mutations C69S and C71S and found that in both cases the polymeric form did not increase with 0.5 mm GSSG for 2 h at room temperature (Fig. 3D). The results indicate that Cys69 and Cys71 cooperate to form intermolecular disulfide bonds.
To probe if CRHSP-24 forms oligomer in vivo, we have also carried out an immunoprecipitation experiment in which HeLa cells were transiently transfected to express wild-type FLAG-CRHSP-24 and its C69S/C71S mutant. We used anti-FLAG tag antibodies to immunoprecipitate soluble CRHSP-24 from the lysates pretreated with 0.5 mm arsenite for 0.5 and 1 h. Thus, dl-dithiothreitol is absent in the entire process to preserve the reaction in the oxidative environment. As shown in Fig. 3E, disulfide bond was formed between FLAG-CRHSP-24 in HeLa cells treated with arsenite for 30 min (Fig. 3E). Interestingly, disulfide bond formation of CRHSP-24 decreased with extended periods of arsenite treatment (e.g. 1 h). Consistent with our hypothesis, replacement of Cys with Ser (C69S/C71S) abolished this disulfide bond formation (Fig. 3E), indicating that CRHSP-24 forms intermolecular disulfide bonds upon oxidative stress in vivo.
To examine whether the intermolecular interaction of CRHSP-24 bears physiological relevance, HeLa cells were transiently transfected to express FLAG-CRHSP-24 and GFP-CRHSP-24. Twenty-four hours after the transfection, cell lysates were prepared and subjected to anti-FLAG immunoprecipitation. To eliminate the involvement of disulfide bond-mediated dimerization, 2 mm dl-dithiothreitol was added in cell lysate preparation and immunoprecipitation. As shown in Fig. 3F, GFP-CRHSP-24 was pulled down by FLAG-CRHSP-24, suggesting that intermolecular association between CRHSP-24 occurs in the reducing environment. We reason that intermolecular association of CRHSP-24 is primarily mediated by hydrophobic interaction under physiological reducing conditions, and Cys69 and Cys71 involve further polymerization of CRHSP-24 dimers in oxidative stress.
The Ligand Binding of CRHSP-24
By superimposing the CSD region of human CRHSP-24 and the CSP protein portion of the Bs-CspB·dT6 complex, aromatic residues His76, Phe78, Phe89, and His91 and polar residues Lys68, Asp87, and Gln122 which are marked in dark green in CRHSP-24, have positions and orientations very similar to those of the corresponding residues that are marked in light green in Bs-CspB (Fig. 1B). All these corresponding residues in Bs-CspB·dT6 are necessary in continual thymine (T) binding (13) and are highly conserved in CRHSP-24 (Fig. 1A). This part of the preformed site in Bs-CspB interacts with T2–4 of hexathymidine (dT6) through base stacking and hydrogen bonds (Fig. 1B). Based on the high conservation of residues involved in T binding, we propose that CRHSP-24 should also interact with thymine (T)- or uracil (U)-rich sequences containing at least three tandem thymine bases.
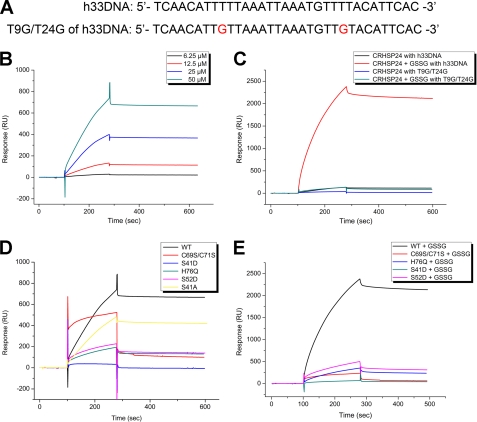
CRHSP-24 is primarily distributed in the cytoplasm (16), and PIPPin, the brain-specific paralog of CRHSP-24, has been shown to bind to the 3′-untranslated region of replacement histone H1° and H3.3 mRNAs (20). Thus, we were interested in determining whether CRHSP-24, which shares a high degree of homology with PIPPin, can also bind to mRNA in physiological environments. Because of the ability of CSD to bind ssDNA and ssRNA (1), we selected a sequence that we called h33DNA, a 33-nt-length ssDNA with a T-rich sequence corresponding to the 3′-untranslated region of H3.3 mRNA except for the substitution of U by T (Fig. 4A). SPR experiments were performed to examine the ligand binding ability of CRHSP-24 in vitro, with a resulting equilibrium dissociation constant (KD) between human CRHSP-24 and h33DNA of 8.87 μm (Fig. 4B). Attempts to crystallize the complex of CRHSP-24 and thymine-rich h33DNA under different conditions have failed due to poor diffraction of the crystals regardless of multiple optimization protocols (data not shown).
FIGURE 4.
Biochemical characterization of the bindings of wild-type and mutant CRHSP-24 proteins to immobilized nucleotides. A, shown are the two ssDNA sequences used in the SPR experiments. h33DNA (NM_005324.3 1085∼1117) represents the poly thymine probe. T9G/T24G of h33DNA is a double mutant of the h33DNA that breaks two polythymine fragments. B, various concentrations of CRHSP-24 were injected over the flow cell with h33DNA and reference flow cell. The sensogram shows the relative response in resonance units (RU) after background subtraction versus time in seconds. The concentrations of CRHSP-24 are marked by numbers, and the dissociation constant (Kd) is calculated. C, curves are comparisons between h33DNA and T9G/T24G of h33DNA after 12.5 μm CRHSP-24 was injected, and then 12.5 μm CRHSP-24 in the presence of 0.5 mm GSSG for 2 h at room temperature was injected. The T9G/T24G double mutant of h33DNA resulted in a significant decrease of binding signal. All of the curves are marked above. D, 50 μm native CRHSP-24, C69S/C71S, S41D, S52D, H76Q, and S41A were injected with h33DNA. The native and mutant proteins are marked in the corresponding curve. E, 12.5 μm native CRHSP-24, C69S/C71S, S41D, S52D, and H76Q in the presence of 0.5 mm GSSG for 2 h at room temperature were injected with h33DNA. The native and mutant proteins are shown in the corresponding curve.
To further explore the mode of binding, we performed mutagenesis analysis using the interaction system above. S41D and H76Q both exhibit similar peak profiles as the wild-type CRHSP-24 from size exclusion chromatography (Fig. 3C), indicating that they have the correct fold. SPR results showed that the H76Q mutant of human CRHSP-24 and the T9G/T24G double mutant of h33DNA (Fig. 4A), which breaks two poly thymine fragments, resulted in a significant decrease of binding affinity (Figs. 4, C and D). These findings support our structure-based hypothesis that human CRHSP-24 has a preference for binding thymine-rich sequences via its preformed binding site. We also verify that polymer formation in oxidative solution does not prevent human CRHSP-24 from ligand binding (Fig. 4E). This is consistent with the crystal packing containing intermolecular disulfide linkages which show that the ligand binding subsite of CSD is still solvent-exposed. Interestingly, in contrast to the wild-type CRHSP-24, the DNA association and dissociation kinetics curve of C69S/C71S double mutant changed. Residues 69 and 71 are close to the ligand binding subsite in the structure, and the corresponding residues in Bs-CspB can also participate in binding ssDNA (13). We, therefore, proposed that hydroxyl substitution of the C69S/C71S double mutant for sulfhydryl would change the DNA binding mode. Moreover, the highly conserved aromatic residues His76, Phe78, Phe89, and His91 of CSD play an important role in the correct folding of CRHSP-24. Unlike the high expression and solubility of wild-type human CRHSP-24, substitutions of these aromatic residues by alanine resulted in low protein expression and acute precipitation, as demonstrated by the different peak profiles in size exclusion chromatography (Fig. 3B).
Further molecular docking analysis showed that residues 41–43 are partially ordered, with the side chain of Leu43 in molecules A and C forming van der Waals contacts with Phe76, Phe89, and His91, and it occupies a similar site as T2 of dT6 in the Bs-CspB·dT6 complex (Fig. 1C). Ser41 is just visible in the crystal structure of molecule A, indicating that residues 41–43 pack loosely into the ligand binding subsite of human CRHSP-24. Ser41 is known to be phosphorylated by a DYRK isoform in serum-fed or -starved cells (21). A phosphomimetic mutation of Ser41 to Asp resulted in complete loss of human CRHSP-24 binding ability whether in the oxidative state or not (Figs. 4, D and E). Combined with the structural observation, these results suggest a mechanism in which the phosphorylation of Ser41 together with the side chain of Leu43 fix clasp-like onto the ligand binding subsite of CSD and regulate CRHSP-24 binding to mRNA.
CRHSP-24 Localizes Stress Granules Possibly via an Interaction with mRNA in SG
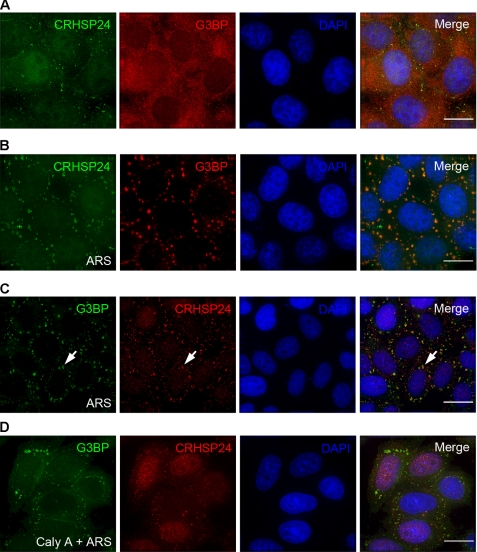
SG assembly is promoted by a subset of RNA-binding proteins, many of which are able to aggregate as well as bind RNA (27). Based on the crystal structure of CRHSP-24, particularly the polymeric state under oxidative conditions in vivo and the nucleotide binding property in vitro, we speculate that CRHSP-24 may be involved in response to environmental stress by binding to stress granules. To probe for the cellular location of CRHSP-24, the cells were exposed to arsenite for 50 min followed by immunofluorescence staining with an anti-CRHSP-24 goat antibody and an anti-G3BP mouse antibody to mark stress granules. As shown in Fig. 5B, antibodies reacting with CRHSP-24 were localized to cytoplasmic dots and co-localized with G3BP-positive granules. To further validate the co-distribution profile between CRHSP-24 and G3BP, HeLa cells were transiently transfected to express GFP-CRHSP-24 followed by dual color imaging. As shown in Fig. 6A, GFP-CRHSP-24 co-localizes with G3BP, indicating that CRHSP-24 is a component of SGs. Notably, SGs stimulated in GFP-CRHSP-24 expression cells were larger in size and lower in quantity compared to the cells stained by CRHSP-24 antibody.
FIGURE 5.
CRHSP-24 localization to SGs is regulated by its dephosphorylation. A, localization of CRHSP-24 in HeLa cells without arsenite treatment is shown. Fixed HeLa cells were stained with anti-CRHSP-24 antibody (green) and anti-G3BP antibody (red). B, shown is a distribution profile of G3BP and CRHSP-24 on SGs. HeLa cells were treated with 0.5 mm arsenite (ARS) for 50 min before being fixed and stained for CRHSP-24 (green) and G3BP (red). DAPI staining (blue) indicates the location of nuclei. C, the cells were exposed to arsenite for 50 min, and anti-CRHSP-24 antibody (red) and anti-G3BP antibody (green) were used to stain HeLa cells. Antibodies directed against CRHSP-24 localized to cytoplasmic dots and co-localized with antibodies directed against G3BP in response to oxidative stress. White arrows indicate the location of representative SGs. DAPI staining (blue) indicates the location of nuclei. D, phosphatase inhibitor calyculin A (100 nm) was added to the culture medium of HeLa cells for 30 min before arsenite treatment for 50 min. Treated cells were fixed and stained with anti-CRHSP-24 antibody (red) and anti-G3BP antibody (green). CRHSP-24 localized to cytoplasmic dots with a minimal overlapping with G3BP. The scale bar represents 5 μm.
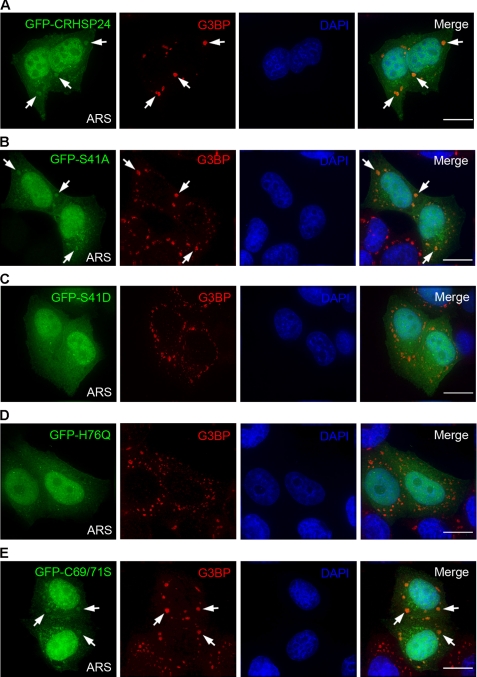
FIGURE 6.
Regulation of CRHSP-24 distribution to stress granules. A, GFP-CRHSP-24 was transfected into HeLa cells, and the cells were exposed to arsenite (ARS) for 50 min. In transfected cells, GFP-CRHSP-24 (green) co-localized with G3BP (red) in SGs. White arrows indicate the presence of SGs in transfected cells. DAPI staining (blue) indicates the location of nuclei. B, shown is non-phosphorylate-able GFP-S41A mutant (green) localized to SGs (red). C, phospho-mimicking GFP-S41D mutant (green) failed to localize to SGs (red). D, GFP-H76Q mutant (green) was not detected in SGs (red). E, disulfide bond-forming-deficient mutant C69S/C71S (green) localized to SGs (red). The scale bar represents 5 μm.
To pinpoint the structural elements responsible for localizing CRHSP-24 to SGs, HeLa cells were transiently transfected to express GFP-CRHSP-24 and its mutants. Interestingly, neither phosphomimetic S41D nor H76Q mutant, which breaks the base-stacking interaction with thymine-rich sequences in vitro, localizes to SGs in response to oxidative stress (Figs. 6, C and D). However, the C69S/C71S mutant with disrupted intermolecular disulfide bond formation still localized to SGs (Fig. 6E), suggesting that polymerization of CRHSP-24 seems not to be essential for localization to SGs. Incidentally, CRHSP-24 could not interact with G3BP by immunoprecipitation, and we could not find interaction of CRHSP-24 with any other proteins previously reported to be components of SGs (data not shown).
S41D and H76Q also had an obvious defect in binding with h33DNA as described previously, indicating that CRHSP-24 may participate in SGs via interaction with mRNA in stress granules. Additionally, the C69S/C71S mutant did localize to stress granules, which lends further support to the hypothesis that CRHSP-24 can localize to stress granules via interaction with mRNA, as the C69S/C71S could still bind to h33DNA in vitro.
Phosphorylation Regulates CRHSP-24 Localization to Stress Granules
CRHSP-24 is phosphorylated on at least four serine sites under basal conditions (16), and phosphomimetic mutant S41D did not localize to SGs, suggesting that dephosphorylation is required of CRHSP-24 localizing to SGs. To verify this hypothesis, HeLa cells were treated with 100 nm calyculin A, a PP4/PP2A inhibitor, for 30 min before exposure to arsenite for 50 min. After the treatments, the cells were then fixed and stained with anti-CRHSP-24 antibody and counter-stained for stress granules marker G3BP. As shown in Fig. 5D, calyculin A treatment inhibited the localization of CRHSP-24 on the SGs. However, as a control, antibodies directed against CRHSP-24 co-localize with antibodies against G3BP without calyculin A treatment (Fig. 5C), suggesting that phosphorylation may be a negative regulator for controlling CRHSP-24 localization to the stress granules. The calyculin A-regulated CRHSP-24 dynamics also suggest that phosphorylation of Ser41 may represent a potential target for phosphatases PP2A or PP4 (22).
Interestingly, CRHSP-24 remains localized to SGs in cyclosporine A-treated cells, suggesting that calcineurin is not responsible for the dynamics of CRHSP-24 phosphorylation and stress granule localization (supplemental Fig. S5). In fact, it was reported that Ser30 and Ser32 are the primary sites responsible for calcineurin dephosphorylation (22), and we propose that Ser41 is the candidate phosphorylation-regulated site responsible for CRHSP-24 localization to SGs, whereas Ser30 and Ser32 are not involved.
To test this hypothesis, we created phospho-mimicking and non-phosphorylate-able mutants of CRHSP-24 and expressed them in HeLa cells by transient transfection. As shown in Figs. 4, D and E, the phosphomimetic mutant CRHSP-24S41D (change Ser to Asp) resulted in a complete loss of human CRHSP-24 binding ability with h33DNA in vitro regardless of the oxidative or reducing state (supplemental Figs. S3 and S4). In contrast, the non-phosphorylate-able mutant CRHSP-24S41A to remains localized to the SGs (Fig. 6B). Combined with observations from the structure, the results suggest a mechanism whereby phosphorylation of Ser41 in the ligand binding subsite of CSD can prevent CRHSP-24 binding to mRNA and then block its localization to SGs in vivo.
To examine whether CRHSP-24 is important for SG assembly, siRNA oligonucleotide duplexes to CRHSP-24 were transfected into HeLa cells to suppress CRHSP-24 protein expression. Trial experiments revealed that treatment of HeLa cells with 150-nm siRNA for 48 h produced an optimal suppression of CRHSP-24 (supplemental Fig. S1). This suppression is relatively specific, as it did not alter the levels of other proteins such as tubulin. Interestingly, suppression of CRHSP-24 does not alter the assembly of SGs judged by G3BP labeling, suggesting that CRHSP-24 might not be necessary for assembly of SGs (data not shown).
CRHSP-24 Associates with mRNA P-bodies
CSPs in prokaryotes are small in size (7–10 kDa) and have been shown to be sufficient for nucleic acid binding and cold shock response by functioning as an RNA chaperone (28). The CSD, with high similarity to CSP, comprises 65–75 amino acid residues and is capable of binding RNA, single-stranded DNA, and double-stranded DNA in eukaryotes (29). Consistent with mRNA binding activity, CRHSP-24 contains a cold-shock domain flanked on each side by RNA binding motifs (amino acids 73–91), suggesting that the protein may act as a translational regulatory molecule (30). Moreover, CSD-containing eukaryotic Y-box protein 1 had also been found to localize to P-bodies and SGs (31).
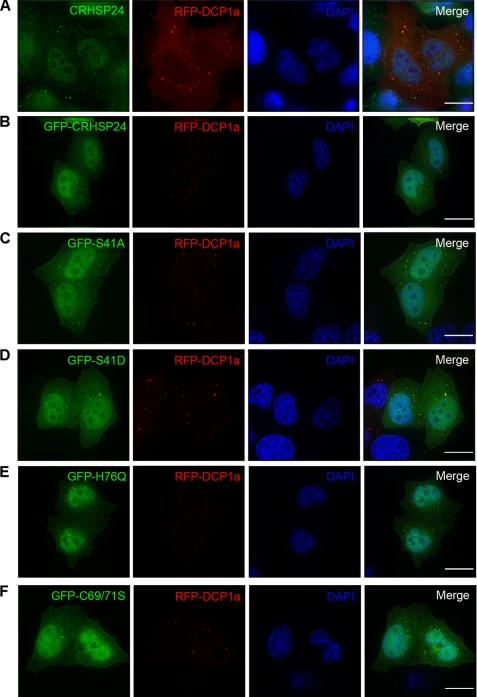
To examine the cellular location of CRHSP-24, DCP1a, which has widely been used as a marker of mRNA P-bodies, was transfected into HeLa cells, and the cells were stained with anti-CRHSP-24 antibody. In cells expressing RFP-DCP1a, antibodies reacting with CRHSP-24 appeared as cytoplasmic dots that were co-localized with RFP-DCP1a (Fig. 7A). In addition, overexpression of GFP-CRHSP-24 was found to localize to mRNA P-bodies (Fig. 7B).
FIGURE 7.
Structure-functional analyses of CRHSP-24 localization to the P-body. A, shown is endogenous CRHSP-24 marked in green localized to discrete, dot-like structures labeled with RFP-DCP1a (red). B, shown is GFP-CRHSP-24-WT localized to cytoplasmic dots (green) superimposed onto that of P-bodies labeled by RFP-DCP1a (red). DAPI staining (blue) indicates the location of nuclei. C, non-phosphorylate-able GFP-S41A (green) localizes to P-bodies (red). D, phosphomimetic mutant GFP-S41D (green) also localizes to mRNA processing bodies (red). E, H76Q, which breaks the base stacks interaction with thymine-rich sequence (green), co-localizes with P-bodies (red). F, disulfide bond-deficient mutant C69S/C71S (green), resulting in the loss of polymeric form, remains localized to P-bodies (red). DAPI staining (blue) indicates the location of nuclei. The scale bar represents 5 μm.
To pinpoint the structural elements essential for CRHSP-24 location to the P-bodies, HeLa cells were transiently transfected to express GFP-CRHSP-24 and its mutants together with RFP-DCP1a. As shown in Figs. 7, D and E, the GFP-CRHSP-24 S41D and H76Q mutants did co-localize with RFP-DCP1a in the physiological state. In addition, C69S/C71S mutant was found localized to P-bodies (Fig. 7F). These data show that the phosphorylation and nucleotide binding ability of CRHSP-24 are not necessary for localization to P-bodies, indicating that CRHSP-24 should localize to P-bodies via another mechanism other than binding mRNA, such as interacting with particular proteins preassembled in the P-bodies.
To identify the potential impact of CRHSP-24 on the assembly of P-bodies, we attempted to suppress CRHSP-24 protein level by transfecting siRNA oligonucleotide duplexes of CRHSP-24 into HeLa cells. Although the siRNA induced a remarkable suppression of CRHSP-24 protein synthesis, P-body assembly and organization exhibits no apparent changes, suggesting that CRHSP-24 is not essential for the P-body core complex assembly (data not shown).
DISCUSSION
The CSD is an evolutionarily conserved structure module that exhibits nucleic acid binding activity including RNA, ssDNA, and dsDNA. Our crystallographic analyses have revealed the atomic structure of CRHSP-24 and mapped the nucleic acid binding activity. Interestingly, phosphorylation of Ser41 regulates CRHSP-24 trafficking between SGs and P bodies. This new class of phosphorylation-regulated interaction between the CSD and nucleic acid is unique in SG plasticity. In fact, computational analyses indicate that CRHSP-24 contains a unique CXCXC motif that is absent from other CSD proteins. The CXCXC motif orchestrates intermolecular disulfide bonding essential for CRHSP-24 oligomerization and subsequent stress granule assembly process. We reason that CRHSP-24 is a dynamic regulator for SGs and P bodies.
To date many structures of ssRNA binding domain complexes have been determined, and the RNA binding surfaces of ssRNA binding domain are almost always located on one surface. In general, ssRNA binding domain-RNA binding interfaces are in close proximity for binding RNA, e.g. the structures of Trypanosoma brucei MRP1/MRP2 guide the RNA binding complex (32) (PDB code 2gje) and sex-lethal proteins complexed with single-stranded RNA (33). However, the two CSD domains of CRHSP-24 are located in the outer layer in the dimer that exists in solution (Fig. 2C). In other words, once ssDNA or ssRNA binds with this CRHSP-24 dimer, it may “bend” over to interact with the protein-DNA/RNA binding surface across the whole dimer.
Interestingly, among the four molecules in one asymmetric unit in the crystal structure, Leu43 in molecules A and C occupies a similar site to T2 of dT6 in the Bs-CspB·dT6 complex (Fig. 1C), whereas Leu43 in molecules B and D does not (supplemental Fig. S2). Therefore, we can infer that molecules B and D may bind with nucleotides, whereas molecules A and C may not. At the same time, the phosphorylation of Ser41 regulates nucleic acid binding by molecules B and D.
CRHSP-24 is a paralog of the brain-specific protein, PIPPin (Fig. 1A), which has been shown to bind to the 3′-untranslated region of replacement histone H1° and H3.3 mRNAs (20) and inhibit their translation (18). Our results show that CRHSP-24 can bind to the h33DNA, a 33-nt-length ssDNA that completely corresponds to the 3′-untranslated region of H3.3 mRNA and that features a thymine (T)-rich sequence via its cold shock domain. Furthermore, a subset of PIPPin found in the nucleus may play a role during the early stages of mRNA synthesis within chromatin structures (34). However, CRHSP-24 distributes in the cytoplasm and acts on SGs to store mRNAs, which possibly have uracil (U)-rich sequences, during environmental stress and P-bodies to regulate mRNA in physiological surroundings. Thus CRHSP-24 and PIPPin can form a nucleo-cytoplasmic shuttle to regulate mRNA.
P-bodies and SGs are distinct subcellular structures but act in concert in response to stress responses in mammalian cells (35). Furthermore, mRNAs may be delivered from SGs to P-bodies for decay (36). Thus, SGs act as intermediates between polysomes and P-bodies and may sort and modulate the increased flow of untranslated mRNA that accompanies stress (37). CRHSP-24 localizes to both SGs and P-bodies, and its mRNA binding ability is essential for localization to SGs but not to P-bodies. Therefore, CRHSP-24 may possibly flow between SGs and P-bodies to accomplish the protection and degradation of mRNAs in a phosphorylation-dependent manner.
SG assembly is promoted by a subset of several RNA-binding proteins, many of which are able to aggregate as well as bind RNA (27). For example, a prion-like aggregation has been demonstrated for TIA-1 (38) and cytoplasmic polyadenylation element-binding protein (39), whereas G3BP possesses an oligomerization domain that is regulated by phosphorylation (40). Survival motor neuron protein (41) self-aggregation appears mediated by its Y-G box, whereas the aggregation of FMRP (42) family members appears mediated by a coiled-coil domain. Interestingly, CRHSP-24 exhibits a polymeric form under oxidative conditions both in vivo and in vitro and localizes to stress granules via intermolecular disulfide bonds. Thus, CRHSP-24 has a brand new aggregated form to participate in stress granule assembly.
Our current data show that CRHSP-24 is a component of stress granules, which are involved in mRNA binding and stabilization in response to stress. The protected mRNA can soon be recovered after the removal of stress. In fact, a recent publication appeared online during our revision that supports our working model (44). We speculate that oligomerization of CRHSP-24 facilitates the assembly of stress granules and perhaps mRNA stability protection. To this end, it would be of great interest and challenge to visualize mRNA dynamics and stability in the regulated setting of CRHSP-24 plasticity and dynamics. This can be achieved with supra-resolution imaging techniques in real time.
Taken together, our study revealed atomic structure and novel function of CRHSP-24 in the trafficking between SGs and P-bodies. There are many other unresolved and interesting issues with regard to its precise function in SG dynamics and plasticity. It will be important, perhaps by following single CRHSP-24 and mRNA molecules, to determine the pathways that CRHSP-24 and mRNA follow between these different subcellular compartments and how those transitions are modulated in an mRNA- and CRHSP-24-specific manner to affect either translation or degradation of the mRNA. Finally, understanding the nature of the CRHSP-24 complexes that form within SGs and P-bodies is likely to provide great insight into a precise structure-function relationship of CRHSP-24 and the novel class of cargo adaptor trafficking between SGs and P-bodies.
Supplementary Material
Acknowledgments
We thank Dr. Thomas Mayo for kindly providing the DCP1a plasmid. We are grateful to staff members of the Structural Biology Core Facility in the Institute of Biophysics, Chinese Academy of Sciences, for excellent technical assistance.
This work was supported by Chinese Natural Science Foundation Grants 30900497 (to F. W.) and 90913016 (to X. Y.), Chinese Academy of Science Grants KSCX1-YW-R-65, KSCX2-YW-H-10, and KSCX2-YW-R-195, and Chinese 973 Project Grants 2007CB914503 and 2010CB912103 (to X. Y.), 2007CB914304 (to Z. R.), and 2009ZX10603 (to Z. R.).
The atomic coordinates and structure factors (code 3AQQ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures and Figs. S1–S5.
- CSD
- cold shock domain
- CSP
- cold-shock protein
- SPR
- surface plasmon resonance
- SG
- stress granule.
REFERENCES
- 1. Graumann P., Marahiel M. A. (1996) Bioessays 18, 309–315 [DOI] [PubMed] [Google Scholar]
- 2. Jiang W., Hou Y., Inouye M. (1997) J. Biol. Chem. 272, 196–202 [DOI] [PubMed] [Google Scholar]
- 3. Bae W., Xia B., Inouye M., Severinov K. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7784–7789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Davies H. G., Giorgini F., Fajardo M. A., Braun R. E. (2000) Dev. Biol. 221, 87–100 [DOI] [PubMed] [Google Scholar]
- 5. Lu Z. H., Books J. T., Ley T. J. (2005) Mol. Cell. Biol. 25, 4625–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Evdokimova V., Ruzanov P., Imataka H., Raught B., Svitkin Y., Ovchinnikov L. P., Sonenberg N. (2001) EMBO J. 20, 5491–5502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nekrasov M. P., Ivshina M. P., Chernov K. G., Kovrigina E. A., Evdokimova V. M., Thomas A. A., Hershey J. W., Ovchinnikov L. P. (2003) J. Biol. Chem. 278, 13936–13943 [DOI] [PubMed] [Google Scholar]
- 8. Jurchott K., Bergmann S., Stein U., Walther W., Janz M., Manni I., Piaggio G., Fietze E., Dietel M., Royer H. D. (2003) J. Biol. Chem. 278, 27988–27996 [DOI] [PubMed] [Google Scholar]
- 9. Sorokin A. V., Selyutina A. A., Skabkin M. A., Guryanov S. G., Nazimov I. V., Richard C., Th'ng J., Yau J., Sorensen P. H., Ovchinnikov L. P., Evdokimova V. (2005) EMBO J. 24, 3602–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kohno K., Izumi H., Uchiumi T., Ashizuka M., Kuwano M. (2003) Bioessays 25, 691–698 [DOI] [PubMed] [Google Scholar]
- 11. Lopez M. M., Yutani K., Makhatadze G. I. (1999) J. Biol. Chem. 274, 33601–33608 [DOI] [PubMed] [Google Scholar]
- 12. Zeeb M., Balbach J. (2003) Protein Sci. 12, 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Max K. E., Zeeb M., Bienert R., Balbach J., Heinemann U. (2006) J. Mol. Biol. 360, 702–714 [DOI] [PubMed] [Google Scholar]
- 14. Tafuri S. R., Wolffe A. P. (1992) New Biol. 4, 349–359 [PubMed] [Google Scholar]
- 15. Kloks C. P., Spronk C. A., Lasonder E., Hoffmann A., Vuister G. W., Grzesiek S., Hilbers C. W. (2002) J. Mol. Biol. 316, 317–326 [DOI] [PubMed] [Google Scholar]
- 16. Groblewski G. E., Yoshida M., Bragado M. J., Ernst S. A., Leykam J., Williams J. A. (1998) J. Biol. Chem. 273, 22738–22744 [DOI] [PubMed] [Google Scholar]
- 17. Wishart M. J., Dixon J. E. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2112–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Castiglia D., Scaturro M., Nastasi T., Cestelli A., Di Liegro I. (1996) Biochem. Biophys. Res. Commun. 218, 390–394 [DOI] [PubMed] [Google Scholar]
- 19. Nastasi T., Muzi P., Beccari S., Bellafiore M., Dolo V., Bologna M., Cestelli A., Di Liegro I. (2000) Neuroreport 11, 2233–2236 [DOI] [PubMed] [Google Scholar]
- 20. Nastasi T., Scaturro M., Bellafiore M., Raimondi L., Beccari S., Cestelli A., di Liegro I. (1999) J. Biol. Chem. 274, 24087–24093 [DOI] [PubMed] [Google Scholar]
- 21. Auld G. C., Campbell D. G., Morrice N., Cohen P. (2005) Biochem. J. 389, 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee S., Wishart M. J., Williams J. A. (2009) Biochem. Biophys. Res. Commun. 385, 413–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jönsson U., Fägerstam L., Ivarsson B., Johnsson B., Karlsson R., Lundh K., Löfås S., Persson B., Roos H., Rönnberg I. (1991) Biotechniques 11, 620–627 [PubMed] [Google Scholar]
- 24. Laskowski R. A., Macarthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 25. Yao X., Abrieu A., Zheng Y., Sullivan K. F., Cleveland D. W. (2000) Nat. Cell Biol. 2, 484–491 [DOI] [PubMed] [Google Scholar]
- 26. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 27. Kedersha N., Anderson P. (2007) Methods Enzymol. 431, 61–81 [DOI] [PubMed] [Google Scholar]
- 28. Graumann P. L., Marahiel M. A. (1998) Trends Biochem. Sci. 23, 286–290 [DOI] [PubMed] [Google Scholar]
- 29. Lorković Z. J., Barta A. (2002) Nucleic Acids Res. 30, 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schäfer C., Steffen H., Krzykowski K. J., Göke B., Groblewski G. E. (2003) Am. J. Physiol. Gastrointest Liver Physiol. 285, G726–G734 [DOI] [PubMed] [Google Scholar]
- 31. Yang W. H., Bloch D. B. (2007) RNA 13, 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schumacher M. A., Karamooz E., Zíková A., Trantírek L., Lukes J. (2006) Cell 126, 701–711 [DOI] [PubMed] [Google Scholar]
- 33. Handa N., Nureki O., Kurimoto K., Kim I., Sakamoto H., Shimura Y., Muto Y., Yokoyama S. (1999) Nature 398, 579–585 [DOI] [PubMed] [Google Scholar]
- 34. Bono E., Compagno V., Proia P., Raimondi L., Schiera G., Favaloro V., Campo V., Donatelli M., Di Liegro I. (2007) Endocrinology 148, 252–257 [DOI] [PubMed] [Google Scholar]
- 35. Anderson P., Kedersha N. (2006) J. Cell Biol. 172, 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Anderson P., Kedersha N. (2009) Nat. Rev. Mol. Cell Biol. 10, 430–436 [DOI] [PubMed] [Google Scholar]
- 37. Coller J., Parker R. (2005) Cell 122, 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L. M., Anderson P. (2004) Mol. Biol. Cell 15, 5383–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wilczynska A., Aigueperse C., Kress M., Dautry F., Weil D. (2005) J. Cell Sci. 118, 981–992 [DOI] [PubMed] [Google Scholar]
- 40. Tourrière H., Chebli K., Zekri L., Courselaud B., Blanchard J. M., Bertrand E., Tazi J. (2003) J. Cell Biol. 160, 823–831 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41. Hua Y., Zhou J. (2004) FEBS Lett. 572, 69–74 [DOI] [PubMed] [Google Scholar]
- 42. Mazroui R., Huot M. E., Tremblay S., Filion C., Labelle Y., Khandjian E. W. (2002) Hum Mol. Genet 11, 3007–3017 [DOI] [PubMed] [Google Scholar]
- 43. Diederichs K., Karplus P. A. (1997) Nat. Struct. Biol. 4, 269–275 [DOI] [PubMed] [Google Scholar]
- 44. Pfeiffer J. R., McAvoy B. L., Fecteau R. E., Deleault K. M., Brooks S. A. (2011) Mol. Cell. Biol. 31, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.