Abstract
Toll-like receptors (TLRs) direct a proinflammatory program in macrophages. One mediator whose generation is induced by TLR ligation is prostaglandin E2 (PGE2), which is well known to increase intracellular cAMP upon G protein-coupled receptor ligation. How PGE2/cAMP shapes the nascent TLR response and the mechanisms by which it acts remain poorly understood. Here we explored PGE2/cAMP regulation of NO production in primary rat alveolar macrophages stimulated with the TLR4 ligand LPS. Endogenous PGE2 synthesis accounted for nearly half of the increment in NO production in response to LPS. The enhancing effect of PGE2 on LPS-stimulated NO was mediated via cAMP, generated mainly upon ligation of the E prostanoid 2 receptor and acting via protein kinase A (PKA) rather than via the exchange protein activated by cAMP. Isoenzyme-selective cAMP agonists and peptide disruptors of protein kinase A anchoring proteins (AKAPs) implicated PKA regulatory subunit type I (RI) interacting with an AKAP in this process. Gene knockdown of potential RI-interacting AKAPs expressed in alveolar macrophages revealed that AKAP10 was required for PGE2 potentiation of LPS-induced NO synthesis. AKAP10 also mediated PGE2 potentiation of the expression of cytokines IL-10 and IL-6, whereas PGE2 suppression of TNF-α was mediated by AKAP8-anchored PKA-RII. Our data illustrate the pleiotropic manner in which G protein-coupled receptor-derived cAMP signaling can influence TLR responses in primary macrophages and suggest that AKAP10 may coordinate increases in gene expression.
Keywords: Cyclic AMP (cAMP), Cytokines Induction, Nitric-oxide Synthase, Prostaglandins, Toll-like Receptors (TLR), A-kinase Anchoring Protein
Introduction
Tissue macrophages play a pivotal role in innate immune responses. Via recognition receptors such as Toll-like receptors (TLRs),3 these sentinel cells sense pathogens and other danger signals and then initiate a coordinated inflammatory response (1). The best-studied example is the recognition by TLR4 of LPS of Gram-negative bacterial cell walls. TLR-mediated inflammation is orchestrated by the activation of transcription factors including NFκB (2). NFκB-dependent genes that are critical to antimicrobial defense include proinflammatory cytokines and iNOS (3). NO is an important signaling molecule involved in regulating a wide range of biological activities in the neural, vascular, and immune systems. NO and its metabolites mediate a number of host defense functions of activated macrophages, including antimicrobial and tumoricidal activities, but are also implicated in the pathogenesis of tissue damage associated with acute and chronic inflammation (4).
Ligation of TLRs also enhances the expression of COX-2, the enzyme responsible for the inducible generation of prostanoids including prostaglandin E2 (PGE2) (5, 6). PGE2 is produced in abundance at sites of inflammation and infection and itself modulates many aspects of macrophage function (7). The immunomodulatory effects of PGE2 largely result from its ability to increase intracellular cAMP through the stimulatory G protein (Gs)-coupled E prostanoid (EP) receptors EP2 and EP4 (8). Increases in intracellular cAMP are transduced into cellular responses via the activation of two downstream effector molecules, cAMP-dependent protein kinase A (PKA) and the exchange protein directly activated by cAMP-1 (Epac-1) (9, 10). Specificity in PKA signaling exists at both biochemical and spatial levels. Type I and type II isozymes of PKA (PKA-I and -II, respectively) display different biochemical properties in their cAMP binding regulatory (R) subunits. Additionally, cAMP signaling can be compartmentalized by the actions of a family of scaffold proteins termed protein kinase A anchoring proteins (AKAPs), which assemble PKA holoenzymes into multiprotein signaling complexes at specific intracellular sites. Most AKAPs preferentially interact with RII subunits (11).
Increases in intracellular cAMP generally suppress innate immune functions of macrophages, including the generation of inflammatory mediators such as TNF-α and the phagocytosis and killing of microbes (12–17). However, depending on the cell type investigated, the PGE2/EP2-EP4/cAMP/PKA cascade has been shown to enhance, inhibit, or exert no effect on iNOS expression (18, 19). As infections of the lung are associated with greater morbidity, mortality, and economic cost than those of any other organ, it can be argued that no resident macrophage population plays as crucial a role in innate immunity as the alveolar macrophage (AM) (20). In this study we investigated the role of endogenous PGE2-cAMP signaling in LPS-induced NO production in resident rat AMs. We identify an autocrine amplification loop for NO synthesis composed of PGE2, EP2, cAMP, and PKA. Interestingly, whereas PGE2/cAMP inhibits TNF-α via a PKA-II interaction with AKAP8, enhancement of NO generation proceeds via a less common type of interaction between PKA-I and a specific AKAP, namely AKAP10.
EXPERIMENTAL PROCEDURES
Animals and Reagents
Pathogen-free female Wistar rats weighing 125–150 g (Charles River Laboratories) were utilized as a source for AMs and treated according to National Institutes of Health guidelines for the use of experimental animals with the approval of the University of Michigan Committee for the Use and Care of Animals.
RPMI 1640 culture medium and penicillin/streptomycin/amphotericin B solution were purchased from Invitrogen. Escherichia coli (055:B5) LPS and SDS were from Sigma. PKA inhibitors KT5720 and myristoylated PKI peptide (14–22) were purchased from Enzo Life Sciences (Plymouth Meeting, PA). Dibutyryl cAMP (N6,2′-O-dibutyryladenosine 3′:5′-cAMP) was from Calbiochem. The PKA-specific cAMP analog 6-Bnz-cAMP (N6-benzoyladenosine-3′,5′-cyclic monophosphate), Epac-specific cAMP analog 8-pCPT-2-O-Me-cAMP (8–4-chlorophenylthio)-2′-O-methyladenosine-3′,5′-cyclic monophosphate), PKA RI-selective activator 2-Cl-8-MA-cAMP (2-chloro-8-methylaminoadenosine-3′, 5′-cyclic monophosphate), and PKA RII-selective activator 6-MBC-cAMP (N6-mono-t- butylcarbamoyladenosine-3′, 5′-cyclic monophosphate) were purchased from Biolog Life Science Institute (Howard, CA). The selective EP2 receptor agonist butaprost free acid, EP2 receptor antagonist AH6809, prostacyclin (PGI2) agonists iloprost and treprostinil, I prostanoid (IP) antagonists CAY 10449 and CAY 10441, and PGE2 were purchased from Cayman Chemicals (Ann Arbor, MI). EP4 receptor agonist (ONO-AE1-329) and EP4 receptor antagonist (ONO-AE3-208) were generous gifts from Ono Pharmaceutical Co., Ltd. (Osaka, Japan). The RII/AKAP disruptor peptide Ht31 was obtained from Promega (Madison, WI). The RI/AKAP disruptor peptide RIAD and the corresponding control peptide scRIAD were purchased from Anaspec (San Jose, CA). Required dilutions of all compounds were prepared immediately before use, and equivalent quantities of vehicle were added to the appropriate controls. Experimental compounds showed no adverse effects on cell viability as determined by LDH release (data not shown).
Cell Isolation, Culture, and Ligand Treatment
Resident rat AMs were obtained by ex vivo lung lavage, as previously described (21) and resuspended in RPMI 1640. Cells were allowed to adhere for 1 h (37 °C, 5% CO2), and after a single wash with warm RPMI, >99% of adherent cells were identified as AMs by use of a modified Wright-Giemsa stain (Diff-Quik; American Scientific Products, McGraw Park, IL). Cells were cultured overnight in RPMI 1640 containing 10% FBS and 1% penicillin/streptomycin/amphotericin B. Cells were washed twice the next day with warm medium so that nonadherent cells could be removed, and the medium was changed to RPMI without serum. Cells were treated with compounds of interest at the concentrations and times indicated in the figure legends and then cultured for an additional 24 h in the presence or absence of LPS or, in selected experiments, the TLR2 agonist peptidoglycan (1 μg/ml), before harvesting.
Measurement of Nitrite, TNF-α, IL-10, and IL-6
AMs were cultured and stimulated as described above, and after 24 h of incubation, cell-free supernatants were harvested. Aliquots were prepared and frozen until ready for nitrite and cytokine analysis. To evaluate NO production, nitrite concentration in the supernatants of AM cultures was measured using the standard Griess reaction (Cayman Chemicals) (22). TNF-α (eBioscience, San Diego, CA), IL-10, and IL-6 (both from R&D Systems, Minneapolis, MN) were measured by ELISA according to the manufacturers' instructions.
Western Blotting
Freshly harvested AMs were lysed in buffer (50 mm Tris-HCl (pH 7.4), 25 mm KCl, 5 mm MgCl2, and 0.2% Nonidet P-40) supplemented with protease inhibitors (Roche Diagnostics). For immunoblot analysis, protein samples (30 μg) were mixed with loading buffer (50 mm Tris HCl (pH 6.8), 2% SDS, 100 mm DTT, 10% glycerol, and 0.1% bromphenol blue), boiled, applied to 10% SDS-polyacrylamide gels, and subjected to electrophoresis. The separated proteins were transferred to nitrocellulose membranes. After transfer, membranes were blocked in 5% milk TTBS for 1 h and probed with respective primary antibodies (iNOS, 1:1000, Assay Designs; GAPDH, 1:500, Santa Cruz Biotechnology, Santa Cruz, CA). Antibodies recognizing the regulatory and catalytic subunits of PKA (PKA-RIα, PKA-RIIα, PKA-RIIβ, and PKA Cα and Cβ) were from BD Biosciences, whereas that for PKA-RIβ was from Chemicon (Temecula, CA); all were used at 1:500. Antibody incubations were for 2 h at room temperature. Bound primary antibodies were visualized with appropriate secondary antibody conjugated to horseradish peroxidase and developed with ECL reagent (Amersham Biosciences). Relative band densities were determined by densitometric analysis using National Institutes of Health Image software, and the ratio of iNOS to that of GAPDH was calculated. In all instances, density values of bands were corrected by subtraction of the background values.
RNA Isolation and Quantitative Real Time RT-PCR
AMs were plated at 2 × 106 cells/well in 6-well plates as described (12). RNA was solubilized in 1 ml of TRIzol and extracted according to the manufacturer's instructions. RNA was quantitated on a spectrophotometer at a wavelength of 260 nm, transcribed to cDNA, and amplified by quantitative real-time RT-PCR performed on an ABI Prism 7000 Thermocycler (Applied Biosystems, Carlsbad, CA). β-Actin was used as a housekeeping gene. Primer efficiency analysis was performed on all primers and ranged from 97 to 108%. Electrophoretic separation of each RT-PCR product yielded a single fragment of the expected size. (23) The average of the control sample was set to 1 for each experiment, and the relative gene expression for each experimental sample was compared with that of the lowest expressed AKAP, AKAP1.
RNA Interference
RNA interference was performed according to a protocol provided by Dharmacon (Lafayette, CO). Rat AMs were transfected using DharmaFECT 1 reagent with 30 nm nonspecific control or specific ON-TARGET SMARTpool AKAP 8, AKAP 10, and AKAP 11 siRNAs from Dharmacon. After 48 h of transfection, AMs were harvested for mRNA isolation or pretreated with PGE2 for 10 min before LPS challenge for 24 h and subsequent analysis of NO, TNF-α, IL-10, and IL-6 secretion.
Statistical Analysis
Data are represented as the mean ± S.E. Statistical differences between groups were determined by one-way ANOVA followed by the Bonferroni test using GraphPad Prism Software (San Diego, CA). Differences were considered significant if p < 0.05.
RESULTS
Effect of Endogenous and Exogenous PGE2 on TLR4-induced iNOS Expression and NO Production
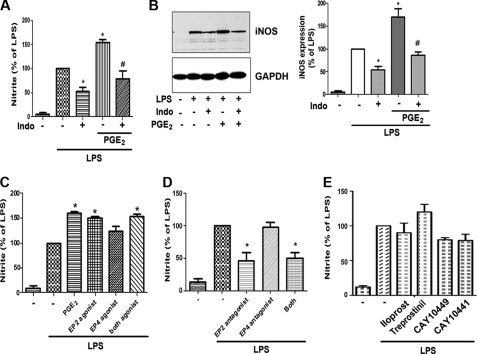
To determine the role of endogenous prostanoids in LPS-induced NO production and iNOS expression in rat AMs, cells were challenged with LPS (100 ng/ml) in the absence or presence of the COX-1/2 inhibitor indomethacin (5 μm). Indomethacin significantly reduced LPS-induced production of NO (measured as nitrite secretion) (Fig. 1A) and expression of iNOS protein (Fig. 1B), implicating some endogenous prostanoid as potentiating NO and iNOS synthesis. The capacity of PGE2 to accomplish this was tested by the addition of exogenous reagent PGE2, which was able to largely restore indomethacin-inhibited responses to the LPS-only level as well as to enhance LPS-induced responses in the absence of indomethacin (Fig. 1, A and B). Of note, iNOS expression was not induced by PGE2 in the absence of LPS (not shown). This suggests that rather than inducing iNOS expression directly, PGE2 potentiates the intracellular signaling events associated with cellular expression of iNOS enzyme in response to LPS. To determine whether the enhancement by PGE2 of LPS-induced NO production was unique for TLR4 stimulation, the TLR2 agonist peptidoglycan was employed. Exogenous PGE2 also modestly enhanced NO production in response to peptidoglycan (Fig. 2E).
FIGURE 1.
PGE2/EP2 signaling enhances LPS-induced NO production in AMs. A, AMs were pretreated with the COX inhibitor indomethacin (Indo, 5 μm) for 30 min followed by PGE2 (1 μm) for 10 min and then challenged with LPS (100 ng/ml) for another 24 h. Supernatants were harvested after 24 h, and nitrite was determined by the Griess reaction. Data are expressed as percent of LPS alone. B, left, AMs were incubated as in A, and cell lysates (30 μg of protein) were subjected to Western blot analysis of iNOS and GAPDH. Results from one experiment of three are shown. Right, relative expression of iNOS was determined by densitometric analysis of immunoblots from three different experiments, normalized for GAPDH expression, and expressed as percent of LPS alone. C, AMs were pretreated with PGE2 (1 μm), the EP2-specifc agonist butaprost free acid (1 μm), the EP4-specific agonist ONO-AE1-329 (1 μm), or both agonists for 10 min followed by LPS for 24 h, and nitrite was measured as in A. D, cells were incubated with the EP2-specific antagonist AH6809 (1 μm) or the EP4-specific antagonist ONO-AE3-208 (100 μm) or both antagonists for 30 min followed by LPS for 24 h, and nitrite was measured as in A. E, cells were pretreated for 30 min with the IP antagonists CAY 10449 and CAY 10441 (both 1 μm) or for 10 min with the PGI2 analogs iloprost and treprostinil (both 1 μm) before incubation for 24 h with LPS; nitrite was subsequently measured as in A. In all circumstances the data are the mean ± S.E. values of three separate experiments, each performed in duplicate. *, p < 0.05 versus LPS alone; #, p < 0.05 versus indomethacin alone.
FIGURE 2.
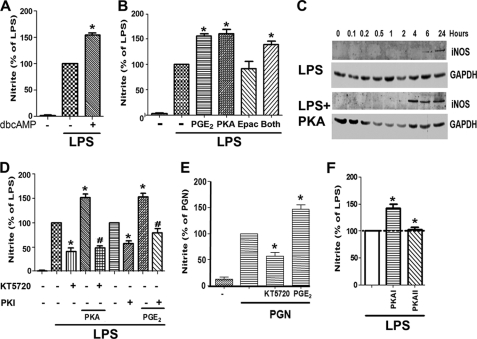
cAMP/PKA axis is responsible for the potentiation by PGE2 of LPS-induced NO production. A, AMs were pretreated with or without dibutyryl cAMP (dbcAMP, 1 μm) for 10 min followed by LPS for 24 h. B, AMs were pretreated for 10 min with PGE2, PKA-specific cAMP analog 6-Bnz-cAMP (500 μm), Epac-specific cAMP analog, 8-pCPT-2-O-Me-cAMP (500 μm), or both analogs followed by 24 h incubation with LPS. C, AMs were pretreated (bottom) or not (top) with the PKA agonist 6-Bnz-cAMP for 10 min followed by incubation with LPS for the indicated time points. Lysates (30 μg of protein) were subjected to Western blot analysis for iNOS and GAPDH. Results from one experiment of three are shown. D, AMs were incubated with the PKA inhibitors KT5720 (1 μm) or PKI amide (10 μm) for 30 min before the addition of PGE2 or 6-Bnz-cAMP for 30 min and subsequent 24 h incubation with LPS. E, AMs were incubated with KT5720 for 30 min or PGE2 for 10 min before treatment with peptidoglycan (PGN) (1 μg/ml) for 24 h. F, cells were incubated for 10 min with site-selective cAMP analogs to activate PKA type I (10 μm 2-Cl-8-MA-cAMP) or PKA type II (10 μm 6-MBC-cAMP) and then incubated with LPS for 24 h. In A, B, D, E, and F, supernatants were harvested after 24 h, and nitrite was determined. The data are the means ± S.E. values of 3–7 separate experiments, each performed in triplicate. *, p < 0.05 versus LPS alone; #, p < 0.05 versus PGE2 or PKA agonist alone.
We have reported that EP2 signaling is required for the inhibitory effect of PGE2 on phagocytosis (14), whereas both EP2 and EP4 mediate suppression of AM microbicidal activity (12). The importance of specific EP receptors in PGE2 modulation of inflammatory mediator production by LPS-stimulated AMs is unknown. We, therefore, challenged AMs with LPS in the presence or absence of an EP2-specific agonist (butaprost free acid) or an EP4-specific agonist (ONO-AE1-329). The EP2 agonist mimicked the effect of PGE2 to enhance LPS-induced NO secretion; the EP4 agonist also had a tendency to enhance LPS-induced NO secretion, but the effect was modest and non-significant (Fig. 1C). These data identified EP2 signaling as being capable of enhancing LPS-induced NO. Importantly, treatment of AMs with the EP2 antagonist AH-6809 attenuated LPS-induced NO secretion to the same extent as did indomethacin (Fig. 1D). By contrast, the EP4 antagonist ONO-AE3-208 had no such effect.
PGI2 is another COX-derived prostanoid that signals via a Gs-coupled receptor, termed IP. As shown in Fig. 1E, two antagonists of IP (CAY 10449 and CAY 10441) failed to significantly attenuate the LPS-induced NO response in the manner achieved by both indomethacin (Fig. 1A) and the EP2 antagonist (Fig. 1D); this argues against an important contribution of endogenous PGI2 in contributing to the LPS response. Moreover, exogenous addition of two stable analogs of PGI2, iloprost and treprostinil, was also tested. Neither had a significant potentiating effect above that of LPS alone (Fig. 1E), in contrast to exogenous PGE2. The small but non-significant increase in NO observed with treprostinil, but not iloprost, can likely be explained by our previous observation (23) that in AMs, treprostinil can ligate not only IP but also EP2; by contrast, iloprost is a pure IP agonist in these cells. These data clearly demonstrate that endogenous and exogenous PGE2 selectively potentiates LPS-induced NO secretion via EP2 receptor signaling.
PGE2 Enhancement of LPS-induced NO Production Is Mediated by PKA
Because EP2 generally signals via Gs-dependent increases in intracellular cAMP (8), we sought to verify that elevation of this second messenger was able to augment NO production. Indeed, the cell membrane-permeable cAMP analog dibutyryl cAMP potentiated LPS-induced NO secretion (Fig. 2A) in a manner similar to PGE2 itself. Modulation of macrophage functions by cAMP can be mediated by either PKA or Epac-1 (24). To delineate which cAMP effector amplifies TLR4 responses, we first employed cell membrane-permeable analogs specific for activation of PKA (6-Bnz-cAMP) or Epac-1 (8-pCPT-2-O-Me-cAMP). As shown in Fig. 2B, the PKA agonist enhanced LPS-induced NO secretion as much as PGE2, whereas the Epac agonist had no effect. No additive effect was observed when the two agonists were used together. The kinetics of iNOS induction by LPS in the absence or presence of the PKA agonist were examined by immunoblot analysis (Fig. 2C). In cells stimulated with LPS alone, iNOS protein was detectable at 6 h and increased at 24 h. When PKA agonist was included, iNOS expression was not only greater in magnitude than with LPS alone but also reached its maximal level at 4 h, indicating a marked increase in the rate of induction as well. To confirm that PKA is indeed responsible for the potentiation of NO generation by endogenous or exogenous PGE2, cells were treated with the PKA inhibitor KT5720 or with the cell-permeable PKA inhibitory peptide PKI. Both inhibitors attenuated LPS-induced NO to an extent similar to that observed with indomethacin or EP2 antagonist and also blocked the potentiating effects of exogenous PGE2 or PKA agonist on LPS-induced NO (Fig. 2D). Indeed, KT5720 also abrogated the increment in NO synthesis elicited by the TLR2 agonist peptidoglycan (Fig. 2E). Taken together, these results suggest that PKA is responsible for the amplification of NO formation by PGE2 in response to ligation of TLR4 and likely other TLRs as well.
Activation of PKA-I Enhances LPS-induced NO Production in an AKAP-dependent Manner
PKA consists of two R subunits, RI and RII, each of which exists in α and β isoforms, as well as two catalytic subunits, Cα and Cβ (11). The profile of PKA subunits in AMs is not known. By Western blotting of resting AMs, we observed robust expression of RIα, RIIα, RIIβ, Cα, and Cβ but only minimal expression of RIβ (supplemental Fig. 1), an expression profile identical to that reported for murine RAW 264.7 macrophages (25). To investigate the ability of PKA isoenzymes to modulate LPS-induced NO production, we employed isoenzyme-selective cAMP agonists to preferentially activate PKA-I (2-Cl-8-MA-cAMP) or PKA-II (6-MBC-cAMP) as described (26). As shown in Fig. 2F, selective activation of PKA-I, but not of PKA-II, enhanced LPS-induced NO production.
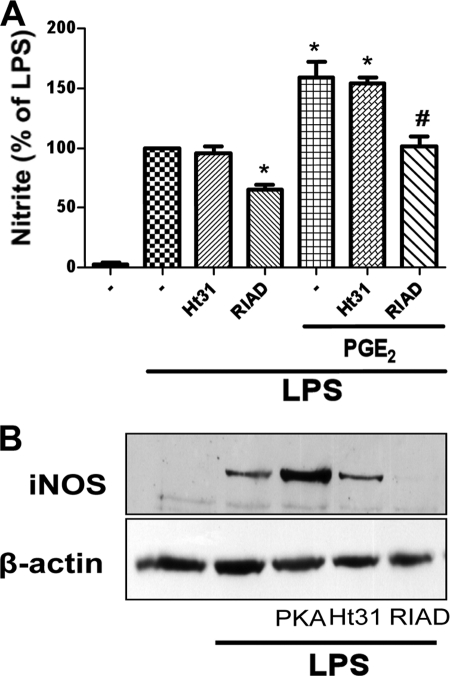
AKAPs serve as scaffold proteins to target PKA to specific microdomains and thereby enhance proximity to particular substrates (27). To determine whether AKAPs are involved in the PKA-dependent potentiation of LPS-induced NO production, we disrupted potential PKA RI-AKAP interactions with RIAD peptide (28) and potential PKA RII-AKAP interactions with Ht31 peptide (25, 29). Ht31 had no effect on LPS-induced NO production (Fig. 3A) or iNOS expression (Fig. 3B) or on PGE2 enhancement of LPS-induced NO (Fig. 3A). By contrast, RIAD attenuated LPS-induced NO production (Fig. 3A) as well as LPS-induced iNOS expression (Fig. 3B). It also prevented the enhancing effect of PGE2 on LPS-induced NO production (Fig. 3A). The RIAD control peptide scRIAD had no such effects (data not shown). These data confirm that the enhanced production of NO by endogenous and exogenous PGE2 depends on PKA-I and implicate an interaction between RI and an AKAP in this effect.
FIGURE 3.
PKA RI/AKAP interaction is required for PGE2 effects on LPS-induced iNOS expression and NO secretion. A, cells were pretreated for 20 min with or without the AKAP/PKA RII-specific disruptor peptide Ht31 (25 μm) or the AKAP/PKA RI-specific disruptor RIAD (25 μm) followed by PGE2 or vehicle for 10 min and then incubated with LPS for 24 h. Supernatants were harvested, and nitrite was determined. Data represent the mean ± S.E. values of 3–5 separate experiments, each performed in duplicate. *, p < 0.05 versus LPS alone; #, p < 0.05 versus PGE2 alone. B, AMs were pretreated for 20 min with Ht31, RIAD, or the PKA agonist 6-Bnz-cAMP followed by LPS for 24 h. Lysates were subjected to Western blot analysis of iNOS and β-actin. Results from one experiment of three are shown.
AKAP10 Is Necessary for PGE2 Enhancement of TLR4-induced NO Production
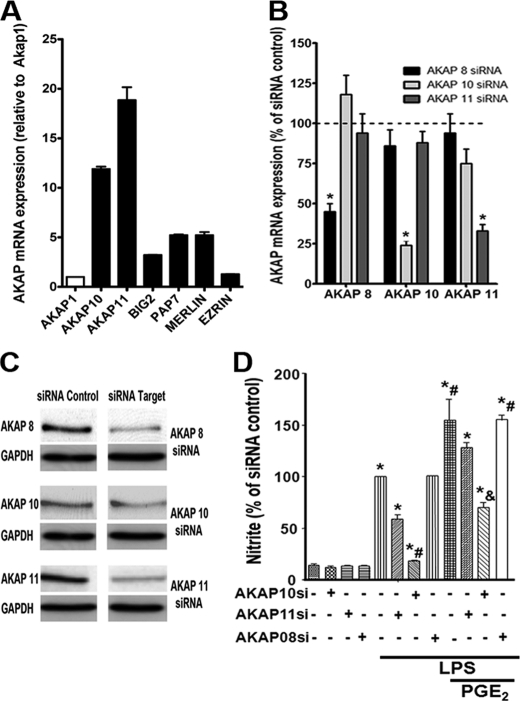
Because most AKAPs preferentially bind RII rather than RI, the suggestion from data in Fig. 3 that PGE2 potentiation of TLR4-mediated NO production is mediated by AKAP-anchored RI was unexpected, and we, therefore, sought to identify the AKAP responsible for this effect. Little is known about AKAP expression in macrophages. Wall et al. (25) identified 10 AKAPs expressed in the RAW264.7 murine macrophage cell line, but only two, AKAP1 (also known as d-AKAP1) and AKAP10 (also known as d-AKAP2), are dual function AKAPs known to interact with RI subunits. We utilized real time RT-PCR to assess the mRNA expression in AMs of these two AKAPs along with several others capable of interacting with RI. As shown in Fig. 4A, transcripts for AKAP10 and AKAP11 (also known as AKAP220) were more abundant than those encoding AKAP1, BIG2, PAP7, and Merlin, whereas that encoding Ezrin was barely detected. To determine the functional importance of the most abundant RI-interacting AKAPs in mediating PGE2 effects on LPS responses, we utilized gene silencing of AKAPs 10 and 11. As a control, we silenced AKAP8, a RII-interacting scaffold protein that has previously been implicated in mediating PKA suppression of TNF-α expression in murine RAW 264.7 macrophages (25). Treatment of AMs for 48 h with siRNA targeting AKAPs 10 and 11 reduced their mRNA expression by ∼65 and 75%, respectively, relative to a non-targeting control siRNA, whereas AKAP8 siRNA reduced its expression by 55% (Fig. 4B). Confirmation of >50% knockdown at the protein level was confirmed for each of these AKAPs (Fig. 4C). The impact of pretreatment with siRNAs on NO production was evaluated (Fig. 4D). LPS-induced NO generation was unaffected by AKAP8 siRNA pretreatment but was inhibited by ∼90 and ∼40% after pretreatment with AKAP10 and AKAP11 siRNA, respectively. Likewise, the increment in LPS-induced NO generation elicited by exogenous PGE2 was unaffected by AKAP8 siRNA, slightly abrogated by AKAP11 siRNA, and entirely abrogated by AKAP10 siRNA. These data implicate AKAP10 in the potentiation by PGE2 of LPS-induced NO synthesis, with a possible minor contribution of AKAP11.
FIGURE 4.
AKAP 10 is required for potentiation of LPS-induced NO production by endogenous and exogenous PGE2. A, shown is mRNA expression of RI-interacting AKAPs in AMs, as determined by real time RT-PCR; data represent the mean ± S.E. from three individual experiments, each performed in triplicate, and values are expressed as -fold change relative to β-actin with AKAP1 set as 1. B, AMs were treated for 48 h with AKAP 8, 10, and 11 siRNAs or control siRNA (indicated by the dashed line), and the expression of the indicated AKAP mRNA was determined by real time RT-PCR. C, AMs were treated with target or control siRNAs as in B, and expression of the AKAP protein and of the housekeeping protein GAPDH was determined by Western blot analysis. D, cells were treated with AKAP or control siRNAs as in B followed by PGE2 for 10 min and then LPS for another 24 h. Supernatants were harvested, and nitrite was determined. The data are the mean ± S.E. values of three separate experiments, each performed in duplicate. *, p < 0.05 versus control siRNA; #, p < 0.05 versus LPS alone; &, p < 0.05 versus PGE2.
Role of AKAPs in PGE2 Regulation of TLR4-induced Synthesis of IL-10, IL-6, and TNF-α
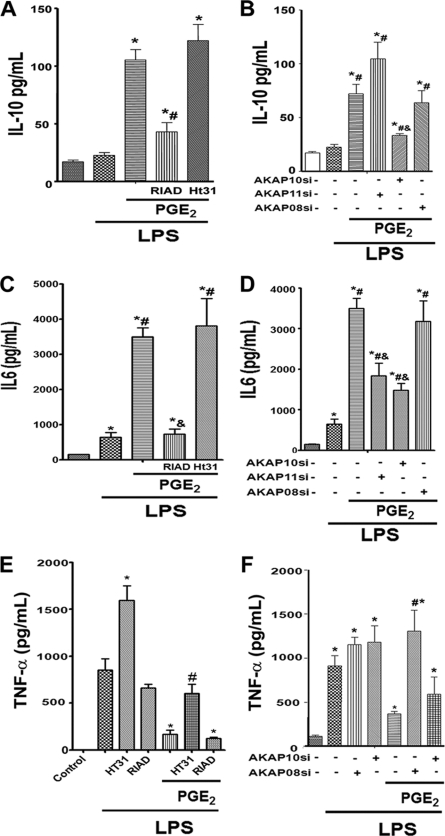
We next assessed the role of AKAPs in regulating the secretion of the important immunomodulatory cytokines IL-10, IL-6, and TNF-α. As reported previously (13), rat AMs fail to secrete appreciable IL-10 in response to LPS alone but they do when treated with LPS + PGE2 (Fig. 5A). This IL-10 response was markedly abrogated by the AKAP/RI-disrupting peptide RIAD but unaffected by the AKAP/RII-disrupting peptide Ht31 (Fig. 5A). The role of specific AKAPs in this potentiation of LPS-induced IL-10 by PGE2 was determined using gene silencing. Although knockdown of AKAP8 appeared to have no effect on this additive response and that of AKAP11 actually enhanced it, silencing of AKAP10 significantly abrogated IL-10 secretion under these circumstances (Fig. 5B). Taken together, these data suggest that RI-AKAP10 interactions play a critical role in potentiation of IL-10 just as in the potentiation of NO synthesis. PGE2 enhancement of LPS-induced IL-6 production likewise depended on RI-AKAP interactions, as determined by the striking inhibitory effects of RIAD (Fig. 5C). As expected, knockdown of AKAP8 appeared to have no impact on this response, whereas that of AKAP10 and AKAP11 each partially abrogated PGE2 potentiation (Fig. 5D). The specificity of these enhancing effects of AKAP10 on TLR4 responses was explored by assessing its impact on LPS-induced generation of TNF-α. PGE2 has been reported to inhibit TNF-α generation in RAW 264.7 macrophages via PKA RII interactions with AKAP8 (25); although we have previously confirmed that PGE2 inhibits TLR4-induced TNF-α via PKA in AMs (13), the role of AKAPs in this AM response was not previously determined. The ability of PGE2 to suppress LPS-induced TNF-α secretion was attenuated by Ht31 but not RIAD (Fig. 5E). Interestingly, LPS induced TNF-α secretion was itself enhanced by Ht31. These results indicate that PKA RII-AKAP interactions down-regulate production of this cytokine during TLR4 activation. Knockdown of AKAP10 had no significant effect on the inhibitory influence of PGE2, whereas that of AKAP8 completely abolished this effect (Fig. 5F). Taken together, these findings suggest that AKAP10-anchored PKA RI is required for PGE2 enhancement of NO, IL-10, and IL-6 generated in response to TLR4 ligation. AKAP11-anchored PKA RI is capable of contributing to PGE2 potentiation of IL-6 and, to a lesser extent, NO. By contrast, AKAP8-anchored PKA RII is required for PGE2 inhibition of TLR4-induced elaboration of the proinflammatory mediator TNF-α.
FIGURE 5.
Role of specific AKAPs in PGE2 modulation of LPS-induced cytokine secretion. Cells were pretreated with Ht31 or RIAD (A, C, and E) as described in Fig. 3A or treated with control or AKAP siRNAs (B, D, and F) as described in Fig. 4B followed by PGE2 for 10 min and then LPS for another 24 h. Supernatants were harvested, and IL-10 (A and B), IL-6 (C and D), and TNF-α (E and F) were determined by ELISA. The data are the mean ± S.E. values of three separate experiments, each performed in duplicate. *, p < 0.05 versus control siRNA without LPS; #, p < 0.05 versus control siRNA plus LPS; &, p < 0.05 versus control siRNA plus LPS plus PGE2.
DISCUSSION
Both TLR and G protein-coupled receptor classes of receptors play key roles in directing macrophage responses to specific components of their local milieu. Synthesis of PGE2 typically accompanies the induction of other NFκB-dependent gene products including iNOS and cytokines. Indeed, PGE2 likely represents the most abundant ligand of Gs-coupled G protein-coupled receptors at sites of inflammation and infection (7). The increases in cAMP that follow, therefore, serve to shape and to modulate the emerging macrophage response to TLR ligation. In this report we examined the mechanisms by which PGE2 modulates iNOS induction and NO generation in resident rat AMs stimulated with the TLR4 ligand LPS. We identified an important role for AKAP10-anchored PKA-I in PGE2 potentiation of elaboration of NO as well as cytokines IL-10 and IL-6. By contrast, PGE2 inhibition of TNF-α in the context of LPS stimulation required AKAP8-anchored PKA RII. Our results help to explain how PGE2, an autocrine modulator whose generation is integral to TLR responses, can exert pleiotropic effects on specific gene products that comprise this response program. In particular, they illustrate how AKAP scaffold proteins can coordinate and specify diverse outcomes from a single second messenger molecule.
Chen et al. (30) identified an autocrine loop consisting of COX-2 induction, cAMP elevation, and PKA activation that contributes to iNOS expression and NO synthesis in the RAW 264.7 macrophage cell line. Data showing that a COX-2 inhibitor attenuated LPS-induced iNOS/NO are consistent with this autocrine loop being operative in rat AMs as well (31). Neither of these reports, however, established that PGE2 was the prostanoid responsible for potentiating LPS-induced NO. We identified PGE2 as the responsible prostanoid and EP2 rather than EP4 as the Gs receptor responsible for its action. Although we previously reported that rat AMs have a lower capacity for COX-2 induction and PGE2 synthesis than resident peritoneal macrophages (32), these data nevertheless demonstrate that PGE2 synthesis and signaling in these cells are sufficient to account for nearly half of the total amount of iNOS/NO synthesized in response to LPS. We have previously reported (33) that murine AMs deficient in the EP3 receptor, which signals via Gi-mediated decreases in cAMP, generate increased NO in response to challenge with the important respiratory pathogen Streptococcus pneumoniae. Such data support the present finding of a potentiating effect on AM NO generation by cAMP.
Cyclic AMP can act via two effectors, PKA and Epac-1 (34). Our previous work demonstrated that each can mediate specific differential effects on macrophage function, with PKA identified as modulating synthesis of inflammatory mediators including cytokines and leukotriene B4 (13, 24). Our present results with both (i) effector-selective cAMP analogs as well as (ii) small molecule and peptide inhibitors of PKA indicate that iNOS induction and NO synthesis in LPS-stimulated AMs is likewise mediated by PKA rather than Epac-1. These results confirm the findings of Moon and colleagues (35) in murine microglial cells. Effects of cAMP on iNOS/NO have been shown to vary depending on the cell type (3, 18), yet conflicting results on the directionality of cAMP modulation of iNOS have been reported even in macrophages. In contrast to our results and those noted above (30, 31), other reports have described a suppressive effect of cAMP on LPS-stimulated iNOS/NO in murine J774 macrophages (36), murine peritoneal macrophages (37, 38), and RAW 264.7 macrophages (37). It has been suggested that disparate types of regulation by cAMP of iNOS transcription may be a function of the specific complement of pertinent transcription factors active in a given cell and circumstance, with suppression being attributable to inhibition of NFκB (39) and potentiation being attributable to the activation of CREB (39). Although we did not comprehensively explore the transcriptional control of iNOS by cAMP in our experimental system, in preliminary studies the small molecule CREB inhibitor KG-501 (40) did abrogate the potentiating actions of PGE2 (data not shown). Additional work will be necessary to clarify this issue in the future.
Anchoring by AKAPs of specific molecular pools of PKA to particular organelles and membranes provides a means for spatial and substrate specificity of PKA signaling. Most AKAPs selectively or preferentially bind RII subunits, dictating that most PKA type I actions reflect the functions of soluble, rather than anchored, holoenzymes (27, 41). Experiments with PKA subtype-selective cAMP agonists (Fig. 2F) suggested that potentiation of LPS-induced NO generation reflected the actions of PKA-I. However, the ability of the RIAD peptide, which selectively disrupts interactions between PKA RI and AKAPs, to attenuate PGE2 potentiation of iNOS and NO (Fig. 3) surprisingly points to AKAP-anchored PKA-I in this action.
There is little information available about AKAPs in macrophages. Wall et al. (25) determined that the RII-interacting AKAP8 (also known as AKAP95) mediated PGE2 suppression of LPS-induced TNF-α in RAW 264.7 macrophages. We confirmed this role of AKAP8-anchored PKA-II in AMs, finding that PGE2 suppression of LPS-induced TNF-α was largely reversed by the RII-delocalizing peptide Ht31 (Fig. 5E) and by siRNA targeting AKAP8 (Fig. 5F). Wall et al. (25) also reported that PGE2 potentiated LPS-induced production of granulocyte colony-stimulating factor and suggested on the basis of inhibition by RIAD that a RI-AKAP complex accounted for this effect. However, they did not identify the RI-interacting AKAP responsible. We identified mRNAs encoding six AKAPs capable of binding RI subunits in AMs, with the most highly expressed being AKAPs 10 and 11. We then employed siRNA to assess the functional contributions of each of these to PGE2 potentiation of LPS-induced mediator production. Although target mRNA knockdown was incomplete, results suggested that AKAP10 was necessary for PGE2 enhancement of NO, IL-10, and IL-6 generated in response to TLR4 ligation, whereas AKAP11 contributed to potentiation of IL-6 and, to a lesser extent, NO. To our knowledge, these data represent the first demonstration of a functional role for a specific RI-interacting AKAP in any macrophage population. We did not examine granulocyte colony-stimulating factor production in our AM cultures and, therefore, cannot evaluate the possibility that AKAP10, which was also expressed in RAW 264.7 cells, might likewise mediate PKA-I-dependent potentiation of this cytokine by PGE2 in AMs. However, an important role for AKAP10 in mediating potentiation of TLR4-induced synthesis of iNOS/NO, IL-10, and IL-6 suggests that this scaffold protein may subserve the general function of mediating transcriptional enhancement by cAMP/PKA during TLR stimulation. As not only iNOS (39) but also IL-6 (42) and IL-10 (43) are transcriptionally up-regulated by CREB in the context of NFκB activation, we speculate that AKAP-10-anchored PKA-I may accomplish this by either directly or indirectly activating CREB; future work will be necessary to characterize this interaction.
In conclusion, our studies suggest that AKAP10 coordinates PGE2/EP2/cAMP/PKA enhancement of NFκB-dependent gene products, including iNOS, IL-10, and IL-6, generated upon TLR stimulation of resident AMs. Such cross-talk between G protein-coupled receptor and TLR responses would be anticipated to typify and help to shape innate immune responses to infections in the lung.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants HL58897 (to M. P.-G.) and HD057176 (to D. M. A.). This work was also supported by American Lung Association Senior Postdoctoral Research Fellowships (to C. H. S. and K. O.) and a Uehara Memorial Foundation research fellowship (to K. O.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- TLR
- Toll-like receptor
- AKAP
- A-kinase anchoring protein
- AM
- alveolar macrophage
- 6-Bnz-cAMP
- N6-benzoyladenosine-3′,5′-cyclic monophosphate
- C
- catalytic subunit of PKA
- 2-Cl-8-MA-cAMP
- 2-chloro-8-methylaminoadenosine-3′,5′-cyclic monophosphate
- EP
- E prostanoid receptor
- Epac
- exchange protein directly activated by cAMP
- iNOS
- inducible nitric-oxide synthase
- 6-MBC-cAMP
- N6-mono-t-butylcarbamoyladenosine- 3′,5′-cyclic monophosphate
- 8-pCPT-2-O-Me-cAMP
- 8–4-chlorophenylthio-2′-O-methyladenosine-3′,5′-cyclic monophosphate
- PGE2
- prostaglandin E2
- R
- regulatory subunit of PKA
- IP
- I prostanoid receptor
- CREB
- cAMP-response element-binding protein
- RIAD
- RI anchoring disruptor.
REFERENCES
- 1. Medzhitov R., Horng T. (2009) Nat. Rev. Immunol. 9, 692–703 [DOI] [PubMed] [Google Scholar]
- 2. Verstrepen L., Bekaert T., Chau T. L., Tavernier J., Chariot A., Beyaert R. (2008) Cell. Mol. Life Sci. 65, 2964–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pautz A., Art J., Hahn S., Nowag S., Voss C., Kleinert H. (2010) Nitric Oxide 23, 75–93 [DOI] [PubMed] [Google Scholar]
- 4. Gaston B., Drazen J. M., Loscalzo J., Stamler J. S. (1994) Am. J. Respir. Crit. Care Med. 149, 538–551 [DOI] [PubMed] [Google Scholar]
- 5. Smith W. L., DeWitt D. L., Garavito R. M. (2000) Annu. Rev. Biochem. 69, 145–182 [DOI] [PubMed] [Google Scholar]
- 6. Aderem A. A., Cohen D. S., Wright S. D., Cohn Z. A. (1986) J. Exp. Med. 164, 165–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vancheri C., Mastruzzo C., Sortino M. A., Crimi N. (2004) Trends Immunol. 25, 40–46 [DOI] [PubMed] [Google Scholar]
- 8. Sugimoto Y., Narumiya S. (2007) J. Biol. Chem. 282, 11613–11617 [DOI] [PubMed] [Google Scholar]
- 9. Serezani C. H., Ballinger M. N., Aronoff D. M., Peters-Golden M. (2008) Am. J. Respir. Cell Mol. Biol. 39, 127–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bos J. L. (2003) Nat. Rev. Mol. Cell Biol. 4, 733–738 [DOI] [PubMed] [Google Scholar]
- 11. Jarnaess E., Taskén K. (2007) Biochem. Soc. Trans. 35, 931–937 [DOI] [PubMed] [Google Scholar]
- 12. Serezani C. H., Chung J., Ballinger M. N., Moore B. B., Aronoff D. M., Peters-Golden M. (2007) Am. J. Respir. Cell Mol. Biol. 37, 562–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aronoff D. M., Carstens J. K., Chen G. H., Toews G. B., Peters-Golden M. (2006) J. Interferon Cytokine Res. 26, 827–833 [DOI] [PubMed] [Google Scholar]
- 14. Aronoff D. M., Canetti C., Peters-Golden M. (2004) J. Immunol. 173, 559–565 [DOI] [PubMed] [Google Scholar]
- 15. Fernández-Repollet E., Mittler R. S., Tiffany S., Schwartz A. (1982) J. Histochem. Cytochem. 30, 466–470 [DOI] [PubMed] [Google Scholar]
- 16. Konopski Z., Seljelid R., Eskeland T. (1993) Scand. J. Immunol. 37, 587–592 [DOI] [PubMed] [Google Scholar]
- 17. Kunkel S. L., Spengler M., May M. A., Spengler R., Larrick J., Remick D. (1988) J. Biol. Chem. 263, 5380–5384 [PubMed] [Google Scholar]
- 18. Kleinert H., Pautz A., Linker K., Schwarz P. M. (2004) Eur. J. Pharmacol. 500, 255–266 [DOI] [PubMed] [Google Scholar]
- 19. Kleinert H., Schwarz P. M., Förstermann U. (2003) Biol. Chem. 384, 1343–1364 [DOI] [PubMed] [Google Scholar]
- 20. Welsh D. A., Mason C. M. (2001) Med. Clin. North Am. 85, 1329–1347 [DOI] [PubMed] [Google Scholar]
- 21. Peters-Golden M., Thebert P. (1987) Am. Rev. Respir. Dis. 135, 1020–1026 [DOI] [PubMed] [Google Scholar]
- 22. Serezani C. H., Perrela J. H., Russo M., Peters-Golden M., Jancar S. (2006) J. Immunol. 177, 3201–3208 [DOI] [PubMed] [Google Scholar]
- 23. Aronoff D. M., Peres C. M., Serezani C. H., Ballinger M. N., Carstens J. K., Coleman N., Moore B. B., Peebles R. S., Faccioli L. H., Peters-Golden M. (2007) J. Immunol. 178, 1628–1634 [DOI] [PubMed] [Google Scholar]
- 24. Aronoff D. M., Canetti C., Serezani C. H., Luo M., Peters-Golden M. (2005) J. Immunol. 174, 595–599 [DOI] [PubMed] [Google Scholar]
- 25. Wall E. A., Zavzavadjian J. R., Chang M. S., Randhawa B., Zhu X., Hsueh R. C., Liu J., Driver A., Bao X. R., Sternweis P. C., Simon M. I., Fraser I. D. (2009) Sci. Signal. 2, ra28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ogreid D., Ekanger R., Suva R. H., Miller J. P., Døskeland S. O. (1989) Eur. J. Biochem. 181, 19–31 [DOI] [PubMed] [Google Scholar]
- 27. Wong W., Scott J. D. (2004) Nat. Rev. Mol. Cell Biol. 5, 959–970 [DOI] [PubMed] [Google Scholar]
- 28. Carlson C. R., Lygren B., Berge T., Hoshi N., Wong W., Taskén K., Scott J. D. (2006) J. Biol. Chem. 281, 21535–21545 [DOI] [PubMed] [Google Scholar]
- 29. Stokka A. J., Gesellchen F., Carlson C. R., Scott J. D., Herberg F. W., Taskén K. (2006) Biochem. J. 400, 493–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen C. C., Chiu K. T., Sun Y. T., Chen W. C. (1999) J. Biol. Chem. 274, 31559–31564 [DOI] [PubMed] [Google Scholar]
- 31. Khanduja K. L., Sohi K. K., Pathak C. M., Kaushik G. (2006) Life Sci. 78, 1662–1669 [DOI] [PubMed] [Google Scholar]
- 32. Wilborn J., DeWitt D. L., Peters-Golden M. (1995) Am. J. Physiol. 268, L294–L301 [DOI] [PubMed] [Google Scholar]
- 33. Aronoff D. M., Lewis C., Serezani C. H., Eaton K. A., Goel D., Phipps J. C., Peters-Golden M., Mancuso P. (2009) J. Immunol. 183, 2642–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bos J. L. (2006) Trends Biochem. Sci. 31, 680–686 [DOI] [PubMed] [Google Scholar]
- 35. Moon E. Y., Oh S. Y., Han G. H., Lee C. S., Park S. K. (2005) J. Neurosci. Res. 81, 38–44 [DOI] [PubMed] [Google Scholar]
- 36. Pang L., Hoult J. R. (1997) Biochem. Pharmacol. 53, 493–500 [DOI] [PubMed] [Google Scholar]
- 37. Harbrecht B. G., Kim Y. M., Wirant E. A., Simmons R. L., Billiar T. R. (1997) J. Leukoc. Biol. 61, 712–720 [DOI] [PubMed] [Google Scholar]
- 38. Raddassi K., Petit J. F., Lemaire G. (1993) Cell. Immunol. 149, 50–64 [DOI] [PubMed] [Google Scholar]
- 39. Galea E., Feinstein D. L. (1999) FASEB J. 13, 2125–2137 [DOI] [PubMed] [Google Scholar]
- 40. Best J. L., Amezcua C. A., Mayr B., Flechner L., Murawsky C. M., Emerson B., Zor T., Gardner K. H., Montminy M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17622–17627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dodge-Kafka K. L., Soughayer J., Pare G. C., Carlisle Michel J. J., Langeberg L. K., Kapiloff M. S., Scott J. D. (2005) Nature 437, 574–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Spooren A., Kooijman R., Lintermans B., Van Craenenbroeck K., Vermeulen L., Haegeman G., Gerlo S. (2010) Cell. Signal. 22, 871–881 [DOI] [PubMed] [Google Scholar]
- 43. Avni D., Ernst O., Philosoph A., Zor T. (2010) Mol. Immunol. 47, 1396–1403 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.