Abstract
Autophagy is a key regulator of cellular homeostasis that can be activated by pathogen-associated molecules and recently has been shown to influence IL-1β secretion by macrophages. However, the mechanisms behind this are unclear. Here, we describe a novel role for autophagy in regulating the production of IL-1β in antigen-presenting cells. After treatment of macrophages with Toll-like receptor ligands, pro-IL-1β was specifically sequestered into autophagosomes, whereas further activation of autophagy with rapamycin induced the degradation of pro-IL-1β and blocked secretion of the mature cytokine. Inhibition of autophagy promoted the processing and secretion of IL-1β by antigen-presenting cells in an NLRP3- and TRIF-dependent manner. This effect was reduced by inhibition of reactive oxygen species but was independent of NOX2. Induction of autophagy in mice in vivo with rapamycin reduced serum levels of IL-1β in response to challenge with LPS. These data demonstrate that autophagy controls the production of IL-1β through at least two separate mechanisms: by targeting pro-IL-1β for lysosomal degradation and by regulating activation of the NLRP3 inflammasome.
Keywords: Dendritic Cell, Inflammation, Interleukin, Lipopolysaccharide (LPS), Macrophage, Toll-like Receptors (TLR), 3-Methyladenine, Rapamycin
Introduction
IL-1β is an important proinflammatory cytokine, released at sites of infection or injury, that regulates diverse physiological responses, including cellular recruitment, appetite, sleep, and body temperature (1). IL-1β is first produced as a proform in response to inflammatory stimuli, such as TLR ligands. This inactive precursor is cleaved into the bioactive (p17) molecule by caspase 1, following the activation of an inflammasome (2). Inflammasomes are molecular scaffolds that trigger the activation of caspase 1 and subsequent maturation of IL-1β and IL-18. Typically, inflammasomes are formed from at least one member of the cytosolic innate immune sensor family, the NOD-like receptors (NLRs), including NLRP1, NLRP3, and NLRC4, coupled with the adaptor apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC or PYCARD) and caspase 1 (2). The NLRP3 inflammasome is the best characterized to date and is activated by a number of endogenous and exogenous signals.
Most studies in vitro employ TLR ligands, particularly LPS, to induce pro-IL-1β formation, but in many cases, this is not enough to stimulate inflammasome activation and secretion of the mature cytokine. Instead, a second signal is commonly required, and this can come from a number of endogenous and exogenous sources, including ATP and particulates, including uric acid crystals, amyloid-β, silica, asbestos, synthetic microparticles, and alum (3–8). Extracellular ATP triggers the P2X7 ATP-gated ion channel, leading to K+ efflux and induces recruitment of the pannexin-1 membrane pore (9). This may then allow extracellular NLRP3 agonists to enter the cell and activate inflammasome assembly (9). Particulates have been proposed to act through one of two mechanisms. Uptake of particulates by phagocytes may lead to lysosomal damage and release of lysosomal products into the cytosol, which activates NLRP3 (4, 10). In particular, the lysosomal protease cathepsin B has been implicated, due to the inhibitory effects of the cathepsin B inhibitor CA-074 methyl ester on IL-1β secretion (4, 8, 10). Alternatively, others have proposed that NLRP3 agonists induce inflammasome assembly by stimulating the production of reactive oxygen species (ROS)5 (11–13).
Autophagy is a highly conserved homeostatic process for the sequestration and degradation of cytosolic macromolecules, excess, or damaged organelles and some pathogens (14–16). Controlled by the products of autophagy-related genes (Atg), autophagy occurs under normal physiological conditions but can be up-regulated by numerous factors, including nutrient deprivation and pharmacological inhibitors of the mammalian target of rapamycin (mTOR). In addition, autophagy is regulated by cytokines; it is up-regulated by IFN-γ and TNF-α (17, 18) and inhibited by IL-4 and IL-13 (19). It was previously reported that inhibition of autophagy in macrophages by knockdown of Atg7 or Atg16L1 promoted the secretion of IL-1β in response to LPS (20). The mechanisms behind this observation, or the inflammasome responsible, have not been fully determined, although the process was found to be dependent on signaling via TIR (Toll/IL-1 receptor) domain-containing adaptor inducing IFN-β (TRIF) and ROS (20).
The aim of this study was to determine the mechanisms behind autophagy-dependent regulation of IL-1β secretion. Using bone marrow-derived macrophages (iBMM) and DC (BMDC) from knock-out mice we found that autophagy regulates the NLRP3 inflammasome. Moreover, we found that after TLR ligation in macrophages IL-1β was specifically sequestered into autophagosomes. Furthermore, induction of autophagy using rapamycin resulted in the loss of intracellular pro-IL-1β and inhibited secretion of mature IL-1β in cells stimulated with LPS and alum, chitosan or ATP and reduced IL-1β in the blood of mice challenged with LPS.
EXPERIMENTAL PROCEDURES
Materials and Reagents
Unless otherwise stated, reagents were from Sigma. Culture media and FCS were from Biosera (Ringmer, East Sussex, UK). The PI3Kα inhibitor 2, 3-[4-(4-morpholinyl)thieno[3,2-d]pyrimidin-2-yl]-phenol, was from Cayman Chemicals (Tallinn, Estonia). The PI3Kδ inhibitor, IC87114, was from Symansis (Washdyke, NZ). Wortmannin, 3-methyladenine, and Akt inhibitor were all from Sigma. All TLR agonists were obtained from Invivogen. The cathepsin B inhibitor CA-074 methyl ester and Akt inhibitor were from Calbiochem. Rabbit polyclonal antibodies against IL-1β and caspase 1 were from Abcam (codes ab9722 and ab1872, respectively). Rat monoclonal antibody against murine IL-1β (for Western blotting) was from R&D Systems. Alexa Fluor 568-conjugated goat anti-rabbit IgG was from Invitrogen. Alum (Alhydrogel, 2.0% Al(OH)3) was from Brenntag Biosector (Frederikssund, Denmark). Chitosan CL213 was obtained from Novamatrix.
Cell Culture
Female C57BL/6 or BALB/c mice were obtained from Harlan Olac (Bicester, UK) and were used at 8–16 weeks of age. Animals were maintained according to the regulations of the European Union and the Irish Department of Health. BMDCs were generated as described previously (21) and grown in RPMI 1640 with 10% FCS, l-glutamine (2 mm), and penicillin and streptomycin (50 units/ml and 50 μg/ml, respectively) (complete medium). The media was supplemented with granulocyte macrophage-colony-stimulating factor (40 ng/ml). Cells were plated out on day 10 and stimulated with particulates and TLR agonists on day 11.
Immortalized BMM Cell Lines
Immortalized bone marrow-derived macrophages (iBMM) from wild type C57BL/6 and gp91phox (NOX2)−/− mice (a generous gift from P. Newburger) were generated using J2 transforming retroviruses (expressing Raf and Myc), as described previously for other knock-out cells (4). iBMM stably expressing EGFP-LC3 (GFP-LC3) were generated by taking EGFP-LC3 out of the pEGFP-C1 vector (22) and inserting it into the multiple cloning site of the retroviral pRP vector (23) using restriction endonucleases NotI and AgeI. Retrovirus carrying the EGFP-LC3 construct was made in HEK cells by transfection with EGFP-LC3::pRP, along with plasmids expressing Gag/Pol and VSV-G using TransIT® transfection reagent (Mirus, Madison, WI). Three days later, retrovirus was isolated from supernatant of the HEK cells and used to transduce the EGFP-LC3 construct into wild type C57BL/6 immortalized WT macrophages. 48 h post-transduction, cells expressing the plasmid were selected for with medium containing 5 μg/ml puromycin. One week later, colonies expressing the construct were isolated based on GFP fluorescence by flow cytometry and expanded under constant selection with 10 μg/ml puromycin. All iBMM were grown in complete medium.
LPS Challenge in Vivo
Female BALB/c mice were injected intraperitoneally with 200 μl PBS, LPS (10 μg/mouse in PBS), or LPS with rapamycin (1.5 mg/kg in PBS). After 4 h, mice were bled, and ELISA analysis was performed on plasma samples.
Propidium Iodide Incorporation Assay
BMDC were grown and stimulated in 96-well U-bottomed tissue culture plates. After treatment with LPS and 3-MA or alum, cells were incubated with propidium iodide (PI, 1 μg/ml) for 5 min. A CyAN (Dako) flow cytometer was used to analyze fluorescence and the data were analyzed using FlowJo software (Tree Star, Ashland, OR).
Macrophage Transfection
iBMM were transfected by nucleoporation with an Amaxa Nucleofector Device (Lonza, Wokingham, UK) as described previously (19). iBMM were harvested after 2–3 days in culture and resuspended in 100 μl of the appropriate electroporation buffer (Lonza AG) with 10 nm siGENOME SMARTpool siRNA against beclin 1 or siGENOME non-targeting control siRNA (Thermo Scientific, Lafayette, CO). Transfected cells were incubated overnight in complete medium before stimulation with LPS for 24 h. Targeting efficiency was analyzed by Western blot for beclin 1.
ELISA
Cytokine secretion in cell culture supernatants was measured by ELISA, according to the manufacturer's standard protocols (R&D Systems). Absorbance was read on a Multiscan FC plate reader and analyzed with SkanIt for Multiscan FC software (Thermo Scientific).
Confocal Microscopy
Immortalized GFP-LC3+ iBMM were grown and stimulated on coverslips (19-mm diameter) in 12-well plates. Cells were fixed with 2% paraformaldehyde for 30 min at room temperature and permeabilized for 10 min with Triton X-100 (0.1% in PBS). All subsequent steps were conducted at room temperature. The cells were blocked with 10% goat serum in PBS with 1% BSA for 1 h then stained with primary antibody (1/200 in blocking buffer) for 1 h. After washing with PBS (×5), the cells were stained with Alexa Fluor 568-conjugated goat anti-rabbit IgG (1/500 in PBS) for 1 h. The cells were stained with bisbenzimide H 33258 (5 μg/ml) for 5 min then washed five times with PBS. The coverslips were mounted onto glass slides with fluorescent Mounting medium (Dako, Ireland) and analyzed on an Olympus FV1000 laser scanning confocal microscope.
Immunoblotting
BMDC and immortalized iBMM were grown in 12-well tissue culture plates and centrifuged at 3,000 rpm, and supernatants were kept for protein isolation and analysis of secreted IL-1β. Cells were washed twice with PBS and lysed according to a protocol described previously (24). Proteins from supernatants were isolated by methanol/chloroform precipitation; 500 μl of supernatant was mixed with 500 μl of methanol and 100 μl of chloroform and centrifuged at 13,000 rpm for 5 min. The supernatant was removed, and the protein pellet was washed with 500 μl methanol (13,000 rpm for 5 min). The protein pellet was air dried and resuspended in 50 μl of Laemmli buffer. Samples were loaded and separated on a 15% SDS-polyacrylamide gel and transferred to a nitrocellulose membrane. The membrane was blocked overnight with 5% milk in PBS/Tween 20 (0.1%) at 4 °C. After washing with PBS/Tween, the membrane was probed with primary antibody (1/500 in 3% BSA) for 1 h at room temperature, washed three times with PBS/Tween and probed with HRP-conjugated secondary antibody (1/2000 in PBS) for 1 h at room temperature. The membrane was washed three times with PBS/Tween and developed with freshly prepared luminol-based detection solution.
Statistical Analysis
Data sets were analyzed for statistical significance (p < 0.05) using one-way ANOVA, followed by Tukey's honestly significantly different (HSD) test.
RESULTS
Inhibition of Autophagy Stimulates LPS-induced Inflammasome Activation
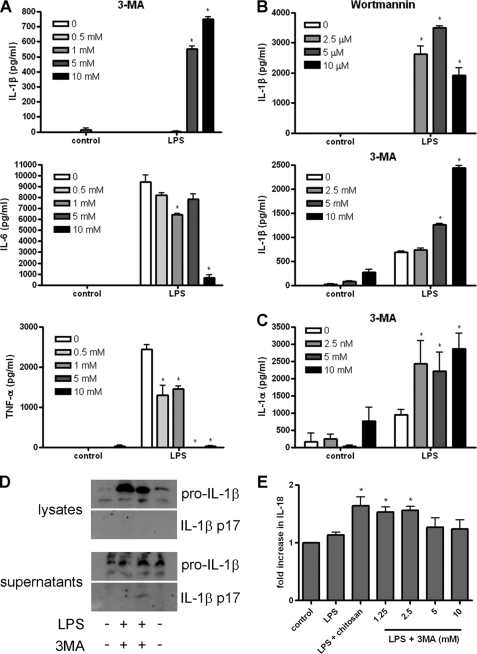
A previous study demonstrated that LPS can induce inflammasome activation and IL-1β secretion in embryonic liver macrophages from Atg16L−/− mice, suggesting a role for autophagy in dampening the inflammatory response to endotoxin (20). Here, we investigated the response of iBMM and BMDC to LPS after inhibition of autophagy. When treated with the PI3K inhibitors 3-MA or wortmannin, LPS-stimulated iBMM and BMDC secreted increased levels of IL-1β, whereas cells treated with LPS alone secreted low or undetectable amounts of the cytokine (Fig. 1, A and B). At the highest concentration used (10 mm), 3-MA strongly induced IL-1β but inhibited IL-6 secretion from BMDC (Fig. 1B and supplemental Fig. S1, A and B) and both IL-6 and TNF-α from iBMM (Fig. 1A). 3-MA, in combination with LPS, also increased IL-1α secretion by BMDC (Fig. 1C). Moreover, mature IL-1β was detected in the supernatants of BMDC treated with 3-MA and LPS, indicating that pro-IL-1β was being processed into the mature, bioactive form of the cytokine (Fig. 1D). In addition, iBMM treated with 3-MA and LPS secreted higher levels of IL-18 than LPS alone (Fig. 1E). Similar IL-1β responses were also observed in human peripheral blood monocyte-derived macrophages from two donors treated with LPS and 3-MA (supplemental Fig. S1C).
FIGURE 1.
The autophagy inhibitors 3-MA and wortmannin induce endotoxin-dependent inflammasome activation. A, iBMM were treated with LPS (100 ng/ml) and 3-MA at the concentrations indicated for 24 h, and supernatants were analyzed for IL-1β, IL-6, and TNF-α by ELISA. B, BMDC were treated with LPS (10 ng/ml) and wortmannin or 3-MA at the concentrations indicated for 24 h, and supernatants were analyzed for IL-1β by ELISA. C, BMDC were treated with LPS (10 ng/ml) and 3-MA at the concentrations indicated for 24 h, and supernatants were analyzed for IL-1α by ELISA. D, Western blot analysis of IL-1β in cell lysates and supernatants from BMDC stimulated with LPS (10 ng/ml) and 3-MA (10 mm) for 24 h (representative of three separate experiments). E, ELISA analysis of IL-18 from supernatants of iBMM stimulated with LPS (100 ng/ml) and the indicated concentrations of 3MA for 24 h. Data are means ± S.D. from one experiment representative of 2 (A, C, and E) or 3 (B). *, p < 0.05, significant difference from cells treated with LPS alone (one-way ANOVA, followed by Tukey's HSD test).
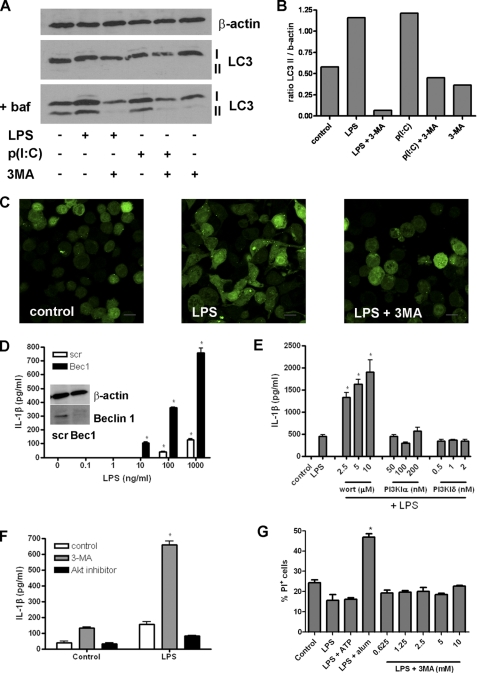
To confirm that inhibition of autophagy is responsible for the enhancing effects on IL-1β secretion, we looked at autophagy induction in iBMM in response to LPS and the TLR3 agonist poly(I:C) with and without 3-MA. Western blot analysis of LC3 showed that conversion of the cytosolic LC3 I to the lipidated LC3 II, which is found on autophagosome membranes, was increased in response to both LPS and poly(I:C) but reduced when cells were treated with 3-MA (Fig. 2, A and B). These assays were conducted in the presence of bafilomycin A to prevent loss of LC3 II due to lysosomal degradation and recycling of LC3 II to LC3 I after fusion of autophagosomes with lysosomes (25). Similarly, using confocal microscopy, LPS induced an increase in LC3+ vesicles in iBMM stably transfected with GFP-LC3, and this was inhibited by treatment with 3-MA (Fig. 2C). We also inhibited autophagy by transfection of iBMM with siRNA against beclin 1 (Atg6), which has been shown to inhibit autophagy in macrophages (19, 26). Cells transfected with siRNA against beclin 1 secreted higher levels of IL-1β, in a dose-dependent manner, compared with those transfected with control (“scrambled”) siRNA (Fig. 2D).
FIGURE 2.
3-MA activates the inflammasome through inhibition of type III PI3K and autophagy. A, Western blot analysis of LC3 in cell lysates from iBMM treated with LPS (100 ng/ml) or poly(I:C) (p(I:C), 10 μg/ml) and 3-MA (10 nm) for 6 h (representative blot from three separate experiments). Cells were treated with or without bafilomycin A (baf; 100 nm). B, densitometric measurement of the ratio of LC3 II:β-actin staining in A. C, confocal analysis of GFP-LC3-expressing iBMM treated with LPS (100 ng/ml) and 3-MA (10 mm) for 6 h. D, ELISA analysis of IL-1β in the supernatants of iBMM transfected with siRNA against beclin 1 (Bec1) or control (scr) siRNA and treated with different concentrations of LPS. Bec1 protein knockdown was confirmed by Western blot (inset). E, BMDC were treated with LPS (10 ng/ml) and either wortmannin (wort), the PI3KIα inhibitor 2,3-[4-(4-morpholinyl)thieno[3,2-d]pyrimidin-2-yl]-phenol or the PI3kIδ inhibitor IC87114 for 24 h, and supernatants were analyzed for IL-1β by ELISA. F, BMDC were treated with LPS (10 ng/ml) and Akt inhibitor (10 μm) for 24 h, and IL-1β was measured in the supernatants by ELISA. G, BMDC were treated with LPS (10 ng/ml) with ATP (5 mm), alum (5 mm), or 3-MA for 24 h. Cells were stained with PI and analyzed by flow cytometry. Data are means ± S.D. from one experiment representative of two (D, E, and F) or mean ± S.E. from three (G) experiments. *, p < 0.05, significant difference from cells treated with TLR agonist alone (one-way ANOVA, followed by Tukey's HSD test).
Both 3-MA and wortmannin suppress autophagy through the inhibition of the type III PI3K vps34 (27). However, these molecules also inhibit other PI3K, so we investigated the effects of alternative PI3K inhibitors. Inhibitors of PI3Kα and PI3Kδ had no effect on IL-1β secretion by BMDC in response to LPS (Fig. 2E). In addition, inhibition of Akt, a downstream effector of type I PI3K activation, did not stimulate IL-1β secretion in response to LPS (Fig. 2F).
As many stimulators of inflammasome activation also induce cell death (28), we measured cell death by incorporation of PI in BMDC. Cell death was increased in BMDC treated with the NLRP3 inflammasome activator alum, in combination with LPS for 24 h, but not in cells treated with 3-MA and LPS. (Fig. 2G).
Inhibition of Autophagy Induces Activation of NLRP3 Inflammasome
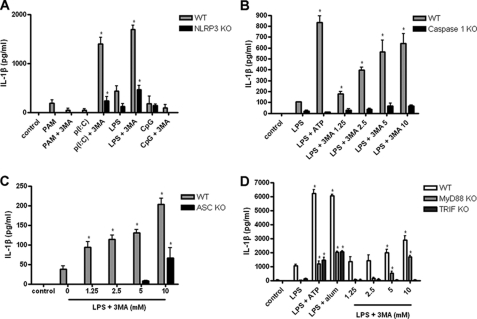
Activation of caspase 1 and processing of pro-IL-1β has been shown to require the assembly of an inflammasome complex, containing an NLR, such as NLRP3 or NLRC4. We looked at the response of BMDC from WT and NLRP3−/− mice to 3-MA in combination with different TLR agonists. IL-1β secretion was significantly increased in BMDC from WT mice in response to 3-MA in combination with LPS or the TLR3 agonist poly(I:C), but not in response to 3-MA with the TLR2 agonist PAM3CysK4 or the TLR9 agonist CpG (Fig. 3A). Moreover, BMDC from NLRP3−/− mice secreted significantly less IL-1β in response to LPS or poly(I:C) and 3-MA (Fig. 3A). In addition, IL-1β secretion in response to 3-MA and LPS was reduced in caspase 1−/− and ASC−/− iBMM (Fig. 3, B and C). Secretion of IL-1β by BMDC deficient in NLRC4 in response to 3-MA and LPS was comparable with WT cells (supplemental Fig. S2A). These data suggest that IL-1β secretion following the inhibition of autophagy is at least partially dependent on activation of the NLRP3 inflammasome. In contrast, IL-1α secretion in response to 3-MA was independent of NLRP3 and induced in the presence of PAM3CySK4, poly(I:C), LPS, and CpG, suggesting it is also not solely dependent on TRIF signaling (supplemental Fig. S2B).
FIGURE 3.
3-MA-induced inflammasome activation is dependent on NLRP3. A, BMDC from WT C57Bl/6 and NLRP3−/− mice were treated with PAM3CysK4 (PAM; 1 μg/ml), poly(I:C) (p(I:C); 10 μg/ml), LPS (10 ng/ml), or CpG (10 μg/ml) with 3-MA (10 mm) for 24 h, and supernatants were analyzed for IL-1β by ELISA. WT and caspase 1−/− (B) or ASC−/− (C) iBMM were treated with LPS (100 ng/ml) and indicated concentrations of 3MA for 24 h, and supernatants were analyzed by ELISA for IL-1β secretion. D, BMDC from WT C57Bl/6, MyD88−/−, and TRIF−/− mice were treated with LPS (10 ng/ml) plus ATP (5 mm), alum (5 mm), or indicated concentrations of 3-MA for 24 h. Supernatants were analyzed by ELISA for IL-1β. Data are means ± S.D. from one experiment representative of two (B, C, and D) or three (A). *, p < 0.05; significant difference from cells treated with TLR agonist alone (one-way ANOVA, followed by Tukey's HSD test).
It was previously demonstrated that IL-1β secretion in response to LPS in the absence of autophagy was dependent on signaling through TRIF (20). To date, only TLR3 and TLR4 have been shown to signal through TRIF; therefore, our data support this observation (Fig. 3A). We also looked at the effects of 3-MA and a larger range of TLR agonists on IL-1β secretion in BMDC. BMDC secreted significantly increased levels of IL-1β in response to 3-MA in combination with LPS and poly(I:C), but not in response to 3-MA with PAM3CysK4, the TLR7/8 agonists R837 (imiquimod) and R848 (Resiquimod) or CpG (supplemental Fig. S2C). Each of these agonists induced IL-1β secretion in combination with ATP. In addition, we found that IL-1β secretion in response to LPS and 3-MA was absent in BMDC from TRIF−/− mice, whereas it was still present in BMDC from MyD88−/− mice, albeit at a lower level compared with WT controls (Fig. 3D).
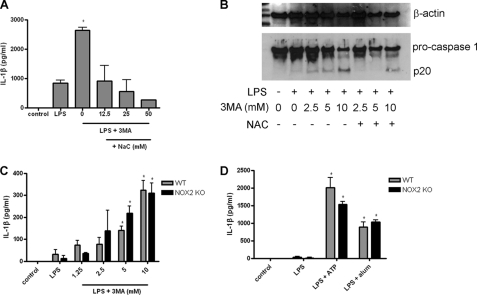
Numerous studies have demonstrated a role for ROS, potassium efflux, and cathepsin B/lysosomal disruption in NLRP3 inflammasome activation in response to various stimuli (4, 8, 10, 11, 13, 20). We found that treatment of cells with the ROS scavenger N-acetyl cysteine (NAc) inhibited IL-1β secretion from BMDC treated with 3-MA and LPS (Fig. 4A). Moreover, treatment of 3-MA/LPS-activated BMDC or iBMM with NAc inhibited caspase 1 processing (Fig. 4B and supplemental Fig. S3). However, NOX2 (gp91phox)−/− iBMM showed no impairment of IL-1β secretion in response to LPS with 3-MA (Fig. 4C) or LPS with ATP or alum (Fig. 4D).
FIGURE 4.
3-MA-induced inflammasome activation is dependent on ROS but not NOX2. A, BMDC were treated with LPS (10 ng/ml) and 3-MA (10 mm) with different concentrations of the reactive oxygen species scavenger NAc for 24 h, and supernatants were analyzed by ELISA for IL-1β. B, Western blot analysis of caspase 1 in cell lysates from iBMM treated with LPS (100 ng/ml) and 3-MA (at concentrations indicated) ± NAc (25 mm) for 24 h (representative of two separate experiments). ELISA analysis of IL-1β in supernatants of WT and NOX2−/− iBMM treated with LPS (100 ng/ml) plus 3MA (C) or with LPS plus ATP (5 mm) or alum (5 mm) (D) for 24 h. Data are means ± S.D. from one experiment representative of two (C and D) or three (A). *, p < 0.05, significant difference from cells treated with LPS alone (one-way ANOVA, followed by Tukey's HSD test).
We found that treatment of BMDC with extracellular KCl to inhibit potassium efflux inhibited IL-1β secretion in response to 3-MA and LPS at high concentrations (50 and 100 mm), but at these concentrations, KCl also inhibited IL-6 secretion from the same cells, suggesting a general inhibitory or toxic effect (supplemental Fig. S2D). Treatment of cells with the cathepsin B inhibitor CA-074 methyl ester at 10 μm had no effect on IL-1β secretion in response to LPS and 3-MA (supplemental Fig. S2E). These data suggest that inflammasome activation in the absence of autophagy is independent of cathepsin B but is reduced by inhibition of ROS.
Induction of Autophagy Leads to Degradation of Pro-IL-1β
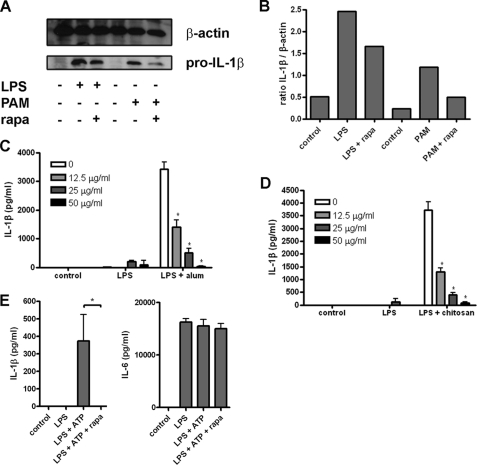
To determine the effect of autophagy induction on IL-1β processing and secretion, we treated iBMM with LPS or PAM3CysK4 for 20 h and then with rapamycin (a known inducer of autophagy) for 4 h and looked at IL-1β in cell lysates by Western blot. Following treatment with rapamycin, a reduction in pro-IL-1β was observed in cells treated with either LPS or PAM3CysK4 (Fig. 5, A and B). We did not detect any processed IL-1β in the supernatants from these cells (data not shown), suggesting that induction of autophagy is inducing degradation of pro-IL-1β, rather than processing and secretion of the mature (p17) form. In addition, we found that rapamycin inhibited IL-1β secretion by BMDC treated with LPS and the particulate inflammasome activators alum or chitosan, both of which have been shown previously to induce activation of the NLRP3 inflammasome and IL-1β secretion (Fig. 5, C and D) (5, 29). Similarly, rapamycin abrogated IL-1β secretion in response to treatment with LPS and ATP (Fig. 5E). The combination of LPS and rapamycin strongly induced autophagosome formation in LC3-GFP iBMM (supplemental Fig. S4). Taken together, these data suggest that autophagy acts to limit the availability of pro-IL-1β within stimulated cells and represents a novel mechanism for self-regulation of inflammatory responses by macrophages and dendritic cells.
FIGURE 5.
Induction of autophagy inhibits IL-1β secretion. A, Western blot analysis of IL-1β in cell lysates from iBMM treated with LPS (100 ng/ml) or PAM3CysK4 (PAM; 1 μg/ml) for 20 h then with rapamycin (rapa; 50 μg/ml) for 4 h (representative of three separate experiments). B, densitometric analysis of the ratio of pro-IL-1β:β-actin staining in A. BMDC were treated with LPS (10 ng/ml) and alum (5 μm) (C) or chitosan (2 μg/ml) (D) with the indicated concentrations of rapamycin and supernatants analyzed for IL-1β by ELISA. E, BMDC were treated with LPS for 20 h, followed by rapamycin (50 μg/ml) for 4 h and ATP (5 mm) for 1 h, and supernatants were analyzed by ELISA for secreted IL-1β and IL-6. Data are means ± S.D. *, p < 0.05, significant difference from cells treated with LPS alone (one-way ANOVA, followed by Tukey's HSD test).
IL-1β Is Targeted by Autophagosomes
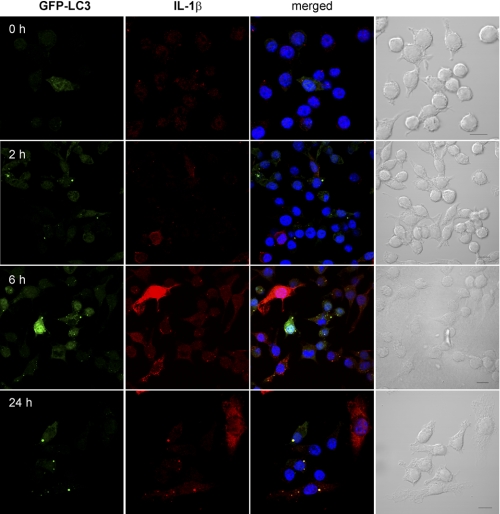
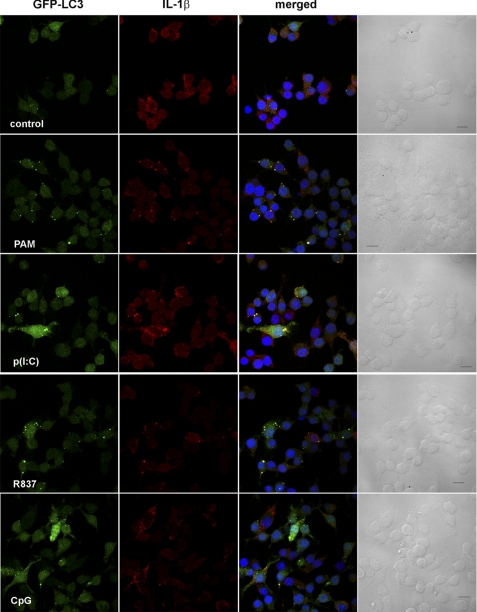
To determine the intracellular localization of IL-1β and how autophagy might regulate its secretion, we stained LPS-stimulated LC3-GFP iBMM with antibody against IL-1β and observed the cells by confocal microscopy. After 2 h, IL-1β staining was low, mostly cytosolic, and comparable with that after 0 h (Fig. 6). Additionally, no increase in autophagosome formation was observed. After treatment with LPS for 6 h, there was an increase in intracellular IL-1β in LC3-GFP iBMM, as well as an increase in autophagosome formation, as evidenced by a greater number of LC3+ vesicles (Fig. 6). Moreover, IL-1β was observed in the autophagosomes of LPS-treated iBMM after 6 h, suggesting that IL-1β is sequestered into autophagosomes following stimulation with LPS (Fig. 6). Similarly, after 24 h, extensive co-localization between LC3 and IL-1β was observed (Fig. 6). We also observed co-localization of IL-1β and LC3 in macrophages after treatment with PAM3CysK4, poly(I:C), R837, and CpG for 6 h (Fig. 7), suggesting that this effect is mediated through both TRIF- and MyD88-dependent pathways. In contrast, we saw no co-localization between caspase 1 and LC3 after treatment with LPS for 0–24 h (supplemental Fig. S5), suggesting that IL-1β sequestered into autophagosomes is effectively segregated from caspase 1. Caspase 1 staining appeared to be localized to vesicles (LC3-negative), rather than cytosolic.
FIGURE 6.
Intracellular IL-1β is targeted by autophagosomes. iBMM stably transfected with GFP-LC3 (green) were treated with LPS (100 ng/ml) for the indicated times, fixed, and stained with antibody against IL-1β (red). Nuclei were stained with bisbenzimide H 33258 (blue). Cells were analyzed by confocal microscopy. Scale bar, 10 μm.
FIGURE 7.
TLR agonists induce targeting of IL-1β by autophagosomes. iBMM stably transfected with GFP-LC3 (green) were treated for 6 h with PAM3CysK4 (PAM; 1 μg/ml), poly(I:C) (p(I:C); 10 μg/ml), R837 (10 μg/ml), or CpG (10 μg/ml). Cells were fixed and stained with antibody against IL-1β (red). Nuclei are stained with bisbenzimide H 33258 (blue). Cells were analyzed by confocal microscopy. Scale bar, 10 μm.
Induction of Autophagy Reduces IL-1β in Vivo
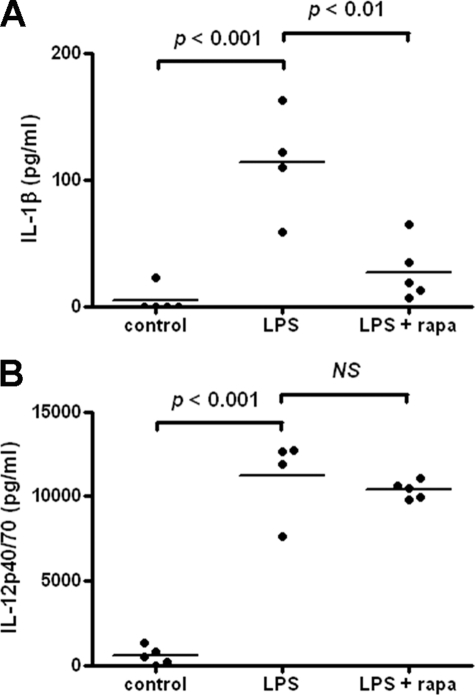
To determine the role of autophagy in controlling IL-1β secretion in vivo, we injected BALB/c mice intraperitoneally with either LPS alone or LPS together with rapamycin and took blood samples for analysis after 4 h. Control mice were given PBS. In mice treated with LPS alone, IL-1β in the blood was raised significantly, compared with PBS-treated controls (Fig. 8A). However, IL-1β was significantly reduced in mice treated with LPS and rapamycin together (Fig. 8A). In contrast, serum levels of IL-12p40, which were raised in response to LPS, were not affected by rapamycin treatment (Fig. 8B). IL-6 and TNF-α were not detectable in the serum of any of the mice (data not shown). Similarly, IL-1β secretion in the peritoneal cavity of the same mice was increased in response to LPS treatment but not in response to LPS plus rapamycin, whereas IL-6 levels were unaffected by the inclusion of rapamycin (supplemental Fig. S6). These data demonstrate that treatment of mice with the autophagy-inducing drug rapamycin can specifically inhibit LPS-induced IL-1β secretion in vivo.
FIGURE 8.
Induction of autophagy reduces IL-1β secretion in vivo. BALB/c mice were injected intraperitoneally with PBS, LPS (10 μg/mouse), or LPS with rapamycin (1.5 mg/kg) and blood taken after 4 h for analysis of IL-1β (A) or IL-12p40 (B) by ELISA. Data are means ± S.E. from a single experiment and analyzed by one-way ANOVA, followed by Tukey's HSD test (n = 5 (PBS and LPS/rapamycin) and n = 4 (LPS alone)).
DISCUSSION
The link between autophagy and innate and adaptive immunity is now well established. In particular, autophagy has been demonstrated to play a role in the maturation of pathogen-containing phagosomes and MHC class II presentation of viral antigens (17, 30). Moreover, autophagy is itself regulated by cytokines and may be an effector of Th1/Th2 polarization; it is induced by IFN-γ and TNF-α (17, 18) and inhibited by IL-4 and IL-13 (19). In addition, inflammatory stimuli, including LPS and other TLR agonists, have been shown to induce autophagy (31–33). A recent study has suggested that autophagy may also be a regulator of inflammation, modulating inflammasome activation and release of IL-1β (20). Here, we show that the inhibition of autophagy in LPS-activated BMDC and iBMM leads to activation of the NLRP3 inflammasome and secretion of IL-1β and IL-18. This is dependent on caspase 1, ASC, and TRIF and may involve the generation of ROS.
Although it is clear from this study and that of Saitoh et al. (20) that activation of the inflammasome by LPS in the absence of autophagy is dependent on TRIF signaling, the mechanism behind this is not clear. Both TRIF and MyD88 can mediate TLR-induced production of pro-IL-1β and activation of the NLRP3 inflammasome, as demonstrated by the fact that all the TLR agonists we tested can induce production of pro-IL-1β and secretion of mature IL-1β, when given in combination with ATP. Moreover, our data show that although both TRIF−/− and MyD88−/− BMDC show equally reduced secretion of IL-1β in response to LPS with ATP or LPS with alum, compared with WT cells, both still secrete significantly more IL-1β than cells treated with LPS alone. This suggests that, in the case of LPS, one can compensate for the other, at least to a degree, but the full response is dependent on both pathways. Indeed, BMDC from TRIF−/− MyD88−/− double knock-out mice do not respond at all to LPS with ATP or alum (data not shown). However, the response of BMDC to LPS with 3-MA is completely abrogated in TRIF−/− cells but is still present in MyD88−/− cells. This would suggest that, unlike LPS with ATP or alum, the role of TRIF in response to LPS and 3-MA cannot be compensated for by the presence of MyD88. Interestingly, TLR agonists can induce autophagy in macrophages through both TRIF- and MyD88-dependent pathways (31–33), although we have found that LPS-induced autophagy in BMDM requires MyD88, but not TRIF, signaling.6
Numerous studies have demonstrated a role for ROS, particularly peroxynitrite, in the processing and secretion of IL-1β (11, 12, 20, 34), although other studies have suggested that ROS are not involved in IL-1β processing or may in fact dampen IL-1β-induced inflammation (35, 36). Saitoh et al. (20) found that Atg16L1-deficient macrophages generated higher levels of ROS in response to LPS compared with WT or Atg16L1/TRIF double-deficient cells, suggesting that autophagy is an important regulator of intracellular ROS. Although we did not see an increase in LPS-induced ROS generation in 3-MA-treated BMDC or iBMM (data not shown), we did find that the ROS scavenger NAc inhibited IL-1β secretion in response to LPS in combination with either wortmannin or 3-MA. However, we found that IL-1β secretion by iBMM deficient in the phagocytic NADPH oxidase, NOX2 (gp91phox), was not impaired in response to LPS with 3-MA or LPS with ATP or alum. This would suggest either that ROS from an alternative source are involved in activation of the NLRP3 inflammasome or that NAc inhibits inflammasome activation via a ROS-independent mechanism. One possible alternative source of ROS is from altered or damaged mitochondria and peroxisomes. Given that autophagy is involved in the removal of these organelles (15, 16), this could represent a mechanism for the inflammasome-activating action of autophagy inhibitors. Indeed, a recent study has demonstrated that ROS from mitochondria can activate the NLRP3 inflammasome in human THP-1 cells and that autophagy of mitochondria (mitophagy) inhibits this (37). Moreover, ROS derived from both mitochondria and NADPH oxidases stimulate autophagy (38, 39).
Various studies have demonstrated a role for potassium efflux and/or lysosomal disruption/cathepsin B in the NLRP3-dependent processing and secretion of IL-1β (4, 8, 10, 13). Moreover, Saitoh et al. (20) found that inhibition of potassium efflux with extracellular KCl prevented LPS-induced IL-1β secretion in macrophages deficient in Atg16L1, which is involved in the formation of autophagosomes. In agreement with this, we found that KCl inhibited LPS-induced IL-1β secretion in macrophages treated with 3-MA at high concentrations. However, at these concentrations, IL-6 production was also abrogated, which may suggest a general inhibitory or toxic effect.
Our finding that IL-1β is sequestered into autophagosomes suggests a previously unidentified role for autophagy in controlling inflammatory responses in macrophages and dendritic cells. Moreover, the fact that further induction of autophagy with rapamycin in LPS-treated cells leads to the loss of intracellular pro-IL-1β and inhibition of IL-1β secretion after treatment with LPS and alum, chitosan, or ATP strongly suggests that autophagosomes specifically target pro-IL-1β for degradation. Moreover, caspase 1 was not seen in autophagosomes, suggesting that autophagosomes do not act as a platform for inflammasome assembly and processing of IL-1β, but rather segregate pro-IL-1β from caspase 1. This sequestration of IL-1β by autophagosomes might explain why inhibition of autophagy leads to increased secretion of IL-1β in response to LPS; more pro-IL-1β is available in the cytosol. However, LPS-induced inflammasome activation and secretion of IL-1β by cells deficient in autophagy is dependent on TRIF, while both MyD88- and TRIF-dependent mechanisms lead to the generation of pro-IL-1β and the sequestration of IL-1β into autophagosomes, as demonstrated by our experiments with other TLR agonists, including poly(I:C), which signals through TRIF and PAM3CysK4, R837, and CpG, which signal through MyD88 (40). Thus, these data suggest that inhibition of autophagy increases IL-1β secretion through two different mechanisms. First, inhibiting autophagy prevents degradation of pro-IL-1β, leaving more of the cytokine available in the cytosol for processing and secretion. Second, inhibition of autophagy leads to activation of the NLRP3 inflammasome and caspase 1-dependent processing of IL-1β through a ROS/TRIF-dependent mechanism.
Previous studies have implicated the autophagy gene Atg16L 1 as a candidate gene for susceptibility to Crohn disease (41–43), and Saitoh et al. (20) demonstrated that loss of ATG16L1 in mouse hematopoietic cells renders the animals highly susceptible to dextran sulfate sodium-induced acute colitis. Using an in vivo LPS challenge model, we have demonstrated that rapamycin, a potent activator of autophagy, reduces serum levels of IL-1β in mice, compared with those treated with LPS alone. These data indicate that autophagy has an important role to play in regulating acute inflammation in vivo. Conversely, a recent study has demonstrated that serum IL-1β and IL-18 levels in MAPLC3−/− mice are raised compared with WT in response to LPS challenge (44). Thus, short-term induction of autophagy may represent a novel therapeutic strategy for sepsis and fever, whereas targeted administration of rapamycin or alternative autophagy-inducing compounds might offer new remedies for inflammatory conditions such as Crohn disease.
In summary, we have demonstrated that the inhibition of autophagy in the presence of TLR3 or TLR4 signaling leads to activation of the NLRP3 inflammasome and secretion of IL-1β and IL-18. This is dependent on caspase 1 and on signaling via the TRIF pathway. In addition, we found that inhibitors of ROS and potassium efflux inhibited IL-1β secretion in response to autophagy blockade. Stimulation of cells with LPS induces autophagy and results in IL-1β being sequestered by autophagosomes, suggesting that autophagy may control inflammation through the degradation of pro-IL-1β. Furthermore, the induction of autophagy in mice using rapamycin reduced LPS-induced elevation of serum IL-1β. Thus, we present a novel mechanism by which autophagy can regulate inflammatory responses in antigen-presenting cells.
Supplementary Material
Acknowledgments
We thank Lisa Waggoner, Andres Mori, Graham Tynan, Edel McNeela, Anne McNaughton, Orla Hanrahan, and Ewa Oleszycka for technical support.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
J. Harris, A. O'Shea, and E. C. Lavelle, unpublished observations.
- ROS
- reactive oxygen species
- iBMM
- immortalized bone marrow-derived macrophage(s)
- TLR
- Toll-like receptor
- BMDC
- bone marrow-derived dendritic cell(s)
- PI
- propidium iodide
- ANOVA
- analysis of variance
- 3-MA
- 3-methyladenine.
REFERENCES
- 1. Dinarello C. A. (2010) Eur. J. Immunol. 40, 599–606 [DOI] [PubMed] [Google Scholar]
- 2. Schroder K., Tschopp J. (2010) Cell 140, 821–832 [DOI] [PubMed] [Google Scholar]
- 3. Eisenbarth S. C., Colegio O. R., O'Connor W., Sutterwala F. S., Flavell R. A. (2008) Nature 453, 1122–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hornung V., Bauernfeind F., Halle A., Samstad E. O., Kono H., Rock K. L., Fitzgerald K. A., Latz E. (2008) Nat. Immunol. 9, 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li H., Nookala S., Re F. (2007) J. Immunol. 178, 5271–5276 [DOI] [PubMed] [Google Scholar]
- 6. Martinon F., Pétrilli V., Mayor A., Tardivel A., Tschopp J. (2006) Nature 440, 237–241 [DOI] [PubMed] [Google Scholar]
- 7. Masters S. L., Dunne A., Subramanian S. L., Hull R. L., Tannahill G. M., Sharp F. A., Becker C., Franchi L., Yoshihara E., Chen Z., Mullooly N., Mielke L. A., Harris J., Coll R. C., Mills K. H., Mok K. H., Newsholme P., Nuñez G., Yodoi J., Kahn S. E., Lavelle E. C., O'Neill L. A. (2010) Nat. Immunol. 11, 897–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sharp F. A., Ruane D., Claass B., Creagh E., Harris J., Malyala P., Singh M., O'Hagan D. T., Pétrilli V., Tschopp J., O'Neill L. A., Lavelle E. C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 870–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kanneganti T. D., Lamkanfi M., Kim Y. G., Chen G., Park J. H., Franchi L., Vandenabeele P., Núñez G. (2007) Immunity 26, 433–443 [DOI] [PubMed] [Google Scholar]
- 10. Halle A., Hornung V., Petzold G. C., Stewart C. R., Monks B. G., Reinheckel T., Fitzgerald K. A., Latz E., Moore K. J., Golenbock D. T. (2008) Nat. Immunol. 9, 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cruz C. M., Rinna A., Forman H. J., Ventura A. L., Persechini P. M., Ojcius D. M. (2007) J. Biol. Chem. 282, 2871–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dostert C., Pétrilli V., Van Bruggen R., Steele C., Mossman B. T., Tschopp J. (2008) Science 320, 674–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pétrilli V., Papin S., Dostert C., Mayor A., Martinon F., Tschopp J. (2007) Cell Death Differ. 14, 1583–1589 [DOI] [PubMed] [Google Scholar]
- 14. Deretic V. (2010) Curr. Opin. Cell Biol. 22, 252–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Monastyrska I., Klionsky D. J. (2006) Mol. Aspects Med. 27, 483–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moreau K., Luo S., Rubinsztein D. C. (2010) Curr. Opin. Cell Biol. 22, 206–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gutierrez M. G., Master S. S., Singh S. B., Taylor G. A., Colombo M. I., Deretic V. (2004) Cell 119, 753–766 [DOI] [PubMed] [Google Scholar]
- 18. Harris J., Keane J. (2010) Clin. Exp. Immunol. 161, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harris J., De Haro S. A., Master S. S., Keane J., Roberts E. A., Delgado M., Deretic V. (2007) Immunity 27, 505–517 [DOI] [PubMed] [Google Scholar]
- 20. Saitoh T., Fujita N., Jang M. H., Uematsu S., Yang B. G., Satoh T., Omori H., Noda T., Yamamoto N., Komatsu M., Tanaka K., Kawai T., Tsujimura T., Takeuchi O., Yoshimori T., Akira S. (2008) Nature 456, 264–268 [DOI] [PubMed] [Google Scholar]
- 21. Lutz M. B., Kukutsch N., Ogilvie A. L., Rössner S., Koch F., Romani N., Schuler G. (1999) J. Immunol. Methods 223, 77–92 [DOI] [PubMed] [Google Scholar]
- 22. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bauernfeind F. G., Horvath G., Stutz A., Alnemri E. S., MacDonald K., Speert D., Fernandes-Alnemri T., Wu J., Monks B. G., Fitzgerald K. A., Hornung V., Latz E. (2009) J. Immunol. 183, 787–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Harris J., Hanrahan O., De Haro S. A. (2009) Curr. Protoc. Immunol. [DOI] [PubMed] [Google Scholar]
- 25. Mizushima N., Yoshimori T. (2007) Autophagy 3, 542–545 [DOI] [PubMed] [Google Scholar]
- 26. Singh S. B., Davis A. S., Taylor G. A., Deretic V. (2006) Science 313, 1438–1441 [DOI] [PubMed] [Google Scholar]
- 27. Petiot A., Ogier-Denis E., Blommaart E. F., Meijer A. J., Codogno P. (2000) J. Biol. Chem. 275, 992–998 [DOI] [PubMed] [Google Scholar]
- 28. Kepp O., Galluzzi L., Zitvogel L., Kroemer G. (2010) Eur. J. Immunol. 40, 627–630 [DOI] [PubMed] [Google Scholar]
- 29. Li H., Willingham S. B., Ting J. P., Re F. (2008) J. Immunol. 181, 17–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Paludan C., Schmid D., Landthaler M., Vockerodt M., Kube D., Tuschl T., Münz C. (2005) Science 307, 593–596 [DOI] [PubMed] [Google Scholar]
- 31. Delgado M. A., Elmaoued R. A., Davis A. S., Kyei G., Deretic V. (2008) EMBO J. 27, 1110–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shi C. S., Kehrl J. H. (2008) J. Biol. Chem. 283, 33175–33182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu Y., Jagannath C., Liu X. D., Sharafkhaneh A., Kolodziejska K. E., Eissa N. T. (2007) Immunity 27, 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hewinson J., Moore S. F., Glover C., Watts A. G., MacKenzie A. B. (2008) J. Immunol. 180, 8410–8420 [DOI] [PubMed] [Google Scholar]
- 35. Romani L., Fallarino F., De Luca A., Montagnoli C., D'Angelo C., Zelante T., Vacca C., Bistoni F., Fioretti M. C., Grohmann U., Segal B. H., Puccetti P. (2008) Nature 451, 211–215 [DOI] [PubMed] [Google Scholar]
- 36. van de Veerdonk F. L., Smeekens S. P., Joosten L. A., Kullberg B. J., Dinarello C. A., van der Meer J. W., Netea M. G. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 3030–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou R., Yazdi A. S., Menu P., Tschopp J. (2010) Nature [DOI] [PubMed] [Google Scholar]
- 38. Chen Y., Azad M. B., Gibson S. B. (2009) Cell Death Differ. 16, 1040–1052 [DOI] [PubMed] [Google Scholar]
- 39. Huang J., Canadien V., Lam G. Y., Steinberg B. E., Dinauer M. C., Magalhaes M. A., Glogauer M., Grinstein S., Brumell J. H. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6226–6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. O'Neill L. A., Bowie A. G. (2007) Nat. Rev. Immunol. 7, 353–364 [DOI] [PubMed] [Google Scholar]
- 41. Hampe J., Franke A., Rosenstiel P., Till A., Teuber M., Huse K., Albrecht M., Mayr G., De La Vega F. M., Briggs J., Günther S., Prescott N. J., Onnie C. M., Häsler R., Sipos B., Fölsch U. R., Lengauer T., Platzer M., Mathew C. G., Krawczak M., Schreiber S. (2007) Nat. Genet. 39, 207–211 [DOI] [PubMed] [Google Scholar]
- 42. Prescott N. J., Fisher S. A., Franke A., Hampe J., Onnie C. M., Soars D., Bagnall R., Mirza M. M., Sanderson J., Forbes A., Mansfield J. C., Lewis C. M., Schreiber S., Mathew C. G. (2007) Gastroenterology 132, 1665–1671 [DOI] [PubMed] [Google Scholar]
- 43. Rioux J. D., Xavier R. J., Taylor K. D., Silverberg M. S., Goyette P., Huett A., Green T., Kuballa P., Barmada M. M., Datta L. W., Shugart Y. Y., Griffiths A. M., Targan S. R., Ippoliti A. F., Bernard E. J., Mei L., Nicolae D. L., Regueiro M., Schumm L. P., Steinhart A. H., Rotter J. I., Duerr R. H., Cho J. H., Daly M. J., Brant S. R. (2007) Nat. Genet. 39, 596–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nakahira K., Haspel J. A., Rathinam V. A., Lee S. J., Dolinay T., Lam H. C., Englert J. A., Rabinovitch M., Cernadas M., Kim H. P., Fitzgerald K. A., Ryter S. W., Choi A. M. (2010) Nat. Immunol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.