Abstract
The RNA-binding protein Y14 heterodimerizes with Mago as the core of the exon junction complex during precursor mRNA splicing and plays a role in mRNA surveillance in the cytoplasm. Using the Y14/Magoh heterodimer as bait in a screening for its interacting partners, we identified the protein-arginine methyltransferase PRMT5 as a candidate. We show that Y14 and Magoh, but not other factors of the exon junction complex, interact with the cytoplasmic PRMT5-containing methylosome. We further provide evidence that Y14 promoted the activity of PRMT5 in methylation of Sm proteins of the small nuclear ribonucleoprotein core, whereas knockdown of Y14 reduced their methylation level. Moreover, Y14 overexpression induced the formation of a large, active, and small nuclear ribonucleoprotein (snRNP)-associated methylosome complex. However, Y14 may only transiently associate with the snRNP assembly complex in the cytoplasm. Together, our results suggest that Y14 facilitates Sm protein methylation probably by its activity in promoting the formation or stability of the methylosome-containing complex. We hypothesize that Y14 provides a regulatory link between pre-mRNA splicing and snRNP biogenesis.
Keywords: Protein Methylation, Protein-Protein Interactions, RNA, RNA-binding Protein, RNA-Protein Interaction, Sm Proteins, Arginine Methylation, Exon Junction Complex, Pre-mRNA Splicing, snRNPs
Introduction
The RNA-binding protein Y14 is evolutionally conserved in metazoans and participates in mRNA biogenesis (1–4). Y14 contains an RNA recognition motif in the central region, which is involved in the interaction with its stable partner Magoh (5). The C-terminal region of Y14 harboring two consecutive arginine-serine (RS) dipeptides and several arginine and glycine residues is predicted to be less structured but can be post-translationally modified (6). In Drosophila, the Y14/Mago homolog participates in transport and translation control of posterior mRNAs during oogenesis (7, 8). In vertebrates, Y14/Mago, together with another heterodimeric factor eIF4AIII/MLN51, constitutes the core of the exon junction complex (EJC),2 which is a multiprotein complex assembled on spliced mRNAs in a splicing-dependent manner (9, 10). In the EJC, Y14 directly interacts with several other factors, including RNA export factor (REF/Aly), RNPS1, and Upf3 (11). The EJC serves as a platform for binding of the mRNA export receptor Tip-associated protein (TAP) and factors involved in nonsense-mediated mRNA decay (4, 10, 12). Y14 indeed plays an important role in nonsense-mediated mRNA decay (4).
We have reported previously that the RG-rich sequences in the C-terminal domain of human Y14 can be methylated, and the RS dipeptides can be phosphorylated (6). In this study, our initial attempt to search for the enzymes or regulators responsible for post-translational modification of Y14 led to the identification of the protein-arginine methyltransferase PRMT5 as a potential interacting factor of Y14. PRMT5 is a type II protein methyltransferase that catalyzes both monomethylation and symmetrical dimethylation (13–15). PRMT5 localizes to both the nucleus and the cytoplasm (14, 16). In the nucleus, PRMT5 methylates transcriptional regulatory factors and may thereby affect their activity in transcriptional control (17, 18). In the cytoplasm, PRMT5 associates with chaperone protein chloride conductance regulatory protein ICln (pICln) and WD repeat-containing protein MEP50 to form the 20 S methylosome complex (13, 19–21). The methylosome essentially participates in the symmetrical dimethylation of Sm proteins of the spliceosomal small nuclear ribonucleoproteins (snRNPs) and is thus important for snRNP biogenesis (22–24).
In vertebrate cells, newly synthesized snRNAs are exported to the cytoplasm by a nuclear transporter CRM1-containing complex (25). In the cytoplasm, assembly of seven common spliceosomal snRNP Sm proteins with a single-stranded region of each snRNA occurs in a highly ordered manner (26, 27). Initially, the chaperone protein pICln binds SmD1/D2 and SmD3/B, respectively, and then SmE/F/G proteins join to preformed pICln/D1/D2 (24). pICln also stimulates the activity of PRMT5 to methylate Sm proteins (13, 19, 21, 24). Subsequently, the survival motor neuron protein (SMN) complex recruits methylated Sm proteins and catalyzes formation of the closed ring structure of Sm proteins on the snRNA (23, 28–31). Upon association with the Sm core, the cap of snRNAs undergoes hypermethylation, which allows its recognition by the nuclear import receptor snurportin1 (SPN1). Finally, the heterodimeric transport SPN1-importin-β complex delivers snRNPs to the nucleus (32, 33).
In this report, we provide evidence suggesting an unprecedented role for Y14 in facilitating Sm protein methylation via its interaction with the methylosome. Therefore, Y14 is involved in the biogenesis of both mRNAs and snRNAs.
EXPERIMENTAL PROCEDURES
Plasmid Construction
The C-terminally truncated Y14 (Y14ΔC; amino acid residues 1–151), C-terminal Y14 (Y14C; amino acid residues 152–174), and SmD1 cDNAs were each cloned into pGEX-5X (GE Healthcare) to create the vector for expressing GST fusion proteins in bacteria. The bacterial vectors for expression of GST-Y14 and PRMT1 and His-tagged Y14, Magoh, and SRPK1 were as described previously (6). The mammalian expression vector for pcDNA-FLAG-Y14, REF, and Upf3b were kind gifts from Jens Lykke-Andersen (University of Colorado, Boulder, CO). The cDNAs encoding Magoh, SMN, PRMT5, MLN51, and eIF4AIII were PCR-amplified and cloned into pcDNA3.1 (Invitrogen) to generate the vectors for expression of FLAG-tagged proteins. The cDNAs for Y14 and SmB were PCR-amplified and cloned into pCEP4 (Invitrogen) to generate the vectors for expression of HA-tagged protein.
Recombinant Protein Purification
GST fusion proteins, including Y14, Y14ΔC, Y14C, PRMT1, and SmD1, were overexpressed in Escherichia coli strain BLR (DE3), purified using glutathione-Sepharose 4B (GE Healthcare), and dialyzed against buffer D (20 mm HEPES, pH 7.9, 50 mm KCl, 0.2 mm EDTA, 0.5 mm DTT, 0.5 mm PMSF, and 20% glycerol). Non-phosphorylated and phosphorylated Y14/Magoh heterodimers were prepared as described (6).
Antibodies
The monoclonal antibodies used were against each of the following: PRMT5 (Sigma), pICln (BD Biosciences), MEP50 (Abnova), SMN (Abnova), CRM1 (Abnova), SPN1 (Abcam), PARP1 (Santa Cruz Biotechnology), α-tubulin (NeoMarkers), Gemin3 (Sigma), transportin (Sigma), Sm (Y12; a gift from Joan A. Steitz, Yale University, New Haven, CT), and actin (Chemicon). The polyclonal antibodies used included anti-HA (Covance), anti-FLAG (Sigma), anti-small nuclear ribonucleoprotein B (SNRPB) (Abcam), and SYM10 that recognizes symmetrical dimethylarginine (Upstate). Polyclonal anti-Y14 was prepared as described (6).
In Vitro Pulldown and Mass Spectrometry
Recombinant GST-Y14/His-Magoh heterodimer was prepared as described previously (6). For in vitro pulldown, 5 μg of GST, GST-Y14/His-Magoh, or any other GST fusion proteins used in this study was incubated with 25 μl of HeLa cell nuclear or cytoplasmic extract in a 50-μl mixture for 30 min at 30 °C followed by affinity selection with glutathione-Sepharose as described (6). Bound proteins were analyzed by silver staining or immunoblotting. For MS analysis, the pulldown reaction was scaled up by 3-fold. After gel electrophoresis, samples were stained with SYPRO Ruby (Bio-Rad) and visualized using a Typhoon 9410 (GE Healthcare). The bands of interest were excised and subjected to in-gel trypsinization followed by liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) (LTQ XL, ThermoFinnigan).
Cell Culture, Transient Transfection, and Establishment of Stable Cell Lines
Culture and transient transfection of HEK293 cells were essentially as described (6). To establish FLAG-tagged Y14 or DDX3-expressing stable cell lines, HEK293 cells were transfected with the corresponding expression vector and cultured under G418 (400 μg/ml; Clontech) selection for 2 weeks. Resistant colonies were picked, and selection continued for additional 2 weeks. The surviving cells were screened for stable expression of FLAG-tagged protein by immunoblotting. To knock down PRMT5, 200 nm PRMT5-targeting small interfering RNA (siRNA) (5′-aaguccggaaguugugccauu; Dharmacon) was transiently transfected into FLAG-Y14-expressing HEK293 cells. Moreover, HEK293 cells were transfected with 100 nm luciferase- (5′-ggauuucgagucgucuuaauguaua; Invitrogen) or Y14-targeting siRNA (5′-agagaauccagccuucaacagagcg; Invitrogen).
Preparation of Nuclear and Cytoplasmic Extracts of HeLa Cells
HeLa cell (S3 strain) culture and extract preparation were carried out as described (34). The nuclear and cytoplasmic extracts were in buffer D and at a concentration of ∼8 and ∼20 mg/ml, respectively.
In Vitro Methylation Assay
FLAG-PRMT5 was transiently expressed in HEK293 cells and immunopurified as described (13). For in vitro methylation, 5 μg of recombinant GST-Y14/His-Magoh or GST-SmD1 was incubated with 200 ng of FLAG-PRMT5 immunoprecipitate, 2.5 μg of purified GST-PRMT1 (6), or additionally with different amounts of purified GST-Y14/His-Magoh (0.3, 0.6, 1.25, or 2.5 μg). Detection of 3H-labeled proteins was performed using EN3HANCE (PerkinElmer Life Sciences) except for Fig. 3B. In vitro methylation of GST-SmD1 was also performed in 500 μl of sucrose gradient fractions (see below); after methylation, GST-SmD1 was affinity-selected by glutathione-Sepharose 4B.
FIGURE 3.
Y14 is not methylated by PRMT5 but may modulate its methylation activity in vitro. A, GST-Y14/His-Magoh (YM, non-phosphorylated; pYM, phosphorylated) and GST-SmD1 were subjected to the in vitro methylation assay using recombinant GST-PRMT1 or immunopurified PRMT5 in the presence of 3H-labeled methyl donor and then detected by autoradiography (Auto-radio.) and Coomassie (Cooma.) Brilliant Blue staining. The lower panel shows Coomassie Blue staining of purified GST-PRMT1 (lane 1) and immunopurified FLAG-PRMT5 (lane 2). Lane 3 shows immunoblotting (IB) of the FLAG-PRMT5 co-precipitate using antibodies against PRMT5, pICln, and MEP50. The asterisk in lane 1 indicates a truncated Y14 fragment. B, in vitro methylation of GST-SmD1 using FLAG-PRMT5 immunoprecipitates was performed as in A; different amounts of GST-Y14/His-Magoh were used. Upper panel, autoradiogram; lower panel, Coomassie Blue staining.
Sucrose Density Gradient Sedimentation
Cytoplasmic extracts were fractionated on linear 5–20% sucrose gradients according to Friesen et al. (13). Centrifugation was performed in an SW41 rotor (Beckman) at 33,000 rpm for 15 h and 20 min at 4 °C. Twenty fractions of 500 μl each were taken sequentially from the top of each gradient.
Immunoprecipitation
For immunoprecipitation, transfected cells were cultured for 48 h before harvest. Collected cells were washed with ice-cold PBS and then lysed in hypotonic buffer containing 10 mm Tris-HCl, pH 7.5, 10 mm NaCl, 10 mm EDTA, 0.5% Triton X-100, and protease inhibitor mixture (Roche Applied Science) on ice for 10 min. Subsequently, additional NaCl was added to the lysate to a final concentration of 150 mm. After centrifugation at 13,400 × g for 5 min at 4 °C, cell debris were removed, and the lysate was treated with RNase A (20 μg/ml) for 10 min on ice. Immunoprecipitation was carried out essentially as described (35). Anti-FLAG M2 affinity gel (Sigma) and anti-2,2,7-trimethylguanosine-conjugated agarose (Calbiochem) were used to immunoprecipitate FLAG-tagged proteins and spliceosomal snRNPs, respectively. For immunoprecipitation of Sm proteins, sucrose gradient fractions were incubated with Y12-conjugated protein A-Sepharose (Amersham Biosciences).
Northern Blot Analysis
To obtain the cytoplasmic and nuclear fractions, cells were resuspended in buffer A containing 10 mm HEPES, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 0.5 mm DTT, 5% glycerol, and protease inhibitor mixture. After incubation on ice for 15 min, Triton X-100 was added to a final concentration of 0.01%. After incubation on ice for 2 min, extracts were centrifuged at 2300 × g for 5 min at 4 °C. The supernatants were saved as the cytoplasmic fractions. The pellets were resuspended in buffer C containing 20 mm HEPES, pH 7.9, 450 mm NaCl, 1.5 mm MgCl2, 0.2 nm EDTA, 0.5 mm DTT, 25% glycerol, and protease inhibitor mixture, incubated on ice for 30 min, and then centrifuged at 13,400 × g for 10 min at 4 °C to yield the nuclear fraction. RNA was extracted from immunoprecipitates using TRIzol reagent (Invitrogen) and subsequently subjected to northern blotting. RNA probes complementary to snRNAs (U1, U2, U4, and U5) were produced by in vitro transcription according to previous reports (36). DNA oligonucleotide probe for U6 (cttcacgaatttgcgtgtcatccttg) was labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs).
Chemical Cross-linking
Sucrose gradient fractions were incubated with 0.01% glutaraldehyde (Sigma) for 15 min at room temperature.
RESULTS
Y14 Interacts with PRMT5
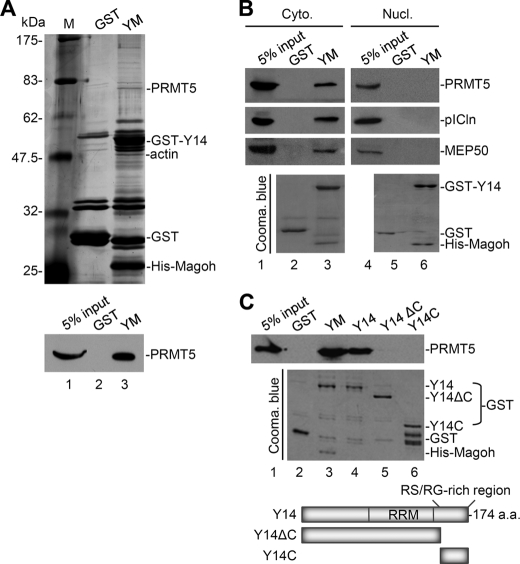
We attempted to identify the interacting partners or post-translational modifiers of Y14 from the cytoplasmic extract of HeLa cells. Because Y14 binds tightly to Magoh in cells, we used the preformed heterodimer of recombinant GST-Y14 and His-Magoh as bait in our in vitro pulldown assay. After selection with glutathione-Sepharose resin, Y14/Magoh-interacting proteins were subjected to gel electrophoresis and silver staining (Fig. 1A, upper panel), and their identity was determined by LC-MS/MS. The result indicated that the ∼72-kDa protein was probably the type II protein-arginine methyltransferase PRMT5. By using immunoblotting with anti-PRMT5, we confirmed that PRMT5 interacted with Y14/Magoh (Fig. 1A, lower panel).
FIGURE 1.
Y14 interacts with PRMT5 and the cytoplasmic methylosome. A, HeLa cell cytoplasmic extract was subjected to a pulldown assay using GST or GST-Y14/His-Magoh heterodimer (YM) as bait. Pulled down proteins were detected by silver staining of SDS-polyacrylamide gels (upper panel) and immunoblotting with anti-PRMT5 (lower panel). Lane M, molecular mass markers. B, HeLa cell cytoplasmic (Cyto.) or nuclear (Nucl.) extract was subjected to GST pulldown assays as in A. Immunoblotting was performed using antibodies against PRMT5, pICln, and MEP50. C, the pulldown assay was performed as in A. The bait proteins were as indicated above the gel. For both B and C, the bait proteins were detected by Coomassie (Cooma.) Brilliant Blue staining (lower panels). A schematic diagram shows the domain structure of Y14 and its truncated versions. RRM, RNA recognition motif; a.a., amino acids.
Y14 Associates with the Cytoplasmic Methylosome
Y14 is predominantly present in the nucleus, although it continuously shuttles between the nucleus and the cytoplasm (37). In contrast, PRMT5 is primarily localized in the cytoplasm but is still detectable in the nucleus (14). To investigate the cellular compartment in which Y14 may interact with PRMT5, we performed an in vitro pulldown assay using HeLa cell nuclear and cytoplasmic extracts. The result showed that Y14/Magoh preferentially pulled down the cytoplasmic but not nuclear PRMT5 (Fig. 1B) even in the presence of RNase (data not shown). Moreover, cytoplasmic pICln and MEP50 were also detected in the Y14/Magoh pulldown (Fig. 1B), indicating that Y14 may associate with the entire methylosome in the cytoplasm.
Next, we tested whether Magoh is essential for being a partner of Y14 to interact with PRMT5. Using uncomplexed GST-Y14 as bait, we found that Y14 itself was sufficient to pull down PRMT5 from the cytoplasmic extract (Fig. 1C, lane 4). RNase treatment did not disrupt such Y14-PRMT5 interaction (data not shown), suggesting that the interaction was not RNA-mediated. However, neither C-terminally truncated Y14 nor the C-terminal peptide alone could pull down PRMT5 (Fig. 1C, lanes 5 and 6). Structural studies have recently shown that two Y14/Mago-interacting proteins, PYM and importin-13, individually interact with the common surface of the Y14-Mago complex, and such interactions do not involve the most C-terminal region of Y14 (38, 39). Therefore, unlike PYM and importin-13, PRMT5 or the entire methylosome could interact with both Mago-associated and singular Y14 and requires the C-terminal RS/RG-rich region of Y14 for such interaction(s). However, whether the structural features of Y14/Magoh and Y14 alone in the interaction with PRMT5 are distinct still needs further investigation.
Y14, but Not Other EJC Factors, Interacts with the Methylosome
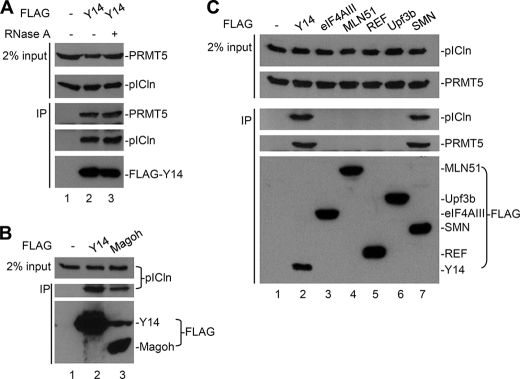
We next examined whether Y14 indeed interacts with the methylosome in vivo. FLAG-tagged Y14 was transiently expressed in HEK293 cells and subjected to immunoprecipitation with anti-FLAG antibody. Immunoblotting analysis showed that endogenous PRMT5 and pICln were co-precipitated with FLAG-Y14 even in the presence of RNase A (Fig. 2A). FLAG-Magoh also interacted with pICln (Fig. 2B). Perhaps Magoh interacted with the methylosome via Y14. Because the Y14/Magoh heterodimer is a part of the EJC core (9), we examined whether any other EJC components associate with the methylosome. Immunoprecipitation was performed with FLAG-tagged EJC factors followed by immunodetection of endogenous pICln or PRMT5 (Fig. 2C). However, except for Y14, neither the EJC nor its associated factors tested in this study, including eIF4AIII, MLN51, Upf3b, REF (Fig. 2C), RNPS1, TAP, and SRm160 (data not shown), significantly interacted with the methylosome components. Nevertheless, FLAG-tagged SMN co-precipitated pICln (Fig. 2C), indicating that pICln was indeed an integral component of the SMN-containing snRNP assembly complex. Therefore, our results indicated that Y14 or Y14/Magoh alone, rather than the entire EJC complex is associated with the methylosome.
FIGURE 2.
Y14, but not other EJC factors, interacts with the methylosome. In all panels, HEK293 cells were transiently transfected with the empty vector (−) or the expression vector for each FLAG-tagged protein as indicated above the gels. Cell lysates (input) and anti-FLAG immunoprecipitates (IP) were analyzed by immunoblotting using antibodies against PRMT5, pICln, and FLAG.
Y14 Is Not a Substrate of PRMT5
The interaction of PRMT5 with Y14 prompted us to examine whether Y14 can be methylated by PRMT5 or the methylosome. FLAG-tagged PRMT5 was transiently expressed in HEK293 cells and then immunoprecipitated from cell lysates. Anti-FLAG primarily precipitated FLAG-PRMT5, but the co-precipitate still contained endogenous MEP50 and pICln as evidenced by immunoblotting (Fig. 3A, lower panel). Immunopurified PRMT5 could effectively methylate a recombinant snRNP protein, SmD1, in vitro but not Y14 or Magoh (Fig. 3A, upper panel, lanes 4–6). Nevertheless, Y14 was able to be methylated by recombinant PRMT1 (Fig. 3A, upper panel, lane 1) as reported previously (6). We concluded that Y14 might not be a substrate of PRMT5.
Y14 Promotes the Methylation Activity of PRMT5
The above result that PRMT5 failed to methylate Y14 prompted us to examine whether Y14 plays a role in modulating the activity of the methylosome. We titrated purified GST-Y14/His-Magoh proteins in the in vitro methylation assay using GST-SmD1 as substrate. No detectable methylation activity was copurified with GST-Y14/His-Magoh (data not shown). However, we observed that the activity of immunopurified PRMT5 in SmD1 methylation was enhanced by Y14/Magoh in a concentration-dependent manner (Fig. 3B).
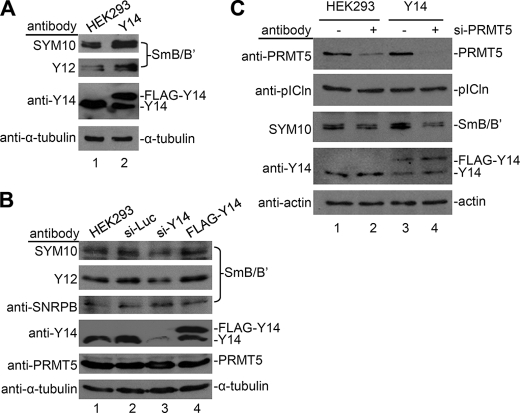
We next examined whether Y14 could modulate Sm protein methylation in vivo. Using the antibody SYM10, which recognizes symmetrically dimethylated arginines, we observed that the signal of SmB/B′ in HEK293 cells that stably overexpressed Y14 was enhanced (Fig. 4A). A similar result was detected with the anti-Sm antibody Y12, which recognizes symmetrical dimethylarginine in arginine/glycine-rich repeats of the Sm proteins (Fig. 4A). Nevertheless, overexpression of Y14 did not significantly affect the total amount of SmB/B′ proteins (Fig. 4B, lane 4). Moreover, using siRNA, we depleted Y14 in HEK293 cells to examine whether it is essential for Sm protein methylation. In such cells, Y14 expression was reduced to ∼25% of the control level, and accordingly, the methylation level of SmB/B′ detected by SYM10 and Y12 was reduced by ∼50% (Fig. 4B, lane 3). Therefore, this result provided another piece of evidence supporting the role of Y14 in facilitating methylation of Sm proteins.
FIGURE 4.
Y14 modulates methylation activity of PRMT5 in vivo. Total cell lysates were prepared from the following cells: mock or FLAG-Y14-expressing HEK293 cells (A), HEK293 cells that were mock-transfected or transfected with luciferase (Luc), Y14-targeting siRNAs, or the FLAG-Y14 expression vector (B), and mock or FLAG-Y14-expressing HEK293 cells that were either additionally transfected or not with an siRNA targeting PRMT5 (si-PRMT5) (C). Antibodies used for immunoblotting were as indicated to the left of each gel. The SYM10 and Y12 antibodies recognize symmetrical dimethyl-arginine.
Next, we evaluated whether Y14-induced Sm protein methylation involves PRMT5. When PRMT5 expression was down-regulated by siRNA, Y14-induced SYM10 signal was largely diminished (Fig. 4C, compare lane 4 with lane 3). Therefore, Y14 may modulate Sm protein methylation via the activity of PRMT5.
Y14 Overexpression Induces Formation of a Larger PRMT5-containing Complex
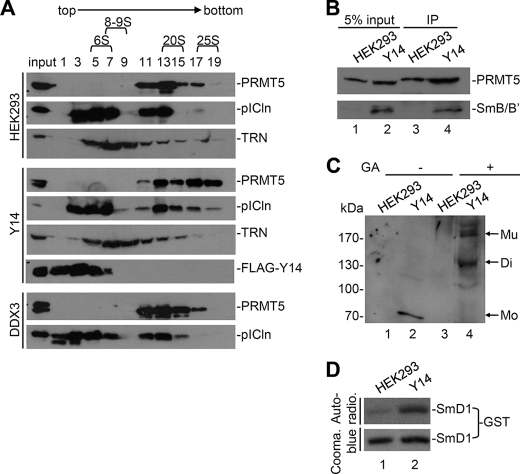
To explore how Y14 might facilitate Sm protein methylation via PRMT5 in vivo, we first examined the integrity of PRMT5-containing complexes in FLAG-Y14-overexpressing cells by using density sedimentation through sucrose gradients. The cytoplasmic extract prepared from FLAG-Y14-expressing HEK293 cells was subjected to sedimentation analysis on a 5–20% sucrose gradient. The vast majority of FLAG-Y14 was detected in low density fractions that, however, also contained a part of pICln (Fig. 5A, fractions 1–5). Cofractionation of overexpressed Y14 with pICln was likely due to their direct interaction (Fig. 2), and its implication is discussed below. Meanwhile, we used a stable HEK293 clone expressing an unrelated DEAD box-containing RNA helicase, DDX3, as a negative control. As compared with mock cells and FLAG-DDX3-expressing cells, ectopic overexpression of Y14 significantly moved both PRMT5 and pICln from the fractions of ∼20 S (fractions 13–15) to those of ∼25 S (fractions 17–19) (Fig. 5A). Under the same conditions, the fractionation pattern of a negative control protein, transportin, was not changed (Fig. 5A, TRN). In the 25 S fractions obtained from Y14-overexpressing cells, SmB/B′ was also detectable (Fig. 5B, lane 2) and indeed interacted with PRMT5 (lane 4), suggesting that Y14-induced high order PRMT5 complexes participate in Sm protein biogenesis.
FIGURE 5.
Y14 promotes formation of larger PRMT5-containing complex. A, cytoplasmic extracts prepared from mock, FLAG-Y14-, or DDX3-expressing stable HEK293 cells were fractionated on a 5–20% sucrose gradient. Odd-numbered fractions were analyzed by immunoblotting with antibodies against PRMT5, pICln, transportin (TRN), or FLAG epitope. B, pooled 25 S fractions from mock cell extract (lanes 1 and 3) or FLAG-Y14-expressing extract (lanes 2 and 4) were subjected to immunoprecipitation (IP) by using Y12 antibody. Total proteins (lanes 1 and 2) and Y12 co-precipitates (lanes 3 and 4) were immunoblotted using anti-PRMT5 and anti-SNRPB. C, as in B, the 25 S fractions were subjected to cross-linking in the absence (−) or presence (+) of glutaraldehyde (GA) followed by immunoblotting using anti-PRMT5. D, in vitro methylation of GST-SmD1 was performed in the 25 S fractions using 3H-labeled methyl donor. GST-SmD1 was recovered and shown by autoradiography (Auto-radio.) (upper panel) and Coomassie (Cooma.) Blue staining (lower panel). Mo, monomer; Mu, multimer; Di, dimer.
Because a previous report has shown that PRMT5 may form homo-oligomers that possess higher methylation activity (14), we tested whether Y14 could promote PRMT5 multimerization by using glutaraldehyde-mediated intramolecular cross-linking. As shown in Fig. 5C, Y14 indeed induced dimer and multimer forms of PRMT5 (lane 4); a similar result was obtained by using disuccinimidyl suberate as cross-linking reagent (data not shown). Accordingly, the fractions containing multimerized PRMT5 had a higher activity in methylation of recombinant SmD1 than the mock control (Fig. 5D). Therefore, Y14 might induce the formation of a large but active PRMT5 complex(es) and promote its association with Sm proteins.
Y14 May Participate in snRNP Core Assembly
The above data indicated that Y14/Magoh interacted with the cytoplasmic methylosome and may thus facilitate Sm protein methylation. To examine whether Y14 also associates with cytoplasmic snRNP assembly complexes, we co-expressed epitope-tagged Y14 and SmB, an snRNP core protein, in HEK293 cells. Immunoprecipitation of FLAG-Y14 from the cytoplasmic extract of transfected HEK293 cells showed that it associated with HA-SmB, but their interaction was partially disrupted by RNase A treatment (Fig. 6A). Moreover, FLAG-Y14 also co-precipitated snRNAs U1, U2, U4, and U5 but not U6 (Fig. 6B, lane 8). The association of Y14 with nuclear U6 probably occurred because Y14 is present in the spliceosome (40, 41) (Fig. 6B, lane 6). To evaluate the specificity of Y14 association with cytoplasmic snRNAs, we expressed FLAG-tagged REF, another EJC component, and Y14 in HEK293 cells. The antibody against the 5′-trimethylguanosine cap of mature snRNAs only precipitated FLAG-Y14 but not FLAG-REF (Fig. 6C). Therefore, Y14, but not REF, was likely a part of the core snRNPs and likely remained associated with these snRNPs at least until their respective snRNA underwent 5′ cap hypermethylation.
FIGURE 6.
Y14 associates with snRNP assembly complex. HEK293 cells were mock-transfected or transiently transfected with the expression vector FLAG-Y14 alone (B, D, and F) or together with the vector of HA-SmB (A). Anti-FLAG immunoprecipitates (IP) were subjected to immunodetection of tagged proteins (A) or endogenous proteins as indicted (D and F) or Northern blotting using antisense RNA probes complementary to U1, U2, U4, U5, and U6 (B). In B, subcellular fractionation was evaluated by immunoblotting using anti-PARP1 and anti-α-tubulin (lower panel). N and C represent nuclear and cytoplasmic fractions, respectively. C, the FLAG-REF and -Y14 expression vectors were transiently transfected into HEK293 cells. Proteins that co-precipitated with anti-trimethylguanosine were immunoblotted with anti-FLAG. In D, anti-Gemin3 cross-reacted to a protein band indicated by the asterisk. E, HEK293 cells were mock-transfected (lane 1) or were transfected with vector expressing FLAG-PRMT5 (lane 2), HA-Y14 alone (lane 4), or both protein-expressing vectors (lane 3). Immunoprecipitation was performed using anti-FLAG followed by immunoblotting using antibodies as indicated.
Because Y14 interacted with the methylosome and cytoplasmic snRNPs, we assumed that Y14 also associates with the SMN complex. By immunoprecipitation of FLAG-Y14 from HEK293 cells, we observed that Y14 interacted with two components of the SMN complex, SMN and Gemin3, although its interaction with Gemin3 was weak and RNA-dependent (Fig. 6D, lanes 5 and 6). Moreover, upon co-expression of HA-tagged Y14, FLAG-PRMT5 could co-immunoprecipitate a higher amount of SMN (Fig. 6E, lane 3). Therefore, Y14 not only interacted with the SMN complex but also promoted PRMT5 association with SMN. Finally, we assessed whether Y14 may escort snRNAs during their transport through nuclear pores. By using antibodies against CRM1 and SPN1 for immunoblotting of FLAG-Y14 co-precipitates, we were unable to detect significant interactions between Y14 and these two snRNA transporters (Fig. 6F). Taken together, our results revealed that Y14 joined the snRNP core assembly complex in the cytoplasm to promote Sm protein methylation but dissociated from the snRNP assembly complex before reimport of the snRNPs into the nucleus.
DISCUSSION
This study unveils a previously undefined role for Y14 in cytoplasmic snRNP biogenesis. We provide evidence that 1) Y14/Magoh directly interacts with the methylosome and participates in snRNP core protein assembly and that 2) Y14 promotes Sm protein methylation perhaps by inducing the formation of a larger complex containing active PRMT5 and the snRNP assembly complex.
We report for the first time that Y14/Magoh, but not any other EJC factors, directly associates with the cytoplasmic methylosome (Figs. 1 and 2). We have previously shown that the C-terminal domain of Y14 is methylated in cells (6). However, this study does not support a role of PRMT5 in Y14 methylation (Fig. 3). As compared with the Sm protein substrates of PRMT5, the C-terminal region of Y14 is relatively deficient in glycine and, particularly, lacks the glycine-arginine-glycine motif that comprises the optimal methylation site for PRMT5 (17, 22, 42). Therefore, the result that Y14 was not methylated by PRMT5 is not completely unexpected.
Nevertheless, both our in vitro and in vivo data suggested that Y14 instead plays a role in modulating the activity of the methylosome. We provided evidence showing that the recombinant Y14/Magoh heterodimer could stimulate the activity of PRMT5 on SmD1 methylation (Fig. 3). Consistently, overexpression of Y14 in cells also enhanced Sm protein methylation, which was likely mediated by PRMT5 (Fig. 4). The result that knockdown of Y14 reduced the Sm protein methylation level further supported the role of Y14 in modulating the methylation activity of the methylosome (Fig. 4). Moreover, we observed that overexpressed Y14 cofractionated with a portion of pICln although not with the methylosome on sucrose gradients (Fig. 5). Nevertheless, ectopic expression of Y14 could induce the formation of a larger PRMT5-containing complex(es) (Fig. 5), which indeed had a high activity for Sm protein methylation (Fig. 5). Indeed, overexpressed Y14 could interact with the SMN complex and promote SMN association with PRMT5 (Fig. 6). Therefore, we assume that a small portion of Y14/Magoh functions transiently with the methylosome and leaves its imprint on the dynamics or activity of the methylosome-snRNP assembly complexes; however, this hypothesis needs further study.
It has been reported that PRMT5 may form a multimer with higher methylation activity (14). However, it is so far unclear whether such a PRMT5 multimer is present in the methylosome and how it may participate in Sm protein methylation. Here, we report that Y14 could promote the formation of a larger and active PRMT5 complex that may contain multimerized PRMT5. Such a PRMT5-containing complex indeed interacted with its substrates, Sm proteins, and the SMN complex (Fig. 6). The enhanced Sm-SMN association might result from methylation of Sm proteins induced by Y14. Nevertheless, the detailed mechanism by which Y14 modulates the methylation activity (or other functions) of PRMT5 still needs further investigation.
During snRNP biogenesis, newly synthesized snRNA precursors are exported to the cytoplasm by CRM1 and are subsequently recruited to the SMN complex for Sm core assembly and snRNA modifications (25, 43). Here, our data show that Y14 had no detectable interaction with CRM1 but associated with SMN as well as trimethyl-capped snRNAs. Therefore, we deduced that Y14 might first interact with the methylosome and then join the snRNP assembly complexes. Upon completion of Sm protein assembly and snRNA cap modification, the core snRNPs are imported to the nucleus by the SPN1/importin-β transporter (32, 33). Y14 might dissociate from the core snRNPs before their import into the nucleus. We thus concluded that Y14 may transiently but functionally associate with snRNP assembly complexes in the cytoplasm (Fig. 7).
FIGURE 7.
Model for function of Y14/Magoh complex in snRNP core methylation and assembly. The Y14/Magoh complex directly interacts with the methylosome to facilitate its activity in methylation of Sm proteins of the spliceosomal snRNPs and also promotes association of the SMN complex with the methylosome for snRNP core assembly. Subsequently, Y14/Magoh dissociates from the snRNPs before their import into the nucleus.
A recent report has indicated that severe reduction of the SMN level results in defective snRNP assembly and hence aberrant splicing, suggesting that snRNP biogenesis is not merely a constitutive process but instead a regulatable process (44). Here, we show for the first time that Y14 may play a role in assisting or regulating snRNP core methylation and hence assembly. Therefore, Y14 probably participates in not only mRNA but also snRNP biogenesis pathways. Finally, it would also be interesting to test whether Y14 serves to link pre-mRNA splicing and perhaps other cytoplasmic mRNA biogenesis activities to snRNP production.
Acknowledgments
We thank Drs. Jens Lykke-Andersen and Joan A. Steitz for generously providing plasmids and the Y12 antibody, respectively. We also thank colleagues M.-C. Lai, R.-M. Lu, and K.-M. Lee for technical assistance.
This work was supported by an Academia Sinica investigator award (to W.-Y. T.).
- EJC
- exon junction complex
- REF
- RNA export factor
- TAP
- Tip-associated protein
- pICln
- chloride conductance regulatory protein ICln
- SMN
- survival motor neuron protein
- snRNP
- small nuclear ribonucleoprotein
- SPN1
- snurportin1
- LC-MS/MS
- liquid chromatography coupled with tandem mass spectrometry.
REFERENCES
- 1. Gehring N. H., Lamprinaki S., Kulozik A. E., Hentze M. W. (2009) Cell 137, 536–548 [DOI] [PubMed] [Google Scholar]
- 2. Degot S., Le Hir H., Alpy F., Kedinger V., Stoll I., Wendling C., Seraphin B., Rio M. C., Tomasetto C. (2004) J. Biol. Chem. 279, 33702–33715 [DOI] [PubMed] [Google Scholar]
- 3. Palacios I. M., Gatfield D., St Johnston D., Izaurralde E. (2004) Nature 427, 753–757 [DOI] [PubMed] [Google Scholar]
- 4. Gehring N. H., Neu-Yilik G., Schell T., Hentze M. W., Kulozik A. E. (2003) Mol. Cell 11, 939–949 [DOI] [PubMed] [Google Scholar]
- 5. Fribourg S., Gatfield D., Izaurralde E., Conti E. (2003) Nat. Struct. Biol. 10, 433–439 [DOI] [PubMed] [Google Scholar]
- 6. Hsu I. W., Hsu M., Li C., Chuang T. W., Lin R. I., Tarn W. Y. (2005) J. Biol. Chem. 280, 34507–34512 [DOI] [PubMed] [Google Scholar]
- 7. Hachet O., Ephrussi A. (2001) Curr. Biol. 11, 1666–1674 [DOI] [PubMed] [Google Scholar]
- 8. Mohr S. E., Dillon S. T., Boswell R. E. (2001) Genes Dev. 15, 2886–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tange T. Ø., Shibuya T., Jurica M. S., Moore M. J. (2005) RNA 11, 1869–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dreyfuss G., Kim V. N., Kataoka N. (2002) Nat. Rev. Mol. Cell Biol. 3, 195–205 [DOI] [PubMed] [Google Scholar]
- 11. Shi H., Xu R. M. (2003) Genes Dev. 17, 971–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kashima I., Yamashita A., Izumi N., Kataoka N., Morishita R., Hoshino S., Ohno M., Dreyfuss G., Ohno S. (2006) Genes Dev. 20, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Friesen W. J., Paushkin S., Wyce A., Massenet S., Pesiridis G. S., Van Duyne G., Rappsilber J., Mann M., Dreyfuss G. (2001) Mol. Cell. Biol. 21, 8289–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rho J., Choi S., Seong Y. R., Cho W. K., Kim S. H., Im D. S. (2001) J. Biol. Chem. 276, 11393–11401 [DOI] [PubMed] [Google Scholar]
- 15. Branscombe T. L., Frankel A., Lee J. H., Cook J. R., Yang Z., Pestka S., Clarke S. (2001) J. Biol. Chem. 276, 32971–32976 [DOI] [PubMed] [Google Scholar]
- 16. Pal S., Baiocchi R. A., Byrd J. C., Grever M. R., Jacob S. T., Sif S. (2007) EMBO J. 26, 3558–3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kwak Y. T., Guo J., Prajapati S., Park K. J., Surabhi R. M., Miller B., Gehrig P., Gaynor R. B. (2003) Mol. Cell 11, 1055–1066 [DOI] [PubMed] [Google Scholar]
- 18. Pal S., Yun R., Datta A., Lacomis L., Erdjument-Bromage H., Kumar J., Tempst P., Sif S. (2003) Mol. Cell. Biol. 23, 7475–7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meister G., Eggert C., Bühler D., Brahms H., Kambach C., Fischer U. (2001) Curr. Biol. 11, 1990–1994 [DOI] [PubMed] [Google Scholar]
- 20. Friesen W. J., Wyce A., Paushkin S., Abel L., Rappsilber J., Mann M., Dreyfuss G. (2002) J. Biol. Chem. 277, 8243–8247 [DOI] [PubMed] [Google Scholar]
- 21. Pesiridis G. S., Diamond E., Van Duyne G. D. (2009) J. Biol. Chem. 284, 21347–21359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brahms H., Meheus L., de Brabandere V., Fischer U., Lührmann R. (2001) RNA 7, 1531–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meister G., Fischer U. (2002) EMBO J. 21, 5853–5863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chari A., Golas M. M., Klingenhäger M., Neuenkirchen N., Sander B., Englbrecht C., Sickmann A., Stark H., Fischer U. (2008) Cell 135, 497–509 [DOI] [PubMed] [Google Scholar]
- 25. Fornerod M., Ohno M., Yoshida M., Mattaj I. W. (1997) Cell 90, 1051–1060 [DOI] [PubMed] [Google Scholar]
- 26. Urlaub H., Raker V. A., Kostka S., Lührmann R. (2001) EMBO J. 20, 187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kambach C., Walke S., Young R., Avis J. M., de la Fortelle E., Raker V. A., Lührmann R., Li J., Nagai K. (1999) Cell 96, 375–387 [DOI] [PubMed] [Google Scholar]
- 28. Paushkin S., Gubitz A. K., Massenet S., Dreyfuss G. (2002) Curr. Opin. Cell Biol. 14, 305–312 [DOI] [PubMed] [Google Scholar]
- 29. Raker V. A., Plessel G., Lührmann R. (1996) EMBO J. 15, 2256–2269 [PMC free article] [PubMed] [Google Scholar]
- 30. Pellizzoni L., Yong J., Dreyfuss G. (2002) Science 298, 1775–1779 [DOI] [PubMed] [Google Scholar]
- 31. Friesen W. J., Massenet S., Paushkin S., Wyce A., Dreyfuss G. (2001) Mol. Cell 7, 1111–1117 [DOI] [PubMed] [Google Scholar]
- 32. Palacios I., Hetzer M., Adam S. A., Mattaj I. W. (1997) EMBO J. 16, 6783–6792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huber J., Cronshagen U., Kadokura M., Marshallsay C., Wada T., Sekine M., Lührmann R. (1998) EMBO J. 17, 4114–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lai M. C., Lin R. I., Huang S. Y., Tsai C. W., Tarn W. Y. (2000) J. Biol. Chem. 275, 7950–7957 [DOI] [PubMed] [Google Scholar]
- 35. Li C., Lin R. I., Lai M. C., Ouyang P., Tarn W. Y. (2003) Mol. Cell. Biol. 23, 7363–7376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Black D. L., Pinto A. L. (1989) Mol. Cell. Biol. 9, 3350–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Diem M. D., Chan C. C., Younis I., Dreyfuss G. (2007) Nat. Struct. Mol. Biol. 14, 1173–1179 [DOI] [PubMed] [Google Scholar]
- 38. Bono F., Ebert J., Unterholzner L., Güttler T., Izaurralde E., Conti E. (2004) EMBO Rep. 5, 304–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bono F., Cook A. G., Grünwald M., Ebert J., Conti E. (2010) Mol. Cell 37, 211–222 [DOI] [PubMed] [Google Scholar]
- 40. Kataoka N., Yong J., Kim V. N., Velazquez F., Perkinson R. A., Wang F., Dreyfuss G. (2000) Mol. Cell 6, 673–682 [DOI] [PubMed] [Google Scholar]
- 41. Reichert V. L., Le Hir H., Jurica M. S., Moore M. J. (2002) Genes Dev. 16, 2778–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Richard S., Morel M., Cléroux P. (2005) Biochem. J. 388, 379–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mattaj I. W. (1986) Cell 46, 905–911 [DOI] [PubMed] [Google Scholar]
- 44. Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., Dreyfuss G. (2008) Cell 133, 585–600 [DOI] [PMC free article] [PubMed] [Google Scholar]