Abstract
Activated epidermal growth factor receptor (EGFR) continues to signal in the early endosome, but how this signaling process is regulated is less well understood. Here we describe a protein complex consisting of TIP30, endophilin B1, and acyl-CoA synthetase long chain family member 4 (ACSL4) that interacts with Rab5a and regulates EGFR endocytosis and signaling. These proteins are required for the proper endocytic trafficking of EGF-EGFR. Knockdown of TIP30, ACSL4, endophilin B1, or Rab5a in human liver cancer cells or genetic knock-out of Tip30 in mouse primary hepatocytes results in the trapping of EGF-EGFR complexes in early endosomes, leading to delayed EGFR degradation and prolonged EGFR signaling. Furthermore, we show that Rab5a colocalizes with vacuolar (H+)-ATPases (V-ATPases) on transport vesicles. The TIP30 complex facilitates trafficking of Rab5a and V-ATPases to EEA1-positve endosomes in response to EGF. Together, these results suggest that this TIP30 complex regulates EGFR endocytosis by facilitating the transport of V-ATPases from trans-Golgi network to early endosomes.
Keywords: Endocytosis, Hepatocyte, Signal Transduction, Trafficking, Tumor Suppressor, Vesicles, EGFR, Endophilin B1, Rab5a, TIP30
Introduction
Receptor-mediated endocytosis is a mechanism utilized by eukaryotic cells to rapidly take up specific nutrients and reduce receptor signaling at the plasma membrane. Internalized ligand-receptor complexes are enclosed in early endosomes, also called sorting endosomes, where they are either recycled or delivered to lysosomes for destruction (1, 2). Signaling receptors continue to activate certain downstream pathways from early endosomes (3–6) until ligand-receptor complexes dissociate due to lower luminal pH created by vacuolar V-ATPases,2 the major proton pump responsible for endosomal and lysosomal acidification (2, 7–9). Inactivation of V-ATPases blocks the transition from early to late endosomes (10). Therefore, the proper endosomal targeting and activity of V-ATPases contribute to the tight regulation of both endocytic trafficking and receptor endosomal signaling.
Acidic luminal pH in early endosomes is the driving force for receptors to release their ligands, such as insulin and low density lipoprotein (LDL) (1, 2). On the basis of individually tracking EGF and EGFR, it has long been considered that EGFR travels together with EGF until they reach lysosomes (11, 12). However, it is unclear whether they remain bound to each other on the way to lysosomes. Recent studies have suggested that EGFR is inactivated before being degraded and that EGF dissociates from EGFR prior to lysosomal transfer (6, 13–15).
Rab5a is a small GTPase that regulates early endosome fusion in vitro (16), motility of early endosomes on microtubules (17), and the traffic between endosomes and lysosomes (18). Deletion of Rab5a in cells inhibits the transport of EGFR from early endosomes to lysosomes and consequently causes sustained EGFR signaling and delayed EGFR degradation (19). Despite its importance to endocytic transport, how Rab5a mediates down-regulation of receptor signaling remains unclear.
TIP30, also known as HTATIP2 or CC3 (20, 21), is a tumor suppressor that has been demonstrated to act as a transcription cofactor to repress transcription in the nucleus (22, 23) and to localize at the nuclear envelope to block nuclear importing (24). However, TIP30 also localizes in the cytoplasm, where its function is not known (25–28).
Here we report that a newly identified protein complex containing TIP30, ACSL4, and Endo B1 drives EGF-EGFR complex endocytic trafficking by facilitating the localization of Rab5a and V-ATPases to early endosomes. Rab5a and V-ATPase reside in vesicles devoid of the early endosomal marker EEA1 and the recycling endosomal marker transferrin receptor (TfR), suggesting that these vesicles are post-trans-Golgi network vesicles responsible for the transport of integral membrane protein V-ATPases. Our data suggest a mechanism by which Rab5a in cooperation with other proteins in the TIP30 complex transports V-ATPases to early endosomes and induces the dissociation of EGF from EGFR and the termination of EGFR endosomal signaling.
EXPERIMENTAL PROCEDURES
Cell Culture
PLC/PRF/5 and HepG2 cell lines were purchased from ATCC. Cells were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum and penicillin/streptomycin (Invitrogen) at 37 °C in 5% CO2.
DNA Constructs and shRNAs
The pSin-EF2 vector (29) was converted to destination vectors by cloning the Gateway cassette RfA (reading frame A, Invitrogen) with either N-terminal or C-terminal HA tag, CFP, EYFP, or DsRed fluorescent proteins into blunted SpeI and EcoRI sites. Human Rab5a, ACSL4, and EndoB1 were amplified using RT-PCR from mRNA isolated from PLC/PRF/5 cells. TIP30 was subcloned from pFlag7-TIP30 and pFlag7-TIP30M (30). For bimolecular fluorescence complementation assays, VC155 and VN173 (31) were cloned into pCDNA3.1 and pSin-EF2, respectively, and both were also converted to destination vectors. Lentiviral plasmids producing shRNAs against TIP30, Rab5a, and ACSL4 were from Sigma-Aldrich. Lentiviral plasmids for shRNAs against Endo B1 were from Open Biosystems.
Antibodies
HA (HA-7), β-actin (AC-15), and Endo B1 antibodies were from Sigma-Aldrich. AKT, AKT-pS473, EEA1, and Rab5a antibodies were from Cell Signaling. EGFR-pY845, TfR, Alexa Fluor 546 goat anti-mouse, and Alexa Fluor 594 goat anti-rabbit antibodies were from Invitrogen. Anti-EGFR antibody was from Millipore. ATP6V1H antibody was from Santa Cruz Biotechnology. LAMP1 antibody was purchased from The Developmental Studies Hybridoma Bank at University of Iowa.
EGFR Internalization and Immunofluorescence
PLC/PRF/5 cells were infected by lentiviruses producing shRNA against indicated genes. Cells were pooled after being selected for 4 days with 2 μg/ml puromycin. At least two confirmed knockdown pools for each targeted gene were used for the experiments in Figs. 4–8. Control and knockdown cells were cultured on cover glass and were serum-starved for 24 h in DMEM. Wild type and Tip30−/− primary hepatocytes were starved for 3 h. Cells were incubated with 100 ng/ml Alexa-488-conjugated EGF (Alexa488-EGF) (Invitrogen) and 20 μg/ml cycloheximide on ice for 1 h and then were washed four times with cold PBS and incubated in DMEM with 20 μg/ml cycloheximide at 37 °C for different time periods. Cells were fixed in 4% paraformaldehyde in PBS for 15 min, permeabilized with 0.1% Triton X-100 for 2 min, and stained for the indicated proteins. Images were obtained with a Zeiss LSM 510 Meta confocal microscope (Carl Zeiss) using Plan-Apochromat 63×/1.40 oil objective. Pinhole size was set to 1 airy unit for all channels. All images are representative single optical sections. To determine EGFR stability upon EGF treatment, cells were cultured in 6-cm dishes and treated as above except that unlabeled EGF was used, and the cells were collected for immunoblot at various time points.
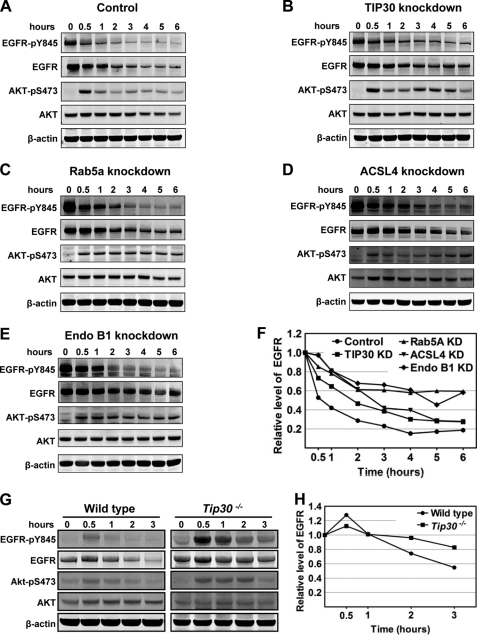
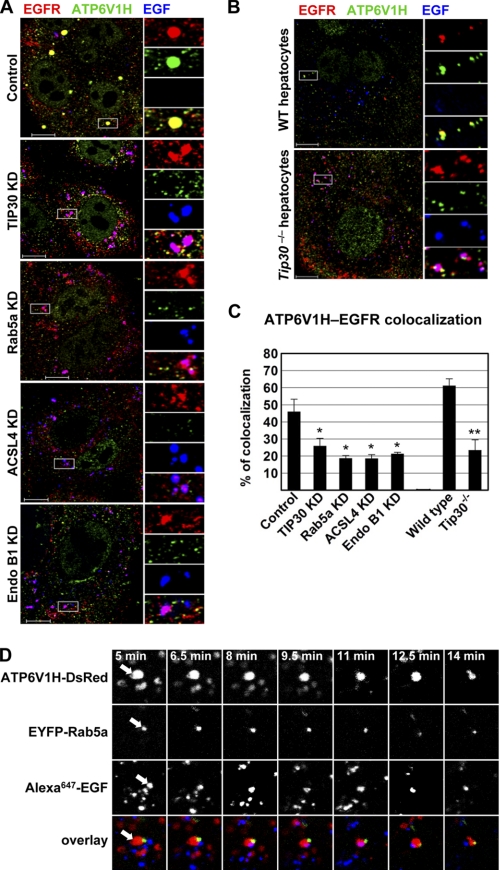
FIGURE 4.
TIP30, Rab5a, ACSL4, and Endo B1 promote the endocytic down-regulation of EGFR. A–E, knockdown of TIP30, Rab5a, ACSL4, or Endo B1 results in delayed EGFR degradation and sustained EGFR signaling. Control PLC/PRF/5 cells (A), TIP30 (B), Rab5a (C), Endo B1 (D), and ACSL4 (E) knockdown cells were incubated with 100 ng/ml EGF after being serum-starved for 24 h and then were washed with cold PBS and incubated with cycloheximide at 37 °C. Cells were collected at various time points after EGF internalization and subjected to immunoblot analysis with the indicated antibodies. Results are typical and representative of experiments on cells from two different shRNAs. F, quantification of EGFR protein levels in A–E using Odyssey 2.1 software. KD, knockdown. G, deletion of Tip30 in mouse primary hepatocytes leads to delayed EGFR degradation and sustained EGFR signaling. Primary hepatocytes were isolated from wild type and Tip30−/− mice. Endocytosis-induced EGFR degradation was analyzed as described in A–E. H, quantification of EGFR protein levels in G using Odyssey 2.1 software.
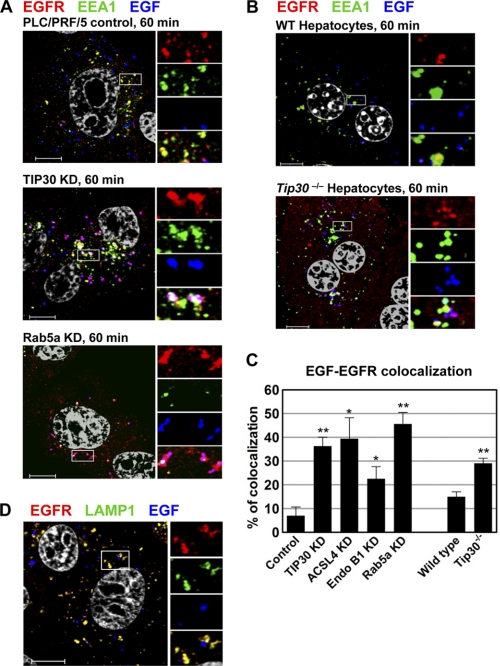
FIGURE 5.
TIP30 and Rab5a depletion delays the exit of EGF from early endosomes. A, knockdown (KD) of TIP30 or Rab5a causes delayed exit of Alexa488-EGF from early endosomes. Cells expressing control shRNA or shRNA against TIP30 or Rab5a were subjected to EGFR internalization analysis. After internalization of Alexa488-EGF (blue) for 60 min, cells were fixed and double-immunostained for EGFR (red) and EEA1 (green). Nucleus was stained by DAPI (gray). Magenta results from overlap between red and blue. Results are representative of at least three independent experiments on cells expressing two different shRNAs. The boxed areas are magnified. Scale bars, 10 μm. B, deletion of Tip30 in mouse primary hepatocytes leads to trapping of EGF in early endosomes. Wild type and Tip30−/− primary hepatocytes were subjected to the same EGFR internalization analysis as described in A. The boxed areas are magnified. Scale bars, 10 μm. C, quantitative analysis of EGF-EGFR colocalization 60 min after EGF internalization. 60 cells represented by Fig. 5, A and B, and Fig. 6A in each group were analyzed using MBF_ImageJ. Pearson's colocalization coefficients were calculated and converted to percentages. Data represent means ± S.E. *, p < 0.05, **, p < 0.01, relative to control or wild type cells; Student's t test. D, EGF exits endosomes in LAMP1-negative vesicles. Cells were stained for EGFR and LAMP1 60 min after EGF internalization. Scale bar, 10 μm.
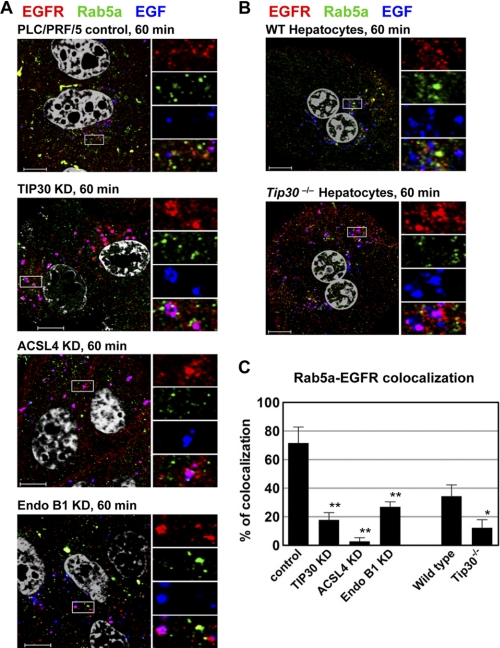
FIGURE 6.
TIP30, ACSL4, and Endo B1 are required for the localization of Rab5a to early endosomes. A, knockdown (KD) of TIP30, ACSL4, or Endo B1 inhibits the localization of Rab5a to early endosomes. Control cells and TIP30, ACSL4, or Endo B1 knockdown cells were subjected to EGFR internalization analysis. 60 min after internalization of Alexa488-EGF (blue), cells were immunostained for EGFR (red) and Rab5a (green). Nucleus is stained by DAPI (gray). Results are typical and representative of three experiments on cells from two different shRNAs. The boxed areas are magnified. Scale bars, 10 μm. B, Tip30 deletion in primary hepatocytes inhibits the recruitment of Rab5a to early endosomes. Primary hepatocytes were stained for EGFR (red) and Rab5a (green) after 60 min of EGF (blue) internalization. The boxed areas are magnified. Scale bars, 10 μm. C, quantitative analysis of Rab5a-EGFR colocalization after 60 min of EGF internalization was performed as described in the legend for Fig. 2. Data represent means ± S.E. *, p < 0.05, **, p < 0.01, relative to control or wild type cells; Student's t test.
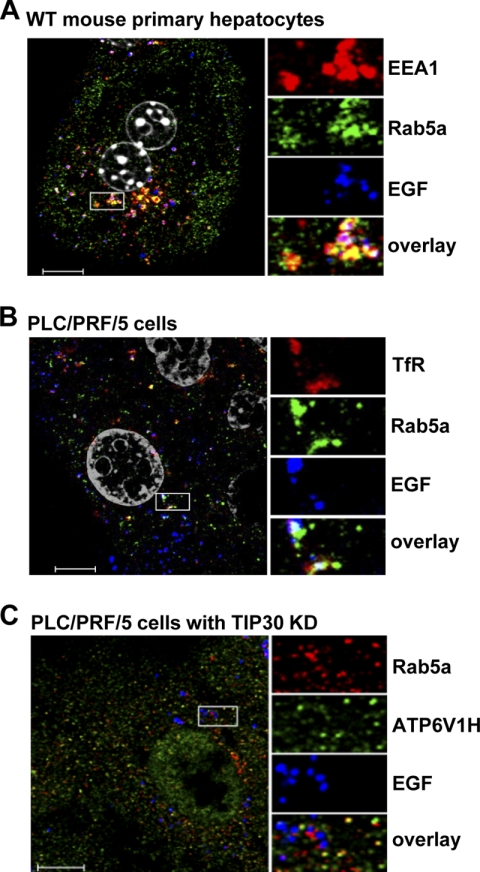
FIGURE 7.
Rab5a vesicles carry V-ATPases and are devoid of EEA1 and TfR. A, EGFR-negative Rab5a vesicles are not early endosomes. Wild type mouse primary hepatocytes were immunostained for EEA1 (red) and Rab5a (green) after 30 min of EGF (blue) internalization. Scale bar, 10 μm. B, TfR partially colocalizes with Rab5a. PLC/PRF/5 cells were immunostained for TfR (red) and Rab5a (green) after 10 min of EGF (blue) internalization. Scale bar, 10 μm. C, V-ATPases colocalize with Rab5a in transport vesicles. TIP30 knockdown (KD) cells were immunostained for V-ATPase (red) and Rab5a (green) after 30 min of EGF (blue) internalization. Scale bar, 10 μm.
FIGURE 8.
Rab5a vesicles transport V-ATPases to early endosomes. A, depletion of TIP30, ACSL4, or Endo B1 inhibits the loading of V-ATPases on endocytic vesicles. Cells were immunostained for EGFR (red) and ATP6V1H (green) after 60 min of EGF (blue) internalization. Results are typical and representative of three experiments on cells from two different shRNAs. The boxed areas are magnified. Scale bars, 10 μm. KD, knockdown. B, deletion of Tip30 in mouse primary hepatocytes results in mislocalization of V-ATPases. Immunostaining was performed as described in A. Scale bars, 10 μm. C, colocalization between V-ATPases and EGFR was analyzed using MBF_ImageJ. Pearson's colocalization coefficients were calculated and converted to percentages. *, p < 0.05, **, p < 0.01, relative to control or wild type cells; Student's t test. D, Alexa647-EGF is released after EGF endocytic vesicles merge with Rab5a vesicles. Live cells expressing ATP6V1H-DsRed (red) and EYFP-Rab5a (green) were imaged by confocal microscopy at the indicated times after Alexa647-EGF internalization, and images of a single focal plane were acquired. A typical EGF endocytic vesicle movement was shown. Arrows point toward the two vesicles undergoing merge and EGF release.
Immunoprecipitation
PLC/PRF/5 cells were transfected with indicated constructs. Whole cell extracts were prepared from pooled stable clones as described previously with modifications (32). Briefly, cells were homogenized by 20 strokes in two packed cell pellet volumes of buffer A (10 mm Hepes, pH 7.9, 10 mm KCl, 0.5 mm DTT, protease inhibitor mixture) using a Kontes homogenizer (B pestle). Another 20 strokes were applied after adding 1.5 cell pellet volumes of buffer B (50 mm Hepes, pH 7.9, 0.6 mm EDTA, 1.5 mm DTT, 1.26 m NaCl, 75% glycerol) followed by centrifugation at 100,000 × g for 1 h. The supernatant was dialyzed against BC300 (20 mm Hepes, pH 7.9, 20% glycerol, 0.2 mm EDTA, 0.5 mm DTT, 0.3 m KCl) (33) and centrifuged at 15,000 rpm for 20 min followed by rotating with anti-HA agarose beads (Roche Diagnostics) overnight at 4 °C. The beads were centrifuged and extensively washed using BC300 buffer. Immunoprecipitates were eluted with HA peptides (Roche Diagnostics), denatured, resolved on SDS-PAGE, and subjected to silver stain, immunoblot, or LC-MS/MS spectral analyses (The MSU Proteomics Facility, Michigan State University).
Mouse Hepatocyte Isolation
Primary hepatocytes were isolated from 8-week-old wild type and Tip30−/− mice as described (34) with modifications. Briefly, the inferior vena cava was cannulated, and the liver was first perfused in situ with an oxygenated Krebs-Ringer buffer (115 mm NaCl, 5.9 mm KCl, 25 mm NaHCO3, 10 mm glucose, 20 mm Hepes, pH 7.4) with 0.1 mm EGTA at 37 °C followed by perfusion with oxygenated Krebs-Ringer buffer containing 0.25 mm CaCl2 and 20 μg/ml Liberase Blendzyme 3 (Roche Applied Science). Liver was removed and then gently minced in ice-cold Krebs-Ringer buffer. Liver cell suspension was then filtered with Falcon cell strainers (BD Biosciences) and washed three times by centrifugation at 50 × g for 2 min at 4 °C. Cell viability was determined by trypan blue exclusion. Cells were cultured in DMEM (with 10% FBS and 1× penicillin/streptomycin) at 37 °C with 5% CO2.
RESULTS
TIP30 Forms a Complex with Rab5a, Endo B1, and ACSL4
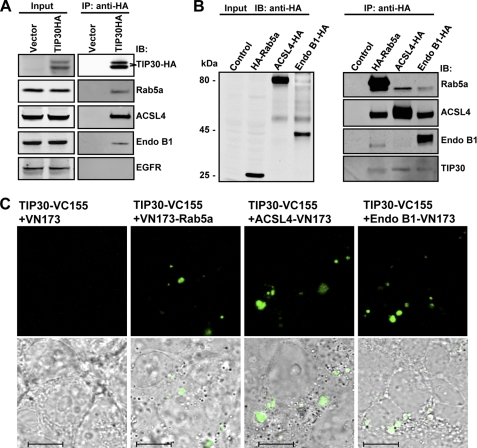
To identify cytosolic proteins that interact with TIP30, we stably expressed TIP30 protein with an HA tag fused to its C-terminal end (TIP30-HA) in human hepatocellular carcinoma cells (PLC/PRF/5). Co-immunoprecipitation assays were performed using whole cell extracts generated from cells expressing TIP30-HA or control vector. Mass spectrometric analysis identified Rab5a, ACSL4, and EndoB1 (also known as Bif-1) in the immunoprecipitates. Association of these proteins with TIP30-HA was confirmed by immunoblot analysis using specific antibodies (Fig. 1A). The interactions were further confirmed by reciprocal immunoprecipitation. When Rab5a, Endo B1, or ACSL4 were HA-tagged, each could be specifically co-immunoprecipitated with endogenous TIP30 and the other three endogenous proteins (Fig. 1B). Although Endo B1 failed to be immunoprecipitated with ACSL4-HA, possibly due to the interference of HA at its C terminus, endogenous ACSL4 was detected in Endo B1-HA immunoprecipitates. Notably, Rab5a was co-immunoprecipitated with these proteins in the presence of 0.2 mm EDTA, implying that the interaction is independent of its nucleotide binding status. We next used bimolecular fluorescence complementation analysis (31) to visualize the association of TIP30 with these proteins in living cells. We observed that co-expression of TIP30-VC155 with VN173-Rab5a, ACSL4-VN173, or Endo B1-VN173 in cells reconstituted fluorescence, whereas co-expression of TIP30-VC155 and control VN173 did not (Fig. 1C), indicating that TIP30 directly or indirectly interacts with these proteins in living cells.
FIGURE 1.
TIP30 interacts with Rab5a, ACSL4, and Endo B1. A, immunoblot confirmed the association of Rab5a, ACSL4, and Endo B1 with TIP30. The immunoprecipitates were resolved on SDS-PAGE and analyzed by immunoblotting (IB) with the indicated antibodies. The two polypeptides that reacted with anti-HA antibodies might result from posttranslational modifications on TIP30. Input was 10% (equals 70 μg of protein) of the volume of lysates used in the immunoprecipitation (IP) assays. The EGFR blot serves as a negative control. B, reciprocal co-immunoprecipitation. Whole cell extracts made from cells expressing HA-Rab5a, ACSL4-HA, Endo B1-HA, or empty vector were subjected to co-immunoprecipitation with α-HA-agarose beads. The immunoprecipitates were subjected to immunoblot analysis with the indicated antibodies. Input was 10% of the volume of lysates used in the immunoprecipitation assays. C, bimolecular fluorescence complementation analysis was performed on 293T cells by co-expressing TIP30-VC155 with VN173, VN173-Rab5a, ACSL4-VN173, or Endo B1-VN173. Green indicates fluorescent signal from Venus, a GFP variant. Bottom panels show overlays of differential interference contrast and representative confocal microscope images. Scale bars, 10 μm.
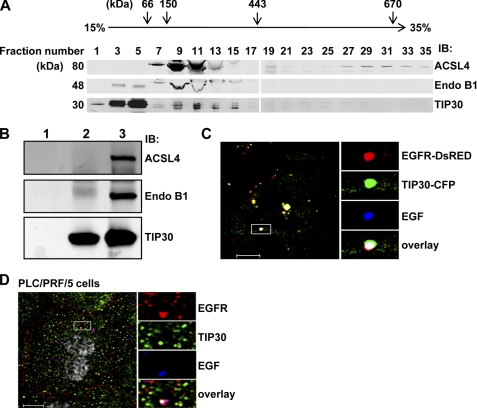
To determine whether endogenous TIP30 associates with endogenous ACSL4 and EndoB1, we did glycerol gradient sedimentation and observed that these proteins co-sedimented at a position between the 150- and 443-kDa marker proteins in a 15–35% glycerol gradient (Fig. 2A). The majority of TIP30 and part of Endo B1 appeared in fractions containing proteins smaller than 66 kDa, suggesting that these two proteins may exist mainly as a heterodimer or as separated homodimers. An in vitro binding assay showed that bacterially expressed recombinant ACSL4 and Endo B1 were able to directly bind purified baculovirus-expressed recombinant TIP30 (Fig. 2B). These results indicate that TIP30, ACSL4, and Endo B1 may form a protein complex to interact with Rab5a.
FIGURE 2.
Endogenous TIP30, ACSL4, and Endo B1 associate together. A, endogenous TIP30, ACSL4, and Endo B1 were co-sedimented in a glycerol gradient. Whole cell extracts of PLC/PRF/5 cells and protein markers were loaded on the top of linear 10–35% (v/v) glycerol gradients (10 ml) formed from the bottom of 12-ml Beckman tubes. The buffer throughout the gradients was 20 mm Hepes-KOH (pH 7.0), 100 mm KCl, 1 mm dithiothreitol. The gradients were centrifuged at 200,000 × g in a SW41 Ti rotor (Beckman) at 4 °C for 20 h. After centrifugation, fractions were collected, concentrated, and subjected to immunoblot (IB) analysis. Albumin (66 kDa), alcohol dehydrogenase (150 kDa) apoferritin (443 kDa), thyroglobulin (670 kDa), and blue dextran (2,000 kDa) were run in parallel as molecular mass indicators. B, direct interaction of TIP30, ACSL4, and Endo B1. GST-ACSL4 and His-Endo B1 (lane 1), FLAG-TIP30 (lane 2), or GST-ACSL4, His-Endo B1, and FLAG-TIP30 (lane 3) were subjected to anti-FLAG M2 immunoprecipitation. 100 ng of each protein was used. Aliquots of precipitates were resolved on SDS-PAGE and analyzed by immunoblot with the indicated antibodies. Purified FLAG-TIP30 was described previously (30). Bacterially expressed GST-ACSL4 was purchased from Abnova. His-Endo B1 was expressed in BL21. C, TIP30 and EGF-EGFR are colocalized in endosomes in response to EGF treatment. HepG2 cells co-expressing TIP30-CFP (green) and EGFR-DsRed (red) were treated with 10 ng/ml Alexa647-EGF (blue) for 10 min and then analyzed by confocal microscopy. A typical image is shown. The boxed areas are magnified. Scale bar, 10 μm. D, TIP30 is localized to EGFR-positive endosomes. PLC/PRF/5 cells were immunostained with antibodies for EGFR (red) and TIP30 (green) after 30 min of Alexa488-EGF (blue) internalization. The boxed areas are magnified. Scale bar, 10 μm.
Rab5a colocalizes with EGFR in endosomes in response to EGF (35, 36). To test whether TIP30 is also targeted to endosomes, we co-expressed TIP30-CFP and EGFR-DsRed fusion proteins and examined their localization in HepG2 cells lacking detectable endogenous TIP30 and EGFR. Confocal microscopy analysis showed that TIP30-CFP partially colocalized with EGF and EGFR-DsRed in endosomes 10 min after cells were treated with Alexa-647-conjugated EGF (Alexa647-EGF; Fig. 2C), suggesting that TIP30 was also targeted to endosomes. Consistently, immunostaining of endogenous TIP30 and EGFR revealed that TIP30 was partially localized to EGF-EGFR-positive endosomes (Fig. 2D). Taken together, our protein-protein interaction and colocalization studies suggest that TIP30, ACSL4, and Endo B1 form a protein complex and may function together with Rab5a on the endocytic pathway.
Inhibition of TIP30, ACSL4, or Endo B1 Leads to Delayed EGFR Degradation and Sustained EGFR Signaling
Knockdown of Rab5a expression in HeLa cells retards EGFR transport from early endosomes to late endosomes and delays the degradation of EGFR (19). The interaction of TIP30, ACSL4, and Endo B1 with Rab5a raises the possibility that these proteins may also function in endocytic pathways. We performed EGFR internalization analysis to investigate whether the inhibition of TIP30, ACSL4, or Endo B1 affects the endocytic down-regulation of EGFR. Instead of continuous incubation with EGF, serum-starved cells were first incubated with EGF on ice to allow for binding of ligand to receptors, and then they were washed to remove unbound EGF before being moved to 37 °C for internalization. Nascent protein synthesis was blocked by cycloheximide in the culture medium. This approach eliminates the interference from continuous ligand entrance and new receptor synthesis, thus enabling us to monitor both the traffic and the fate of EGF-EGFR complexes. We found that knockdown of TIP30, Rab5a, ACSL4, or Endo B1 in PLC/PRF/5 cells (Fig. 3, A–D) resulted in a slower reduction of total EGFR protein levels (Fig. 4, A–F). To corroborate these findings, we further examined whether phosphorylation of EGFR at Tyr-845 (EGFR-pY845) and phosphorylation of AKT at Ser-473 (AKT-pS473), a downstream target of EGFR signaling, were affected. Remarkably, the levels of EGFR-pY845 and AKT-pS473 were sustained much longer in TIP30, ACSL4, Endo B1, or Rab5a knockdown cells than in control cells (Fig. 4, A–F). Of note, Akt phosphorylation was rapidly increased followed by a decline and then increased again after a 3-h internalization (Fig. 4, A–D). Although the exact mechanism to explain this phenomenon remains to be tested, one can envisage that it may reflect the autonomic balance of kinase and phosphatase activities for Akt in cells.
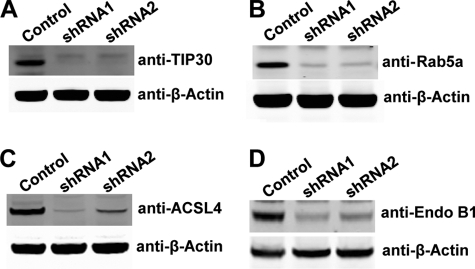
FIGURE 3.
Knockdown of TIP30, Rab5a, ACSL4, and Endo B1 in PLC/PRF/5 cells. A–D, immunoblot analyses show knockdown of TIP30, Rab5a, ACSL4, or Endo B1 in PLC/PRF/5 cell. Whole cell lysates were made from cells expressing a scramble shRNA or shRNA against TIP30 (A), Rab5a (B), ACSL4 (C), or Endo B1 (D). Two shRNAs against each gene were used, and equal amounts of protein were loaded per lane.
To assess the effect of TIP30 deletion on EGFR endocytic down-regulation in normal cells and to exclude off-target effects of shRNAs, we performed EGFR internalization analysis using primary hepatocytes isolated from wild type and Tip30 knock-out mouse littermates. Deletion of Tip30 in primary hepatocytes delayed EGF-induced EGFR degradation (Fig. 4, G and H), and the phosphorylation of EGFR at Tyr-845 and Akt at Ser-473 was higher and sustained longer in Tip30−/− hepatocytes than in wild type hepatocytes. Together, these data provide strong evidence that TIP30, ACSL4, Endo B1, and Rab5a not only physically associate but also function together in promoting the endocytic down-regulation of EGFR protein level and signaling.
TIP30, ACSL4, Endo B1, and Rab5a Are Required for EGF-EGFR Endocytic Trafficking
An early study showed that EGF dissociates from EGFR at later stages of endocytosis (37). Moreover, it has been suggested that EGFR is inactivated before being transferred to lysosomes and degraded (6, 13–15). To test whether depletion of TIP30, ACSL4, Endo B1, or Rab5a affects EGF dissociation from EGFR, we simultaneously tracked Alexa488-EGF and EGFR at different time points after internalization using confocal microscopy. We found that fluorescent EGF partially colocalizes with EGFR and Rab5a in control cells after 15 and 30 min of internalization (supplemental Fig. S1A). Interestingly, Alexa488-EGF exited in membrane-bound vesicles from EGFR-positive endosomes in control cells after 60 min of internalization (Fig. 5A). In contrast, the majority of EGF remained colocalized with EGFR in TIP30 or Rab5a knockdown cells (control cells, 7 ± 3%; TIP30 knockdown cells, 36 ± 4%; Rab5a knockdown cells, 46 ± 5%; n = 60, p < 0.01 versus control cells; Fig. 5, A and C). The EGF vesicles were devoid of early endosomal markers EEA1 and Rab5a, as well as late endosomal and lysosomal marker LAMP1 (Fig. 5D), indicating that they are neither early/late endosomes nor lysosomes.
We next performed EGFR internalization analysis using wild type and Tip30−/− primary hepatocytes. Consistently, Tip30 deletion increased the colocalization of EGF and EGFR nearly 2-fold (wild type primary hepatocytes, 15 ± 2%; Tip30−/− primary hepatocytes, 29 ± 2%; n = 20, p < 0.01; Fig. 5, B and C).
Likewise, knockdown of ACSL4 or Endo B1 expression also increased the colocalization of EGF and EGFR (ACSL4 knockdown cells, 40 ± 8%; Endo B1 knockdown cells, 23 ± 5%; n = 60, p < 0.05 versus control cells; Figs. 5C and 6A). The majority of EGF colocalized with EGFR even after 120 min of internalization in knockdown and Tip30−/− cells (supplemental Fig. S1, A–C). Taken together, these results suggest that TIP30, ACSL4, Endo B1, and Rab5a are involved in promoting EGF dissociation from EGFR during endocytic trafficking.
TIP30, ACSL4, and Endo B1 Promote the Loading of Rab5a on Early Endosomes
Endocytic vesicles gain Rab5a dynamically mostly by membrane fusion with Rab5-positive endosomes in the course of cargo transport (38). This is further supported by the observation that Rab5a is mainly localized on numerous vesicles in the perinuclear region of cells under steady state condition and is rapidly recruited to early endocytic vesicles at the cell periphery in response to EGF (35, 36). The similar effects of TIP30 and Rab5a knockdown on EGFR endocytosis prompted us to test whether TIP30 is involved in the loading of Rab5a on early endosomes. In control cells, Rab5a appeared in EGFR-positive endosomes from which EGF had exited after 60 min of EGF internalization (Fig. 6A). By contrast, the colocalization of Rab5a and EGFR was significantly decreased in TIP30 knockdown cells (control cells, 72 ± 11%; TIP30 knockdown cells, 18 ± 5%; n = 60, p < 0.01; Fig. 6, A and C) and Tip30−/− primary hepatocytes (wild type hepatocytes, 34 ± 8%; Tip30−/− hepatocytes, 12 ± 5%; n = 20, p < 0.05; Fig. 6, B and C). Similar results were obtained with ACSL4 or Endo B1 knockdown cells (ACSL4 knockdown cells, 3 ± 2%; Endo B1 knockdown cells, 27 ± 4%; n = 60, p < 0.01 versus control cells; Fig. 6, A and C). Interestingly, there were a few Rab5a-positive early endosomes in Endo B1 knockdown cells, which was probably the result of incomplete knockdown. However, these Rab5a-positive endosomes appeared different from those in control cells. They did not release EGF even after 120 min of internalization (supplemental Fig. S1B), indicating that Endo B1 has an additional function after Rab5a recruitment. This provides a possible explanation for the observation that Endo B1 knockdown cells have less EGF-EGFR colocalization and more Rab5a-EGFR overlap but have longer lasting EGFR stability when compared with TIP30 and ACSL4 knockdown cells (Figs. 4, A–F, 5C, and 6C). Together, these results indicate that TIP30, ACSL4, and Endo B1 promote efficient Rab5a localization to early endosomes.
Rab5a Vesicles Transport V-ATPases to Early Endosomes
Intriguingly, we noted that Rab5a appeared in vesicles when it was not localized to EGFR-positive endosomes (Fig. 6, A and B). To further characterize those EGFR-negative Rab5a vesicles, we co-stained EEA1 and Rab5a in wild type mouse primary hepatocytes 30 min after EGF internalization. The EGFR-negative Rab5a vesicles were also negative for EEA1 and TfR (Fig. 7, A and B), suggesting that they are neither plasma membrane-derived endocytic vesicles nor recycling endosomes, but likely transporting vesicles that originate from the trans-Golgi network.
Dissociation of ligand-receptor complexes inside endosomes is caused by the low luminal pH created by V-ATPases. Our previously presented results showed that lack of Rab5a in early endosomes was concomitant with delayed EGF-EGFR dissociation induced by loss of TIP30 or its interacting proteins. Rab5a did not coexist with EGF in EGFR-positive endosomes, suggesting that Rab5a vesicles may deliver V-ATPases to early endosomes to drive EGF-EGFR dissociation. To test this hypothesis, we examined the intracellular localization of V-ATPases by staining for the regulatory subunit H (ATP6V1H). ATP6V1H-positive staining was observed in Rab5a vesicles lacking EGF and EGFR in TIP30 knockdown cells (Fig. 7C). Significant reduction of ATP6V1H localization to EGFR-positive endosomes was observed in TIP30, Rab5a, ACSL4, or Endo B1 knockdown cells (control cells, 46 ± 7%; TIP30 knockdown cells, 25 ± 4%; Rab5a knockdown cells, 19 ± 1%; ACSL4 knockdown cells, 19 ± 2%; Endo B1 knockdown cells, 21 ± 1%; n = 60, p < 0.05 versus control cells; Fig. 8, A and C) and in Tip30−/− primary hepatocytes (wild type hepatocytes, 61 ± 4%; Tip30−/− hepatocytes, 23 ± 6%; n = 20, p < 0.01; Fig. 8, B and C). Moreover, in live PLC/PRF/5 cells co-expressing EYFP-Rab5a and ATP6V1H-DsRed, EGF-positive endosomes fused with Rab5a-ATP6V1H vesicles at 11 min after EGF internalization and released Alexa647-EGF 3 min after the merge (Fig. 8D).
To determine whether mislocalization of V-ATPases affects endosomal acidification, we monitored endosomal pH after pHrodo-EGF:Alexa647-EGF (7:3) internalization (39). Indeed, TIP30 knockdown resulted in significantly less acidic endosomes (supplemental Fig. S2, A and B). Taken together, these data indicate that Rab5a contribute to endosomal acidification by transporting V-ATPases to endocytic vesicles and that TIP30, ACSL4, and Endo B1 are required for efficient transport.
DISCUSSION
The present study describes a novel protein complex consisting of TIP30, ACSL4, and Endo B1 that interacts with Rab5a and regulates EGFR endocytic trafficking. Down-regulation of these proteins results in trapping of EGF-EGFR complexes in early endosomes, which enabled us to further dissect the EGFR endocytic pathway. We uncovered two consecutive events following EGF-EGFR entrance into the early endosomes because these two events became obviously detectable when the function of the TIP30 complex was inhibited. First, newly formed endosomes are devoid of V-ATPases and Rab5a. V-ATPases are delivered to early endosomes by the Rab5a vesicles. Second, localization of Rab5a and V-ATPases to the early endosomes results in the termination of EGFR endosomal signaling. Clearly, inhibiting the function of the TIP30 complex can block the transport of V-ATPases to endosomes, leading to EGF-bound EGFR detention in endosomes, delayed EGFR endocytic degradation, and sustained activation of Akt. Given that EGF-bound EGFR enclosed in endosomes is sufficient to activate downstream molecules and to induce cell survival (40), our results indicate that enhanced EGFR signaling by inhibition of TIP30, ACSL4 and EndoB1 may contribute to the initiation and progression of cancers. Indeed, we have recently reported that Tip30 deletion in MMTV-Neu mice leads to enhanced EGFR signaling and development of estrogen receptor-positive and progesterone receptor-negative mammary tumors (41). Thus, the regulation of EGFR-mediated endocytosis by the TIP30 complex may serve as a general mechanism to govern EGFR signaling and suppress tumorigenesis.
Endo B1 belongs to a family of proteins containing N-terminal Bin-Amphiphysin-Rvs (BAR) domains and is involved in apoptosis, autophagy, mitochondrial fission, and endocytic trafficking (42). The role of Endo B1 in EGFR endocytosis is consistent with a previous report that Endo B1 colocalizes with EEA1 in the early endosome of neural cells in response to nerve growth factor (43). It is noteworthy that Endo B1 knock-out mice exhibited spontaneous development of lymphomas, hepatocellular carcinomas, and mammary tumors (44). These phenotypes resemble the phenotypes of Tip30 knock-out mice (25).3 Thus, we speculate that TIP30 and Endo B1 may act in concert to suppress tumorigenesis, at least in part, through the regulation of EGFR-mediated endocytosis. With the identification of the interaction between TIP30 and Endo B1, this hypothesis can now be addressed.
Rab family proteins have been well known to be essential for vesicle targeting (45). Rab5a is recruited to the early endosomes, where it promotes the endocytic down-regulation of EGFR and transition from endosome to lysosome. Before entering endosomes, Rab5a was found in smaller vesicles localized in the perinuclear region (36, 46). These Rab5 vesicles are redistributed to the cell periphery region, where they colocalize with EGFR in early endosomes in response to EGF treatment (17, 35, 36). Notably, newly formed endocytic vesicles containing transferrin, LDL, EGF, or influenza virus were devoid of Rab5a and acquired Rab5a through merging with Rab5a vesicles (35). Our data support these observations and clearly show that besides the EEA1- and EGFR-positive endocytic vesicles, there is a population of Rab5a-positive vesicles that are negative for EEA1 and EGFR but positive for V-ATPases. Given that intracellular trafficking of EGF-EGFR or entry of dengue and West Nile viruses into HeLa cells by clathrin-mediated endocytosis requires vacuolar acidic pH and Rab5a (47), we propose that Rab5a is involved in post-trans-Golgi network transport of integral membrane proteins and helps create acidic endosomal pH by targeting V-ATPases to endosomes, thereby providing the driving force for EGFR endocytic trafficking and viral entry into host cells (Fig. 9).
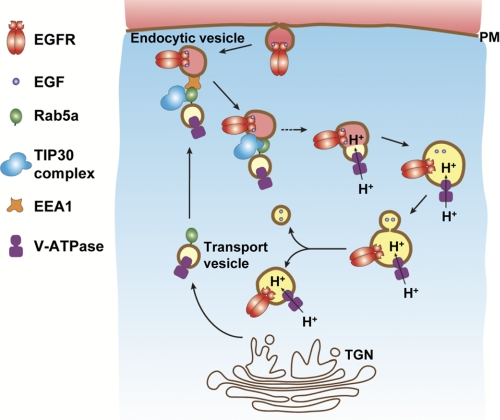
FIGURE 9.
Proposed model shows the regulation of Rab5a vesicle-mediated transport of V-ATPases to endocytic vesicles in EGFR endocytic trafficking. Rab5a directs the trans-Golgi network (TGN)-originated vesicles carrying V-ATPases toward the cell peripheral and recognizes endocytic vesicles presumably by interacting with EEA1 (48, 49). Then, the TIP30 complex facilitates the fusion of the two vesicles to help load V-ATPases, which can start pump H+ into endosomes. EGF dissociates from EGFR due to lower pH created by V-ATPases and might exit endosomes in membrane-bound vesicles.
We were surprised to see that Alexa488-EGF vesicles pinched off from endosomes shortly after internalization. This event was observed in both human liver cancer cells and mouse primary hepatocytes. The Alexa488-EGF vesicles are previously uncharacterized vesicles that are devoid of early endosomal, recycling endosomal, and lysosomal markers, implying that EGF might exit endosomes after the dissociation with EGFR. Nevertheless, it will be important to determine whether Alexa488-EGF in these vesicles is intact or degraded.
Supplementary Material
Acknowledgments
We thank Dr. Stephen Prescott for generously sharing anti-ACSL4 antibody, Dr. Chang-deng Hu for generously sharing bimolecular fluorescence complementation plasmids, and Drs. Richard Schlegel and Frank Suprynowicz for generously sharing pHrodo-EGF. We are grateful to Drs. Sandra Haslam, Hans Cheng, Jerry Dodgson, Karen Friderici, and Richard Schwartz for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 DK066110-01 and W81XWH-08-1-0377 from the NIDDK (to H. X.). This work was also supported by a grant from the Department of Defense.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S2.
A. Li, J. Pecha, and H. Xiao, unpublished data.
- V-ATPase
- vacuolar (H+)-ATPase
- EGFR
- epidermal growth factor receptor
- Endo B1
- endophilin B1
- TfR
- transferrin receptor.
REFERENCES
- 1. Mellman I. (1996) Annu. Rev. Cell Dev. Biol. 12, 575–625 [DOI] [PubMed] [Google Scholar]
- 2. Maxfield F. R., McGraw T. E. (2004) Nat. Rev. Mol. Cell Biol. 5, 121–132 [DOI] [PubMed] [Google Scholar]
- 3. Miaczynska M., Pelkmans L., Zerial M. (2004) Curr. Opin. Cell Biol. 16, 400–406 [DOI] [PubMed] [Google Scholar]
- 4. Polo S., Di Fiore P. P. (2006) Cell 124, 897–900 [DOI] [PubMed] [Google Scholar]
- 5. Sorkin A., von Zastrow M. (2009) Nat. Rev. Mol. Cell Biol. 10, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Murphy J. E., Padilla B. E., Hasdemir B., Cottrell G. S., Bunnett N. W. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17615–17622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yamashiro D. J., Tycko B., Fluss S. R., Maxfield F. R. (1984) Cell 37, 789–800 [DOI] [PubMed] [Google Scholar]
- 8. Backer J. M., Kahn C. R., White M. F. (1990) J. Biol. Chem. 265, 14828–14835 [PubMed] [Google Scholar]
- 9. Nishi T., Forgac M. (2002) Nat. Rev. Mol. Cell Biol. 3, 94–103 [DOI] [PubMed] [Google Scholar]
- 10. Clague M. J., Urbé S., Aniento F., Gruenberg J. (1994) J. Biol. Chem. 269, 21–24 [PubMed] [Google Scholar]
- 11. Carpenter G., Cohen S. (1979) Annu. Rev. Biochem. 48, 193–216 [DOI] [PubMed] [Google Scholar]
- 12. Carpenter G., Cohen S. (1976) J. Cell Biol. 71, 159–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Authier F., Chauvet G. (1999) FEBS Lett. 461, 25–31 [DOI] [PubMed] [Google Scholar]
- 14. Burke P., Schooler K., Wiley H. S. (2001) Mol. Biol. Cell 12, 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Umebayashi K., Stenmark H., Yoshimori T. (2008) Mol. Biol. Cell 19, 3454–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gorvel J. P., Chavrier P., Zerial M., Gruenberg J. (1991) Cell 64, 915–925 [DOI] [PubMed] [Google Scholar]
- 17. Nielsen E., Severin F., Backer J. M., Hyman A. A., Zerial M. (1999) Nat. Cell Biol. 1, 376–382 [DOI] [PubMed] [Google Scholar]
- 18. Rosenfeld J. L., Moore R. H., Zimmer K. P., Alpizar-Foster E., Dai W., Zarka M. N., Knoll B. J. (2001) J. Cell Sci. 114, 4499–4508 [DOI] [PubMed] [Google Scholar]
- 19. Chen P. I., Kong C., Su X., Stahl P. D. (2009) J. Biol. Chem. 284, 30328–30338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shtivelman E. (1997) Oncogene 14, 2167–2173 [DOI] [PubMed] [Google Scholar]
- 21. Xiao H., Tao Y., Greenblatt J., Roeder R. G. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2146–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiang C., Ito M., Piening V., Bruck K., Roeder R. G., Xiao H. (2004) J. Biol. Chem. 279, 27781–27789 [DOI] [PubMed] [Google Scholar]
- 23. Zhao J., Lu B., Xu H., Tong X., Wu G., Zhang X., Liang A., Cong W., Dai J., Wang H., Wu M., Guo Y. (2008) Hepatology 48, 265–275 [DOI] [PubMed] [Google Scholar]
- 24. King F. W., Shtivelman E. (2004) Mol. Cell. Biol. 24, 7091–7101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ito M., Jiang C., Krumm K., Zhang X., Pecha J., Zhao J., Guo Y., Roeder R. G., Xiao H. (2003) Cancer Res. 63, 8763–8767 [PubMed] [Google Scholar]
- 26. Lee L. W., Zhang D. H., Lee K. T., Koay E. S., Hewitt R. E. (2004) Ann. Acad. Med. Singapore 33, S30–32 [PubMed] [Google Scholar]
- 27. Tong X., Li K., Luo Z., Lu B., Liu X., Wang T., Pang M., Liang B., Tan M., Wu M., Zhao J., Guo Y. (2009) Am. J. Pathol. 174, 1931–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao J., Ni H., Ma Y., Dong L., Dai J., Zhao F., Yan X., Lu B., Xu H., Guo Y. (2007) Hum. Pathol. 38, 293–298 [DOI] [PubMed] [Google Scholar]
- 29. Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin I. I., Thomson J. A. (2007) Science 318, 1917–1920 [DOI] [PubMed] [Google Scholar]
- 30. Xiao H., Palhan V., Yang Y., Roeder R. G. (2000) EMBO J. 19, 956–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hu C. D., Grinberg A. V., Kerppola T. K. (2005) in Current Protocols in Protein Science (Coligan J. E., Dunn B. M., Speicher D. W., Wingfield P. T. eds) pp. 19.10.1–19.10.2, John Wiley & Sons, Inc., Somerset, NJ: [DOI] [PubMed] [Google Scholar]
- 32. Stringer K. F., Ingles C. J., Greenblatt J. (1990) Nature 345, 783–786 [DOI] [PubMed] [Google Scholar]
- 33. Chiang C. M., Roeder R. G. (1995) Science 267, 531–536 [DOI] [PubMed] [Google Scholar]
- 34. Seglen P. O. (1972) Exp. Cell Res. 74, 450–454 [DOI] [PubMed] [Google Scholar]
- 35. Lakadamyali M., Rust M. J., Zhuang X. (2006) Cell 124, 997–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Leonard D., Hayakawa A., Lawe D., Lambright D., Bellve K. D., Standley C., Lifshitz L. M., Fogarty K. E., Corvera S. (2008) J. Cell Sci. 121, 3445–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Carter R. E., Sorkin A. (1998) J. Biol. Chem. 273, 35000–35007 [DOI] [PubMed] [Google Scholar]
- 38. Rink J., Ghigo E., Kalaidzidis Y., Zerial M. (2005) Cell 122, 735–749 [DOI] [PubMed] [Google Scholar]
- 39. Suprynowicz F. A., Krawczyk E., Hebert J. D., Sudarshan S. R., Simic V., Kamonjoh C. M., Schlegel R. (2010) J. Virol. 84, 10619–10629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang Y., Pennock S., Chen X., Wang Z. (2002) Mol. Cell. Biol. 22, 7279–7290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang C., Mori M., Gao S., Li A., Hoshino I., Aupperlee M. D., Haslam S. Z., Xiao H. (2010) Cancer Res. 70, 10224–10233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Takahashi Y., Meyerkord C. L., Wang H. G. (2009) Cell Death Differ. 16, 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wan J., Cheung A. Y., Fu W. Y., Wu C., Zhang M., Mobley W. C., Cheung Z. H., Ip N. Y. (2008) J. Neurosci. 28, 9002–9012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Takahashi Y., Meyerkord C. L., Wang H. G. (2008) Autophagy 4, 121–124 [DOI] [PubMed] [Google Scholar]
- 45. Pfeffer S. R. (2001) Trends Cell Biol. 11, 487–491 [DOI] [PubMed] [Google Scholar]
- 46. Bucci C., Parton R. G., Mather I. H., Stunnenberg H., Simons K., Hoflack B., Zerial M. (1992) Cell 70, 715–728 [DOI] [PubMed] [Google Scholar]
- 47. Krishnan M. N., Sukumaran B., Pal U., Agaisse H., Murray J. L., Hodge T. W., Fikrig E. (2007) J. Virol. 81, 4881–4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lawe D. C., Chawla A., Merithew E., Dumas J., Carrington W., Fogarty K., Lifshitz L., Tuft R., Lambright D., Corvera S. (2002) J. Biol. Chem. 277, 8611–8617 [DOI] [PubMed] [Google Scholar]
- 49. Simonsen A., Lippé R., Christoforidis S., Gaullier J. M., Brech A., Callaghan J., Toh B. H., Murphy C., Zerial M., Stenmark H. (1998) Nature 394, 494–498 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.