Abstract
Protein S-palmitoylation, the reversible thioester linkage of a 16-carbon palmitate lipid to an intracellular cysteine residue, is rapidly emerging as a fundamental, dynamic, and widespread post-translational mechanism to control the properties and function of ligand- and voltage-gated ion channels. Palmitoylation controls multiple stages in the ion channel life cycle, from maturation to trafficking and regulation. An emerging concept is that palmitoylation is an important determinant of channel regulation by other signaling pathways. The elucidation of enzymes controlling palmitoylation and developments in proteomics tools now promise to revolutionize our understanding of this fundamental post-translational mechanism in regulating ion channel physiology.
Keywords: Calcium Channels, Ion Channels, Membrane Trafficking, Post-translational Modification, Potassium Channels, Signal Transduction, Sodium Channels, Acylation, Ligand-gated Ion Channel, Palmitoylation
Introduction
Protein S-palmitoylation, the most common form of protein S-acylation identified over 30 years ago (1), involves the addition of a 16-carbon chain palmitic acid, via a hydroxylamine-sensitive thioester linkage, to intracellular cysteine residues. Importantly, protein palmitoylation is a reversible post-translational modification (Fig. 1a) and, unlike other irreversible lipid modifications such as myristoylation and prenylation, thus represents a dynamic mechanism to spatiotemporally control protein function and interactions (for reviews, see Refs. 2–10). Indeed, dysregulation of protein palmitoylation results in a number of major disorders, including cancer, X-linked mental retardation, and schizophrenia (11–17), emphasizing the importance of this post-translational modification (PTM)2 in normal physiology and disease. Recent advances in proteomics approaches to characterize protein palmitoylation and the identification of the enzymes that control palmitoylation (palmitoyl acyltransferases and zDHHCs (zinc finger- and DHHC domain-containing proteins)) and depalmitoylation (palmitoyl thioesterases) have revealed the central role of palmitoylation in controlling a diverse array of proteins, including an ever-expanding number of ion channels (see Tables 1–3).
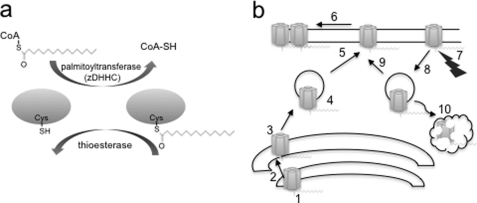
FIGURE 1.
Reversible protein palmitoylation and regulation of the ion channel life cycle. a, schematic illustrating reversible protein palmitoylation. Palmitoylation is controlled by a family of acyl palmitoyltransferases (zDHHCs), and depalmitoylation is controlled by a limited number of thioesterases. b, palmitoylation controls multiple steps in the life cycle of an ion channel that include assembly (step 1), maturation (step 2), control of Golgi exit/sorting (step 3) and trafficking (step 4), insertion in the plasma membrane (step 5), clustering and localization in membrane microdomains (step 6), determination of activity regulation by other signaling pathways (step 7), internalization (step 8), recycling (step 9), and degradation (step 10).
TABLE 1.
Ligand gated-ion channels
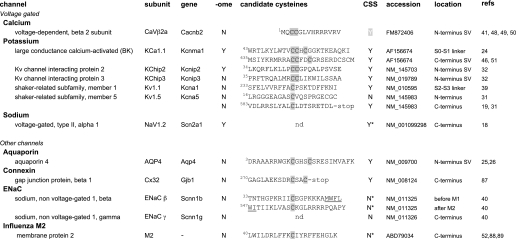
Common channel abbreviations and subunit as well as gene names are given. “-ome” indicates that that subunit has also been identified in mammalian palmitoylome screens (73, 75, 79, 85, 86). “candidate cysteine” indicates experimentally determined cysteine residues (shaded boxes) with flanking 10 amino acids. The predicted membrane domain is underlined. Amino acid numbering corresponds to the NCBI murine accession number given for consistency. “Y” indicates that at least one cysteine within the corresponding candidate cysteine sequence is predicted using the CSS-Palm 2.04 algorithm (67) at high threshold using the corresponding accession number of the full-length murine channel-coding sequence (also validated for cognate species used in reference). “N” indicates that cysteines in the candidate sequence are not predicted. An asterisk indicates that alternative cysteines are predicted in the coding sequence. “location” indicates the predicted location in the channel subunit. “after” or “before” the membrane domain indicates palmitoylated cysteine within 10 amino acids of a membrane domain. nAChR, nicotinic acetylcholine.
TABLE 2.
Voltage-gated and other ion channels
Common channel names and subunit as well as gene names are given. “-ome” indicates that that subunit has also been identified in mammalian palmitoylome screens (73, 75, 79, 85, 86). “candidate cysteine” indicates experimentally determined cysteine residues (shaded boxes) with flanking 10 amino acids. The predicted transmembrane domain is underlined. Amino acid numbering corresponds to the NCBI murine accession number given for consistency. “Y” indicates that at least one cysteine within the corresponding candidate cysteine sequence is predicted using the CSS-Palm 2.04 algorithm (67) at high threshold using the corresponding accession number of the full-length murine channel-coding sequence (also validated for cognate species used in reference). “N” indicates that cysteines in the candidate sequence are not predicted. An asterisk indicates that alternative cysteines are predicted in the coding sequence. “location” indicates the predicted location in the channel subunit. “after” or “before” the membrane domain (S or M) indicates palmitoylated cysteine within 10 amino acids of a membrane domain. SV, splice variant.
TABLE 3.
Other channels identified in mammalian palmitoylome screens
The channels listed were identified in S-acylation screens from rat brain (79), Jurkat cells (73, 85), dendritic cells (75), and prostate cancer cells (86) and are not independently characterized as in Tables 1 and 2. Common channel and gene names are given. Y indicates that at least one cysteine is predicted with the CSS-Palm 2.04 algorithm (67) at high threshold using the corresponding NCBI accession number of the full-length murine channel-coding sequence (also validated for cognate species used in reference). N indicates that cysteines in the coding region are not predicted.
| Channel | Gene | CSS-Palm | Accession no. |
|---|---|---|---|
| Anion | |||
| Chloride channel 6 | Clcn6 | Y | NM_008166 |
| Chloride intracellular channel 1 | Clic1 | N | NM_033444 |
| Chloride intracellular channel 4 | Clic4 | N | NM_013885 |
| Tweety homolog 1 | Ttyh1 | Y | NM_001001454 |
| Tweety homolog 3 | Ttyh3 | Y | NM_175274 |
| Voltage-dependent anion channel 1 | Vdac1 | N | NM_011694 |
| Voltage-dependent anion channel 2 | Vdac2 | Y | NM_011695 |
| Voltage-dependent anion channel 3 | Vdac3 | Y | NM_001198998 |
| Calcium | |||
| Voltage-dependent, gamma subunit 8 | Cacng8 | Y | NM_133190 |
| Cation | |||
| Amiloride-sensitive cation channel 2 | Accn2 | Y | NM_009597 |
| Glutamate | |||
| Ionotropic, delta 1 | Grid1 | Y | NM_008166 |
| Perforin | |||
| Perforin-1 | Prf1 | Y | NM_011073 |
| Potassium | |||
| Voltage-gated channel, subfamily Q, member 2 | Kcnq2 | Y | NM_010611 |
| Sodium | |||
| Voltage-gated, type I, alpha | Scn1a | Y | NM_018733 |
| Voltage-gated, type III, alpha | Scn3a | Y | NM_018732 |
| Voltage-gated, type IX, alpha | Scn9a | Y | NM_018852 |
| Transient receptor potential | |||
| Cation channel, subfamily V, member 2 | Trpv2 | N | NM_011706 |
| Cation channel, subfamily M, member 7 | Trpm7 | Y | NM_021450 |
The addition of palmitic acid increases the hydrophobicity of a protein and thus may affect ion channel function in multiple ways. Evidence suggests that protein S-palmitoylation controls many stages in the life cycle of ion channels (Fig. 1b).
Palmitoylation-dependent Control of Ion Channels: From Assembly to Regulation
A diverse array (>40) of pore-forming and regulatory subunits of ion channels have been experimentally identified as being palmitoylated, ranging from the virally encoded M2 channel to voltage- and ligand-gated ion channels involved in complex behaviors (Tables 1–3). Both surface expression and intrinsic activity/regulation of ion channels can be controlled by palmitoylation of cognate pore-forming and/or regulatory subunits. Although the focus of this minireview is palmitoylation-dependent regulation of ion channel subunits per se, it is important to consider that palmitoylation also controls many adaptor and cell signaling proteins (for example, PSD-95, AKAP18, G-proteins, etc. (5)) that control macromolecular ion channel complexes.
Control of Channel Cell-surface Expression and Spatial Organization
The surface expression of a channel is dependent upon the delicate balance between channel synthesis, forward trafficking to the membrane and subsequent internalization, recycling, and degradation. Palmitoylation plays a significant role in controlling channel cell-surface expression and clustering by acting at distinct stages of the trafficking pathway. For several channels, including voltage-gated sodium (Nav1.2) (18) and potassium (Kv1.5) (19) channels, palmitoylation is thought to occur very early in the biosynthetic pathway and to regulate channel maturation/quality control. Indeed, palmitoylation regulates the formation of ligand-binding sites in nicotinic acetylcholine receptors (20, 21). Palmitoylation of a cluster of C-terminal cysteines in the pore-forming NR2A subunit of NMDA receptors (22) and a cysteine residue juxtaposed to the M2 membrane domain in the GluR1 subunit of AMPA receptors (23) controls Golgi retention of the respective channel, whereas palmitoylation of the intracellular N-terminal S0-S1 loop of large conductance calcium- and voltage-activated potassium (BK) channels modulates but is not essential for cell-surface delivery (24). Palmitoylation also controls the spatial organization of channels once at the plasma membrane as illustrated by the formation of orthogonal arrays of AQP4 (aquaporin-4) channels controlled by palmitoylation of two C-terminal cysteine residues (25, 26), whereas C-terminal palmitoylation promotes association of P2X7 (P2X purinoceptor 7) receptors with cholesterol-rich microdomains (“lipid rafts”) (27). Furthermore, synaptic clustering of GABAA receptors is controlled by palmitoylation of an intracellular loop of the γ2 subunit (28). Agonist-induced internalization of AMPA receptors is determined by palmitoylation of a single cysteine residue in GluR1 and GluR2 subunits, distinct from that controlling Golgi retention (23, 29, 30). Palmitoylation also controls internalization of Kv1.5 channels, as palmitoylation-deficient Kv1.5 channels display a reduced internalization rate with consequently higher surface expression (19, 31).
Palmitoylation of regulatory subunits and adaptor proteins is also an important determinant of channel surface delivery and stability. For example, surface expression of the voltage-gated potassium channel Kv4.3 is intimately controlled by palmitoylation of the regulatory KChip2 and KChip3 subunits (32), whereas NMDA and AMPA receptor trafficking is controlled by palmitoylation of a number of scaffolding proteins such as PSD-95 (33–35), Delphilin (36), and GRIP1b (37, 38).
Control of Channel Activity at the Plasma Membrane: Cross-talk with Other PTMs
Relatively few studies have revealed the effects of palmitoylation on the intrinsic activity and/or gating kinetics of ion channels. Palmitoylation of the voltage-sensitive potassium channel Kv1.1 (39), within the intracellular linker between transmembrane domains 2 and 3, increases the intrinsic voltage sensitivity of the channel. Palmitoylation of the β subunit of epithelial sodium channels also affects channel gating (40), whereas the palmitoylated regulatory β2a subunit of N-type calcium channels controls voltage-dependent inactivation (41, 42).
In contrast, an emerging concept is that palmitoylation is an important determinant of ion channel regulation by other PTMs. Indeed, 15 years ago, palmitoylation of the GluR6 subunit of kainate receptors was reported to reduce channel phosphorylation by PKC (43). Similarly, palmitoylation inhibits PKC-mediated phosphorylation of the GluR1 subunit of AMPA receptors, which is important for receptor insertion (29). In both cases, the mechanism likely results from steric hindrance, as the palmitoylated cysteine is immediately adjacent to the consensus phosphorylation site analogous to that hypothesized for palmitoylation-dependent regulation of β2-adrenergic receptor phosphorylation (44, 45). However, palmitoylation can also promote channel phosphorylation. Fyn-dependent tyrosine phosphorylation of the NR2A subunit of NMDA receptors, at a site juxtaposed between the M4 membrane-spanning helix and a cluster of palmitoylated cysteine residues, is abrogated upon site-directed mutation of the palmitoylated cysteines (22). This palmitoylation-dependent enhancement of tyrosine phosphorylation inhibits internalization of the NMDA receptor (22).
A novel mechanism for cross-talk between palmitoylation and phosphorylation, through regulation of membrane association of an intracellular channel domain, has been revealed in BK channels (46). In this system, palmitoylation of a dicysteine motif in the alternatively spliced C-terminal STREX (stress-regulated exon) insert promotes association of the STREX domain with the plasma membrane. PKA-dependent phosphorylation of a serine residue immediately upstream of the palmitoylated cysteines results in dissociation of the STREX domain from the plasma membrane, leading to channel inhibition. The reciprocal control of membrane association of a protein domain by these PTMs likely represents a common mechanism in other signaling proteins. For example, the PDE10A (phosphodiesterase 10A) splice variant is targeted to the plasma membrane via palmitoylation of an alternatively spliced N-terminal insert. PKA phosphorylation of the spliced insert, adjacent to the palmitoylated cysteine residue, results in dissociation of the PDE10A variant from the plasma membrane (47).
Palmitoylation also determines regulation by other signaling pathways. G-protein (Gq)-mediated stimulation of N-type calcium channels is conferred by the palmitoylated N terminus of the regulatory β2a subunit splice variant acting as a steric inhibitor of an arachidonic acid-binding domain (48–50). In the presence of non-palmitoylated regulatory β subunits, Gq-mediated signaling, via arachidonic acid, inhibits calcium channel activity. A potentially important but as yet unexplored mechanism for cross-talk is palmitoylation-dependent control of cysteine reactivity per se upon thioester addition of palmitate to a target cysteine. Indeed, cysteine residues often form disulfide cross-bridges, or their free sulfhydryl groups are targets for both redox as well as S-nitrosylation pathways, all of which are important ion channel regulators.
Location of Palmitoylated Cysteine Is Important for Function
The functional effect of palmitoylation is dependent on both the channel type and the location of the palmitoylated cysteine residue within a given channel subunit. In both ligand-gated (e.g. AMPA and NMDA) and voltage-gated (e.g. BK) channels, palmitoylation of discrete sites on the same channel subunit can exert fundamentally distinct effects. For example, palmitoylation of the C-terminal cluster of cysteine residues in NR2A and NR2B controls Golgi retention, whereas palmitoylation of the cysteine cluster proximal to the M4 transmembrane domain controls channel internalization (22). In BK channels, palmitoylation of the N-terminal intracellular S0-S1 linker controls surface expression (24), whereas inclusion of an alternatively spliced insert (STREX) in the cytosolic C terminus determines channel regulation by PKA-dependent phosphorylation (46). Such data raise several fundamental questions regarding both the mechanistic basis for regulation of channel properties by palmitoylation and how distinct sites on the same protein may be differentially palmitoylated.
Mechanistically, it is generally assumed that palmitoylation facilitates membrane association. However, although exploitation of fluorescent fusion proteins encoding the entire cytosolic C terminus of BK channels supports such a mechanism (46), direct experimental evidence is lacking for most channels. Importantly, the addition of palmitic acid can modify protein hydrophobicity. Thus, palmitoylation may also control structural conformation, as well as protein-protein interactions, independently of membrane association especially where the palmitoylated cysteine is juxtaposed to a membrane domain (2–10). Little is known about which of the potential palmitoyl acyltransferases (gene family zDHHC) control ion channel palmitoylation, although analysis of both NMDA (22) and BK (51) channels has suggested that distinct palmitoylated domains on the same polypeptide can be regulated by different zDHHCs.
Mechanisms Controlling and Tools Available to Investigate Ion Channel Palmitoylation
It has been more than 20 years since the first palmitoylated ion channels were discovered (18, 52). The recent development of new bioinformatics and proteomics tools, together with the identification of zDHHCs, is starting to revolutionize our understanding of ion channel palmitoylation.
Identification of Palmitoyl Acyltransferases and Acyl Thioesterases
Although autoacylation of some cysteine-containing peptides/proteins in the presence of palmitoyl-CoA has been reported (9), the recent discovery of a large family (at least 23 members in mammals) of candidate palmitoyl acyltransferases suggests that protein palmitoylation is predominantly an enzymatic process from yeast to man (2, 3, 5–10). These predicted transmembrane zinc finger-containing proteins include a conserved Asp-His-His-Cys (DHHC) signature sequence within a cysteine-rich stretch of ∼50 amino acids that is critical for catalytic activity (53), with palmitoylation proceeding via a two-step mechanism (54). Although no “consensus” motif for protein palmitoylation has been identified, increasing evidence suggests that zDHHCs can display substrate specificity for proteins, including ion channels (22, 55), although the mechanisms involved are poorly characterized. In this regard, although many zDHHCs are thought to be localized to the endoplasmic reticulum/Golgi, largely based on overexpression studies, the subcellular localization of both native zDHHCs and ion channel palmitoylation is poorly characterized. Cysteine palmitoylation is dependent on three main factors: (i) the local concentration of fatty acyl-CoA that can be increased near hydrophobic environments (56), (ii) localization with the zDHHCs found on membranes (5, 53), and (iii) cysteine reactivity that can typically be enhanced by proximity to basic or hydrophobic residues (56–58). Thus, an initial membrane association signal is likely required to allow efficient palmitoylation. In ∼30% of identified channel subunits, the palmitoylated cysteine is within 10 amino acids of a membrane domain (Tables 1 and 2). However, in the majority of channels, this is not the case, suggesting that additional initiating membrane association signals are required adjacent to the site of palmitoylation. Likely candidates include regions of basic charge (59), observed in ∼30% of palmitoylated channels such as the STREX variant of the BK channel (46), hydrophobic domains, and other lipid anchors.
The regulation of zDHHC activity is poorly characterized. Activity-dependent redistribution of zDHHC2 in neurons (60) controls palmitoylation of the postsynaptic scaffolding protein PSD-95, regulating NMDA receptor function. Furthermore, zDHHCs express a range of protein-protein interaction domains and potential sites for PTMs, including phosphorylation and palmitoylation, suggesting multiple mechanisms for zDHHC regulation (5). Intriguingly, as ion channels themselves determine cellular excitability, this may provide a local feedback mechanism to regulate palmitoylation status. zDHHCs are also reported to assemble selectively with a number of ion channels, including GABAA (61) and BK (51) channels, suggesting that channels and palmitoylating enzymes may exist within multimolecular signaling complexes.
To date, enzymes responsible for depalmitoylation of ion channels, as for most other palmitoylated membrane proteins, have not been clearly defined. The cytosolic protein APT1 (acyl-protein thioesterase-1) (62, 63) and the recently characterized protein APT2 (64), as well as PPT1 (palmitoyl-protein thioesterase-1) and PPT2, are likely candidates. However, PPT1 is expressed predominantly in lysosomes (65) and is thus most likely responsible for depalmitoylation of channels undergoing degradation. The development of specific APT1 inhibitors (66) should begin to reveal insights into the role of depalmitoylation in controlling ion channels.
Palmitoylation Site Prediction
In contrast to many PTMs, no canonical consensus site for protein palmitoylation has been characterized. However, several recent freely available predictive tools have proved successful in characterizing potential new palmitoylation targets (e.g. Refs. 67 and 68). For example, the multi-platform CSS-Palm 2.0 tool (67) exploits a clustering and scoring strategy by comparing the surrounding amino acid sequence similarity with that of a set of >340 experimentally determined palmitoylation sites. CSS-Palm predicts >75% of the experimentally identified ion channel palmitoylation sites (Tables 1–3) and suggests that >50% of human channel subunits may be palmitoylated.3 Recent advances in biochemical analysis of palmitoylation should facilitate testing of this latter prediction.
Biochemical Identification of Protein Palmitoylation
Metabolic Labeling (Palmitate-centric Assays)
Traditionally, protein palmitoylation has been characterized using metabolic labeling of cells with radioactive palmitate (e.g. [3H]palmitate) and subsequent immunoprecipitation and identification of candidate proteins by autoradiography/fluorography. However, this approach typically requires extensive (several weeks) exposure of autoradiographs and is not readily amenable for global analysis to enrich and identify palmitoylated proteins (7, 69, 70). The recent development of biorthogonal lipid probes (for review, see Ref. 7), modified fatty acids with reactive groups such as an azide or alkyne group, allows labeled proteins to be conjugated to biotin or fluorophores via the azide or alkyne group using Staudinger ligation or “click” chemistry. In particular, the development of a family of ω-alkynyl fatty acid probes of different chain lengths (such as Alk-C16 and Alk-C18) has been exploited for proteomics profiling or cellular imaging and has identified candidate palmitoylated channels in a number of mammalian cell lines (71–75). Although such metabolic labeling approaches are most suited to analysis of isolated cells rather than tissues, they can provide information on dynamic palmitoylation of proteins during the labeling period.
Cysteine Accessibility Assays (Cysteine-centric Assays) and Acyl-biotin Exchange (ABE)
By coupling hydroxylamine cleavage (at neutral pH) of the cysteine-palmitoyl thioester linkage with subsequent labeling of the newly exposed cysteine thiol with cysteine-reactive biotin groups (such as 1-biotin-amido-4-(4′-(maleimidoethylcyclohexane)carboxamido)-butane or N-(6-(biotinamido)hexyl)-3′-(2′-pyridyldithio)-propionamide), S-acylated proteins can be specifically labeled with biotin and can thus be purified and subsequently identified by mass spectrometry (69, 76–78). This unique approach allows S-acylation of native endogenous proteins to be assayed without the requirement for prior cell isolation and metabolic labeling. The ABE approach has been exploited to determine the “palmitoylome” in a number of species (78, 79), although it must be remembered that it detects S-acylation and does not define palmitoylation per se. In particular, analysis of rat brain identified both previously characterized as well as novel palmitoylated ion channels (Tables 1–3). The ABE approach determines the net amount of pre-existing S-acylated proteins; however, caution is required to eliminate false positives, in particular, the requirement to fully block all reactive cysteines prior to hydroxylamine cleavage. A recent development of this approach, exploiting resin-assisted capture using thiopropyl-Sepharose instead of biotin analogs, has been reported to improve detection of higher molecular weight palmitoylated proteins (80) and thus may prove valuable for ion channel analysis.
The metabolic labeling and ABE methodologies are thus complementary, and exploitation of these approaches should provide very significant insight into the role and regulation of ion channel palmitoylation. However, it is important to note that palmitic acid can also be incorporated into free N-terminal cysteines of proteins via an amide linkage (N-palmitoylation) or the addition of the monounsaturated palmitoleic acid via an oxyester linkage to a serine residue (7, 9). Furthermore, although these modifications can be discriminated by their insensitivity to hydroxylamine cleavage of the S-palmitoylation thioester linkage, other S-linked fatty acids such as arachidonic, oleate, and stearic acids have also been reported (7, 9). Thus, whether other S-linked fatty acids and/or N- or O-linked palmitoylation controls ion channel function remains to be explored. In most cases, candidate palmitoylated cysteine residues have been characterized using mutagenic strategies combined with ABE or [3H]palmitate incorporation assays to screen for loss of palmitoylation. As such, direct biochemical demonstration of native cysteine palmitoylation (e.g. using mass spectrometry) is lacking in most ion channels (and other proteins).
Genetic Manipulation of zDHHCs and Thioesterases
With the identification of the family of palmitoyl acyltransferases, genetic tools have been exploited to identify candidate zDHHCs that control channel palmitoylation and function. To date, most studies have exploited overexpression of candidate zDHHCs and analyzed increases in [3H]palmitate incorporation to define zDHHC channel specificity (e.g. see Refs. 22, 23, and 28). Although this is a powerful approach, caution is required to determine whether this in fact replicates endogenous regulation. For example, overexpression of some zDHHCs results in palmitoylation of a cysteine residue not endogenously palmitoylated in BK channels when expressed in HEK293 cells (51). Few studies have exploited knockdown of endogenous zDHHCs to interrogate palmitoylation of ion channels by native zDHHCs, although such an approach allowed the first systematic characterization of zDHHCs required for palmitoylation of any ion channel (51). This approach revealed that multiple distinct zDHHCs control palmitoylation of the BK channel C terminus. As some zDHHCs are themselves palmitoylated, the functional effect of overexpressing or knocking down individual zDHHCs on the localization and activity of other zDHHCs needs to be determined. Furthermore, as many signaling and cytoskeletal elements are controlled by palmitoylation, direct effects on channel palmitoylation per se need to be evaluated.
Pharmacological Manipulation of Palmitoylation
The most widely employed S-acylation inhibitor for cellular studies is the palmitate analog 2-bromopalmitate (81). However, 2-bromopalmitate does not display selectivity toward specific zDHHC proteins (82), and at high concentrations, this compound has many pleiotropic effects on cells, including cytotoxicity (81). Other less commonly used inhibitors include cerulenin and tunicamycin; however, cerulenin affects many aspects of lipid metabolism (81), and tunicamycin is an established inhibitor of N-linked glycosylation (81). There are no known specific activators of zDHHCs. In contrast to the lack of pharmacological tools for zDHHCs, small molecule inhibitors of the major cytosolic acyl thioesterase APT1 (66) have been developed, although, to date, these have not been exploited to interrogate ion channel depalmitoylation. Clearly, a major goal for the field is to identify novel S-acylation inhibitors that display both specificity and zDHHC selectivity (76, 82, 83).
Conclusions and Perspectives
The last 5 years have seen a major resurgence in the protein palmitoylation field due, in large part, to the development of new proteomics tools and characterization of the major palmitoylating enzymes. To date, >40 different ion channel subunits have been shown experimentally to be S-acylated, and the development of new tools is now beginning to reveal both mechanistic and functional insights into the effect palmitoylation exerts on these important signaling molecules.
Although it is fully expected that the ion channel palmitoylation “catalog” will grow significantly in the next few years, the goal of the field must be to develop fundamental understanding of the mechanistic role of palmitoylation in controlling diverse aspects of ion channel properties and function. Furthermore, a major goal is to elucidate the physiological consequence of ion channel palmitoylation from the single channel to body systems level. Several major challenges and questions must be addressed if we are to tackle these aims, elucidation of which will have major impacts on both the ion channel and wider signaling fields.
The first challenge is the development of improved tools to allow real-time analysis of the palmitoylation status of ion channels and analysis of channel palmitoylation from the single molecule to cellular and tissue function. This includes the development of selective inhibitors (76, 82, 83) and/or activators of zDHHCs combined with improved proteomics tools (e.g. palmitoylation-specific antibodies) and imaging probes. Second, although palmitoylation is widely accepted to be reversible, the spatiotemporal regulation of palmitoylation on any channel is very poorly understood. Imperative in this regard is understanding the physiological and pathological signals that can regulate the activity and localization of the palmitoylating and depalmitoylating enzymes. Furthermore, mechanistic insight into the target selectivity of zDHHCs will allow us to understand the coordinated regulation of ion channels by palmitoylation signaling cascades and to decipher how distinct domains on the same channel may be differentially regulated by palmitoylation. Third, for most ion channels, the mechanism by which palmitoylation modifies trafficking, gating, or regulation is unknown, although it is widely assumed that effects are determined by a simple “membrane anchor” model. Obtaining structural insights into how palmitoylation controls protein architecture, interactions, and properties will be a major challenge. Fourth, as with any PTM, palmitoylation cannot be considered in isolation. Clear evidence from the ion channel field demonstrates significant levels of functional cross-talk of palmitoylation with other major signaling pathways, including phosphorylation. Mechanistic insight into the fundamental rules controlling palmitoylation cross-talk will have an enormous impact far beyond the realms of ion channel biology. Finally, although an increasing number of ion channels are known to be regulated by palmitoylation, very little is known about the functional consequence at the cellular and organismal levels. Although it is clear that disruption of palmitoylation can lead to major disorders, ranging from cancer to affective disorders, the importance of ion channel palmitoylation in (patho)physiology is largely unexplored. For example, will we begin to uncover channel “palmitoylopathies” that result from dysregulation of ion channel palmitoylation analogous to channel phosphorylopathies (84) that have been discovered? Understanding the control and functional consequence of ion channel palmitoylation from the level of the single channel to the whole organism may lead to novel therapeutic strategies to control diverse physiological processes. The next few years represent a challenging and exciting time that promises to reveal fundamental new insights into both ion channel physiology and protein palmitoylation.
Supplementary Material
This work was supported by the Wellcome Trust. This minireview will be reprinted in the 2011 Minireview Compendium, which will be available in January, 2012.
This Minireview was originally commissioned by the late Dale Benos, who served as an associate editor of the Journal of Biological Chemistry until his untimely death on October 7, 2010. It is with great pleasure that I dedicate this contribution to his memory.
M. J. Shipston, unpublished data.
- PTM
- post-translational modification
- ABE
- acyl-biotin exchange.
REFERENCES
- 1. Schmidt M. F., Schlesinger M. J. (1979) Cell 17, 813–819 [DOI] [PubMed] [Google Scholar]
- 2. Baekkeskov S., Kanaani J. (2009) Mol. Membr. Biol. 26, 42–54 [DOI] [PubMed] [Google Scholar]
- 3. Charollais J., Van Der Goot F. (2009) Mol. Membr. Biol. 26, 55–66 [DOI] [PubMed] [Google Scholar]
- 4. el-Husseini Ael-D., Bredt D. S. (2002) Nat. Rev. Neurosci. 3, 791–802 [DOI] [PubMed] [Google Scholar]
- 5. Fukata Y., Fukata M. (2010) Nat. Rev. Neurosci. 11, 161–175 [DOI] [PubMed] [Google Scholar]
- 6. Greaves J., Prescott G. R., Gorleku O. A., Chamberlain L. H. (2009) Mol. Membr. Biol. 26, 67–79 [DOI] [PubMed] [Google Scholar]
- 7. Hannoush R. N., Sun J. (2010) Nat. Chem. Biol. 6, 498–506 [DOI] [PubMed] [Google Scholar]
- 8. Iwanaga T., Tsutsumi R., Noritake J., Fukata Y., Fukata M. (2009) Prog. Lipid Res. 48, 117–127 [DOI] [PubMed] [Google Scholar]
- 9. Linder M. E., Deschenes R. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 74–84 [DOI] [PubMed] [Google Scholar]
- 10. Resh M. D. (2006) Sci. STKE 2006, re14. [DOI] [PubMed] [Google Scholar]
- 11. Kim S. J., Zhang Z., Sarkar C., Tsai P. C., Lee Y. C., Dye L., Mukherjee A. B. (2008) J. Clin. Invest. 118, 3075–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mansouri M. R., Marklund L., Gustavsson P., Davey E., Carlsson B., Larsson C., White I., Gustavson K. H., Dahl N. (2005) Eur. J. Hum. Genet. 13, 970–977 [DOI] [PubMed] [Google Scholar]
- 13. Mukai J., Dhilla A., Drew L. J., Stark K. L., Cao L., MacDermott A. B., Karayiorgou M., Gogos J. A. (2008) Nat. Neurosci. 11, 1302–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mukai J., Liu H., Burt R. A., Swor D. E., Lai W. S., Karayiorgou M., Gogos J. A. (2004) Nat. Genet. 36, 725–731 [DOI] [PubMed] [Google Scholar]
- 15. Oyama T., Miyoshi Y., Koyama K., Nakagawa H., Yamori T., Ito T., Matsuda H., Arakawa H., Nakamura Y. (2000) Genes Chromosomes Cancer 29, 9–15 [DOI] [PubMed] [Google Scholar]
- 16. Raymond F. L., Tarpey P. S., Edkins S., Tofts C., O'Meara S., Teague J., Butler A., Stevens C., Barthorpe S., Buck G., Cole J., Dicks E., Gray K., Halliday K., Hills K., Hinton J., Jones D., Menzies A., Perry J., Raine K., Shepherd R., Small A., Varian J., Widaa S., Mallya U., Moon J., Luo Y., Shaw M., Boyle J., Kerr B., Turner G., Quarrell O., Cole T., Easton D. F., Wooster R., Bobrow M., Schwartz C. E., Gecz J., Stratton M. R., Futreal P. A. (2007) Am. J. Hum. Genet. 80, 982–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang Z., Lee Y. C., Kim S. J., Choi M. S., Tsai P. C., Xu Y., Xiao Y. J., Zhang P., Heffer A., Mukherjee A. B. (2006) Hum. Mol. Genet. 15, 337–346 [DOI] [PubMed] [Google Scholar]
- 18. Schmidt J. W., Catterall W. A. (1987) J. Biol. Chem. 262, 13713–13723 [PubMed] [Google Scholar]
- 19. Zhang L., Foster K., Li Q., Martens J. R. (2007) Am. J. Physiol. Cell Physiol. 293, C152–C161 [DOI] [PubMed] [Google Scholar]
- 20. Alexander J. K., Govind A. P., Drisdel R. C., Blanton M. P., Vallejo Y., Lam T. T., Green W. N. (2010) J. Mol. Neurosci. 40, 12–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Drisdel R. C., Manzana E., Green W. N. (2004) J. Neurosci. 24, 10502–10510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hayashi T., Thomas G. M., Huganir R. L. (2009) Neuron 64, 213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hayashi T., Rumbaugh G., Huganir R. L. (2005) Neuron 47, 709–723 [DOI] [PubMed] [Google Scholar]
- 24. Jeffries O., Geiger N., Rowe I. C., Tian L., McClafferty H., Chen L., Bi D., Knaus H. G., Ruth P., Shipston M. J. (2010) J. Biol. Chem. 285, 33307–33314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Crane J. M., Verkman A. S. (2009) Biophys. J. 97, 3010–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Suzuki H., Nishikawa K., Hiroaki Y., Fujiyoshi Y. (2008) Biochim. Biophys. Acta 1778, 1181–1189 [DOI] [PubMed] [Google Scholar]
- 27. Gonnord P., Delarasse C., Auger R., Benihoud K., Prigent M., Cuif M. H., Lamaze C., Kanellopoulos J. M. (2009) FASEB J. 23, 795–805 [DOI] [PubMed] [Google Scholar]
- 28. Rathenberg J., Kittler J. T., Moss S. J. (2004) Mol. Cell. Neurosci. 26, 251–257 [DOI] [PubMed] [Google Scholar]
- 29. Lin D. T., Makino Y., Sharma K., Hayashi T., Neve R., Takamiya K., Huganir R. L. (2009) Nat. Neurosci. 12, 879–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang G., Xiong W., Kojic L., Cynader M. S. (2009) Eur. J. Neurosci. 30, 35–46 [DOI] [PubMed] [Google Scholar]
- 31. Jindal H. K., Folco E. J., Liu G. X., Koren G. (2008) Am. J. Physiol. Heart Circ. Physiol. 294, H2012–H2021 [DOI] [PubMed] [Google Scholar]
- 32. Takimoto K., Yang E. K., Conforti L. (2002) J. Biol. Chem. 277, 26904–26911 [DOI] [PubMed] [Google Scholar]
- 33. El-Husseini A. E., Craven S. E., Chetkovich D. M., Firestein B. L., Schnell E., Aoki C., Bredt D. S. (2000) J. Cell Biol. 148, 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. El-Husseini Ael-D., Schnell E., Dakoji S., Sweeney N., Zhou Q., Prange O., Gauthier-Campbell C., Aguilera-Moreno A., Nicoll R. A., Bredt D. S. (2002) Cell 108, 849–863 [DOI] [PubMed] [Google Scholar]
- 35. Topinka J. R., Bredt D. S. (1998) Neuron 20, 125–134 [DOI] [PubMed] [Google Scholar]
- 36. Matsuda K., Matsuda S., Gladding C. M., Yuzaki M. (2006) J. Biol. Chem. 281, 25577–25587 [DOI] [PubMed] [Google Scholar]
- 37. Hanley L. J., Henley J. M. (2010) Neurosci. Lett. 485, 167–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yamazaki M., Fukaya M., Abe M., Ikeno K., Kakizaki T., Watanabe M., Sakimura K. (2001) Neurosci. Lett. 304, 81–84 [DOI] [PubMed] [Google Scholar]
- 39. Gubitosi-Klug R. A., Mancuso D. J., Gross R. W. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5964–5968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mueller G. M., Maarouf A. B., Kinlough C. L., Sheng N., Kashlan O. B., Okumura S., Luthy S., Kleyman T. R., Hughey R. P. (2010) J. Biol. Chem. 285, 30453–30462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stephens G. J., Page K. M., Bogdanov Y., Dolphin A. C. (2000) J. Physiol. 525, 377–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Qin N., Platano D., Olcese R., Costantin J. L., Stefani E., Birnbaumer L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4690–4695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pickering D. S., Taverna F. A., Salter M. W., Hampson D. R. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 12090–12094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mouillac B., Caron M., Bonin H., Dennis M., Bouvier M. (1992) J. Biol. Chem. 267, 21733–21737 [PubMed] [Google Scholar]
- 45. Moffett S., Mouillac B., Bonin H., Bouvier M. (1993) EMBO J. 12, 349–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tian L., Jeffries O., McClafferty H., Molyvdas A., Rowe I. C., Saleem F., Chen L., Greaves J., Chamberlain L. H., Knaus H. G., Ruth P., Shipston M. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 21006–21011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Charych E. I., Jiang L. X., Lo F., Sullivan K., Brandon N. J. (2010) J. Neurosci. 30, 9027–9037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chien A. J., Carr K. M., Shirokov R. E., Rios E., Hosey M. M. (1996) J. Biol. Chem. 271, 26465–26468 [DOI] [PubMed] [Google Scholar]
- 49. Heneghan J. F., Mitra-Ganguli T., Stanish L. F., Liu L., Zhao R., Rittenhouse A. R. (2009) J. Gen. Physiol. 134, 369–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mitra-Ganguli T., Vitko I., Perez-Reyes E., Rittenhouse A. R. (2009) J. Gen. Physiol. 134, 385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tian L., McClafferty H., Jeffries O., Shipston M. J. (2010) J. Biol. Chem. 285, 23954–23962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sugrue R. J., Belshe R. B., Hay A. J. (1990) Virology 179, 51–56 [DOI] [PubMed] [Google Scholar]
- 53. Fukata M., Fukata Y., Adesnik H., Nicoll R. A., Bredt D. S. (2004) Neuron 44, 987–996 [DOI] [PubMed] [Google Scholar]
- 54. Mitchell D. A., Mitchell G., Ling Y., Budde C., Deschenes R. J. (2010) J. Biol. Chem. 285, 38104–38114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hou H., John Peter A. T., Meiringer C., Subramanian K., Ungermann C. (2009) Traffic 10, 1061–1073 [DOI] [PubMed] [Google Scholar]
- 56. Bélanger C., Ansanay H., Qanbar R., Bouvier M. (2001) FEBS Lett. 499, 59–64 [DOI] [PubMed] [Google Scholar]
- 57. Britto P. J., Knipling L., Wolff J. (2002) J. Biol. Chem. 277, 29018–29027 [DOI] [PubMed] [Google Scholar]
- 58. Kümmel D., Heinemann U., Veit M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12701–12706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Heo W. D., Inoue T., Park W. S., Kim M. L., Park B. O., Wandless T. J., Meyer T. (2006) Science 314, 1458–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Noritake J., Fukata Y., Iwanaga T., Hosomi N., Tsutsumi R., Matsuda N., Tani H., Iwanari H., Mochizuki Y., Kodama T., Matsuura Y., Bredt D. S., Hamakubo T., Fukata M. (2009) J. Cell Biol. 186, 147–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fang C., Deng L., Keller C. A., Fukata M., Fukata Y., Chen G., Lüscher B. (2006) J. Neurosci. 26, 12758–12768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Devedjiev Y., Dauter Z., Kuznetsov S. R., Jones T. L., Derewenda Z. S. (2000) Structure 8, 1137–1146 [DOI] [PubMed] [Google Scholar]
- 63. Yeh D. C., Duncan J. A., Yamashita S., Michel T. (1999) J. Biol. Chem. 274, 33148–33154 [DOI] [PubMed] [Google Scholar]
- 64. Tomatis V. M., Trenchi A., Gomez G. A., Daniotti J. L. (2010) PLoS ONE 5, e15045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Verkruyse L. A., Hofmann S. L. (1996) J. Biol. Chem. 271, 15831–15836 [DOI] [PubMed] [Google Scholar]
- 66. Dekker F. J., Rocks O., Vartak N., Menninger S., Hedberg C., Balamurugan R., Wetzel S., Renner S., Gerauer M., Schölermann B., Rusch M., Kramer J. W., Rauh D., Coates G. W., Brunsveld L., Bastiaens P. I., Waldmann H. (2010) Nat. Chem. Biol. 6, 449–456 [DOI] [PubMed] [Google Scholar]
- 67. Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. (2008) Protein Eng. Des. Sel. 21, 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang X. B., Wu L. Y., Wang Y. C., Deng N. Y. (2009) Protein Eng. Des. Sel. 22, 707–712 [DOI] [PubMed] [Google Scholar]
- 69. Drisdel R. C., Green W. N. (2004) BioTechniques 36, 276–285 [DOI] [PubMed] [Google Scholar]
- 70. Veit M., Ponimaskin E., Schmidt M. F. G. (2008) Methods Mol. Biol. 446, 163–182 [DOI] [PubMed] [Google Scholar]
- 71. Charron G., Wilson J., Hang H. C. (2009) Curr. Opin. Chem. Biol. 13, 382–391 [DOI] [PubMed] [Google Scholar]
- 72. Hannoush R. N., Arenas-Ramirez N. (2009) ACS Chem. Biol. 4, 581–587 [DOI] [PubMed] [Google Scholar]
- 73. Martin B. R., Cravatt B. F. (2009) Nat. Methods 6, 135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yap M. C., Kostiuk M. A., Martin D. D., Perinpanayagam M. A., Hak P. G., Siddam A., Majjigapu J. R., Rajaiah G., Keller B. O., Prescher J. A., Wu P., Bertozzi C. R., Falck J. R., Berthiaume L. G. (2010) J. Lipid Res. 51, 1566–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yount J. S., Moltedo B., Yang Y. Y., Charron G., Moran T. M., López C. B., Hang H. C. (2010) Nat. Chem. Biol. 6, 610–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Draper J. M., Smith C. D. (2009) Mol. Membr. Biol. 26, 5–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Drisdel R. C., Alexander J. K., Sayeed A., Green W. N. (2006) Methods 40, 127–134 [DOI] [PubMed] [Google Scholar]
- 78. Wan J., Roth A. F., Bailey A. O., Davis N. G. (2007) Nat. Protoc. 2, 1573–1584 [DOI] [PubMed] [Google Scholar]
- 79. Kang R., Wan J., Arstikaitis P., Takahashi H., Huang K., Bailey A. O., Thompson J. X., Roth A. F., Drisdel R. C., Mastro R., Green W. N., Yates J. R., 3rd, Davis N. G., El-Husseini A. (2008) Nature 456, 904–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Forrester M. T., Hess D. T., Thompson J. W., Hultman R., Moseley M. A., Stamler J. S., Casey P. J. (2011) J. Lipid Res. 52, 393–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Resh M. D. (2006) Methods 40, 191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Jennings B. C., Nadolski M. J., Ling Y., Baker M. B., Harrison M. L., Deschenes R. J., Linder M. E. (2009) J. Lipid Res. 50, 233–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ducker C. E., Griffel L. K., Smith R. A., Keller S. N., Zhuang Y., Xia Z., Diller J. D., Smith C. D. (2006) Mol. Cancer Ther. 5, 1647–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Erxleben C., Liao Y., Gentile S., Chin D., Gomez-Alegria C., Mori Y., Birnbaumer L., Armstrong D. L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3932–3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wilson J. P., Raghavan A. S., Yang Y. Y., Charron G., Hang H. C. (2010) Mol. Cell. Proteomics 10.1074/mcp.M110.001198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yang W., Di Vizio D., Kirchner M., Steen H., Freeman M. R. (2010) Mol. Cell. Proteomics 9, 54–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Locke D., Koreen I. V., Harris A. L. (2006) FASEB J. 20, 1221–1223 [DOI] [PubMed] [Google Scholar]
- 88. Holsinger L. J., Shaughnessy M. A., Micko A., Pinto L. H., Lamb R. A. (1995) J. Virol. 69, 1219–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Veit M., Kretzschmar E., Kuroda K., Garten W., Schmidt M. F., Klenk H. D., Rott R. (1991) J. Virol. 65, 2491–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.