Abstract
Swi6/HP1, an evolutionarily conserved protein, is critical for heterochromatin assembly in fission yeast and higher eukaryotes. In fission yeast, histone deacetylation by histone deacetylases is thought to be followed by H3-Lys-9 methylation by the histone methyltransferase Clr4/Suv39H1. H3-Lys-9-Me2 interacts with the chromodomain of Swi6/HP1. Swi6/HP1 is thought to act downstream of Clr4/Suv39, and further self-association of Swi6/HP1 is assumed to stabilize the heterochromatin structure. Here, we show that the self-association-defective mutant of Swi6 does not interact with Clr4. It not only fails to localize to heterochromatin loci but also interferes with heterochromatic localization of H3-Lys-9-Me2 (and thereby Clr4) and the endogenous Swi6 in a dominant negative manner. Thus, self-association of Swi6/HP1 helps in binding to and recruitment of Clr4 and thereby in establishment and maintenance of heterochromatin by a concerted rather than a sequential mechanism.
Keywords: Chromatin Structure, Epigenetics, Gene Silencing, Histone Deacetylase, Histone Methylation
Introduction
Distinct sets of histone modifications organize chromatin into transcriptionally expressed or repressed structures. As in higher eukaryotes, fission yeast euchromatic regions are associated with hyperacetylated histones, particularly Lys-9 and Lys-14 acetylated and Lys-4 methylated histone H3. Conversely, heterochromatin assembly (silent mating type, centromere, rDNA, and telomere loci) involves deacetylation of histone H3 at Lys-9 and Lys-14 by Clr3, Clr6, and Sir2, followed by H3-Lys-9 methylation by the histone methyltransferase Clr4/Suv39H1. H3-Lys-9-Me2 is recognized by the chromodomain in Swi6/HP1 (1, 2). The property of Swi6 to form multimers is thought to cause folding of chromatin into a transcriptionally inactive structure (3). The RNAi pathway is also involved in heterochromatin assembly. The dcr1Δ, rdp1Δ, and ago1Δ mutants are defective in H3-Lys-9 methylation and Swi6 recruitment at heterochromatin loci (4).
Structural studies show that the chromoshadow domain (CSD)3 in Swi6/HP1 contains a dimerization motif that creates a cleft for binding a pentapeptide (5, 6). Mutations in the conserved residues in the cleft inhibit interaction with the pentapeptide motif in proteins (6, 7). We found that mutation of one such residue (L315E) severely compromises the self-association property of Swi6. The mutant protein does not self-associate in vivo or in vitro or with the endogenous Swi6p. Whereas Swi6+p interacts with Clr4 in vivo and in vitro, the Swi6L315E mutant protein interacts poorly with Clr4p. The mutant swi6L315E is not only defective in complementing the silencing defect in the swi6Δ mutant but also exerts a dominant negative effect on silencing, accompanied by loss of heterochromatic localization not only of itself but also that of native Swi6 and H3-Lys-9-Me2 and, by implication, of Clr4. These results support a concerted rather than a sequential action of Clr4 and Swi6 in establishment and maintenance of heterochromatin.
EXPERIMENTAL PROCEDURES
Strains and Plasmids Used
All strains and plasmids used are listed in supplemental Tables 1 and 2, respectively.
Media Compositions
All fission yeast media were prepared according to Moreno et al. (8). The serial dilution plate assay has been described earlier (9). Iodine staining assays for switching and silencing have been described earlier (supplemental “Experimental Procedures”) (8, 10).
Site-directed Mutagenesis
Leucine (Leu) at position 315 in Swi6 was mutated to glutamate (Glu) or isoleucine (Ile) using the site-directed mutagenesis kit from Stratagene according to the manufacturer's instructions. Mutations were confirmed by sequencing. The mutation was introduced into the nmt1-GFP-Swi6 expression construct (kind gift of Dr. Alison Pidoux). The Swi6 region of the construct was in turn PCR-amplified and cloned as GST-, (His)6-, or maltose-binding protein (MBP)-tagged protein using suitable vectors for recombinant expression in Escherichia coli.
Glutaraldehyde Cross-linking
In a 50-μl cross-linking reaction, ∼35–40 μg of protein extract was used. The reaction was carried out for 30 min at RT in the presence of glutaraldehyde cross-linking buffer (1 mm DTT, 100 mm NaCl, 0.2 mm EDTA, 25 mm HEPES, 10% v/v glycerol, 0.05% v/v Nonidet P-40) and 0.01% glutaraldehyde. The reaction was stopped by adding 6× SDS loading buffer (to a final concentration of 1×) and heating the samples at 95 °C for 5 min. The samples were then resolved by 8% denaturing (SDS) polyacrylamide gel electrophoresis followed by Western blotting with α-Swi6 antibody.
Fluorescence Microscopy
Cells expressing GFP-tagged Swi6 or GFP-tagged Swi6L315E mutant were visualized directly without fixation under a Carl Zeiss LSM 510 Meta Confocal microscope. Cells with distinct GFP-Swi6 spots and those with diffuse GFP fluorescence were counted from three independent cultures and graphically represented (with statistically calculated standard deviation).
Co-immunoprecipitation
Direct in vivo physical interaction between Swi6 and Clr4 was checked by co-immunoprecipitation. Whole cell extracts prepared from the required strains were immunoprecipitated with α-Swi6 or α-Myc antibody coupled to protein A-Sepharose beads (GE Healthcare). Immunoprecipitated fractions are immunoblotted with α-Myc and α-Swi6 antibodies. Inputs are Western blotted with α-Myc and α-Swi6 antibodies, and α-tubulin is taken as a loading control.
Iodine Staining and Detection of Sporulation
Wild-type and swi6Δ cells transformed with nmt1 vector alone, nmt1 GFP-swi6+, and nmt1GFP-swi6L315E plasmids were streaked on plates lacking leucine. After growing for 3–4 days at 30 °C, colonies were stained with iodine for 2 min and photographed under an Olympus stereo zoom microscope. Data were represented graphically with statistically calculated standard deviation.
Chromatin Immunoprecipitation
A ChIP experiment was performed according to Ekwall and Partridge (11). Antibody against H3-Lys-9-Me2 was from Millipore. Primer sequences are given in supplemental Table 3. PCR products were resolved by electrophoresis on 4% polyacrylamide gel, exposed to a magnetic screen, and scanned in a Fuji image processor. The bands were quantified densitometrically from three independent sets of PCR using in-built MultiGauge software and represented graphically with statistically calculated standard deviation.
RESULTS
Residue Leu-315 Is Required for Self-association of Swi6
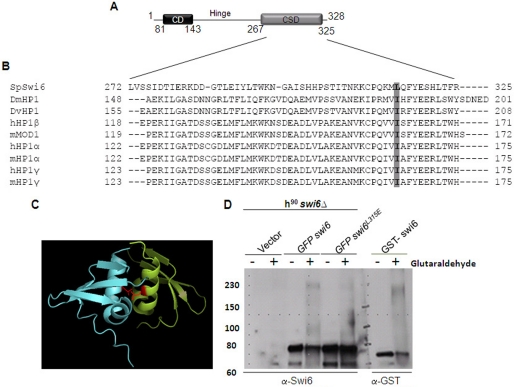
A comparison of the sequences in the CSD of Swi6 (Fig. 1A) with homologues in humans and mice revealed conservation of a hydrophobic amino acid (Leu-315) in Swi6 in place of Ile in the homologues (Fig. 1B). Structure analysis of the CSD of Swi6 has shown that Leu-315 is located at the cleft between the interacting CSD monomers in the dimer (Fig. 1C, 5–6). It is thought that the residue may lead to dimerization of the CSD. Therefore, we sought to study the role of residue Leu-315 in the self-association of Swi6.
FIGURE 1.
L315E mutation in the CSD domain of Swi6 interferes with its self-association. A, schematic showing the domain structure of fission yeast Swi6. B, alignment of sequences of CSDs from different HP1 proteins numbered with respect to MOD1 (adapted from Ref. 7). The hydrophobic residue Leu-315 in Swi6 and Ile in other homologues is conserved and indicated by shading. C, a dimer formed by the association of the CSDs of two monomers. The monomers are shown in different colors. Two Leu-315 residues are shown in magenta in both CSD chains. Their side chains, shown as lines, are believed to play an important role in the association of two CSD monomers of Swi6 (5) (PDB code 1E0B). D, whole cell extracts of the swi6Δ mutant strain harboring empty vector (first and second lanes), GFP-swi6+ (3rd and 4th lanes), and swi6L315E mutant (5th and 6th lanes) genes expressed under the control of the nmt1 promoter and E. coli cells expressing GST-Swi6 (7th and 8th lane) were subjected to glutaraldehyde cross-linking and Western blotted with anti-Swi6 antibody. The GFP-tagged Swi6 monomer migrates at 78 kDa (≈ 28 (GFP) + 50 (Swi6)). Upon cross-linking, the multimer migrates around ∼230–240 kDa (A and B, 4th lane). The level of self-association is reduced in case of Swi6L315E mutant protein (A and B, 6th lane).
Using gel filtration studies, bacterially expressed His-tagged Swi6 has been shown to exist as a tetramer in vitro (3), whereas a recent study using sedimentation analysis suggested that Swi6 may exist as a dimer (12). To directly check whether Swi6 can self-associate and to check the role of the residue Leu-315, glutaraldehyde cross-linking was attempted. Whole cell extracts of swi6Δ cells expressing GFP-tagged swi6+ or swi6L315E mutant genes were cross-linked and analyzed by Western blotting. Results showed that ∼35% of the GFP-Swi6p yielded a cross-linked product of ∼230–240 kDa (Fig. 1D, compare the 3rd and 4th lanes). Swi6 (molecular mass, 37 kDa) migrates at the estimated electrophoretic mobility of 50 kDa (3). The GFP-Swi6 fusion product migrates at ∼78 kDa (50 (Swi6) + 28 kDa (GFP), Fig. 1D, 3rd lane). The migration around 240 kDa suggests the possibility of the cross-linked product being either a trimer or tetramer of GFP-Swi6. (A closer analysis suggests that Swi6 exists most likely as a tetramer, see “Discussion”). However, in case of the GFP-Swi6L315E mutant protein, only a small percentage (∼1.9%) of total protein was cross-linked (Fig. lD, 6th lane), indicating that the L315E mutation drastically reduces the efficiency of self-association of Swi6. This is consistent with the recent gel filtration study showing complete loss of self-association of the recombinant Swi6L315E mutant (12).
Because the cross-linking protocol may not only cause cross-linking of Swi6 monomers but also that of Swi6 with other cellular proteins, we carried out cross-linking of GST-tagged Swi6. A cross-linked product of ∼220 kDa was obtained (Fig. 1D, 8th lane). The close similarity in size of the cross-linked product of recombinant GST-Swi6 with that of GFP-Swi6 expressed in Schizosaccharomyces pombe supports the possibility that the cross-linked product of GFP-Swi6 represents a homopolymeric complex of Swi6 alone.
The Swi6L315E Mutant Fails to Localize to Heterochromatin
GFP-tagged Swi6 has been shown to be localized as distinct spots (1–4) corresponding to heterochromatin loci when transformed either into the swi6Δ mutant or wild-type strains (3, 13). Our results show that the GFP-Swi6L315E mutant protein fails to localize as distinct spots in both genetic backgrounds and rather appears as diffuse fluorescence in the majority (> 95%) of cells (Fig. 2, A (lower panel), B, and C). Thus, the self-association-defective GFP-Swi6L315E mutant protein is defective in localization to the heterochromatic loci.
FIGURE 2.
The Swi6L315E mutant protein fails to localize to heterochromatic loci. A, representative pictures showing defective GFP-Swi6L315E localization in h90 swi6Δ and h90 swi6+ cells. The mutation almost completely deprives Swi6 of its ability to localize to heterochromatic regions; only 1% of the cells show distinct GFP spots in the h90 swi6Δ strain (left panel, lower frame). The mutant protein also fails to localize to heterochromatic regions in the h90 swi6+ strain (right panel, lower frame). B and C, quantitation of the percentage of cells showing distinct spots of GFP-Swi6 fluorescence.
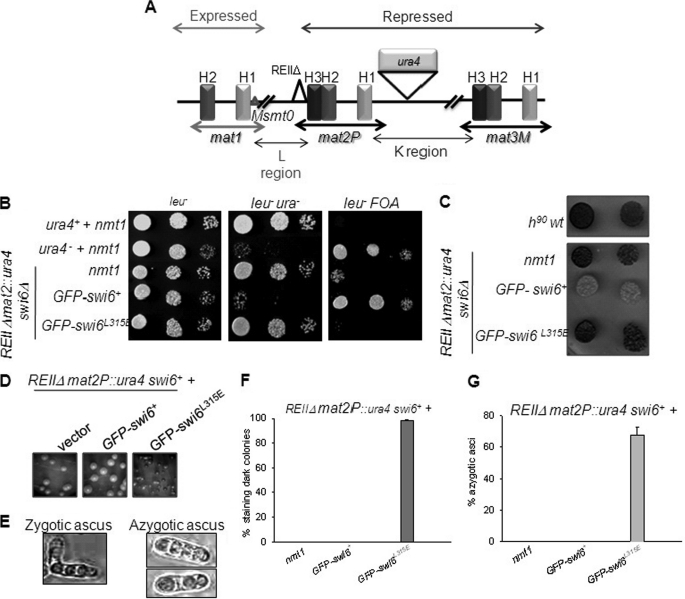
The swi6L315E Mutant Is Defective in Mating-type Switching and Silencing at All Heterochromatin Loci
By regulating the heterochromatin structure spanning the mat2-mat3 interval, Swi6 ensures switching, silencing, and proper donor choice selection (9, 14). Thus, a homothallic (h90) swi6Δ mutant strain switches inefficiently and gives reduced iodine staining (see supplemental Experimental Procedures). Whereas the GFP-tagged swi6+ gene is able to restore switching and thus iodine staining to the swi6Δ mutant (Fig. 3A), the mutant GFP-swi6L315E produced only light staining transformants (A). Interestingly, transformation of the mutant gene into an h90 swi6+ strain produced a significant number of colonies with reduced iodine staining (Fig. 3B). Thus, the mutant is not only defective in complementing swi6Δ mutation but also exerts a dominant negative effect on switching in the WT strain. Overexpression of the GFP-swi6+ gene in the h90 strain had no such effect, as reported earlier (3). Western blot analysis showed similar levels of WT and mutant proteins.4
FIGURE 3.
Self-association of Swi6 is required for efficient switching. A and B, h90 swi6Δ and h90 strains, respectively. The indicated strains were transformed with vector alone, nmt1-GFP-swi6, and nmt1-GFP-swi6L315E plasmids (upper panels). Transformants were streaked for a single colony on a Pombe minimal medium with adenine (PMA) plate lacking leucine (8). Colonies formed after 3–4 days at 30 °C were stained with iodine vapors. Quantitative representation of the percentage of colonies giving dark staining with iodine (lower panels).
The mutation was also checked for its effect on silencing. The strain with the genotype Msmto REIIΔ mat2P::ura4, swi6Δ (10, supplemental Experimental Procedures, and Fig. 4A) showed elevated ura4 expression (Fig. 4B) and dark staining colonies (C), indicating a silencing defect at the mat2P locus and the linked ura4 reporter (see supplemental Experimental Procedures). Expression of GFP-swi6+ restored silencing as indicated by lower expression of the ura4 reporter (reduced growth on plates lacking uracil and enhanced growth on counter-selective fluoroorotic acid plates, Fig. 4B) and loss of iodine staining (C). However, the GFP-swi6L315E gene failed to suppress either the level of ura4 expression (Fig. 4B) or iodine staining (C) phenotypes of the swi6Δ mutant. Thus, the swi6L315E mutant is also defective in silencing at the mating-type locus.
FIGURE 4.
The swi6L315E mutant gene is defective in complementing the silencing defect in swi6Δ mutant and exerts a dominant negative effect on silencing at the mat2 locus. A, diagrammatical representation of the mating-type region (1, 33). Switching the incompetent Msmt0 strain with a wild-type chromosomal copy of swi6 deleted (swi6Δ) and a ura4 reporter cassette inserted on the right side of mat2P, whereas a cis-acting repression element, REII, has been deleted (REIIΔ). The mat1M locus has a deletion of the cis-acting sequence Msmt0; such a strain does not undergo imprinting at mat1 and is thereby unable to switch and retains a stable Minus (M) mating type (1, 8). In combination with swi6Δ mutation, the strain exhibits a silencing defect, increased ura4 expression, and iodine staining (8, 10). B, the above strain was transformed with vector alone, nmt1-GFP-swi6+, and nmt1-GFP-swi6L315E mutant plasmids. Cultures of the transformants were serially diluted and spotted on plates lacking leucine (left panel), lacking leucine and uracil (center panel), or lacking leucine and containing fluoroorotic acid (right panel). Cultures of control ura+ and ura4− strains having the control vector were also spotted. After growth for 3–4 days at 30 °C, the plates were photographed. C, the colonies growing on plates lacking leucine in B were stained with iodine and photographed. D–G, the dominant negative effect of the swi6L315E mutant on silencing at the mat2 locus. D, the effect of the L315E mutant on silencing. The swi6+ strain above was transformed with vector (left panel), nmt1-GFP-swi6+ plasmid (center panel), and nmt1-GFP-swi6L315E plasmid (right panel). The transformants were streaked for single colonies on PMA plates lacking leucine. The colonies formed after 3–4 days of growth at 30 °C were stained with iodine and photographed. E, the haploid meiosis (azygotic ascus) phenotype observed in the right panel of D is shown on the right. F and G show, respectively, the quantitation of the number of iodine staining colonies, and the asci showing haploid meiosis in the transformants are shown in E.
Interestingly, like the dominant negative effect on switching (Fig. 3), expression of the swi6L315E mutant gene in the strain Msmt0 REIIΔmat2P::ura4, swi6+ also elicited loss of silencing, as indicated by 100% of the transformant colonies giving dark iodine staining (Fig. 4, D (right frame) and F) and the haploid meiosis phenotype that accompanies the loss of silencing (Fig. 4, E and G, and supplemental “Experimental Procedures”). A similar phenotype was observed earlier in combination of the above genotype with the swi6–115 mutation (10). Thus, the phenotype elicited by the dominant negative effect of the mutant is similar to that elicited because of the loss of function mutation swi6–115. Therefore, the Swi6L315E mutant protein, although being defective in localization to heterochromatin by itself, may also abrogate the assembly of heterochromatin in a dominant negative manner, possibly by interfering with recruitment of endogenous Swi6. Like the vector control, the swi6+ gene did not produce any transformants giving iodine staining or having haploid meiosis (Fig. 4, D (center panel), F, and G). A similar effect was observed in the case of silencing of the ade6 reporter linked to the mat3 locus.4 Thus, like the effect on switching, the swi6L315E mutant also exerts a dominant negative effect on silencing at the mating-type loci.
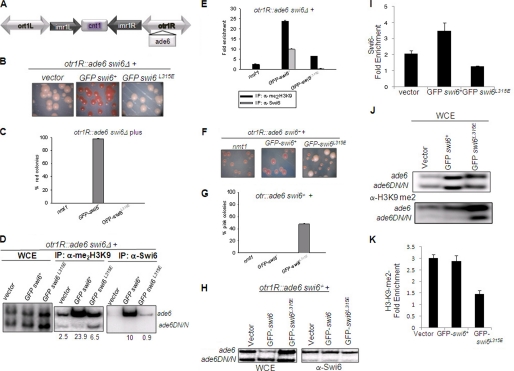
A similar effect was observed in the case of silencing at the outer repeats of the centromere. Unlike the swi6+ gene, the swi6L315E mutant gene was unable to suppress the silencing defect of the ade6 reporter inserted at the otr1 repeat observed in the swi6Δ mutant (Fig. 5, A and B). Pink colonies of the swi6Δ mutant obtained upon growth on adenine-limiting plates (Fig. 5, B and C, vector control) indicate derepression of the ade6 reporter. Whereas expression of the swi6+ gene restored the color to red, indicating restoration of silencing of the ade6 reporter (Fig. 5, B (center panel) and C), expression of the swi6L315E mutant did not (B, right panel). Furthermore, like at mating-type loci, expression of the swi6L315E mutant gene but not the control or swi6+ gene, also exerted a dominant negative effect on silencing at the ade6 reporter at the otr1 repeat in the swi6+ strain, as indicated by the pink colony color of nearly 50% of transformants (Fig. 5, F and G). However, no such effect of the swi6L315E mutant was observed on silencing of the ura4 reporter at the imr repeat of the cen1 locus.4
FIGURE 5.
Non-complementing and dominant negative silencing defect caused by the swi6L315E mutant at the outer repeat-linked ade6 reporter in the cen1 locus. A, diagrammatic representation of the genotype of the strain used. The centromere of each fission yeast chromosome is comprised of a central core (cnt), immediately flanked on both sides by inverted repeats (imr). imr on both sides are flanked by outer repeat regions (otr). The reporter gene ade6 has been inserted into the otr on the right side of chromosome 1 (34). The WT chromosomal copy of the swi6 gene is deleted with the kanr cassette (swi6Δ::kanr). B, this strain was transformed with the nmt1 vector, nmt1-GFP-swi6, and nmt1-GFP-swi6L315E plasmids. Transformants were selected on PMA plates lacking leucine and restreaked on PMA plates lacking leucine and containing a limiting amount of adenine (20 mg/liter). Shown are pictures of colonies obtained upon streaking the transformants. C, quantitative representation of the observed phenotype. Percent of total colonies giving a red color on adenine-limiting plates are plotted for different transformants. D, A ChIP assay was performed for quantitating the localization of Swi6 (lower panel) and H3-Lys-9-Me2 (center panel) among the transformants. PCR was performed for ade6 (heterochromatic) and ade6DN/N (euchromatic) loci, the latter having an internal deletion in the euchromatic ade6 locus (5). WCE represent whole cell extracts (top panel). Enrichment ratios are given in numbers below the lanes or are plotted as a histogram in E. F–G, the dominant negative effect of the swi6L315E mutant on silencing at the otr1-linked ade6 reporter in the cen1 locus. F, the strain was transformed with nmt1, nmt1-GFP-swi6+, and nmt1-GFP-swi6L315E plasmids. The transformants were streaked on plates lacking leucine and a limiting amount of adenine (20 mg/liter), allowed to grow to form colonies at 30 °C for 3–4 days, and then photographed. G, quantitation of the number of pink colonies among the transformants depicted as percentage of total colonies. H–K, the ChIP assay showed reduced localization of Swi6p (H and I) and H3-Lys-9-Me2 (J and K) at the otr1R locus upon overexpression of the swi6L315E mutant gene in the swi6+ strain.
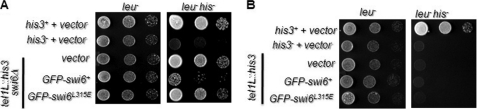
A similar lack of complementation of the silencing defect of the swi6Δ mutant by the swi6L315E mutant gene was observed with respect to the his3 reporter inserted at the telomere locus, indicating a role of self-association of Swi6 in telomere silencing as well (Fig. 6A). However, the swi6L315E mutant gene did not exert a dominant negative effect on silencing at the telomere-linked his3 reporter (Fig. 6B). Thus, the self-association function of Swi6 is important for silencing at all the heterochromatin loci.
FIGURE 6.
The swi6L315E mutant complements the silencing defect of swi6Δ but exerts no dominant negative effect on silencing at the telomere. A and B, swi6Δ (A) and swi6+ (B) strains harboring the his3 gene inserted at the telomere locus and transformed with vector, GFP-swi6+, and GFP-swi6L315E mutant genes were grown to the same cell densities, and 10-fold serial dilutions were spotted on plates lacking leucine (leu−) and plates lacking Leu and His (leu− his−). After 3–4 days of growth at 30 °C, the plates were photographed.
The swi6L315E Mutant Is Defective in Establishment and Maintenance of the Repressed State
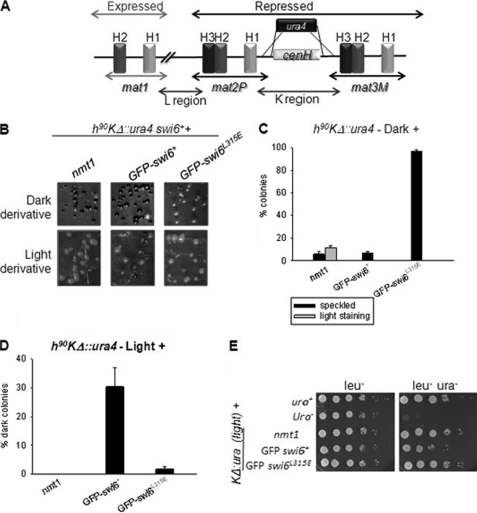
It was shown earlier (15) that replacement of the 7.5-kb region between the mat2 and mat3 loci with the ura4 reporter gene in a homothallic h90 strain (referred to as KΔ::ura4) led to generation of two metastable states: one showing a normal level of switching and staining with iodine (dark) with a low level of expression of the ura4 reporter, and another showing inefficient switching, sporulation, and thereby a lower level of iodine staining (light) along with enhanced expression of the ura4 reporter. Interestingly, these alternative states were inherited as Mendelian alleles (epialleles) during meiosis (15). We tested the effect of overexpression of the swi6+ and swi6L315E genes on the stability of these states. As shown earlier (16), the swi6+ gene converted a significant number of colonies of the “light” derivative of the KΔ::ura4 strain into “dark” (Fig. 7, A, B (lower panel, center), D, and E (with reduced growth on plates lacking uracil). However, the swi6L315E mutant gene was not effective in either restoring iodine staining from light to dark (Fig. 7, B (lower panel, right) and D) or in lowering the level of growth on a plate lacking uracil (E). Thus, the self-association-defective mutant was defective in establishment of silencing.
FIGURE 7.
Swi6 self-association is essential for establishment and propagation of the repressed heterochromatin state. A, diagrammatic representation of the relevant genotype wrt and the mating type region (15, 16, 33). A 7.5-kb fragment of the recombinationally cold spot, called K-region, inclusive of the centromeric non-coding repeat homologous cenH region has been replaced with the 1.8-kb ura4 gene (15, 16). B, the resultant strains called KΔ::ura4 manifested two opposite iodine-staining phenotypes (dark and light) as reported earlier (15). Colonies produced by transforming the dark (upper panel) and light staining derivatives (lower panel) of the KΔ::ura4 strain with vector alone, GFPswi6+, and GFP-tagged swi6L315E genes were grown on selective leu− pates for 3–4 days and stained with iodine. C and D, number of light and dark colonies in B were plotted as the percentage of total transformants of the dark- and light-staining derivatives of the KΔ::ura4 strain. E, serial dilution assay showing suppression of expression of KΔ::ura4 reporter of the light-staining derivative by transformation with the swi6+ but not the swi6L315E mutant gene.
On the other hand, all the transformants of the cells of dark colonies harboring the swi6L315E mutant gene gave light or speckled staining with iodine (Fig. 7, B (upper panel, right) and C), whereas the swi6+ gene had no effect (Fig. 7, B (upper panel, center) and C), indicating that the mutant Swi6 protein is defective in maintenance of the silent state. Thus, the self-association-defective mutant of swi6 is defective in both establishment and maintenance of the repressed heterochromatin state.
The swi6L315E Mutant Abrogates Heterochromatic Localization of H3-Lys-9-Me2 and Itself
Because silencing at the heterochromatin loci is correlated with localization of Me2-Lys-9-H3 and Swi6, a ChIP assay was performed to check the effect of the swi6L315E mutant on heterochromatic localization of Swi6 and H3-Lys-9-Me2. Results show that Swi6 and H3-Lys-9-Me2 (and thereby Clr4) are enriched at the mating-type region (Fig. 8, A–D) and that the ade6 reporter at the otr1 repeat at the cen1 locus (Fig. 5, A–D) in the swi6Δ strain transformed with the swi6+ gene. However, Swi6 and H3-Lys-9-Me2 failed to be localized at both regions when the same strain was transformed with the swi6L315E mutant gene (Fig. 8, A–D and Fig. 5, D and E).
FIGURE 8.
Self-association of Swi6 is required for its own recruitment and setting up the histone H3-Lys-9 dimethylation mark. A, a schematic representation of the K region along with the approximate binding sites of the two sets of oligos used (14, 33). B, whole cell extracts from the swi6Δ mutant strain expressing vector alone, GFP-swi6, and GFP-swi6 L315E mutant plasmids were subjected to ChIP assay with α-Me2-H3-Lys-9 (center panel) and α-Swi6 (right panel) antibodies using specific sets of primers indicated in A. Representative pictures of the amplified products of the chromatin immunoprecipitated samples are shown. C and D, quantitative representation of enrichment of the regions amplified by primer sets 4 and 5 in α-Me2-H3-Lys-9 and α-Swi6 immunoprecipitated samples, respectively. E–H, whole cell extracts from the swi6+ strain expressing vector alone, GFP-tagged swi6, and GFP-tagged swi6 L315E mutant plasmids were subjected to a ChIP assay with α-Swi6 (right panel, E and F) and H3-Lys-9-Me2 (G and H) antibodies using specific sets of primers indicated in A. F and H, quantitation of the data shown in E and G, respectively.
A similar result was obtained when the swi6L315E mutant gene was transformed into the swi6+ strain. Localization of Swi6 and H3-Lys-9-me2 to both the cenH region (Fig. 8, E–H) and otr1R::ade6 (Fig. 5, H–K) was abrogated. The extent of reduction of Swi6 localization suggests that not only was the Swi6L315E protein delocalized from the cenH region (as mentioned above, Fig. 8E), but it interfered with the localization of endogenous Swi6 as well (Fig. 5, H and I and Fig. 8, E and F), indicating a dominant negative effect of the self-association-defective mutant. More interestingly, the swi6L315E mutant exerted a dominant negative effect on localization of H3-Lys-9-Me2 (and thereby Clr4) at the mating-type region (Fig. 8, G and H) as well as the otr1R-linked ade6 reporter (Fig. 5, J and K) when transformed into the swi6+ strain.
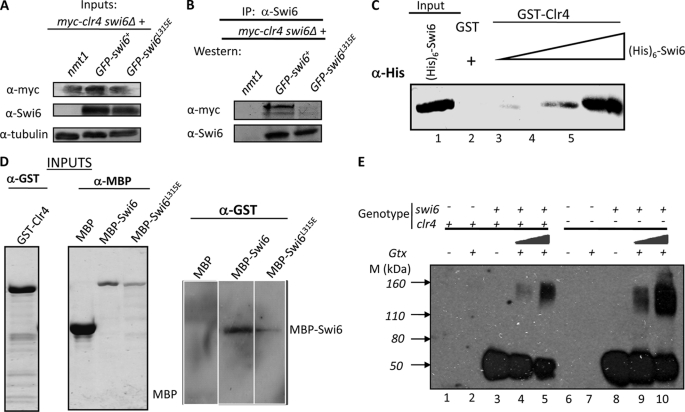
Because Swi6 is thought to act downstream of the Clr4 effect of H3-Lys-9 methylation, the reduced level of H3-Lys-9 methylation at mating-type and centromere loci is quite surprising, suggesting a deleterious effect of the swi6L315E mutation on recruitment of Clr4. Despite the lack of a pentapeptide motif in Clr4p, we checked whether Swi6 and Clr4 can interact with each other. A co-immunoprecipitation experiment showed that Swi6 does indeed interact with Clr4 in vivo and, more importantly, that the mutant Swi6L315E protein is unable to do so (Fig. 9, A and B). Pull-down experiments showed that Clr4p can interact with Swi6p in vitro (Fig. 9C). Furthermore, a far-Western experiment was done to evaluate the relative ability of WT and mutant Swi6 to interact with GST-Clr4 in vitro. Results showed at least a 3–4-fold reduction in the ability of MBP-Swi6L315E to interact with GST-Clr4 as compared with MBP-Swi6+ (Fig. 9D). However, the self-association property of Swi6 was not influenced by Clr4 (Fig. 9E).
FIGURE 9.
The Swi6L315E mutant fails to interact with Clr4. A and B, the Swi6L315E mutant fails to interact with Clr4 in vivo. A, inputs. Whole cell extracts from clr4-Myc, the swi6Δ strain transformed with vector alone, GFP-swi6, and the GFP-swi6L315E mutant gene were immunoblotted with α-Myc (upper panel), α-Swi6 (center panel), and α-tubulin antibody as loading control (lower panel). B, samples in A were immunoprecipitated with α-Swi6 antibody, and the immunoprecipitated samples were immunoblotted with α-Myc (upper panel) and α-Swi6 (lower panel) antibodies. The reverse immunoprecipitation reaction was not performed because of poor extractability of myc-tagged Clr4. C, pull-down assay showing interaction of GST-Clr4 with (His)6-tagged Swi6 in vitro. GST (lane 2) and GST-Clr4 (lanes 3–5) were bound to glutathione-Sepharose beads and incubated with increasing concentrations of extracts of E. coli cells expressing (His)6-Swi6 (lanes 2 and 3-5). Lane 1 contains (His)6-Clr4 at 5% input. After washing, the beads were subjected to SDS-PAGE and immunoblotted with α-His antibody. D, far-Western analysis to study the interaction of WT and the Swi6L315E mutant with GST-Clr4. Equal amounts of MBP, MBP-Swi6, and MBP-Swi6L315E proteins were subjected to SDS-PAGE and electroblotted. The membrane was renatured according to Ref. 35 and incubated with purified GST-Clr4 protein, followed by detection using α-GST antibody. E, self-association of Swi6 is independent of Clr4. A glutaraldehyde cross-linking experiment was done with extracts from swi6+, clr4+ (lanes 3–5), and swi6+, clr4− (lanes 8–10) strains. Lanes 3 and 8 represent mock-treated samples, whereas lanes 4, 5, 9, and 10 show the effect of increasing amounts of glutaraldehyde (Gtx). Lanes 1 and 2 represent extracts from swi6−, clr4+ with and without cross-linking, respectively. Similarly, lanes 6 and 7 represent extracts from the swi6−, clr4− strain without and with cross-linking, respectively.
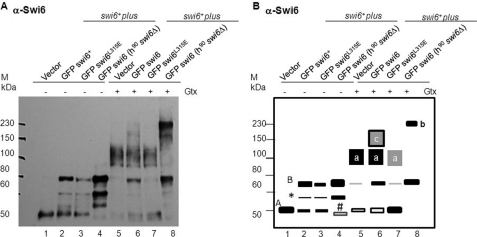
The Swi6L315E Mutant Fails to Interact with and Curtails the Self-association of Native Swi6
Because of the dominant negative effect of the swi6L315E mutant on silencing, we investigated whether it might also exert a dominant negative effect on the self-association of and association with native Swi6. Western blots of the glutaraldehyde cross-linked extracts prepared from the swi6+ strain expressing the GFP-tagged swi6+ and swi6 L315E mutant genes were probed with α-Swi6 antibody. Results showed that the native Swi6p with an apparent molecular mass of ∼50 kDa (Fig. 10, lane 1 (band A) as reported earlier) yielded a cross-linked band of ∼130 kDa (Fig. 10, A and B, lane 5 (band A)). Similarly, cross-linking of GFP-tagged Swi6 expressed in the swi6Δ background, which migrates at ∼78 kDa (50 (Swi6) + 28 kDa (GFP); Fig 10, A and B, lane 4 (band B)), yielded a broad band of ∼230–240 kDa (Fig. 10, A and B, lane 8 (band b)). Extracts of swi6+ cells expressing the GFP-swi6+ gene (Fig. 10A, lane 2) yielded a mixture of cross-linked products of ∼130 kDa (band a representing endogenous native Swi6; Fig 10, lane 6) as well as ∼150 kDa (band c) that reacts with Swi6 antibody (Fig. 10, A and B, lane 6 of the left and right panels), likely representing a stable complex between GFP-Swi6+p and the native Swi6. Further, swi6+ cells expressing GFPSwi6L315E yielded a reduced level of cross-linked product of the endogenous Swi6 (Fig. 10, compare lanes 5 and 7 and band A with band a), observed as reduced relative intensity of band a versus band A in lane 7 as compared with that in lanes 5 and 6, indicating that the mutant Swi6L315E interferes with the efficiency of self-association of native Swi6 itself. Importantly, the complete lack of band c (Fig. 10, A and B, lane 7) indicates the inability of Swi6L315E protein to interact with native endogenous Swi6.
FIGURE 10.
The Swi6L315E mutant protein fails to interact with and interferes with self-association of the native Swi6. A, protein extracts from the swi6+ (lanes 1–3 and 5–7) and swi6Δ (lanes 4 and 8) strains containing empty vector (lanes 1 and 5), the GFP-tagged swi6 gene (lanes 2 and 6), and the GFP tagged swi6L315E mutant gene (lanes 3, 4, 7, and 8) were subjected to glutaraldehyde cross-linking (lanes 5–8), whereas lanes 1–4 represent the input, untreated samples. Western blotting was done with α-Swi6 antibody. B, a schematic representation of the data in A depicting the bands described in the test. The asterisk indicates a possible degradation product of GFP-Swi6 (band B), whereas band A represents endogenous Swi6. Some of the portion of GFP-Swi6 also overlaps with band A, possibly because of truncation of full-length GFP-Swi6. Bands a and b represent the cross-linked products of endogenous native Swi6 and GFP-Swi6, respectively. Band c may represent the putative complex between native Swi6 and GFP-Swi6. # indicates an unknown cross-reacting band in the swi6Δ mutant in lane 4. The relative intensities of bands in A are represented in B qualitatively as thick bars for a higher level to an empty bar to a simple line representing progressively lower levels of the band intensities. Boxes with black and gray shades also depict a decreasing level of cross-linked product.
Thus, apart from being unable to form oligomers by itself, Swi6L315E protein fails not only to associate with native Swi6 but also interferes with the efficiency of self-association of the endogenous Swi6 which, in turn, will interfere with binding to and recruitment of Clr4, thus leading to reduced H3-Lys-9-Me2 and Swi6 recruitment. This turn of events may explain the dominant negative effect of the mutant.
DISCUSSION
Swi6 Exists as a Tetramer in Vivo
Whether Swi6 exists as a tetramer or a dimer has not been fully resolved. In vitro gel filtration studies on recombinant His-tagged Swi6 indicated it to be tetramer (3), whereas a recent gel filtration study suggested that Swi6 may exist as a dimer in vivo with a disproportionately large hydrodynamic volume (12). In SDS page analysis, Swi6 migrates as a 50-kDa protein (3), whereas the estimated molecular mass based on sequence is ∼37 kDa. The difference in electrophoretic mobility cannot be due merely to hydrodynamic volume and may be ascribed to various posttranslational modifications like phosphorylation. In fact, phosphatase treatment enhanced the electrophoretic mobility of Swi6 (17, 18). Our cross-linking data of GFP-Swi6 (78 kDa, Fig. 1D) and native Swi6 (∼50 kDa) yielded products of ∼240 and 130 kDa, respectively, with a gross estimate of a trimer or tetramer for GFP-Swi6 (78 × 3 = 224 kDa, 78 × 4 = 300 kDa) or a dimer or trimer for native Swi6 (50 × 2 = 100 kDa, 50 × 3 = 150 kDa). However, the difference of 110 kDa between the apparent molecular masses of the cross-linked products of native Swi6 (130 kDa) and GFP-Swi6 (240 kDa) corresponds roughly to a tetramer of GFP (28 × 4 = 112 kDa). Because GFP is not reported to exist as dimer or oligomer, the above calculation suggests that Swi6 is more likely to exist as a tetramer in vivo.
Recently, Swi6/HP1 has been shown to be part of several complexes and to interact with a large number of proteins in vivo (19). Keeping this in mind, the cross-linked bands may represent complexes between Swi6 and other proteins. However, the close correspondence between the sizes of the cross-linked product of GST-Swi6 in vitro and that of GFP-Swi6 expressed in S. pombe cells argues in favor of the observed cross-linked complex to be likely to be a homomultimer of Swi6 alone in vivo. It is likely that complexes of Swi6 with other proteins may migrate at much slower electrophoretic mobility and may be less abundant, explaining the lack of visible bands larger than 230–240 kDa.
The Swi6L315E Mutant Is Functional in Vivo
Sequence comparison shows that the Leu-315 residue in S. pombe is replaced by Ile in HP1 homologues (Fig. 1A). Notably, another chromodomain protein, Chp2, in S. pombe contains an Ile residue, which, like the Ile residue in HP1 (3), performs a similar role to that of Swi6 Leu-315 in self-association (12). Mutation of these residues to Glu abolishes their self-association property (12). In the case of Chp2, the silencing function is abrogated and in the case of HP1, the interaction with Suv39 is inhibited (20). Thus, being hydrophobic residues, Ile and Leu may perform a conserved function in self-association of the Swi6/HP1 proteins. In agreement, we find that unlike L315E, the L315I mutant of Swi6 complements the silencing defect of the swi6Δ mutant and fails to exert a dominant negative effect on silencing at the mating-type locus (supplemental Fig. S1, A and B).
Self-association of Swi6 in Binding to and Recruitment of Clr4/Suv39H1
Recently, the swi6L315E mutant has been shown to be defective in self-association as well as silencing at the mating-type locus (12). However, this study investigated the relative contributions of Swi6 and Chp2 in silencing. In this study, we investigated the mechanism by which self-association of Swi6 contributes to silencing. Our results show that self-association of Swi6 is required for complementing the swi6Δ defect at the heterochromatin loci. Furthermore, the self-association-defective mutant exerts a dominant negative effect on silencing at mating-type and centromere loci; thus, self-association is also involved in the maintenance of silencing. Considering the generally accepted model where Swi6 acts downstream of Clr4 (1), an unexpected result of the ChIP assay is a reduced level of H3-Lys-9-Me2, in addition to that of Swi6, at the heterochromatin loci under both conditions; i.e. the swi6Δ and swi6+ strains transformed with the swi6L315E mutant gene (Figs. 5 and 8). Together with the result that Swi6 interacts with Clr4 in vivo whereas the self-association-defective mutant interacts poorly, these results imply that the self-association is important for binding of Swi6 with Clr4 in vivo and Swi6 and Clr4 function as a complex, which ensures both the initial establishment of the heterochromatin mark (that is, H3-Lys-9 methylation and its binding with Swi6) and the cooperative propagation of these events to ensure spreading and subsequent inheritance of the heterochromatin structure.
Recently, the possible role of Clr4 in marking the imprint of H3-Lys-9 methylation through its SET domain and then binding of the imprint by its chromodomain in generating and propagating an epigenetic loop has been mooted (21). This proposal was based in part on the ability of the chromodomain of Clr4 to bind to H3-Lys-9-Me2 in vitro (22). However, the same authors highlight the importance of the presence of both Clr4 and Swi6 in effecting the epigenetic loop (21, 23). Interaction between HP1 and Suv39 has indeed been demonstrated and may involve some other residues in Clr4/Suv39 as well (20). Similarly, human Suv39H1 has been shown to interact with m31 in transgenic flies (24). In Xenopus, targeting of HP1 to chromatin requires a direct protein-protein interaction between SUV39H1 and HP1 (25). There is an apparent difference between our results and previous results, wherein the swi6–115 mutant has been shown to retain a reduced but significant level of H3-Lys-9 methylation (23). However, the same study also showed a significant reduction of H3-Lys-9 methylation in the flanking regions of mating-type loci, suggesting a role of Swi6 in spreading of heterochromatin (a role of Swi6 downstream of Clr4) (23). Further, the swi6–115 mutant does express a drastically reduced albeit detectable level of Swi6 (23), whereas the swi6Δ mutant does not. Thus, earlier results may have had a contribution from the residual Swi6 in initiating the heterochromatin mark by recruiting Clr4, whereas the lack of Swi6 in the present study and our previous study (26) implies a complete lack of Clr4 recruitment in the swi6Δ mutant, leading to a defect in both establishment and maintenance of heterochromatin.
These inferences are indeed supported by the dominant negative effect of the L315E mutant protein not only on self-association of Swi6 itself but also on its binding to (and possibly recruitment of) Clr4, thereby causing reduced H3-Lys-9 methylation. The Swi6 tetramer may require association between the CSD of identical Swi6 monomers. Whereas the lack of Swi6L315E self-association is understandable in terms of the inability of the CSD of the mutant protein to interact, that of interference with association of native Swi6 may be explained by the dilution of the native protein by the mutant protein and unstable association between the WT and mutant Swi6. Thus, both the defects caused by the dominant negative mutant (that is, reduced recruitment of Swi6 and Clr4) may result from lack of a functional Swi6 tetrameric complex which, in turn, fails to bind to and recruit Clr4. In this context, it is surprising to note the lack of a dominant negative effect of the Swi6L315E mutant on telomere silencing. However, it needs to be noted that silencing at the telomere is much more complex as it also involves Taz1 and RNAi/RNA-induced initiation of transcriptional gene silencing in recruitment of heterochromatin machinery (27). Because of this complexity of silencing at telomeres, an analysis of the lack of a dominant negative effect of this mutant on telomere silencing is beyond the scope of the present investigation.
Taken together, both Swi6 and Clr4 may function in unison as integral components of the epigenetic loop in establishing the repressive mark of H3-Lys-9 methylation, its binding by Swi6, and spreading of these landmarks by virtue of their mutually cooperative interaction.
Concerted Rather than Sequential Action of Swi6/HP1 and Clr4/Suv39H1
Thus, the important finding of this study is that the events of histone H3-Lys-9 methylation by Clr4 and binding by Swi6 occur in a concerted rather than a spatiotemporally separated manner. We suggest a model wherein Swi6/HP1 and Clr4/Suv39H1 are recruited as a complex to establish the epigenetic mark of heterochromatin (that is, H3-Lys-9-methylation). A cooperative interaction between them may ensure a stoichiometric recruitment of Clr4, followed by H3-Lys-9 methylation; binding of H3-Lys-9-Me2 by Swi6 may help in spreading. Likewise, at the time of duplication of the heterochromatin structure, the self-association function of Swi6 may be instrumental in recruitment of Clr4 together with Swi6 to ensure reassembly of the heterochromatin structure, thus ensuring the maintenance of heterochromatin. Lack of a self-association property of Swi6 may interfere with both these stages because of its inability to recruit Clr4 (Fig. 11). Our results may apparently seem to be at variance with the recent reports where H3-Lys-9 methylation is thought be a required for H3-Lys-14 deacetylation and Swi6/HP1 binding (19, 28). However, an alternative and plausible scenario could involve concurrent action by Swi6/HP1 (possibly along with SHREC2) and Clr4 (which may be a part of the ClrC complex, 29). Thus, Swi6 may exist in two modes of binding: initial recruitment may be dependent on a cue, possibly from the RNAi pathway coupled with the SHREC2 complex to initiate heterochromatin (30), followed by maintenance, which is ensured by interaction between Swi6 and Clr4. Interestingly, during mouse development, the male pronucleus of the zygote after fertilization first recruits HP1β prior to the appearance of H3-Lys-9-Me2 (31), suggesting a role of HP1β-mediated recruitment of Suv39 in early development. Interestingly, the self-association defect in HP1α has been associated with breast cancer invasiveness (32). Thus, self-association of Swi6 may establish a platform for several physiologically important protein-protein interactions that are critical for development and disease.
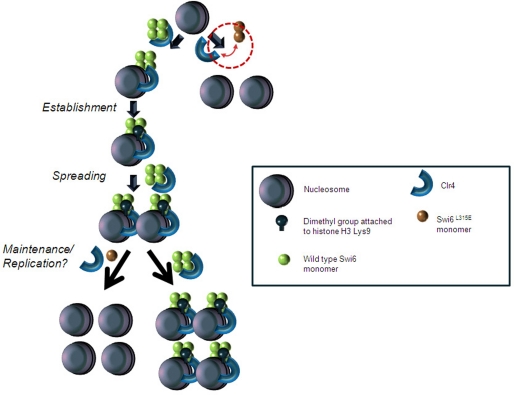
FIGURE 11.
A speculative model depicting the role of self-association of Swi6 in binding to Clr4 and co-recruiting it to establish, spread, and maintain the heterochromatin mark. Clr4 dimethylates histone H3 at the K9 position. Wild-type Swi6, which can self-associate, can thereby interact with Clr4. This interaction most likely facilitates recruitment of Clr4 and subsequent dimethylation of histone H3 on Lys-9. This association may play a dynamic role in establishment of the repressive mark (H3-Lys-9-Me2) and its spreading and maintenance. The Swi6L315E mutant protein, which fails to self-associate, is incapable of interacting with Clr4 and thus recruitment of the latter as well as their mutually cooperative recruitment are adversely affected, which ultimately manifests as reduced localization of H3-Lys-9-Me2 and Swi6 to heterochromatin in the mutant.
Conservation of modular structures of Swi6 and Clr4 (33) suggests that the cooperative action of Swi6/HP1 and Clr4/Suv39H1 in heterochromatin assembly may also be evolutionarily conserved.
Supplementary Material
Acknowledgments
We thank R. Allshire, G. Thon, and A. Klar for the strains; A. Pidoux for the nmt1-GFP-swi6 plasmid; K. Tikoo and M. Pal-Bhadra for anti-H3-Lys-9-Me2 antibody, and S. Ahmed for the protocol of co-immunoprecipitation experiments.
This work was supported by intramural support from the Council of Scientific and Industrial Research, New Delhi, India.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Tables 1–3, references, and Fig. 1.
S. Haldar and J. Singh, unpublished results.
- CSD
- chromoshadow domain
- h90
- homothallic
- MBP
- maltose-binding protein
- PMA
- Pombe minimal medium with adenine.
REFERENCES
- 1. Grewal S. I., Elgin S. C. (2002) Curr. Opin. Genet. Dev. 12, 178–187 [DOI] [PubMed] [Google Scholar]
- 2. Bannister A. J., Zegerman P., Partridge J. F., Miska E. A., Thomas J. O., Allshire R. C., Kouzarides T. (2001) Nature 410, 120–124 [DOI] [PubMed] [Google Scholar]
- 3. Wang G., Ma A., Chow C. M., Horsley D., Brown N. R., Cowell I. G., Singh P. B. (2000) Mol. Cell. Biol. 20, 6970–6983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Volpe T. A., Kidner C., Hall I. M., Teng G., Grewal S. I., Martienssen R. A. (2002) Science 297, 1833–1837 [DOI] [PubMed] [Google Scholar]
- 5. Cowieson N. P., Partridge J. F., Allshire R. C., McLaughlin P. J. (2000) Curr. Biol. 10, 517–525 [DOI] [PubMed] [Google Scholar]
- 6. Smothers J. F., Henikoff S. (2000) Curr. Biol. 10, 27–30 [DOI] [PubMed] [Google Scholar]
- 7. Brasher S. V., Smith B. O., Fogh R. H., Nietlispach D., Thiru A., Nielsen P. R., Broadhurst R. W., Ball L. J., Murzina N. V., Laue E. D. (2000) EMBO J. 19, 1587–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moreno S., Klar A., Nurse P. (1991) Methods Enzymol. 194, 795–823 [DOI] [PubMed] [Google Scholar]
- 9. Thon G., Klar A. J. S. (1993) Genetics 134, 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thon G., Cohen A., Klar A. J. (1994) Genetics 138, 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ekwall K., Partridge J. F. (1999) in Chromosome Structure Analysis: A Practical Approach (Bickmore W. A. ed) pp. 38–57, Oxford University Press, Oxford, UK [Google Scholar]
- 12. Sadaie M., Kawaguchi R., Ohtani Y., Arisaka F., Tanaka K., Shirahige K., Nakayama J. (2008) Mol. Cell. Biol. 28, 6973–6988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pidoux A. L., Uzawa S., Perry P. E., Cande W. Z., Allshire R. C. (2000) J. Cell Sci. 113, 4177–4191 [DOI] [PubMed] [Google Scholar]
- 14. Grewal S. I., Klar A. J. (1997) Genetics 146, 1221–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grewal S. I., Klar A. J. (1996) Cell 86, 95–101 [DOI] [PubMed] [Google Scholar]
- 16. Nakayama J., Klar A. J., Grewal S. I. (2000) Cell 101, 307–317 [DOI] [PubMed] [Google Scholar]
- 17. Bailis J. M., Bernard P., Antonelli R., Allshire R. C., Forsburg S. L. (2003) Nat. Cell Biol. 5, 1111–1116 [DOI] [PubMed] [Google Scholar]
- 18. Shimada A., Dohke K., Sadaie M., Shinmyozu K., Nakayama J., Urano T., Murakami Y. (2009) Genes Dev. 23, 18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Motamedi M. R., Hong E. J., Li X., Gerber S., Denison C., Gygi S., Moazed D. (2008) Mol. Cell 32, 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yamamoto K., Sonoda M. (2003) Biochem. Biophys. Res. Commun. 301, 287–292 [DOI] [PubMed] [Google Scholar]
- 21. Grewal S. I. S. (2010) Curr. Opin. Genet. Dev. 20, 134–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang K., Mosch K., Fischle W., Grewal S. I. S. (2008) Nat. Struct. Mol. Biol. 15, 381–388 [DOI] [PubMed] [Google Scholar]
- 23. Hall I. M., Shankaranarayana G. D., Noma K., Ayoub N., Cohen A., Grewal S. I. (2002) Science 297, 2232–2237 [DOI] [PubMed] [Google Scholar]
- 24. Aagaard L., Laible G., Selenko P., Schmid M., Dorn R., Schotta G., Kuhfittig S., Wolf A., Lebersorger A., Singh P. B., Reuter G., Jenuwein T. (1999) EMBO J. 18, 1923–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stewart M. D., Li J., Wong J. (2005) Mol. Cell. Biol. 25, 2525–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dubey R. N., Nakwal N., Bisht K. K., Saini A., Haldar S., Singh J. (2009) J. Biol. Chem. 284, 7165–7176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kanoh J., Sadaie M., Urano T., Ishikawa F. (2005) Curr. Biol. 15, 1808–1819 [DOI] [PubMed] [Google Scholar]
- 28. Fischer T., Cui B., Dhakshnamoorthy J., Zhou M., Rubin C., Zofall M., Veenstra T. D., Grewal S. I. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8998–9003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sugiyama T., Cam H. P., Sugiyama R., Noma K., Zofall M., Kobayashi R., Grewal S. I. (2007) Cell 128, 491–504 [DOI] [PubMed] [Google Scholar]
- 30. Iida T., Nakayama J., Moazed D. (2008) Mol. Cell 31, 178–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Arney K. L., Bao S., Bannister A. J., Kouzarides T., Surani M. A. (2002) Int. J. Dev. Biol. 46, 317–320 [PubMed] [Google Scholar]
- 32. Norwood L. E., Moss T. J., Margaryan N. V., Cook S. L., Wright L., Seftor E. A., Hendrix M. J., Kirschmann D. A., Wallrath L. L. (2006) J. Biol. Chem. 281, 18668–18676 [DOI] [PubMed] [Google Scholar]
- 33. Klar A. J. (2007) Ann. Rev. Genet. 41, 213–236 [DOI] [PubMed] [Google Scholar]
- 34. Allshire R. C., Javerzat J. P., Redhead N. J., Cranston G. (1994) Cell 76, 157–169 [DOI] [PubMed] [Google Scholar]
- 35. Wu Y., Li Q., Chen X. Z. (2007) Nat. Protoc. 12, 3278–3284 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.