Abstract
The pathogenesis of dengue hemorrhagic fever and dengue shock syndrome (DHF/DSS), both serious complications of dengue virus (DV) infection, remains unclear. In this study, we found that anti-DV NS1 (nonstructural protein 1) polyclonal antibodies cross-reacted with human umbilical vein endothelial cells (HUVECs). We further identified a complex-specific mAb, DB16-1, which could recognize DV NS1 and cross-react with HUVECs and human blood vessels. The target protein of DB16-1 was further purified by immunoaffinity chromatography. LC-MS/MS analysis and co-immunoprecipitation revealed that the target protein of DB16-1 was human LYRIC (lysine-rich CEACAM1 co-isolated). Our newly generated anti-LYRIC mAbs bound to HUVECs in a pattern similar to that of DB16-1. The B-cell epitope of DB16-1 displayed a consensus motif, Lys-X-Trp-Gly (KXWG), which corresponded to amino acid residues 116–119 of DV NS1 and mimicked amino acid residues 334–337 in LYRIC. Moreover, the binding activity of DB16-1 in NS1 of DV-2 and in LYRIC disappeared after the KXWG epitope was deleted in each. In conclusion, DB16-1 targeted the same epitope in DV NS1 and LYRIC protein on human endothelial cells, suggesting that it might play a role in the pathogenesis of DHF/DSS. Future studies on the role of the anti-NS1 antibody in causing vascular permeability will undoubtedly be performed on sera collected from individuals before, during, and after the endothelial cell malfunction phase of a dengue illness.
Keywords: Antibodies, Endothelium, Flavi Viruses, Immunology, RNA Viruses, Dengue Fever (DF), Dengue Hemorrhagic Fever and Dengue Shock Syndrome (DHF/DSS), Lysine-rich CEACAM1 Co-isolated (LYRIC), Molecular Mimicry, Nonstructural Protein 1 (NS1)
Introduction
Dengue virus (DV),2 a flaviviridae, causes diseases ranging from mild dengue fever to severe syndromes, such as DHF and DSS (1, 2). Primary DV infection often leads to a painful but nonfatal dengue fever and protects patients from reinfection of DV of the same serotype. However, secondary infection with DV of a different serotype can trigger the more severe and potentially fatal DHF or DSS (1, 3). The clinical presentations of DHF/DSS include thrombocytopenia, vascular leakage, hemorrhage, and complement activation. Because little is known about the pathogenic mechanisms underlying these disorders, no effective strategy has been developed to prevent their occurrence (4, 5).
Several theories have been proposed to explain the pathogenesis of the DHF/DSS. One of them is antibody-dependent enhancement. It is theorized that upon the second infection by DV of different serotype, monocytes and/or macrophages enhance uptake of complexes of virus with non-neutralizing antibodies, subneutralizing cross-reactive antibodies, or low titer neutralizing antibodies through the Fc receptor (1, 6). Hence, the increased viral load induces the plasma leakage or hemorrhage in DHF/DSS. It has been proposed that host immune reactions, including complement activation, immune cell activation, cytokine production, and immune deviation, are involved in the initiation of DHF/DSS (7–10). Others suggest that viral virulence may play a role in the pathogenesis of DHF/DSS (11, 12). However, although many theories have been put forward, the main mechanism underlying the development of DHF/DSS remains unknown.
Several viruses have similar antigenic determinants that make them able to mimic host proteins (13, 14), a phenomenon known as molecular mimicry. These viruses often initiate the generation of autoantibodies against the host's own tissues (15–17). The presence of cross-reactive antibodies against endothelium after infection by human cytomegalovirus (hCMV), Epstein-Barr virus, and HIV is well documented (17–19). The association of the autoantibody induced by hCMV infections and systemic sclerosis is a good example. The serum antibodies that induce endothelium apoptosis in patients of systemic sclerosis also recognize the late protein UL94 of hCMV (17). The clinical onset of systemic sclerosis is associated with the generation of pathogenetic autoantibodies by chronic infection of hCMV (20, 21). However, vascular permeability and DHF/DSS in dengue is transient. Mouse polyclonal antibodies against DV-2 NS1 have been found able to cross-react with human endothelium (22, 23). Moreover, serum antibodies from dengue fever and DHF patients can bind to HUVECs (23, 24). Once the binding occurs, endothelial cells undergo nitric oxide-mediated apoptosis (23), an effect that can be blocked by recombinant NS1 protein (24). Together, these findings suggest that the endothelial dysfunction caused by the induction of autoantibodies through host-virus interplay may be one of the factors in the pathogenesis of DHF/DSS. Understanding the molecular target of DV autoantibodies may, therefore, be important for diagnosis and the design of a suitable safe vaccine against this viral disease.
In this study, we generated many anti-DV mAbs and successfully identified an anti-DV NS1 mAb, DB16-1, which was found to cross-react with HUVECs and human blood vessels. The protein targeted by DB16-1 was isolated by immunoprecipitation and designated LYRIC (lysine-rich CEACAM1 co-isolated), a finding further confirmed by mass spectrometry. LYRIC protein is also called metadherin (25), 3D3 (26), or AEG-1 (astrocyte-elevated gene-1) (27) and is highly conserved between species (26). However, its biological function remains unclear. Using phage display to identify the B-cell epitope of DB16-1, we found that DV NS1 and LYRIC contained similar epitopes that may induce the production of an autoantibody. These findings suggest that DB16-1 might act as an autoantibody against LYRIC on endothelial cells and lead to the transient vascular leakage in DHF/DSS.
EXPERIMENTAL PROCEDURES
Cells and Viruses
Preparation of the four serotypes of dengue viruses, DV-1 (Hawaii), DV-2 (New Guinea C), DV-3 (H87), and DV-4 (H241), was as previously descried (28). HUVECs were purchased and grown in EGM-2 medium (LONZA).
Preparation of NS1 and Viral Particles of Four Serotypes of DV
DV-infected C6/36 cultured supernatant was applied to DB20-6 (an anti-NS1 mAb)- or 4G2 (an anti-Envelop protein mAb)-coupled protein G-Sepharose 4 Fast Flow gel (GE Healthcare). The antibody-conjugated affinity columns were washed with PBS. The NS1 proteins or viral particles were eluted with elution buffer (Thermo Scientific, Rockford, IL) and neutralized with 1 m Tris-HCl, pH 9.1.
Immunofluorescence
HUVECs were grown on coverslips and then rinsed with PBS and fixed with 2% paraformaldehyde, followed by incubation with mouse anti-NS1 hyperimmune sera (1:500 dilution), DB16-1 (10 μg/ml), 4G2 (10 μg/ml) antibodies, and normal mouse sera (1:500 dilution) or normal mouse IgG (NMIgG) (10 μg/ml) for 1 h at room temperature. The coverslips were incubated with fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG (Jackson ImmunoResearch) for 30 min at room temperature. To verify that DB16-1 cross-reacted with human endothelium, lung cancer tissue sections were cut at 4 μm, deparaffinized in xylene, hydrated in descending alcohol, and rinsed with PBS. Sections were boiled with 0.01 m citrate buffer, pH 6.0, in a high pressure cooker for 5 min to retrieve antigenicity. Then the sections were incubated with DB16-1 and biotinylated Ulex europaeus agglutinin-I (Vector Laboratories, Burlingame, CA) for 1 h at room temperature. The sections were then treated with phycoerythrin-conjugated anti-mouse IgG (Jackson ImmunoResearch) and FITC-conjugated streptavidin (Thermo Scientific, Waltham, MA) for 30 min at room temperature. The slides were counterstained with mounting medium containing Hoechst 33258 (Molecular Probes, Inc., Eugene, OR) and analyzed under a fluorescent microscope.
ELISA
HUVECs were grown on 96-well plates, fixed with 2% paraformaldehyde, and blocked with 1% bovine serum albumin (BSA) in PBS (blocking buffer). Diluted anti-DV NS1 or anti-DV viral particle mouse sera were incubated with HUVECs. The plates were washed with PBS containing 0.1% Tween 20 (PBST0.1) and treated with HRP-conjugated anti-mouse IgG (Jackson ImmunoResearch). After washing with PBST0.1, the plates were incubated with the peroxidase substrate o-phenylenediamine dihydrochloride (Sigma-Aldrich). The reaction was stopped with 3 n HCl, and the plates were read using a microplate reader at 490 nm.
Generation of mAbs
The production of mAbs against D2NS1 and human recombinant LYRIC was generated following a standard procedure (29) with slight modifications (30). Briefly, female BALB/c mice were immunized intraperitoneally with purified D2NS1 or LYRIC four times at 3-week intervals. On day 4 after the final boost, lymphocytes were harvested from the immunized mouse spleen and fused with NSI/1-Ag4–1 myeloma cells by 50% polyethylene glycol (Invitrogen). The fused cells were resuspended in DMEM containing 20% fetal calf serum (FCS), hypoxanthine-aminopterin-thymidine, and hybridoma cloning factor (ICN Biomedicals, Aurora, OH). All animal experiments were performed as per the guidelines of the National Laboratory Animal Center. The protocol was approved by the Committee on the Ethics of Animal Experiments of Academia Sinica. To confirm the specificity of antibodies, cultured hybridoma supernatant was incubated with DV-2 virus-infected C6/36 cells or coated with purified LYRIC (1 μg/ml) protein in 0.1 m NaHCO3, pH 8.6 (coating buffer), as described under “ELISA.” Hybridoma cell lines were grown in DMEM with 10% FCS. Ascites were generated in pristane-primed BALB/c mice and mAbs purified with protein G-Sepharose 4 Fast Flow gel.
Western Blotting
For specificity of DB16-1, the protein extracts prepared from DV-1-, DV-2-, DV-3-, and DV-4-infected C6/36 cell lysates were mixed with an equal volume of the Laemmli sample buffer (Bio-Rad) containing 5% β-mercaptoethanol (Bio-Rad), separated by SDS-PAGE, and transferred to nitrocellulose (NC) membrane (Millipore, Billerica, MA). The membrane was blocked with 5% skimmed milk in PBS and then incubated with DB16-1. For identification of DB16-1 epitope, the NC membrane was incubated with DB16-1 and different concentrations of SP16-1 (LKYSWKTWGKAK, matched to amino acid residues 111–122 of D2NS1) or control peptide P7M (SHRLHNTMPSES) (30). To characterize the anti-LYRIC antibodies, mAbs Lyric 1-7, Lyric 2-1, Lyric 3-13, and Lyric 4-7 were incubated with NC membranes prepared from cell lysates of HUVECs and COLO 205, which is a colon cancer cell line overexpressing LYRIC. DB16-1 and anti-metadherin (anti-Mid) (Zymed Laboratories Inc., San Francisco, CA), which is a commercial anti-LYRIC antibody, served as a positive control. For the competitive inhibition assay, mAbs were incubated with synthetic peptides SP16-1, SP-Ly (NTNGKDWGRS, matched to amino acid residues 330–339 of LYRIC), and SP5-52 (SVSVGMKPSPRP) (31). The anti-V5 antibody (Invitrogen) allows the detection of recombinant proteins containing the V5 epitope. After washing with PBST0.1, the blots were then incubated with HRP-conjugated anti-mouse or anti-rabbit IgG (Jackson ImmunoResearch) and developed with the use of enhanced chemiluminescence reagents (Thermo Scientific, Rockford, IL).
Flow Cytometry
HUVECs were detached with 2 mm EDTA and washed with PBS. The cells (1 × 105) were incubated with the following antibodies: mouse anti-human CD31 (BD Biosciences), DB16-1, Lyric 1-7, Lyric 2-1, Lyric 3-13, and Lyric 4-7 for 1 h at 4 °C. They were then probed with phycoerythrin-conjugated anti-mouse IgG (Jackson ImmunoResearch) for 30 min at 4 °C. After the final wash, the cells were resuspended with 1% FBS in PBS and analyzed by flow cytometry (BD Biosciences).
TUNEL Staining
HUVECs were cultured to ∼80% confluence on coverslips and incubated with DB16-1 or 4G2 (50 μg/ml) in serum-free medium for 24 h at 37 °C in a 5% CO2 incubator. After rinsing with PBS, the cells were fixed with 4% paraformaldehyde for 20 min at 4 °C and permeabilized with 0.2% Triton X-100 for 5 min at room temperature, followed by incubation with TUNEL reaction mixture (Roche Applied Science) for 1 h at 37 °C. The slides were counterstained with DAPI (Vector Laboratories) and visualized under a fluorescent microscope.
Complement-dependent Cytotoxicity
HUVECs or LYRIC-transfected HEK293T cells were harvested (1 × 105 cells/reaction) and incubated with DB16-1 and 4G2 (50 μg/ml) for 1 h at 4 °C. After centrifugation, the supernatant was removed, and cells were washed with 1% FBS in PBS. The HUVECs were then resuspended at a density of 1 × 106 cells/ml in EGM-2 serum-free medium and incubated with Low-Tox-M rabbit complement (1:20 dilution; Cedarlane Laboratories Ltd., Hornby, Canada) for 4 h at 37 °C in a 5% CO2 incubator to facilitate complement-mediated cell lysis. Cell lysis was determined by flow cytometry following propidium iodide staining (1 μg/ml). Propidium iodide-positive cells were defined as a percentage of total cells (32, 33).
Immunoprecipitation
HUVECs (1 × 107) were lysed with lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Nonidet P-40) supplemented with a protease inhibitor mixture tablet (Roche Applied Science) and kept on ice for 30 min. Cell lysate was prepared at 10,000 × g for 15 min at 4 °C. The supernatant was incubated with DB16-1, and then the immunocomplex was precipitated by protein G-Sepharose (GE Healthcare). After washing, the proteins binding to DB16-1 were eluted with 0.2 m glycine, pH 2.5, 150 mm NaCl, and 1% Nonidet P-40, and the eluates were neutralized with 1 m Tris-HCl, pH 9.1. The eluates were fractionated in SDS-PAGE and immunoblotted with DB16-1. The band of interest was cut from the gel; reduced with 50 mm dithioerythreitol in 25 mm ammonium bicarbonate, pH 8.5, for 1 h at 37 °C; and alkylated with 100 mm iodoacetamide in ammonium bicarbonate in the dark for 1 h at room temperature. After washing with 50% acetonitrile in ammonium bicarbonate, the gel was soaked in 100% acetonitrile and incubated with 0.02 μg of trypsin for 16 h at 37 °C. The digested peptides were extracted with 50% acetonitrile in 5% TFA and concentrated using a concentrator (Eppendorf, Hamburg, Germany). The sample was analyzed by LC-MS/MS sequencing in the Core Facility for Proteomics and Structural Biology Research at Academia Sinica.
Co-immunoprecipitation
HUVEC cell lysates were co-immunoprecipitated with anti-Mid (2 μg/ml) and DB16-1 (5 μg/ml) antibodies for 1 h at 4 °C. The immunocomplex was then coupled to protein G-Sepharose (GE Healthcare). Samples were Western blotted with anti-Mid (Zymed Laboratories Inc.) and DB16-1 antibodies following the same procedures as described above under “Western Blotting.”
Phage Display Biopanning
The 8-well module was coated with 100 μg/ml DB16-1 and blocked at 4 °C overnight. A phage-displayed peptide library (New England Biolabs, Inc.) was diluted to 4 × 1010 phages and incubated with the DB16-1-coated well for 50 min at room temperature. After washing with PBS containing 0.5% Tween 20 (PBST0.5), the bound phages were eluted with 0.2 m glycine, pH 2.2. The eluates were neutralized with 1 m Tris-HCl, pH 9.1. The eluted phages were amplified in an ER2738 (New England Biolabs, Inc.) overnight culture, which was vigorously shaken for 4.5 h at 37 °C. The amplified phages were precipitated with 20% polyethylene glycol 8000 in 2.5 m NaCl (PEG/NaCl) at 4 °C overnight. The phages were centrifuged for 20 min at 8,000 × g at 4 °C and suspended with PBS. The phages were reprecipitated with PEG/NaCl, isolated by centrifugation at 4 °C for 10 min, and resuspended in PBS. The amplified phages were titered on LB/isopropyl-β-d-thiogalactoside/X-gal plates. The second round was identical to the first one except for the addition of 2 × 1011 plaque-forming units (pfu) from previously amplified phages. The third round of biopanning was performed once again with 2 × 1011 pfu of second round-amplified phages. The third round-eluted phages were titered on LB/isopropyl-β-d-thiogalactoside/X-gal plates and selected for ELISA.
Identification and Sequencing of Immunopositive Phage Clones
The ELISA plate was coated with 50 μg/ml DB16-1 or NMIgG in coating buffer for 2 h at room temperature and blocked with blocking buffer at 4 °C overnight. The diluted phages were incubated with coated plates for 1 h at room temperature. After washing, the bound phages were probed with HRP-conjugated mouse anti-M13 mAb (GE Healthcare Biosciences) following the same procedures described above under “ELISA.” The immunopositive phage clones were further sequenced with the −96 primer 5′-CCCTCATAGTTAGCGTAACG-3′, which corresponded to the pIII gene sequence of M13 phage. The phage-displayed peptide sequences were translated with the ExPASy Proteomics Server.
Phage Binding Assay
ELISA plates were coated with DB16-1 at a concentration of 10 μg/ml and blocked with blocking buffer at 4 °C overnight. The plates were incubated with PC16-10 and control phage HB47-1 (34), which were serially diluted from 109 to 104 pfu and 0 pfu. After washing with PBST0.5, the plate was incubated with HRP-conjugated anti-M13 mAb (GE Healthcare) for 1 h at room temperature and following the same procedures described under “ELISA.”
Antibody Binding and Competitive Inhibition Assay
For the binding assay, SP16-1, SP-Ly, and arbitrary control peptide SP5-52 were coated at a concentration of 5 μg/ml. DB16-1 was serially 2-fold diluted from 0.5 to 0.007 μg/ml for SP16-1 and 3-fold diluted from 10 to 0.013 μg/ml for SP-Ly and then incubated for 1 h at room temperature. For the competitive inhibition assay, SP-Ly was coated at a concentration of 5 μg/ml and incubated with the mixture of DB16-1 (1 μg/ml) and SP16-1 or control L-peptide (RLLDTNRPLLPY) (35) (2-fold dilution from 50 to 0.05 μg/ml) for 1 h at room temperature. The plates were probed with HRP-conjugated anti-mouse IgG (Jackson ImmunoResearch) following procedures described under “ELISA.”
Construction and Expression of Recombinant Proteins
The full-length D2NS1 (New Guinea C Strain) cDNA was cloned to pcDNA3.1 vector (Invitrogen). The full-length LYRIC cDNA was cloned to pET151-directional TOPO vector (Invitrogen). The deleted forms of D2NS1 (amino acid residues 116–119 deleted) and LYRIC (amino acid residues 334–337 deleted) were established using the site-directed mutagenesis. The pcDNA3.1-D2NS1 and pcDNA3.1-D2NS1-d116–119 plasmids were transfected to BHK21 cells with Lipofectamine 2000 (Invitrogen), and the cell lysates were prepared with radioimmune precipitation assay buffer (150 mm NaCl, 10 mm Tris-HCl, pH 7.5, 5 mm EDTA, 1% Triton X-100, 0.1% SDS, and 1% sodium deoxycholate). The pET151-LYRIC and pET151-LYRIC-d334–337 plasmids were introduced into Escherichia coli BL21 (DE3) (Invitrogen). The LYRIC proteins were induced by 1 mm isopropyl-β-d-thiogalactoside and purified with nickel-Sepharose 6 Fast Flow (GE Healthcare). The full-length and deleted forms of D2NS1 and LYRIC proteins were analyzed with DB16-1 and anti-V5 antibodies following the same procedures described under “Western Blotting.”
Statistical Analysis
Statistical results were tested using unpaired Student's t tests as appropriate. p < 0.05 was considered significant.
RESULTS
Serum Antibody against DV NS1 Cross-reacts with Human Endothelial Cells
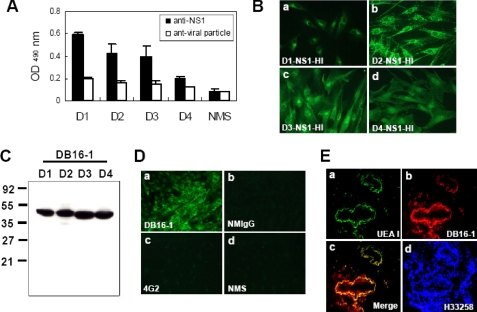
To identify the anti-endothelium autoantibodies, we performed cellular ELISA of HUVECs with hyperimmunized sera from mice challenged by purified NS1 proteins or viral particles of four serotypes of DV. We found that antibodies against DV NS1 proteins bound to HUVECs, whereas antibodies against DV viral particles did not (Fig. 1A). To test whether antibodies against DV NS1 proteins would react with HUVECs, an immunofluorescent assay was performed. We found that sera from mice immunized with four serotypes of NS1 proteins could recognize HUVECs (Fig. 1B). Taken together, it is plausible that anti-DV NS1 antibodies simultaneously cross-reacted with endothelial cells.
FIGURE 1.
Anti-DV NS1 antibodies cross-react with HUVECs and human blood vessels. A, ELISA revealed that anti-DV NS1 antibodies cross-reacted with HUVECs, whereas anti-DV viral particle antibodies did not. B, anti-NS1 hyperimmune sera induced by four serotypes of dengue viruses (a, b, c, and d) contained antibodies bound to HUVECs by an immunofluorescence assay. C, NS1 protein of four serotypes of dengue viruses could be recognized by mAb DB16-1 by Western blot analysis. D, DB16-1 cross-reacted with HUVECs (a), whereas NMIgG (b) 4G2 (c) normal mouse sera (NMS) (d) did not. E, DB16-1 (b) bound to human blood vessels by immunofluorescent double staining (c). U. europaeus agglutinin-I (UEA-I) (a), a lectin expressed on human endothelium, was used as a marker for human vasculature. The nuclear staining was performed by Hoechst 33258 (H33258) (E, d). Cell images were acquired at ×400 magnification (B and E) and ×200 magnification (D). The values were presented as the mean ± S.D.
A mAb Recognizes DV NS1 and Cross-reacts with HUVECs and Human Blood Vessels
It is difficult to use polyclonal antibodies to identify the target proteins of the anti-endothelium autoantibodies. Therefore, we generated more than 100 mAbs and screened the candidate autoantibodies by cellular ELISA. We found a mAb, DB16-1, to have a relatively high reactivity with HUVECs and to be able to recognize the NS1 protein of all four dengue virus subtypes (Fig. 1C). We further validated the reactivity of DB16-1 by an immunofluorescent assay and found that this mAb but not 4G2 (a mAb against the envelope protein of DV), normal mouse IgG (NMIgG) or normal mouse serum could recognize HUVECs (Fig. 1D). We further investigated whether DB16-1 could bind to human blood vessels by double immunostaining. We found that DB16-1 could bind to the blood vessels in surgical specimens of human lung tissues through immunofluorescent localization (Fig. 1E, b). The target protein of DB16-1 was found to be co-localized with an endothelial marker, U. europeus agglutinin-I, in the blood vessels of human lung tissues (Fig. 1E, c). The nuclear staining was performed by Hoechst 33258 (Fig. 1E, d).
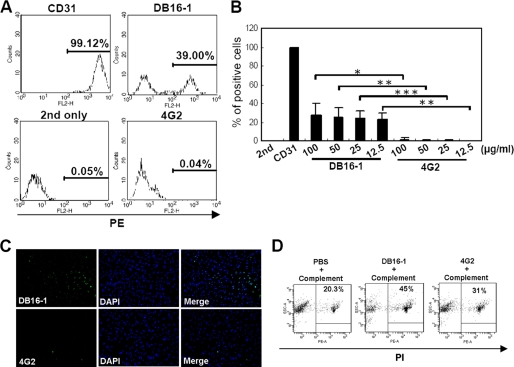
Because DB16-1 could bind to HUVECs in immunofluorescence staining (Fig. 1D), we used flow cytometry to examine the HUVEC binding activity of DB16-1. The presence of a right-shifted peak indicated that the DB16-1 bound to HUVECs (Fig. 2A). Notably, all of the HUVECs were recognized by the endothelial marker CD31, but only some of them (39%) were recognized by DB16-1 (Fig. 2A). Meanwhile, 4G2 did not bind to HUVECs (Fig. 2, A and B, and supplemental Fig. S2). To investigate the hypothesis that dengue NS1 antibodies damage endothelial cells, we tested whether DB16-1 could induce endothelial cell damage by TUNEL and a complement-dependent cytotoxicity assay. We found that DB16-1 but not 4G2 enhanced HUVECs apoptosis by a TUNEL assay (Fig. 2C). Complement-dependent cytotoxicity assay also demonstrated that more HUVECs (45%) underwent lysis after binding to DB16-1 than binding to 4G2 did (31%) (Fig. 2D). These two experiments confirm that endothelial damage occurred after binding of autoantibody DB16-1.
FIGURE 2.
DB16-1 bound to HUVECs and induced cell apoptosis. A, the presence of a right-shifted peak in the flow cytometry indicated DB16-1 bound to HUVECs. Anti-CD31 and 4G2 antibodies were used as the positive and negative control, respectively. B, the percentage of DB16-1 binding to HUVECs was quantified. These data are presented as the mean ± S.D. (error bars) of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001. C, DB16-1 induced apoptosis of HUVECs by TUNEL staining, whereas this phenomenon was not observed in 4G2-treated HUVECs. The nuclear staining was performed by DAPI. Cell images were acquired at ×200 magnification. D, complement-dependent cytotoxicity assay was demonstrated by flow cytometry following staining with propidium iodide in HUVECs treated with complement alone, DB16-1 with complement, or 4G2 with complement. PE, phycoerythrin.
Identification of the Target Protein of DB16-1, LYRIC, in HUVEC Cells
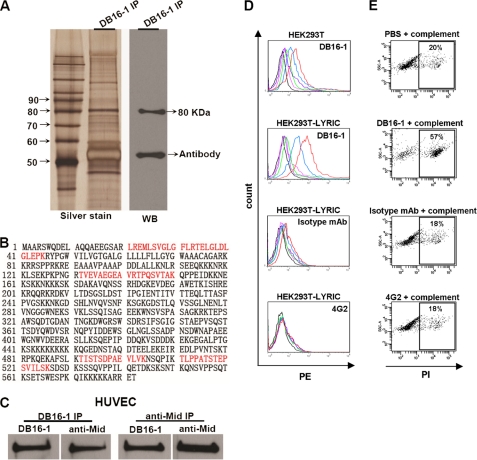
Since molecular mimicry is one of the mechanisms contributing to the link between infection and autoimmunity (36, 37), we were curious to find out if the DB16-1 target protein is expressed on human endothelium. The target proteins were immunoprecipitated from extracts of HUVECs with DB16-1. Western blotting demonstrated that DB16-1 recognized a target protein with a molecular mass of 80 kDa (Fig. 3A). The corresponding band was harvested in silver-stained gel. After in-gel digestion, four peptides were found to have sequences identical to that of the human LYRIC protein by LC-MS/MS sequencing (Fig. 3B). We used immunoaffinity purification to further confirm that human LYRIC protein was the target of DB16-1. Because it was difficult to harvest sufficient HUVECs for immunoaffinity purification of the DB16-1 target, we screened several cancer cell lines and identified COLO 205, a colon cancer cell line that highly expresses the target protein of DB16-1 (data not shown). The purified protein from a DB16-1-conjugated affinity column was further confirmed to be a human LYRIC protein by LC-MS/MS sequencing (supplemental Fig. S1A). The co-immunoprecipitations using DB16-1 and a commercial anti-LYRIC antibody (anti-Mid) revealed that these two antibodies recognized the same target in HUVECs (Fig. 3C) and COLO 205 (supplemental Fig. S1B) cells. To further confirm that DB16-1 could bind to LYRIC-expressed cells, we transfected the LYRIC gene into HEK293T cells and tested the binding activity of DB16-1 by flow cytometry. The results revealed that BD16-1 but not control antibodies recognized LYRIC-expressed HEK293T cells (Fig. 3D). Furthermore, DB16-1 was also demonstrated to enhance the cell undergoing lysis through a complement-dependent cytotoxicity assay. LYRIC-transfected HEK293T cells treated with DB16-1 had a higher percentage of apoptotic cells (57%) than those treated with the isotype mAb (18%) or 4G2 (18%) (Fig. 3E). These data strongly suggest that both NS1 of dengue viruses and human LYRIC protein are targets of DB16-1.
FIGURE 3.
Identification of the target protein of DB16-1 from HUVECs. A, the target protein was immunoprecipitated from HUVEC lysate by DB16-1 and analyzed by silver staining and Western blotting. B, peptide sequences of the target protein through LC-MS/MS analysis are marked. The peptide in red highlights the sequences present in LYRIC protein. C, co-immunoprecipitation (IP) of HUVEC lysates with DB16-1 antibody and a commercial polyclonal anti-LYRIC (anti-Mid) antibody and subsequent immunoblotting with anti-Mid and DB16-1 antibodies. These two antibodies recognized the same target in HUVECs. D, HEK293T cells expressing full-length LYRIC were analyzed by flow cytometry. DB16-1 bound to LYRIC-expressed HEK293T cells in a dose-dependent manner. Isotype mAb and 4G2 antibodies were used as negative controls. Red line, 50 μg/ml; blue line, 12.5 μg/ml; green line, 3.1 μg/ml; purple line, 0.78 μg/ml; pink line, 0.19 μg/ml; black line, 0 μg/ml. E, complement-dependent cytotoxicity assay was performed by flow cytometry in LYRIC-expressed HEK293T cells treated with complement alone, DB16-1 (50 μg/ml) plus complement, isotype mAb (50 μg/ml) plus complement, and 4G2 (50 μg/ml) plus complement.
DB16-1 and Anti-LYRIC Antibodies Recognize LYRIC Protein and HUVECs
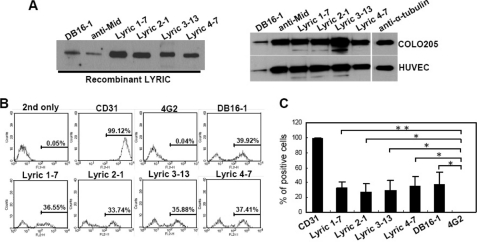
To confirm that DB16-1 recognized LYRIC protein, we cloned and expressed human LYRIC protein and generated mAbs against this protein. Using ELISA and Western blotting screening, we identified four novel monoclonal antibodies against LYRIC protein, Lyric 1-7, Lyric 2-1, Lyric 3-13, and Lyric 4-7. These four anti-LYRIC mAbs, DB16-1, and anti-Mid recognized recombinant LYRIC protein (Fig. 4A, left). LYRIC protein expressed by COLO 205 cells and HUVECs was also recognized by these antibodies (Fig. 4A, right). Flow cytometry further confirmed that HUVECs expressed LYRIC, and this protein could be recognized by DB16-1 and anti-LYRIC antibodies (Fig. 4, B and C). These results indicated that LYRIC was expressed on the cell surface of HUVECs, and anti-DV NS1 antibody cross-reacted with these cells through LYRIC.
FIGURE 4.
LYRIC is the target protein of DB16-1. A, human recombinant LYRIC protein was recognized by DB16-1 and anti-Mid in Western blotting. Similarly, four newly generated anti-LYRIC mAbs, Lyric 1-7, Lyric 2-1, Lyric 3-13, and Lyric 4-7, also bound to the LYRIC protein (left). Expression of LYRIC in COLO 205 and HUVECs was recognized by all of these antibodies (right). B, a right-shifted peak in the flow cytometry indicates DB16-1, Lyric 1-7, Lyric 2-1, Lyric 3-13, and Lyric 4-7 binding to HUVECs. CD31 and 4G2 antibodies were used as a positive and negative control, respectively. C, the binding activities of these mAbs to HUVECs were quantified and are presented as the mean ± S.D. (error bars) of three independent experiments. *, p < 0.05; **, p < 0.01.
Identification and Characterization of the B-cell Epitope of DB16-1
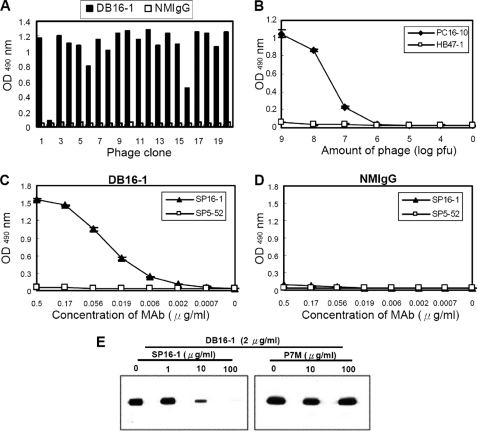
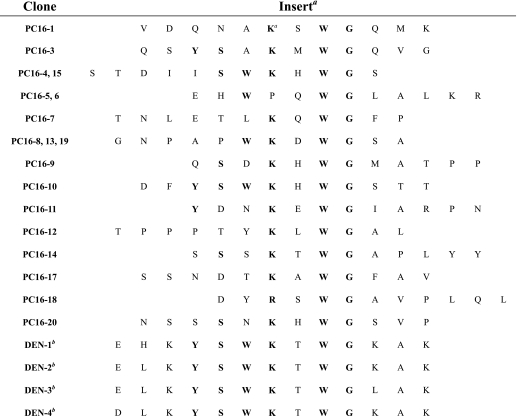
To investigate the molecular mechanism underlying DB16-1 cross-reaction with LYRIC, we identified the precise binding epitope of DB16-1 using a phage-displayed random peptide library. After three rounds of phage display biopanning, 18 of the 20 random selected phage clones were found by ELISA to be significantly reactive to DB16-1 but not to control antibody NMIgG (Fig. 5A). Through alignment of the peptides displayed by selected phage clones, a consensus motif was unraveled as KXWG in most selected phage clones (Table 1). It was interesting to note that this consensus motif matched the amino acid residues 116–119 in NS1 of four dengue virus subtypes (Table 1). To confirm the specificity of the selected phage clones, ELISA plates coated with DB16-1 were incubated with control phage HB47-1 and phage clone PC16-10, the most homologous to DV NS1. We found that PC16-10 bound specifically to DB16-1 in a dose-dependent manner, but control phage did not (Fig. 5B). A synthetic peptide, SP16-1 (LRYSWKTWGKAK), corresponding to amino acid residues 111–122 of D2NS1, was recognized by DB16-1 in a dose-dependent manner. In contrast, control peptide SP5-52 was not recognized by DB16-1 (Fig. 5C). Control antibody NMIgG did not bind to SP16-1 and SP5-52 (Fig. 5D). Furthermore, peptide competition assay indicated that binding activity of DB16-1 to D2NS1 protein was competitively inhibited by SP16-1 in a dose-dependent manner, but an arbitrary control peptide P7M was incapable of inhibiting the binding activity of DB16-1 (Fig. 5E). These results suggested that DB16-1 recognized the epitope corresponding to amino acid residues 111–122 of NS1 of dengue viruses.
FIGURE 5.
Identification and characterization of the B-cell epitope of DB16-1. A, phage clones with high affinity to DB16-1 were selected through ELISA assay. B, binding assay of DB16-1 with phage clones PC16-10 and HB47–1. These two phages were serially diluted from 109 to 104 pfu and 0 pfu. The PC16-10 bound to DB16-1 specifically, but control phage HB47–1 did not. C and D, synthetic peptide corresponding to D2NS1 protein (SP16-1) bound with DB16-1 in a dose-dependent manner. NMIgG served as the negative control. E, competitive inhibition of DB16-1 binding to D2NS1 by SP16-1 was confirmed by Western blotting. Control peptide P7M had no such effect. The values were presented as the mean ± S.D.
TABLE 1.
Alignment of phage-displayed peptide sequences selected by DB16-1
a Phage-displayed consensus amino acids are shown in boldface type.
b Amino acid sequence of residues 110–122 of the NS1 protein of dengue virus.
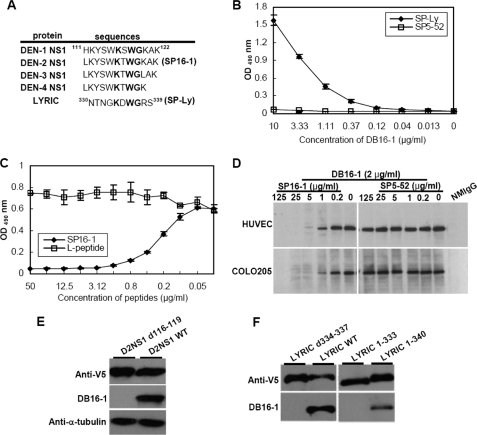
NS1 Protein of Dengue Viruses and LYRIC Protein Contain the Same Binding Motif for DB16-1
When comparing amino acid sequence of DV NS1 and LYRIC proteins, we found a KXWG sequence that corresponded to human LYRIC amino acid residues 334–337 (Fig. 6A). To determine whether the KXWG motif in LYRIC was crucial for the binding of DB16-1, we performed a peptide binding assay. Synthetic peptide SP-Ly (NTNGKDWGRS) representing amino acid residues 330–339 of human LYRIC was recognized by DB16-1 in a concentration-dependent manner (Fig. 6B). The same concentration of control peptide SP5-52 did not react to DB16-1 (Fig. 6B). Moreover, peptide competitive inhibition assay showed that SP16-1 but not control peptide dose-dependently inhibited the binding activity between SP-Ly peptide and DB16-1 (Fig. 6C). Western blot analysis also revealed that SP16-1 but not control peptide SP5-52 could inhibit DB16-1 binding to HUVECs and COLO 205 cells in a dose-dependent manner (Fig. 6D). To confirm that the KXWG motif was important for DB16-1 binding to DV NS1 and LYRIC protein, we expressed D2NS1 and LYRIC deletion mutants without the KXWG motif. Western blot analysis of these recombinant proteins revealed that DB16-1 could recognize D2NS1 (Fig. 6E) and LYRIC (Fig. 6F) proteins. However, DB16-1 did not recognize these proteins when the KXWG motif in D2NS1 or LYRIC was deleted (Fig. 6, E and F). Furthermore, truncated LYRIC 1–340 protein containing the KXWG motif was detectable by DB16-1, whereas truncated LYRIC 1–333 protein without the KXWG motif was not (Fig. 6F). These results strongly suggested that the KXWG consensus motif in NS1 of dengue viruses molecularly mimicked human LYRIC and could be recognized by autoantibody DB16-1.
FIGURE 6.
DV NS1 and LYRIC contain the same B-cell epitope of DB16-1. A, comparison of amino acid sequences of four serotypes of DV NS1 and LYRIC proteins. All of those proteins possessed the KXWG motif. B, DB16-1 recognized synthetic peptide corresponding to LYRIC protein (SP-Ly) in a dose-dependent manner. Synthetic peptide SP5-52 was used as a negative control. C, the binding activity of DB16-1 with SP-Ly was competitively inhibited by SP16-1, whereas control L-peptide had no such effect. D, SP16-1 inhibited DB16-1 binding to LYRIC in HUVECs and COLO 205 cells by Western blot analysis. E, DB16-1 recognized the wild type of D2NS1 protein containing KXWG motif but not the DV-2 NS1 protein with deletion of amino acids 116–119 (D2NS1 d116–119). F, DB16-1 recognized the wild type of LYRIC (LYRIC WT) but not the deletion form of LYRIC (LYRIC d334–337). Truncated LYRIC (LYRIC 1–340) protein containing the KXWG motif was detectable by DB16-1, whereas LYRIC protein with deleted KXWG motif (LYRIC 1–333) lost its binding activity. Anti-V5 antibody was used as an internal control. The values were presented as the mean ± S.D.
DISCUSSION
The molecular mechanisms that cause vascular leakage and hemorrhage after DV infection are not clear. There is some evidence suggesting that autoantibody binding to endothelium might be induced after DV infection (22–24). However, the exact mechanism underlying the molecular mimicry of autoantibodies is unclear. In this study, we found that the antibodies to DV induced in mice were cross-reactive with endothelial cells (Fig. 1, A and B). These results are consistent with previous studies reporting that those polyclonal antibodies against D2NS1 cross-reacted with endothelial cells (22, 23). Anti-NS1 polyclonal antibodies from dengue patients have also been shown to cross-react with endothelial cells and induce endothelial cell apoptosis via a caspase-dependent pathway (24). These results, therefore, suggest the possible involvement of autoantibody reactions in dengue viral infection. To study the mechanism underlying the molecular mimicry of dengue autoantibodies, we generated many mAbs against DV. From these mAbs, we identified a mAb, DB16-1, that could recognize HUVECs and human blood vessels (Fig. 1, D and E). To our knowledge, this is the first report of a mAb against NS1 of DV that can cross-react with human endothelial cells (Figs. 1 (D and E) and 2 and supplemental Fig. S2). Importantly, DB16-1 could induce the apoptosis of HUVECs and cause cell lysis by activating complement-dependent cytotoxicity (Fig. 2, C and D). Together, these findings suggest that DB16-1 may be involved in vascular leakage of endothelium.
The identification of dengue virus-relevant autoantigen is critical when investigating the immunopathogenetic mechanism of DHF/DDS after DV infection. We isolated the target protein of DB16-1 from HUVECs and discovered through LC-MS/MS analysis the target protein to be human LYRIC (Fig. 3, A and B). Antibody-based immunoaffinity chromatography further confirmed the target protein of DB16-1 to be human LYRIC (supplemental Fig. S1B). Co-immunoprecipitation with DB16-1 and anti-Mid antibodies from HUVECs and COLO 205 cells further verified that LYRIC was the target protein of DB16-1 (Fig. 3C and supplemental Fig. S1). To further confirm LYRIC to be the target of DB16-1, we generated LYRIC protein and anti-LYRIC mAbs. Western blotting showed that DB16-1 recognized LYRIC protein as four anti-LYRIC antibodies: Lyric 1-7, Lyric 2-1, Lyric 3-13, and Lyric 4-7 (Fig. 4A). These findings confirmed that DB16-1 was cross-reactive with human LYRIC. Previously, LYRIC has been reported to be a highly conserved protein among species (26), and has been found to be associated with the progression and metastasis of diverse cancers (25, 27, 38), although its cellular function remains unknown.
It is important to define the B-cell epitope of DB16-1 to identify the cross-reactive epitope in the host antigen (17, 39–41). Using phage display, we identified a consensus motif of DB16-1, KXWG, located in amino acid residues 116–119 in all four subtypes of DV NS1 proteins (Table 1). The selected phage PC16-10, not the control phage, bound to DB16-1 in a dose-dependent manner, indicating that PC16-10 interacted with DB16-1 through its displayed peptide rather than another part of the phage particle (Fig. 5B). The synthetic peptide SP16-1 inhibited DB16-1 binding to D2NS1 (Fig. 5E), indicating that amino acid residues 111–122 of the DV NS1 protein made up the B-cell epitope of DB16-1.
To investigate the molecular mimicry of DV NS1 and LYRIC, we compared the amino acid sequences of these two proteins. Interestingly, both proteins contained a KXWG motif (Fig. 6A), and this consensus motif was recognized by autoantibody DB16-1 by the phage display method (Table 1 and Fig. 5). Synthetic peptide SP-Ly, which corresponded to amino acid residues 330–339 of human LYRIC (Fig. 6A), was recognized by DB16-1 in a concentration-dependent manner (Fig. 6B). Furthermore, the ability of DB16-1 to bind to SP-Ly peptide could be competitively inhibited by SP16-1 peptide (Fig. 6C). These results indicated that the KXWG motif located both in amino acid residues 111–122 of the DV NS1 protein and amino acid residues 330–339 of human LYRIC was the B-cell epitope of DB16-1. Moreover, SP16-1 inhibited DB16-1 binding to human LYRIC on HUVECs and COLO 205 cells (Fig. 6D). Western blot analysis further confirmed that DB16-1 bound to D2NS1 and human LYRIC through the KXWG motif (Fig. 6, E and F). The inhibition of autoantibody DB16-1 binding to endothelial cells by synthetic peptide SP16-1 (Fig. 6D) suggests that this peptide and LYRIC protein may be used clinically to prevent damage by autoantibodies.
Systemic sclerosis is an autoimmune disease characterized by immunological abnormalities, vascular damage, and fibroblast proliferation. Autoantibodies against cell surface antigens may be pathogenic because they induce endothelial cell damage (42, 43), considered the primary event in the pathogenesis of the disease. Latent hCMV infection may contribute to progression of systemic sclerosis through its ability to infect endothelial cells (44). A molecular mimicry mechanism links antibodies against the hCMV-derived late protein UL94 to the pathogenesis of systemic sclerosis (17), and increasing levels of antibodies to the HCMV UL94 have been associated with disease severity (20). Anti-UL94 antibodies induce endothelial cell apoptosis and activate dermal fibroblasts and have been linked to two hallmarks of systemic sclerosis, vascular damage and fibrosis (21). Although systemic sclerosis is associated with generation of pathogenetic autoantibodies by chronic infection of hCMV, the vascular permeability in dengue is transient, so the pathogenesis of DHF/DSS is complex and still not well understood. According the clinical course and symptoms of dengue infection, viremia plateaus approximately 2 days before defervescence (45), and patients with high vascular permeability are usually recognized at defervescence and at the end of viremia (3, 46, 47). In this study, we found that DV-induced antibodies were cross-reactive with host LYRIC protein expressed in endothelium. However, because vascular permeability in human dengue infection is transient, future studies should be carried out on groups of human DHF cases at different time points before, during, and after the onset of vascular permeability. More research is needed to elucidate the pathogenic mechanisms underlying the autoantibodies associated with DHF/DSS.
In conclusion, this study established a monoclonal antibody, DB16-1, that recognized DV NS1 and human LYRIC on HUVECs. Our experimental evidence of a motif homology between NS1 of DV and human LYRIC suggests that molecular mimicry could potentially be the mechanism underlying the development of vascular permeability during a dengue infection.
Supplementary Material
Acknowledgments
We thank the Core Facility of the Institute of Cellular and Organismic Biology and the Core Facility for Proteomics and Structural Biology Research, Academia Sinica, for LC-MS/MS sequencing.
This work was supported by Academia Sinica (to H.-C. W.) and National Science Council, Taiwan, Grants NSC-98-3111-B-001-004 and NSC-98-2323-B-001-001 (to H.-C. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- DV
- dengue virus
- DHF
- dengue hemorrhagic fever
- DSS
- dengue shock syndrome
- HUVEC
- human umbilical vein endothelial cell
- hCMV
- human cytomegalovirus
- NC
- nitrocellulose
- anti-Mid
- anti-metadherin
- NMIgG
- normal mouse IgG.
REFERENCES
- 1. Halstead S. B. (1988) Science 239, 476–481 [DOI] [PubMed] [Google Scholar]
- 2. Gubler D. J., Clark G. G. (1995) Emerg. Infect. Dis. 1, 55–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gubler D. J. (1998) Clin. Microbiol. Rev. 11, 480–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bielefeldt-Ohmann H. (1997) Trends Microbiol. 5, 409–413 [DOI] [PubMed] [Google Scholar]
- 5. Halstead S. B. (2007) Lancet 370, 1644–1652 [DOI] [PubMed] [Google Scholar]
- 6. Halstead S. B., Venkateshan C. N., Gentry M. K., Larsen L. K. (1984) J. Immunol. 132, 1529–1532 [PubMed] [Google Scholar]
- 7. Kurane I., Innis B. L., Nisalak A., Hoke C., Nimmannitya S., Meager A., Ennis F. A. (1989) J. Clin. Invest. 83, 506–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Avirutnan P., Malasit P., Seliger B., Bhakdi S., Husmann M. (1998) J. Immunol. 161, 6338–6346 [PubMed] [Google Scholar]
- 9. Gagnon S. J., Ennis F. A., Rothman A. L. (1999) J. Virol. 73, 3623–3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Green S., Vaughn D. W., Kalayanarooj S., Nimmannitya S., Suntayakorn S., Nisalak A., Lew R., Innis B. L., Kurane I., Rothman A. L., Ennis F. A. (1999) J Infect. Dis. 179, 755–762 [DOI] [PubMed] [Google Scholar]
- 11. Rico-Hesse R., Harrison L. M., Salas R. A., Tovar D., Nisalak A., Ramos C., Boshell J., de Mesa M. T., Nogueira R. M., da Rosa A. T. (1997) Virology 230, 244–251 [DOI] [PubMed] [Google Scholar]
- 12. Diamond M. S., Edgil D., Roberts T. G., Lu B., Harris E. (2000) J. Virol. 74, 7814–7823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lane D. P., Hoeffler W. K. (1980) Nature 288, 167–170 [DOI] [PubMed] [Google Scholar]
- 14. Fujinami R. S., Oldstone M. B., Wroblewska Z., Frankel M. E., Koprowski H. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 2346–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Query C. C., Keene J. D. (1987) Cell 51, 211–220 [DOI] [PubMed] [Google Scholar]
- 16. Banki K., Maceda J., Hurley E., Ablonczy E., Mattson D. H., Szegedy L., Hung C., Perl A. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 1939–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lunardi C., Bason C., Navone R., Millo E., Damonte G., Corrocher R., Puccetti A. (2000) Nat. Med. 6, 1183–1186 [DOI] [PubMed] [Google Scholar]
- 18. Kaplan C., Morinet F., Cartron J. (1992) Semin. Hematol. 29, 34–44 [PubMed] [Google Scholar]
- 19. Rhodes G. H., Valbracht J. R., Nguyen M. D., Vaughan J. H. (1997) J. Autoimmun. 10, 447–454 [DOI] [PubMed] [Google Scholar]
- 20. Namboodiri A. M., Rocca K. M., Pandey J. P. (2004) Autoimmunity 37, 241–244 [DOI] [PubMed] [Google Scholar]
- 21. Lunardi C., Dolcino M., Peterlana D., Bason C., Navone R., Tamassia N., Beri R., Corrocher R., Puccetti A. (2006) PLoS Med. 3, e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Falconar A. K. (1997) Arch. Virol. 142, 897–916 [DOI] [PubMed] [Google Scholar]
- 23. Lin C. F., Lei H. Y., Shiau A. L., Liu H. S., Yeh T. M., Chen S. H., Liu C. C., Chiu S. C., Lin Y. S. (2002) J. Immunol. 169, 657–664 [DOI] [PubMed] [Google Scholar]
- 24. Lin C. F., Lei H. Y., Shiau A. L., Liu C. C., Liu H. S., Yeh T. M., Chen S. H., Lin Y. S. (2003) J. Med. Virol. 69, 82–90 [DOI] [PubMed] [Google Scholar]
- 25. Brown D. M., Ruoslahti E. (2004) Cancer Cell. 5, 365–374 [DOI] [PubMed] [Google Scholar]
- 26. Sutherland H. G., Lam Y. W., Briers S., Lamond A. I., Bickmore W. A. (2004) Exp. Cell. Res. 294, 94–105 [DOI] [PubMed] [Google Scholar]
- 27. Kang D. C., Su Z. Z., Sarkar D., Emdad L., Volsky D. J., Fisher P. B. (2005) Gene 353, 8–15 [DOI] [PubMed] [Google Scholar]
- 28. Chen Y. C., Huang H. N., Lin C. T., Chen Y. F., King C. C., Wu H. C. (2007) Clin. Vaccine Immunol. 14, 404–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Köhler G., Milstein C. (1975) Nature 256, 495–497 [DOI] [PubMed] [Google Scholar]
- 30. Wu H. C., Jung M. Y., Chiu C. Y., Chao T. T., Lai S. C., Jan J. T., Shaio M. F. (2003) J. Gen. Virol. 84, 2771–2779 [DOI] [PubMed] [Google Scholar]
- 31. Lee T. Y., Lin C. T., Kuo S. Y., Chang D. K., Wu H. C. (2007) Cancer Res. 67, 10958–10965 [DOI] [PubMed] [Google Scholar]
- 32. Rose A. L., Smith B. E., Maloney D. G. (2002) Blood 100, 1765–1773 [PubMed] [Google Scholar]
- 33. Nechansky A., Szolar O. H., Siegl P., Zinoecker I., Halanek N., Wiederkum S., Kircheis R. (2009) J. Pharm. Biomed. Anal. 49, 1014–1020 [DOI] [PubMed] [Google Scholar]
- 34. Wu H. C., Huang Y. L., Chao T. T., Jan J. T., Huang J. L., Chiang H. Y., King C. C., Shaio M. F. (2001) J. Clin. Microbiol. 39, 977–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee T. Y., Wu H. C., Tseng Y. L., Lin C. T. (2004) Cancer Res. 64, 8002–8008 [DOI] [PubMed] [Google Scholar]
- 36. Di Rosa F., Barnaba V. (1998) Immunol. Rev. 164, 17–27 [DOI] [PubMed] [Google Scholar]
- 37. Zhao Z. S., Granucci F., Yeh L., Schaffer P. A., Cantor H. (1998) Science 279, 1344–1347 [DOI] [PubMed] [Google Scholar]
- 38. Emdad L., Sarkar D., Su Z. Z., Randolph A., Boukerche H., Valerie K., Fisher P. B. (2006) Cancer Res. 66, 1509–1516 [DOI] [PubMed] [Google Scholar]
- 39. Bowditch R. D., Tani P., Fong K. C., McMillan R. (1996) Blood 88, 4579–4584 [PubMed] [Google Scholar]
- 40. Sibille P., Ternynck T., Nato F., Buttin G., Strosberg D., Avrameas A. (1997) Eur. J. Immunol. 27, 1221–1228 [DOI] [PubMed] [Google Scholar]
- 41. Liu I. J., Hsueh P. R., Lin C. T., Chiu C. Y., Kao C. L., Liao M. Y., Wu H. C. (2004) J. Infect. Dis. 190, 797–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Carvalho D., Savage C. O., Black C. M., Pearson J. D. (1996) J. Clin. Invest. 97, 111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bordron A., Dueymes M., Levy Y., Jamin C., Leroy J. P., Piette J. C., Shoenfeld Y., Youinou P. Y. (1998) J. Clin. Invest. 101, 2029–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pandey J. P., LeRoy E. C. (1998) Arthritis Rheum. 41, 10–15 [DOI] [PubMed] [Google Scholar]
- 45. Fink J., Gu F., Vasudevan S. G. (2006) Rev. Med. Virol. 16, 263–275 [DOI] [PubMed] [Google Scholar]
- 46. Nimmannitya S. (1987) Southeast Asian J. Trop. Med. Public Health 18, 392–397 [PubMed] [Google Scholar]
- 47. Carlos C. C., Oishi K., Cinco M. T., Mapua C. A., Inoue S., Cruz D. J., Pancho M. A., Tanig C. Z., Matias R. R., Morita K., Natividad F. F., Igarashi A., Nagatake T. (2005) Am. J. Trop. Med. Hyg. 73, 435–440 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.