Abstract
Vascular endothelial growth factor (VEGF) is vital to physiological as well as pathological angiogenesis, and regulates a variety of cellular functions, largely by activating its 2 receptors, fms-like tyrosine kinase (Flt1) and kinase domain receptor (KDR). KDR plays a critical role in the proliferation of endothelial cells by controlling VEGF-induced phospholipase Cγ-protein kinase C (PLCγ-PKC) signaling. The function of Flt1, however, remains to be clarified. Recent evidence has indicated that Flt1 regulates the VEGF-triggered migration of endothelial cells and macrophages. Here, we show that RACK1, a ubiquitously expressed scaffolding protein, functions as an important regulator of this process. We found that RACK1 (receptor for activated protein kinase C 1) binds to Flt1 in vitro. When the endogenous expression of RACK1 was attenuated by RNA interference, the VEGF-driven migration was remarkably suppressed whereas the proliferation was unaffected in a stable Flt1-expressing cell line, AG1-G1-Flt1. Further, we demonstrated that the VEGF/Flt-mediated migration of AG1-G1-Flt1 cells occurred mainly via the activation of the PI3 kinase (PI3K)/Akt and Rac1 pathways, and that RACK1 plays a crucial regulatory role in promoting PI3K/Akt-Rac1 activation.
Keywords: Akt PKB, Cell Migration, MAP Kinases (MAPKs), PI 3-Kinase, RNA Interference (RNAi), Flt1(VEGFR1), RACK1 (Receptor for Activated C-Kinase 1), Vascular Endothelial Growth Factor (VEGF), Angiogenesis, Signaling
Introduction
Angiogenesis is a critical and complicated process in embryonic vascular development, wound healing, and organ regeneration as well as in multiple pathological conditions, such as rheumatoid arthritis and tumor growth (1). A number of molecules including vascular endothelial growth factor (VEGF), fibroblast growth factor (FGF), and transforming growth factor (TGF)-β have been implicated in angiogenesis. Among known angiogenic factors, VEGF has emerged as a central regulator of vascular development during embryogenesis as well as the formation of blood vessels from the newborn to adult stages (1–4). The VEGF family, consisting of VEGF-A (or simply VEGF), VEGF-B, VEGF-C, and VEGF-D and placenta growth factor (PlGF), directly initiate angiogenic signals by interacting with their receptors (3, 5). VEGF executes various cellular functions largely by binding to 2 homologous tyrosine kinase receptors, fms-like tyrosine kinase (Flt1 or VEGFR1), and kinase domain receptor/fetal liver kinase 1 (KDR2/Flk1 or VEGFR2). Both receptors contain 7 extracellular immunoglobin-like regions, 1 short transmembrane domain, and an intracellular tyrosine-kinase domain that is divided by a kinase-insert sequence (6, 7). Flt1 has high affinity for VEGF (Kd = 2∼10 pm), but weak kinase activity (10-fold less than that of KDR) (8). Gene targeting studies have suggested that the 2 receptors are essential for embryonic development: Flt1-null mutant mice (Flt1−/−) died at E8.5–9.0 due to the excess growth and disorganization of blood vessels, whereas KDR/Flk1−/− mice also died at E-8.5, but due to a lack of blood vessels (9, 10). Accordingly, these studies demonstrate that the 2 receptors utilize distinct signaling cascades to regulate different biological functions. Interestingly, we previously showed that Flt1 tyrosine kinase domain-deficient mice (Flt1 TK−/−) were healthy and had normal blood vessel networks, and thus, the function of Flt1 early in embryogenesis is most likely the trapping of VEGF to reduce its local concentration (11).
VEGF launches receptor-relayed signaling events by binding to the second and third IgG-like domains of Flt1 and KDR, respectively (12, 13). The phosphorylation of Tyr(Y)-1175 on KDR leads to the activation of phospholipase C (PLC)γ, which in turn promotes the intracellular mobilization of calcium and activates a crucial protein kinase C-Raf-mitogen-activated protein kinase (PKC-Raf-MAPK) cascade, the latter regulating endothelial cell proliferation (14–16). The phosphorylation of Tyr(Y)-1169 on Flt1 also provides a binding site for PLCγ and activates a PLCγ-MAPK cascade (17). Moreover, both receptors appear to activate the PI3 kinase (PI3K)-Akt pathway (18, 19). In addition to promoting weak signals for VEGF-deprived cell growth and survival, Flt1 is also involved in regulating cell movement in both endothelial cells and macrophage-lineage cells. Loss of Flt1 expression in endothelial cells led to a decrease in sprout formation and cell migration, which resulted in reduced vascular branching (20). VEGF induces the migration and activation of macrophage-lineage cells into tumor tissue or inflamed areas by binding to Flt1 (11, 21–24). Taken together, these findings suggest that Flt1 plays a key role in regulating VEGF-induced cell migration and cell growth, however, the precise signaling pathway under Flt1 remains to be characterized.
RACK1 (receptor for activated protein kinase C 1), a 36-kDa protein containing 7 internal Trp-Asp 40 (WD40) repeats, is homologous to the G protein β subunit and expressed ubiquitously in both human and animal tissues (25). RACK1 was originally cloned as an anchoring protein for PKCs, and can stabilize the active form of PKC, and permit its translocation to different sites within the cell (26, 27). Studies have implied that RACK1 can associate with a variety of signaling molecules, including members of the Src family, the integrin β subunit, PDE45, and IGF-1 receptors, to regulate cell cycle, survival, adhesion, and migration (25). Such reports imply that RACK1 may function as a scaffolding protein to mediate protein-protein interaction and facilitate tight regulation of cellular function as well as control the cross-talk in different signaling cascades.
Here, we provide evidence that RACK1 plays a regulatory role in VEGF-Flt1-dependent cell migration through direct interaction with Flt1. When the endogenous expression of RACK1 was attenuated by RNA interference (RNAi) in a stable Flt1-expressing cell line, the VEGF-induced migration was remarkably suppressed whereas the proliferation was not affected. Moreover, the activation of PI3K/Akt and small-GTPase Rac1 signaling pathways was clearly inhibited by the RACK1-silencing. Our study indicates a new possible mechanism of VEGF-Flt1-induced migration.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
The recombinant human-VEGF was purchased from R&D Systems (Minneapolis, MN). The anti-RACK1 and anti-phosphotyrosine antibodies were from BD transduction laboratories (San Diego, CA). The antibodies against Akt, phospho-Akt, MAPK, phospho-MAPK, PLCγ, and phospho-PLCγ were obtained from Cell Signaling Technology (Beverly, MA). The anti-Flt1 antibody was from Santa Cruz Technology (Santa Cruz, CA). The Rac1 activation assay Biochem kit TM was bought from Cytoskeleton (Denver, CO). The protein G-Sepharose TM 4 Fast Flow was from GE Healthcare (Piscataway, NJ). The BD BioCoatTM Angiogenesis System-Endothelial Cell Migration 24-well Plate was obtained from BD Bioscience (Bedford, MA). The AP-conjugated anti-mouse and anti-rabbit immunoglobulins were purchased from Promega (Madison, WI). The FuGENE® 6 Transfection Reagent was purchased from Roche (Indianapolis, IN). The LipofectamineTM RNAiMAX reagent was obtained from Invitrogen (Carlsbad, CA).
Two-hybrid Assay
The GAL4-based MATCHMAKER two-hybrid system II (Clontech, Mountain View, CA) was used for the yeast two-hybrid assays. The plasmid vectors pGBT9 and pGAD424 encoding the GAL4 DNA-binding domain and the GAL4-activating domain, respectively, were used to express hybrid proteins. A mixture of embryonic and adult human brain cDNA libraries (both from Clontech) in the GAL4-activating domain vector pACT2 was screened using the intracellular domain of Flt1 (Flt1-IC) cloned into the GAL4 DNA-binding domain vector pGBT9 as bait in the PJ69–4A yeast system. DNAs from positive clones were isolated, and the GAL4-activating domain plasmids were recovered in Escherichia coli strain HB101, and sequenced. The DNA-binding domain or GAL4-activating domain was assayed for selection of drop-out leucine, tryptophan, adenine, and histidine and for galactosidase activity on nitrocellulose filters as described in the Clontech manual.
Generation of AG1-G1-Neo and AG1-G1-Flt1 Cell Lines
AG1-G1-Flt1 cells were established from a human angioma with the permission of the Ethics Committee for scientific research at the Institute of Medical Science, University of Tokyo (28). Briefly, an adult benign angioma was excised surgically, and the pEF1α-SV40 large T antigen plasmid was introduced into the cells. An SV40 large T-positive AG1-G1 cell was isolated, and then pBCMGS-Neo-Flt1 carrying the full-length Flt1 cDNA (29), or the empty vector pBCMGS-Neo was transfected into AG1-G1 cells. A single cell clone expressing Flt1 was isolated and designated as AG1-G1-Flt1.
Cell Culture and Plasmid DNA Transfection
The AG1-G1-Neo and AG1-G1-Flt1 cells were maintained in F-12 medium supplemented with 10% fetal bovine serum (FBS; JRH biosciences, Lenexa, KS) and 200 μg/ml of G-418 at 37 °C in 5% CO2. Human embryonic kidney (HEK) 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% FBS at 37 °C in 5% CO2. All the DNA plasmids were transfected into HEK293T cells using the FuGENE® 6 Transfection Reagent according to instructions.
RNA Interference (RNAi)
The following stealth siRNAs designed based on the human RACK1 cDNA (GenBankTM accession number GNB2L1) were obtained from Invitrogen. The sequences of the two RACK1 siRNAs used in this study are: si1, GCCUCUCGAGAUAAGACCAUCAUCA and si2, GGAACCUGGCUAACUGCAAGCUGAA. AG1-G1-Neo and AG-G1-Flt1 cells were transfected with the RACK1 siRNAs or negative control siRNA using the LipofectamineTM RNAiMAX reagent (Invitrogen) according to the manufacturer's directions. The effect of suppression on RACK1 expression was assessed 60 h post-transfection.
Western Blotting and Immunoprecipitation
Cellular protein extracts were prepared by washing cells with cold PBS and then scraped into lysis buffer (50 mm Tris, pH 7.5, 150 mm NaCl, and 1 mm EDTA, and 1% Triton X-100) supplemented with 2 μg/ml aprotinin, 2 μg/ml leupeptin, 1 mm PMSF, and 1 mm sodium orthovanadate. Cell lysate was briefly centrifuged for 10 min to remove the insoluble debris. The lysate (20–40 μg each) was separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred onto polyvinylidene difluoride (PVDF) membranes. The membranes were soaked in blocking buffer (5% skimmed milk and 1% bovine serum albumin in PBS) for 1 h, and incubated with the primary antibody overnight. The membranes were incubated with a horseradish peroxidase-conjugated secondary antibody for 30 min and visualized using a 5-bromo-4-chloroindolyl phosphate (BCIP) and nitroblue tetrazolium (NBT) detection system (Promega, Madison, WI). For immunoprecipitation, the cell lysate was incubated with antibody for 2 h at 4 °C. Protein A-agarose was then added, and the mixture was further incubated for 1 h. The agarose beads were washed five times with lysis buffer and subjected to immunoblotting.
Migration Assay
The cell migration assay was carried out using a BioCoatTM Angiogenesis System-Endothelial Cell Migration 24-well plate (BD Bioscience, Bedford, MA). Briefly, the cells were trypsinized and suspended at a final concentration of 1 × 106 cells/ml in F-12 medium containing 0.5% FBS. The cell suspension (100 μl) was loaded into each of the upper wells, and fresh F-12 medium (0.5% FBS) containing VEGF (100 ng/ml) was placed in the lower wells. The plate was incubated at 37 °C for 23 h, and the cells were fixed and stained with 4 μg/ml of Calcein AM (Molecular Probes, Eugene, OR). The cells on the top of the filter were removed by wiping with a cotton swab, and chemotaxis was quantified with an immunofluorescence microscope by counting cells that had migrated to the bottom of the filter.
Rac1 Activation Assay
The activation of Rac1 was analyzed by using a Rac1 activation assay kit (Cytoskeleton, Denver, CO) according to the instructions recommended by the manufacturer. The Rac1 activation assay was a direct pull-down experiment performed to measure the binding of GTP-bound Rac1 to a GST-fused form of the p21-binding domain of PAK (GST-PAK-PBD) fixed onto glutathione-agarose beads. The beads were washed four times with lysis buffer, and resuspended in sample buffer. The samples were measured by Western blotting.
Proliferation Assay
Trypsinized cells (0.5 × 104) were seeded into 96-well plates and incubated at 37 °C overnight. The medium was replaced with fresh F-12 medium containing 0.5% FBS and 100 ng/ml of VEGF, and the plates were incubated for 24, 48, and 72 h. The medium was removed, and the cells were fixed with ice-cold 10% (w/v) trichloroacetic acid (TCA). They were then stained with Sulforhodamine B (SRB) in 1% acetic acid for 20 min, and the plates were washed and dried overnight. The dye was solubilized by adding 100 μl of 10 mm Tris base to each well at room temperature for 20 min, and the absorbance (A) at 540 nm was measured using a Asys Hitech Microplate Reader (Biochrom UK).
Immunofluorescence Microscopy
Following fixation in 3.7% formaldehyde in PBS for 15 min, cells treated under different experimental conditions were further permeabilized with 0.2% Triton X-100 in PBS for 10 min, then incubated with the primary antibody for 60 min. They were washed with PBS and incubated with the immunofluorescence-labeled secondary antibodies for 30 min. For the visualization of F-actin, cells were incubated with rhodamine-phalloidin (Molecular Probes, Carlsbad, CA). Finally, the coverslips were washed thoroughly with PBS and mounted on glass slides. Images were captured and analyzed with a Radiance 2000 laser-scanning confocal microscope (Bio-Rad).
RESULTS
Flt1 Mediates VEGF-induced Migration in an Flt1-Expressing Cell Line
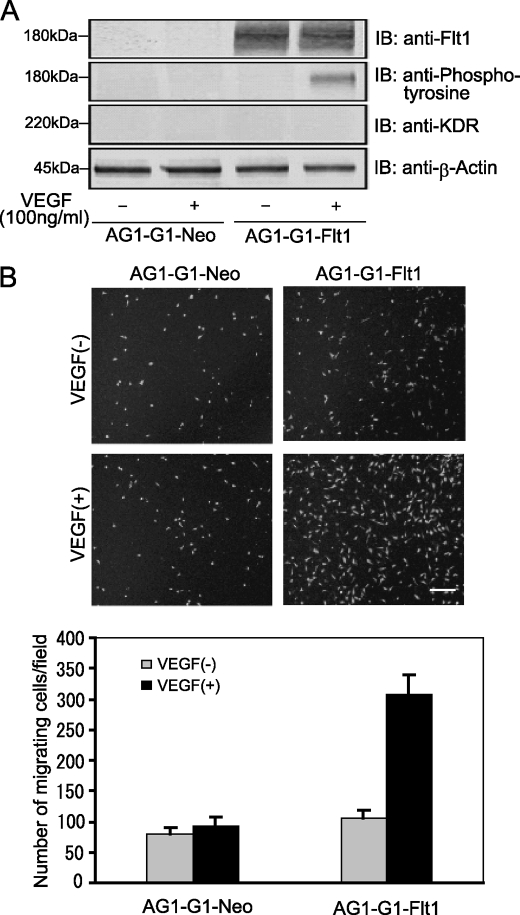
To obtain a model for clarifying the function and signaling under Flt1, we generated a stable Flt1-expressing human cell line, called AG1-G1-Flt1, by introducing Flt1 cDNA into a benign angioma cell line. As shown in Fig. 1A, the Flt1 level in AG1-G1-Flt cells was ∼1000-fold that in AG1-G1-Neo cells, whereas KDR expression was suppressed to an undetectable level. Next, we performed a migration assay using AG1-G1-Neo and AG1-G1-Flt1 cells to assess the effect of Flt1 on the VEGF-induced cell migration. As shown in Fig. 1B, VEGF treatment remarkably increased the migration of AG1-G1-Flt1 cells about 3-fold, whereas the number of migrating AG1-G1-Neo cells was not altered even in the presence of VEGF. These results provide evidence that Flt1 mediates VEGF-driven cell migration.
FIGURE 1.
Flt1 mediates VEGF-induced AG1-G1-Flt1 cell migration. A, serum-deprived AG1-G1-Neo and AG1-G1-Flt1 cells were left untreated or stimulated with VEGF (100 ng/ml) for 5 min. Cell lysate prepared as described under “Experimental Procedures” was immunoblotted with antibody against Flt1, phosphotyrosine, KDR, or β-actin. B, AG1-G1-Neo and AG1-G1-Flt1 cells seeded on a endothelial cell migration 24-well plate were exposed to VEGF (100 ng/ml) in F-12 medium containing 0.5% FBS for 23 h. Migrating cells were stained with calcein AM (4 μg/ml) for 1.5 h and analyzed by fluorescence microscope. A representative image of multiple analogous fields is presented for each condition (upper panels), Scale bar, 250 μm. The numbers of migrating cells were counted (lower panels). VEGF-induced migration was remarkably increased in AG1-G1-Flt1 cells as compared with control AG1-G1-Neo cells.
Flt1 Interacts with RACK1 in Vitro
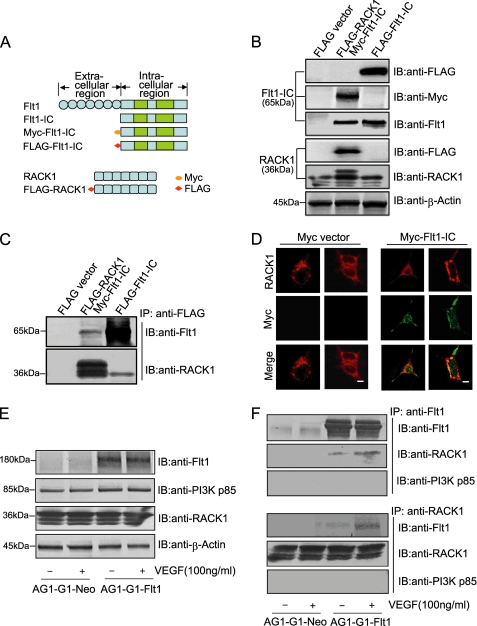
A two-hybrid screening assay to identify the partner of Flt1 uncovered RACK1, a ubiquitously expressed scaffolding protein, which bound Flt1-IC (the truncated intracellular domain of Flt1) as bait. To determine whether the interaction of Flt1 with RACK1 occurs in mammalian cells, several tag-conjugated constructs including FLAG-RACK1, FLAG-Flt1-IC, and Myc-Flt1-IC were individually expressed or co-expressed in HEK293T cells. Schematic diagrams of these constructs and their normal expression patterns are shown in Fig. 2, A and B. The lysates extracted from HEK293T cells expressing the constructs described above were subjected to immunoprecipitation (IP) experiments with the FLAG antibody, the results of which showed that Flt1-IC and RACK1 can bind to each other (Fig. 2C and supplemental Fig. S1). Next, the association was examined using immunofluorescence microscopy. Myc-Flt1-IC co-localized with endogenous RACK1 expressed at the periphery of the cytosol (Fig. 2D). These findings provide evidence that RACK1 can interact with Flt1 within the cell.
FIGURE 2.
Flt1 binds to RACK1 in vitro. A, schematic representation of several tag-conjugated constructs including Myc-Flt1-IC (truncated intracellular domain of Flt1), FLAG-Flt1-IC, and FLAG-RACK1. B, levels of the constructs in HEK293T cells were analyzed by Western blotting using antibody against FLAG, Myc, RACK1, or β-actin. C, HEK293T cells were transfected with plasmids expressing FLAG-Flt1-IC alone, or co-expressing Myc-Flt1-IC and FLAG-RACK1. Cell lysate was subjected to immunoprecipitation (IP) with anti-FLAG antibody, and the FLAG-immunoprecipiates were probed with anti-Flt1 and anti-RACK1 antibodies. D, co-localization of RACK1 and Flt1 in HEK293T cells. HEK293T cells transiently expressing Myc-Flt1-IC were fixed, permeabilized, and stained with anti-Myc (green) or anti-RACK1 (red) antibody. The localization of RACK1 and Myc-Flt1-IC was observed by confocal microscope. Scale bar, 10 μm. E and F, interaction between Flt1 and RACK1 in AG1-G1-Flt1 cells. AG1-G1-Neo and AG1-G1-Flt1 cells were serum-starved and stimulated with VEGF (100 ng/ml) for 5 min. Equal amounts of cell lysate were immunoblotted with antibody against Flt1, PI3K p85, RACK1, or β-actin (E), and then subjected to immunoprecipitation (IP) with anti-Flt1 antibody (upper panels) or anti-RACK1 antibody (lower panels), followed by Western blotting with anti-Flt1, anti-RACK1, or anti-PI3K p85 antibody.
Additionally, the lysates from AG1-G1-Neo and AG1-G1-Flt1 cells exposed to VEGF were immunoprecipitated with anti-Flt1 or anti-RACK1 antibody. As shown in Fig. 2, E and F, although the endogenous RACK1 constitutively associated with Flt1 in the absence of VEGF, the amount of the Flt1-RACK1 complex increased ∼3-fold following stimulation with VEGF. However the interaction was undetected in the AG1-G1-Neo control cell line in which Flt1 is not expressed (Fig. 2F and supplemental Fig. S2). On the other hand, our results also showed that the p85 subunit of PI3K did not bind to either Flt1 or RACK1 at detectable levels (Fig. 2F and supplemental Fig. S3). Taken together, these results indicate that Flt1 can interact with RACK1 directly, and VEGF promotes the formation of a complex between the two proteins, whereas PI3K is not involved.
RACK1-silencing Decreased VEGF-induced Cell Migration
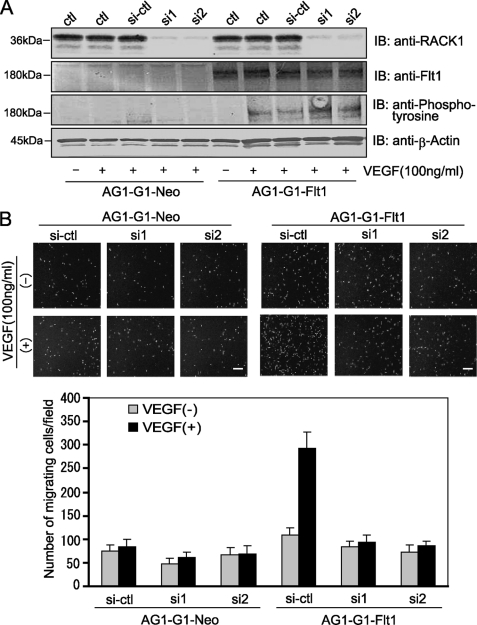
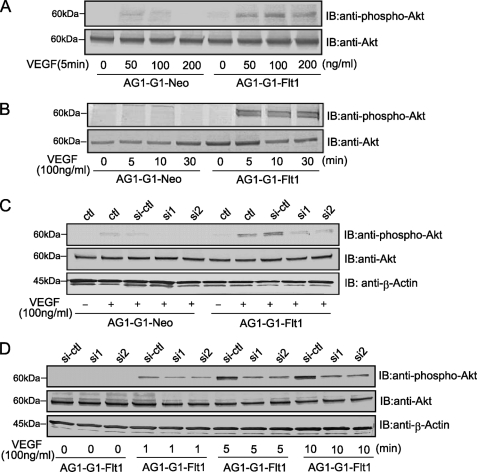
The finding that Flt1 interacts with RACK1 raises the possibility that the multifunctional RACK1 protein is involved in the VEGF/Flt1-mediated cellular functions. We therefore employed RNA interference to investigate the role of RACK1. Two siRNAs (Invitrogen) designed based on the human RACK1 gene showed a very strong suppressing effect with about a 90% reduction in RACK1 expression (Fig. 3A, upper panel). However, the expression and VEGF-induced phosphorylation of Flt1 were not affected significantly under the conditions (Fig. 3A, middle panel). These observations imply that RACK1 does not affect the upstream events of the VEGF-Flt1 signaling pathway.
FIGURE 3.
RACK1-silencing remarkably suppressed VEGF-induced migration of AG1-G1-Flt1 cells. A, silencing of RACK1 by RNAi. AG1-G1-Neo and AG1-G1-Flt1 cells transfected with control siRNA (si-ctl) or RACK1 siRNAs (si1 and si2) were serum-starved and exposed to VEGF (100 ng/ml) for 5 min. (−) indicates no stimulation with VEGF. Cell lysate was subjected to Western blotting with antibody against RACK1, Flt1, phosphotyrosine, or β-actin. The expression and VEGF-induced phosphorylation of Flt1 were not affected by RACK1-silencing. B, AG1-G1-Neo and AG1-G1-Flt1 cells transfected with control siRNA (si-ctl) or RACK1 siRNAs (si1 and si2) were allowed to migrate on an endothelial cell migration 24-well plate. Migrating cells stained with calcein AM were analyzed by fluorescence microscope. A representative image of multiple similar fields is presented for each condition (upper panels), Scale bar, 250 μm. The numbers of migrating cells were counted (lower panels). VEGF-induced migration of AG1-G1-Flt1 cells was significantly inhibited by RACK1-silencing.
We next examined the effect of RACK1-silencing on VEGF/Flt1-mediated cell migration. As compared with control AG1-G1-Neo cells which showed no obvious increase in migration after VEGF treatment, the VEGF-induced migration of AG1-G1-Flt1 cells was remarkably suppressed by RACK1-silencing (Fig. 3B). These results clearly implied that RACK1 plays an important role in the VEGF/Flt1-triggered signaling.
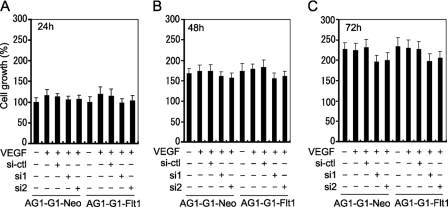
Attenuation of Endogenous RACK1 Expression Does Not Affect VEGF-triggered Cell Proliferation
To evaluate the role of RACK1 in VEGF/Flt1-triggered cell proliferation, the effect of RACK1-silencing on the VEGF-induced growth of AG1-G1-Neo and AG1-G1-Flt1 cells was analyzed. Indeed, even after 72 h, VEGF did not significantly promote proliferation in either cell line (Fig. 4). Certainly, RACK1-silencing did not specifically affect Flt1-mediated proliferation. The growth of AG1-G1-Neo cells unaffected in their response to VEGF is easily understood given the deficiency of the two receptors, whereas the similar effect obtained in AG1-G1-Flt1 cells suggests that Flt1-mediated signaling does not or only very weakly promotes cell growth. The slight inhibitory effect on growth in the two cell lines following RACK-silencing may be due to the intrinsic regulatory role of RACK1 in cell proliferation but independent of the Flt1-relayed signals. Thus, these results indicate that RACK1 plays a more critical role in Flt1-related cell migration than cell proliferation.
FIGURE 4.
RACK1-silencing does not significantly affect AG1-G1-Neo and AG1-G1-Flt1 cell growth. AG1-G1-Neo and AG1-G1-Flt1 cells transfected with control siRNA (si-ctl) or RACK1 siRNAs (si1 and si2) were cultured in 96-well plates in F-12 medium supplemented with 0.5% FBS and 100 ng/ml VEGF for 24 h (A), 48 h (B), and 72 h (C). Cell growth was measured by SRB assay as described under “Experimental Procedures.”
RACK1-silencing Suppresses the VEGF-driven Activation of the PI3K/Akt Cascade
VEGF launches a crucial signal via activation of the PI3K/Akt pathway as well as PLCγ-PKC-MAPK pathway to control cell behavior including the migration and survival of endothelial cells (19, 30, 31). In our model, Akt was activated by VEGF treatment in AG1-G1-Flt1 cells, and the phosphorylation was slightly stronger on stimulation with 100 ng/ml or 200 ng/ml than 50 ng/ml of the growth factor for 5 min (Fig. 5A). This phenotype did not occur in the control AG1-G1-Neo cells. Meanwhile, in a time-course experiment, the phosphorylation of Akt peaked at 5∼10 min and remained high for over 30 min after exposure to VEGF (Fig. 5B). These results clearly show that VEGF activates the PI3K/Akt pathway via Flt1. Next, the potential role of RACK1 in this Flt1-dependent activation of the PI3K/Akt cascade was examined. As shown in Fig. 5, C and D, RACK1-silencing in AG1-G1-Flt1 cells led to a remarkable decrease in Akt phosphorylation. Taken together, these results indicate that RACK1 plays an important role in the downstream signaling of VEGF-Flt1 by regulating activation of Akt.
FIGURE 5.
RACK1-silencing inhibits the VEGF-driven phosphorylation of Akt. A and B, Akt-activation by VEGF in AG1-G1-Flt1 cells. Serum-deprived AG1-G1-Neo and AG1-G1-Flt1 cells were stimulated with 50, 100, or 200 ng/ml VEGF for 5 min (A), or with VEGF (100 ng/ml) for the periods indicated (B). Cell lysates were analyzed by Western blotting with antibody against phospho-Akt or Akt. C and D, effect of RACK1-silencing on Akt phosphorylation. After the serum-starvation, AG1-G1-Neo and AG1-G1-Flt1 cells transfected with control siRNA (si-ctl) or RACK1 siRNAs (si1 and si2) were treated with VEGF (100 ng/ml) for 5 min (C). AG1-G1-Flt1 cells transfected with control siRNA (si-ctl) or RACK1 siRNAs (si1 and si2) were serum-starved and stimulated with VEGF (100 ng/ml) for the periods indicated (D). Cell lysate was analyzed by Western blotting with antibodies against phospho-Akt, Akt, and β-actin.
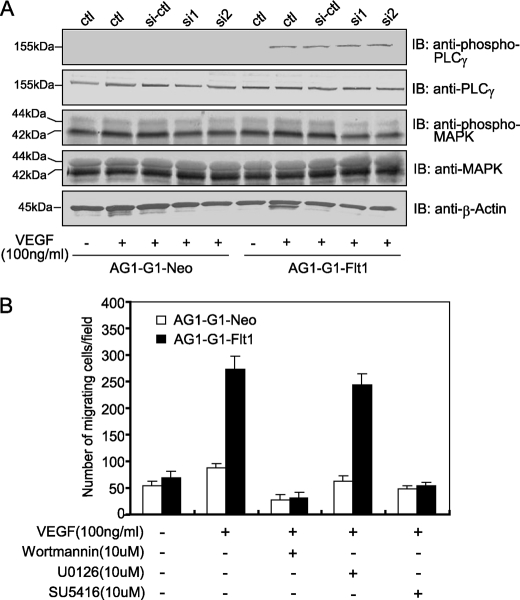
Previous studies revealed that the phosphorylation of 1169-Tyr of Flt1 corresponding to 1175-Tyr of KDR relays a signal to PLCγ and MAPK, leading to the proliferation of endothelial cells (15–17, 32). Hence, the effect of RACK1-silencing on the VEGF-triggered activation of PLCγ and MAPK was examined. VEGF induced modest phosphorylation of PLCγ at 5 min in AG1-G1-Flt1 cells, however, RACK1-silencing did not affect this phosphorylation (Fig. 6A, upper panel). Moreover, the vigorous constitutive activation of MAPK was observed in both AG1-G1-Neo and AG1-G1-Flt1 cells (Fig. 6A, middle panel). We consider that the establishment of immortalized cell lines with T-antigen most likely resulted in this constitutive activation. Therefore, we could not draw a conclusion about the importance of RACK1 in the VEGF-Flt1-MAPK activation pathway.
FIGURE 6.
RACK1-silencing does not affect the VEGF-induced activation of PLCγ and MAPK. A, effect of RACK1-silencing on the VEGF-induced activation of PLCγ and MAPK. AG1-G1-Neo and AG1-G1-Flt1 cells transfected with control siRNA (si-ctl) or RACK1 siRNAs (si1 and si2) were serum-starved and exposed to VEGF (100 ng/ml) for 5 min. Cell extracts were analyzed by Western blotting with antibodies against phospho-PLCγ, PLCγ, phospho-MAPK, MAPK, and β-actin. RACK1-silencing did not significantly affect the VEGF-initiated activation of PLCγ and MAPK. B, AG1-G1-Neo and AG1-G1-Flt1 cells seeded on an endothelial cell migration 24-well plate were left quiescent or stimulated with VEGF (100 ng/ml) alone, or VEGF (100 ng/ml) plus wortmannin, U0126, or SU5416. Migrating cells were stained with calcein AM and observed by fluorescence microscope. The numbers of migrating cells from four random fields were counted. The PI3K- and Flt1-inhibitors but not MAPK-inhibitor substantially attenuated VEGF-induced migration.
PI3K and Flt1 Inhibitors but Not a MAPK Inhibitor Suppresses VEGF-triggered Cell Migration
Several kinase inhibitors including wortmannin (PI3K inhibitor), U0126 (MAPK inhibitor) and SU5416 (VEGFR kinase inhibitor) were used to clarify which of these signaling molecules is involved in regulating the VEGF-induced cell migration. As shown in Fig. 6B, VEGF increased the migration of AG1-G1-Flt1 cells, and this effect was significantly blocked in the presence of wortmannin and SU5416. In contrast, no suppressive effect was observed on treatment with U0126. These results suggest that the Flt1-PI3K signaling pathway is involved in VEGF-triggered cell migration whereas MAPK is not.
RACK1-silencing Inhibited VEGF-induced Small-GTPase Rac1 Activity and Membrane Ruffling
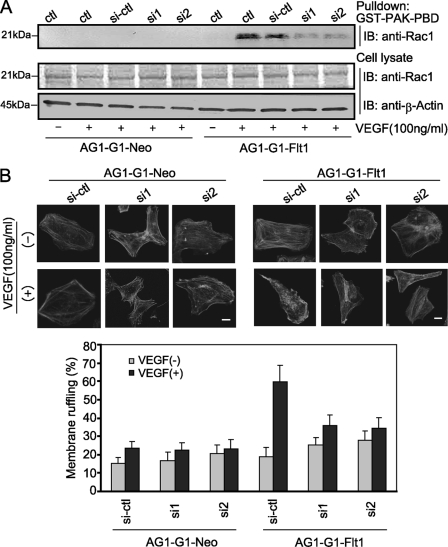
More recently, several groups have demonstrated that small-GTPase Rac1 participates in VEGF-driven endothelial cell migration by regulating the actin reorganization, lamellipodial structures, and focal adhesions (33, 34). To investigate whether the activation of Rac1 is essential for VEGF-induced AG1-G1-Flt1 migration and the potential role of RACK1 in this process, we measured the effect of RACK1-silencing on the Flt1-related activation of Rac1. As shown in Fig. 7A, Rac1 was activated at 3 min after VEGF treatment in AG1-G1-Flt1 cells, and RACK1-silencing intensively suppressed this VEGF-driven activation of Rac1. Because the activated Rac1 induces broad membrane ruffling (35), we further assessed the effect of RACK1-silencing on VEGF-triggered ruffling. As shown in Fig. 7B, stimulation with VEGF increased the membrane ruffling (62%) in AG1-G1-Flt1 cells compared with that in untreated cells (24.4%). RACK-silencing remarkably reduced the VEGF-dependent ruffling (35%). These results indicate that RACK1 plays a regulatory role in VEGF-induced Rac1 activation and membrane ruffling.
FIGURE 7.
RACK1-silencing significantly decreased VEGF-induced small-GTPase Rac1 activation and membrane ruffling. A, AG1-G1-Neo and AG1-G1-Flt1 cells transfected with control siRNA (si-ctl) or RACK1 siRNAs (si1 and si2) were serum-starved and stimulated with VEGF (100 ng/ml) for 3 min. Cell lysate was subjected to a pull-down assay with GST-PAK-PBD (GST-fused GTPase-binding domain) agarose beads, followed by Western blotting with anti-Rac1 antibody. RACK1-silencing remarkably decreased VEGF-induced activation of Rac1. Cell lysates were immunoblotted with antibodies against Rac1, and β-actin as a control. B, AG1-G1-Neo and AG1-G1-Flt1 cells transfected with control siRNA (si-ctl) or RACK1 siRNAs (si1 and si2) were serum-starved and stimulated with VEGF (100 ng/ml) for 5 min. Cells were fixed, permeabilized, and stained with rhodamine-phalloidin (F-actin). Cell morphology was observed by confocal microscope, and representative images are presented (upper panels). Scale bar, 10 μm. The numbers of cells exhibiting membrane ruffling were counted (lower panels). RACK1-silencing significantly inhibited VEGF-induced membrane ruffling.
DISCUSSION
In this study, we attempted to elucidate the downstream signaling pathway of Flt1. By using a two-hybrid approach, we found that RACK1, an important scaffolding protein, can directly interact with Flt1. Inhibition of the endogenous expression of RACK1 suppressed the VEGF-dependent activation of Akt and Rac1, resulting in decreased cell migration in a stable Flt1-expressing cell line where KDR was undetectable.
The cellular function and intracellular signaling of Flt1 remain unclear. Indeed, the lack of an experimental model has severely impeded research in this area. Recently, we generated a cell line, AG1-G1-Flt1, which stably expresses human Flt1. The Flt1 expression level in AG1-G1-Flt1 cells is comparable to that in primary vascular endothelial cells (data not shown). Both AG1-G1-Flt1 and control AG1-G1-Neo cells maintain the unique characteristics of vascular endothelial cells, such as the ability to form blood vessel-like structures and to incorporate acetyl-low-density lipoproteins in vitro.3 Moreover, the stable expression of Flt1 in the AG1-G1-Flt1 cell line also facilitates functional research into Flt1. Importantly, when analyzing cellular function using these cells, we found that the migration, but not growth, of AG1-G1-Flt1 cells was significantly increased by VEGF. Similar findings were reported in several recent papers. For example, the chemotaxis of human monocytes in response to VEGF was mediated via Flt1 because only Flt1, not KDR, was expressed (21, 36). Hiratsuka et al. (37) demonstrated that premetastatic infiltration of macrophages into the lungs is dependent on Flt1 signaling. By utilizing an anti-neutralizing antibody, Kaplan et al. (38) also showed that the antibody to Flt1 but not KDR efficiently suppressed the infiltration of bone marrow-derived VEGFR1-positive progenitor cells into lungs in tumor-bearing mice. Consistent with these studies, the absence of KDR expression in our model implied a specific role for Flt1 in the VEGF-induced cell migration.
Although the phenotype has been reported in several cells, the signaling mechanism underlying Flt1-mediated cell migration remains poorly understood. Recently, by employing a two-hybrid assay, we found that RACK1 can interact with Flt1. Interestingly, in our model, the VEGF-stimulation increased the amount of RACK1 bound to Flt1 as well as triggered AG1-G1-Flt1 cell migration, while the silencing of RACK1 with RNAi significantly suppressed this migration. These results definitely indicate that RACK1 plays a pivotal role in the regulation of Flt1-mediated cell migration. Considering the regulatory effect brought to the individual protein by the Flt1-RACK1 complex, the finding that the activation of Flt1 was unaffected by RACK1-silencing (Fig. 3A) suggests that RACK1 only relays the signal from Flt1 downstream. Most of the signaling molecules underlying a receptor kinase function as (i) a kinase, like PI3K, which after being activated by the receptors, conveys signals by phosphorylating a substrate downstream, or (ii) an adapter protein, facilitating signal transduction by mediating protein-protein interaction. RACK1 is a proposed scaffolding protein that plays an important role in positioning the signaling enzymes at appropriate locations and facilitates the reactions with the substrate proteins (25, 39). Structural analysis showed that RACK1 contains 7 internal Trp-Asp 40 (WD40) repeats, domains involved in protein-protein interaction, but lacks a kinase domain (26, 39). Studies have revealed that RACK1 plays a critical role in membrane-cytoskeletal association by acting as a scaffold to recruit other proteins to focal adhesion complexes and regulate cell chemotaxis (40–43). Accordingly, RACK1 may emerge as an adaptor protein not a kinase and provide docking sites to facilitate the protein-protein interaction required for Flt1-related signaling.
The proteins integrated by RACK1 into Flt1-related signaling remain unknown, however, the PI3K may be a candidate. In our experimental model, Akt was markedly activated by the VEGF-treatment, and the Flt1-mediated AG1-G1-Flt1 cell migration was significantly attenuated by wortmannin, a PI3K inhibitor. Similar findings have been reported previously (36, 44). These results indicate that the activation of the PI3K/Akt pathway is essential for Flt1-mediated cell migration. Interestingly, RACK1-silencing significantly decreased the VEGF-driven phosphorylation of Akt, direct proof that RACK1 is involved in the Flt1-related activation of the PI3K/Akt pathway. The question is how RACK1 facilitates the activation of PI3K by Flt1. In contrast to previous reports that RACK1 can interact with PI3K p85 (45) and both the N- and C-SH2 domains of PI3K p85 bind to Y1213 of Flt1 (46), we found that the p85 subunit of PI3K does not interact with Flt1 or RACK1 at detectable levels. Different signaling pathways or cell-type dependent pathways may result in this difference. It is also possible that RACK1 recruits unidentified proteins to bridge the interaction between Flt1 and PI3K. It is necessary to analyze the RACK1-associated proteins in response to VEGF-treatment in vascular endothelial cells or monocytes to elucidate this regulatory mechanism.
The role and regulation of PLCγ in Flt1-dependent cell migration are unclear. We have previously demonstrated the VEGF-dependent phosphorylation of PLCγ in Flt1-expressing NIH3T3 cells (17). We also found that PLCγ was phosphorylated on VEGF-treatment in AG1-G1-Flt1 cells. However, unlike the PI3K/Akt cascade, the phosphorylation of PLCγ activated by VEGF was unaffected by RACK1-silencing (Fig. 6A). Although the activated PLCγ induces the phosphorylation of MAPK (17), U0126, a MAPK inhibitor, did not inhibit the VEGF-induced migration of AG1-G1-Flt1 cells. Thus, these findings demonstrate a specific regulatory role for RACK1-PI3K/Akt but not PLCγ-MAPK in Flt1-mediated cell migration.
The growth factor-induced cell migration is based on the spatial and temporal coordination of cytoskeletal changes, which are mainly regulated by the Rho family of GTPases. Rac1, a member of this family, is involved in regulating VEGF-driven actin reorganization and cell migration (33, 34, 47). In our model, RACK1-silencing significantly decreased the VEGF-induced activation of Rac1 and membrane ruffling (Fig. 7). We consider that the decreased activity of the PI3K/Akt pathway may, at least partly, give rise to this downstream regulation because the activation of Rac1 depends on PI3K/Akt signaling (48). Meanwhile, it is also possible that RACK1 more directly regulates the Flt1-meidated activation of Rac1 because RACK1 was reported to bind Rac1 (49). Activation of Rac1 may rapidly stimulate the remodeling of actin filaments at the plasma membrane, forming membrane ruffles, and promoting cell migration (35, 50).
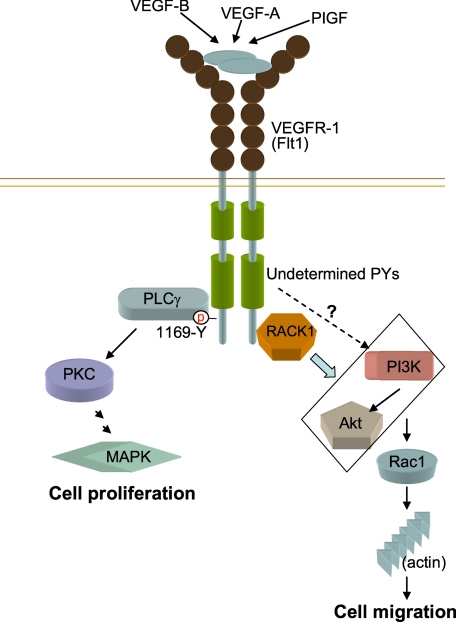
Based on the results described above, we propose a regulatory mechanism for RACK1 in Flt1-PI3K/Akt-Rac1-mediated skeletal dynamics (Fig. 8). In addition, RACK1 has been shown to be involved in the dynamics of focal adhesion (40, 42). Thus, whether and how RACK1 regulates Flt1-related focal adhesion should be investigated further.
FIGURE 8.
The possible role of RACK1 in VEGF/Flt1-induced cell migration. By interacting with Flt1, RACK1 plays an important role in VEGF/Flt1-induced cell migration by regulating the activation of the PI3K/Akt but not PLCγ-MAPK signaling pathway. RACK1 regulates the VEGF/Flt1-induced activation of Rac1 and cytoskeletal dynamics.
Supplementary Material
Acknowledgments
We thank Dr. Y. Yuasa and other members of the laboratory (Department of Molecular Oncology, Tokyo Dental, and Medical University, Tokyo, Japan) for help and advice throughout this study. We also thank Dr. Makoto Watanabe for providing plasmids.
This work was supported by Grant-in-aid Special Project Research on Cancer-Bioscience 17014020 from the Ministry of Education, Culture, Sports, Science and Technology of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
M. Yamauchi and M. Shibuya, unpublished data.
- KDR
- kinase domain receptor
- RACK
- receptor for activated protein kinase C
- BCIP
- 5-bromo-4-chloroindolyl phosphate
- NBT
- nitroblue tetrazolium.
REFERENCES
- 1. Ferrara N., Kerbel R. S. (2005) Nature 438, 967–974 [DOI] [PubMed] [Google Scholar]
- 2. Coultas L., Chawengsaksophak K., Rossant J. (2005) Nature 438, 937–945 [DOI] [PubMed] [Google Scholar]
- 3. Shibuya M., Claesson-Welsh L. (2006) Exp. Cell Res. 312, 549–560 [DOI] [PubMed] [Google Scholar]
- 4. Ferrara N. (2009) Arterioscler. Thromb. Vasc. Biol. 29, 789–791 [DOI] [PubMed] [Google Scholar]
- 5. Lohela M., Bry M., Tammela T., Alitalo K. (2009) Curr. Opin. Cell Biol. 21, 154–165 [DOI] [PubMed] [Google Scholar]
- 6. Shibuya M., Yamaguchi S., Yamane A., Ikeda T., Tojo A., Matsushime H., Sato M. (1990) Oncogene 5, 519–524 [PubMed] [Google Scholar]
- 7. Terman B. I., Carrion M. E., Kovacs E., Rasmussen B. A., Eddy R. L., Shows T. B. (1991) Oncogene 6, 1677–1683 [PubMed] [Google Scholar]
- 8. Sawano A., Takahashi T., Yamaguchi S., Aonuma M., Shibuya M. (1996) Cell Growth & Differ. 7, 213–221 [PubMed] [Google Scholar]
- 9. Fong G. H., Rossant J., Gertsenstein M., Breitman M. L. (1995) Nature 376, 66–70 [DOI] [PubMed] [Google Scholar]
- 10. Shalaby F., Rossant J., Yamaguchi T. P., Gertsenstein M., Wu X. F., Breitman M. L., Schuh A. C. (1995) Nature 376, 62–66 [DOI] [PubMed] [Google Scholar]
- 11. Hiratsuka S., Minowa O., Kuno J., Noda T., Shibuya M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9349–9354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Davis-Smyth T., Chen H., Park J., Presta L. G., Ferrara N. (1996) EMBO J. 15, 4919–4927 [PMC free article] [PubMed] [Google Scholar]
- 13. Fuh G., Li B., Crowley C., Cunningham B., Wells J. A. (1998) J. Biol. Chem. 273, 11197–11204 [DOI] [PubMed] [Google Scholar]
- 14. Xia P., Aiello L. P., Ishii H., Jiang Z. Y., Park D. J., Robinson G. S., Takagi H., Newsome W. P., Jirousek M. R., King G. L. (1996) J. Clin. Invest. 98, 2018–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takahashi T., Yamaguchi S., Chida K., Shibuya M. (2001) EMBO J. 20, 2768–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sakurai Y., Ohgimoto K., Kataoka Y., Yoshida N., Shibuya M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1076–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sawano A., Takahashi T., Yamaguchi S., Shibuya M. (1997) Biochem. Biophys. Res. Commun. 238, 487–491 [DOI] [PubMed] [Google Scholar]
- 18. Lee T. H., Seng S., Sekine M., Hinton C., Fu Y., Avraham H. K., Avraham S. (2007) PLoS Med. 4, e186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Suzuma K., Naruse K., Suzuma I., Takahara N., Ueki K., Aiello L. P., King G. L. (2000) J. Biol. Chem. 275, 40725–40731 [DOI] [PubMed] [Google Scholar]
- 20. Kearney J. B., Kappas N. C., Ellerstrom C., DiPaola F. W., Bautch V. L. (2004) Blood 103, 4527–4535 [DOI] [PubMed] [Google Scholar]
- 21. Barleon B., Sozzani S., Zhou D., Weich H. A., Mantovani A., Marmé D. (1996) Blood 87, 3336–3343 [PubMed] [Google Scholar]
- 22. Krysiak O., Bretschneider A., Zhong E., Webb J., Hopp H., Verlohren S., Fuhr N., Lanowska M., Nonnenmacher A., Vetter R., Jankowski J., Paul M., Schönfelder G. (2005) Circ. Res. 97, 1253–1261 [DOI] [PubMed] [Google Scholar]
- 23. Kerber M., Reiss Y., Wickersheim A., Jugold M., Kiessling F., Heil M., Tchaikovski V., Waltenberger J., Shibuya M., Plate K. H., Machein M. R. (2008) Cancer Res. 68, 7342–7351 [DOI] [PubMed] [Google Scholar]
- 24. Muramatsu M., Yamamoto S., Osawa T., Shibuya M. (2010) Cancer Res. 70, 8211–8221 [DOI] [PubMed] [Google Scholar]
- 25. McCahill A., Warwicker J., Bolger G. B., Houslay M. D., Yarwood S. J. (2002) Mol. Pharmacol. 62, 1261–1273 [DOI] [PubMed] [Google Scholar]
- 26. Ron D., Chen C. H., Caldwell J., Jamieson L., Orr E., Mochly-Rosen D. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 839–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ron D., Mochly-Rosen D. (1994) J. Biol. Chem. 269, 21395–21398 [PubMed] [Google Scholar]
- 28. Ishizaki H., Tsunoda T., Wada S., Yamauchi M., Shibuya M., Tahara H. (2006) Clin. Cancer Res. 12, 5841–5849 [DOI] [PubMed] [Google Scholar]
- 29. Seetharam L., Gotoh N., Maru Y., Neufeld G., Yamaguchi S., Shibuya M. (1995) Oncogene 10, 135–147 [PubMed] [Google Scholar]
- 30. Podar K., Catley L. P., Tai Y. T., Shringarpure R., Carvalho P., Hayashi T., Burger R., Schlossman R. L., Richardson P. G., Pandite L. N., Kumar R., Hideshima T., Chauhan D., Anderson K. C. (2004) Blood 103, 3474–3479 [DOI] [PubMed] [Google Scholar]
- 31. Wu Y., Hooper A. T., Zhong Z., Witte L., Bohlen P., Rafii S., Hicklin D. J. (2006) Int. J. Cancer 119, 1519–1529 [DOI] [PubMed] [Google Scholar]
- 32. Wu L. W., Mayo L. D., Dunbar J. D., Kessler K. M., Baerwald M. R., Jaffe E. A., Wang D., Warren R. S., Donner D. B. (2000) J. Biol. Chem. 275, 5096–5103 [DOI] [PubMed] [Google Scholar]
- 33. Del Valle-Pérez B., Martinez V. G., Lacasa-Salavert C., Figueras A., Shapiro S. S., Takafuta T., Casanovas O., Capellà G., Ventura F., Viñals F. (2010) J. Biol. Chem. 285, 10748–10760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tan W., Palmby T. R., Gavard J., Amornphimoltham P., Zheng Y., Gutkind J. S. (2008) FASEB J. 22, 1829–1838 [DOI] [PubMed] [Google Scholar]
- 35. Ridley A. J., Paterson H. F., Johnston C. L., Diekmann D., Hall A. (1992) Cell 70, 401–410 [DOI] [PubMed] [Google Scholar]
- 36. Tchaikovski V., Fellbrich G., Waltenberger J. (2008) Arterioscler. Thromb. Vasc. Biol. 28, 322–328 [DOI] [PubMed] [Google Scholar]
- 37. Hiratsuka S., Nakamura K., Iwai S., Murakami M., Itoh T., Kijima H., Shipley J. M., Senior R. M., Shibuya M. (2002) Cancer Cell 2, 289–300 [DOI] [PubMed] [Google Scholar]
- 38. Kaplan R. N., Riba R. D., Zacharoulis S., Bramley A. H., Vincent L., Costa C., MacDonald D. D., Jin D. K., Shido K., Kerns S. A., Zhu Z., Hicklin D., Wu Y., Port J. L., Altorki N., Port E. R., Ruggero D., Shmelkov S. V., Jensen K. K., Rafii S., Lyden D. (2005) Nature 438, 820–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sklan E. H., Podoly E., Soreq H. (2006) Prog. Neurobiol. 78, 117–134 [DOI] [PubMed] [Google Scholar]
- 40. Liliental J., Chang D. D. (1998) J. Biol. Chem. 273, 2379–2383 [DOI] [PubMed] [Google Scholar]
- 41. Besson A., Wilson T. L., Yong V. W. (2002) J. Biol. Chem. 277, 22073–22084 [DOI] [PubMed] [Google Scholar]
- 42. Cox E. A., Bennin D., Doan A. T., O'Toole T., Huttenlocher A. (2003) Mol. Biol. Cell 14, 658–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hermanto U., Zong C. S., Li W., Wang L. H. (2002) Mol. Cell. Biol. 22, 2345–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Banerjee S., Mehta S., Haque I., Sengupta K., Dhar K., Kambhampati S., Van Veldhuizen P. J., Banerjee S. K. (2008) Biochemistry 47, 3345–3351 [DOI] [PubMed] [Google Scholar]
- 45. Kiely P. A., Sant A., O'Connor R. (2002) J. Biol. Chem. 277, 22581–22589 [DOI] [PubMed] [Google Scholar]
- 46. Cunningham S. A., Waxham M. N., Arrate P. M., Brock T. A. (1995) J. Biol. Chem. 270, 20254–20257 [DOI] [PubMed] [Google Scholar]
- 47. Garrett T. A., Van Buul J. D., Burridge K. (2007) Exp. Cell Res. 313, 3285–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fukata M., Nakagawa M., Kaibuchi K. (2003) Curr. Opin. Cell Biol. 15, 590–597 [DOI] [PubMed] [Google Scholar]
- 49. Orlichenko L., Geyer R., Yanagisawa M., Khauv D., Radisky E. S., Anastasiadis P. Z., Radisky D. C. (2010) J. Biol. Chem. 285, 19153–19161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Michiels F., Habets G. G., Stam J. C., van der Kammen R. A., Collard J. G. (1995) Nature 375, 338–340 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.