Abstract
Bone integrity is maintained by a dynamic equilibrium between the activities of bone-forming osteoblasts and bone-resorbing osteoclasts. Osteolytic lesions are a painful consequence of metastasis of breast cancer cells to bone in an overwhelming majority of breast cancer patients. Factors secreted by breast cancer cells propel a cascade of events that trigger osteoclastogenesis and elevated bone resorption. In the present study, we show that the Hedgehog (Hh) ligands secreted by breast cancer cells promote osteoclast differentiation and potentiate the activity of mature osteoclasts. Paracrine Hh signaling induced by breast cancer cells mediates a detrimental chain of events by the up-regulation of osteopontin (OPN), which in turn enhances osteoclastic activity by up-regulating cathepsin K and MMP9. Hh signaling is essential for osteoclasts because blocking the Hh pathway using the pharmacological Hh inhibitor, cyclopamine, results in an overall decrease in osteoclastogenesis and resorptive activity. Our studies suggest that inhibiting Hh signaling interferes with the ability of pre-osteoclasts to respond to the stimulatory effects of the breast cancer cells, indicating that Hh signaling is vital to osteoclast activity.
Keywords: Bone, Breast Cancer, Matrix Metalloproteinase, Oncogene, Protease, Hedgehog Pathway, Bone Metastasis, Osteoclastogenesis, Osteolysis, Osteopontin
Introduction
Breast cancer cells preferentially metastasize to the bone. Once within bone, an interaction ensues between breast cancer cells and the cells within the bone microenvironment. Breast cancer cells secrete various factors that stimulate osteoblasts and osteoclasts and other cells within the bone; these in turn secrete factors that stimulate the tumor cells, creating a vicious cycle that nurtures the development and propagation of bone metastases (1). Seminal studies by Kang et al. (2) showed that osteopontin (OPN)2 forms a component of a “bone metastasis signature” of breast cancer cells, i.e. breast cancer cells that metastasized to bone had increased OPN expression. Furthermore, OPN functionally enhanced incidence of bone metastases by breast cancer cells in concert with interleukin-11.
OPN is one of the abundant non-collagenous proteins in bone. It is a bone matrix protein that promotes osteoclast function and is consistently overexpressed in highly metastatic cells. Ultrastructural immunocytochemical studies show that the most prominent accumulation of OPN is seen at cement lines in remodeling bone and at laminae limitantes at bone surfaces (3). It is localized to cell-matrix and matrix-matrix interfaces in mineralized tissue, where it is deposited as the result of osteoclast action. Moreover, OPN appears to be an important component in the communication between osteoclasts and osteoblasts, and there is strong evidence for the involvement of OPN in the formation, migration, and attachment of osteoclasts and in their resorptive activity (4, 5). Importantly, interfering with the adhesion of osteoclasts to osteopontin by RGD peptides abolishes their resorptive activity (6, 7).
Nearly 42% of primary breast tumors express moderate to strong levels of OPN, and 83% of bone metastases resulting from these tumors express OPN (8). OPN expression, specifically within the tumor cells, reciprocally correlates with patient survival. Clinical studies have revealed a correlation between plasma OPN, tumor burden, and prognosis in patients with breast cancer metastasis (9). The levels of OPN in plasma of patients with breast cancer are significantly higher in those with bone metastasis when compared with those who do not have bone metastasis (10). Moreover, the level of OPN increases with progression of the disease. Among these women, those with highest levels of OPN (more than 2000 μg/ml) show poor survival when compared with those with OPN levels between 1000 and 1500 μg/ml. Whether the circulating OPN impacts “homing” of breast cancer cells to bone is still not known. Functionally, OPN expression is vital to the tumorigenic ability of cells (11). Expression of OPN in OPN-negative breast cancer cells increases their adhesion to bone marrow cells (12), and OPN knock-out mice display significantly lower incidence of bone metastases (13, 14). In bone, tumor-derived OPN plays a vital role in the establishment of vasculature by mediating adhesion to endothelial cells, co-operating with vascular endothelial growth factor (VEGF), and preventing apoptosis of the endothelial cells (15).
We have recently reported that the expression of OPN is regulated, in part, by the Hedgehog (Hh) pathway (16). The Hh pathway has been reported to be aberrantly activated in breast cancer (17, 18). In the absence of the ligand, Desert hedgehog (DHH), Indian hedgehog (IHH), Sonic hedgehog (SHH), the Hh signaling pathway is inactive. Ligand molecules bind to the receptor Patched (PTCH), thereby alleviating PTCH-mediated suppression of Smoothened (SMOH), leading to activation of the pathway through the transcription of target genes mediated by the GLI transcription factors (19).
In this study, we have determined that breast cancer cells express Hh ligands. These ligands can mediate a cross-talk directly with osteoclasts and activate expression of OPN in the osteoclasts; this promotes osteoclast maturation and resorptive activity. As such, we have revealed a novel mechanism by which breast cancer cells can directly influence osteoclast development and activity.
EXPERIMENTAL PROCEDURES
Cell Lines
Human metastatic breast cancer cells, MDA-MB-231, were cultured in Dulbecco's modified Eagle's medium (DMEM/F12; Invitrogen), supplemented with 2 mm l-glutamine, 1 mm sodium pyruvate, 0.02 mm non-essential amino acids, 5% fetal bovine serum, FBS (Atlanta Biologicals, Norcross, GA), without antibiotics or antimycotics. SUM1315 and SUM159 cells (20) (Asterand, Detroit, MI) were cultured in DMEM/F12 supplemented with 5 mg/ml insulin, 5% FBS (Atlanta Biologicals), and either 10 ng/ml EGF or 1 ng/ml hydrocortisone, without antibiotics or antimycotics. The SUM1315 cells were derived from a metastasis in a patient with infiltrating ductal carcinoma; SUM159 cells were derived from a primary breast tumor with metaplastic carcinoma. RAW264.7 (ATCC, TIB 71) cells, a murine pre-osteoclastic line capable of differentiation and mineralization in culture (in the presence of RANKL and M-CSF), were grown in DMEM with l-glutamine (ATCC, 30-2002) supplemented with 10% FBS.
A 2× differentiation medium (DM) was formulated for the RAW 264.7 cells (21) comprising RAW264.7 growth medium supplemented with 20% FBS, RANKL (100 ng/ml), and M-CSF (40 ng/ml). Conditioned media were harvested from breast cancer cells and mixed in a 1:1 ratio with double strength differentiation medium to assess the effect on osteoclast differentiation. 1× DM was used as control or wherever SHH (R&D Systems, Minneapolis, MN) or OPN (R&D Systems) was used alone. The medium on the RAW264.7 cells was replenished every 48 h. To assess the effect of secreted Hh ligands, we used the neutralizing 5E1 antibody (22) (Developmental Studies Hybridoma Bank at the University of Iowa, Iowa City, IA). The amount of 5E1 antibody used for the studies was determined following titration of the antibody with respect to its effects on osteoclast differentiation (supplemental Fig. 1A). Medium was supplemented with 5E1 (2.5 μg/ml) and was changed on alternate days until the end of the experiment.
The effect of OPN on osteoclast activity was assessed by transfecting an OPN shRNA-expressing construct (11) on day 6 after induction of differentiation. Fresh DM was added the following day, and cells were allowed to grow for another 12 h before the termination of the experiment. GLI1 expression was silenced in the SUM1315 cells using shRNA targeting GLI1 into pSuperior.gfp+neo (Oligoengine, WA) (16). Silencing of OPN expression was done using OPN-targeting shRNA cloned into pSuper (Oligoengine) (11). All cells were maintained in a humidified 5% CO2 environment at 37 °C. All cultures were confirmed to be negative for Mycoplasma spp. infection using a PCR-based test (TaKaRa, Shiga, Japan).
Osteoclast Differentiation and Activity Assays
Tartrate-resistant acid phosphatase (TRAP) assay was conducted for RAW 264.7 following the manufacturer's protocol (Sigma). This assay was indicative of the extent of differentiation. OAASTM plates (Osteogenic Core Technologies, Choongnam, Republic of Korea) were utilized to measure osteoclastic activity. RAW 264.7 cells (25 × 103) were inoculated in 48-well plates. Cells were treated with serum-free conditioned media from breast cancer cells or 100 nm SHH or 100 ng/ml OPN on the following day. Media were changed every 2 days, and the experiments were either terminated on day 6 or terminated on day 7 for knockdown experiments, with transfections being done on the 6th day. At the completion, cells were detached with 5% sodium hypochlorite, and the wells were observed under a Nikon Eclipse TS 100 microscope at 10× magnification. To test the inhibitory effect of cyclopamine on osteoclast differentiation and activity, RAW264.7 cells were cultured in differentiation medium supplemented with 20 μm cyclopamine (Sigma) dissolved in dimethyl sulfoxide (DMSO) (Sigma). Medium containing cyclopamine was changed every 48 h. The percentage of area resorbed was calculated using the NIS-Elements BR. 3.1 software.
Western Blotting Analysis
Whole cell lysates were collected in Nonidet P-40 buffer (150 mm NaCl, 50 mm Tris, 1% Nonidet P-40). Total protein (30 μg) was resolved by SDS-PAGE gel and transferred to PVDF membranes. Membranes were immunoblotted overnight at 4 °C with antibodies to either SHH (Santa Cruz Biotechnology, Santa Cruz, CA) or IHH (Santa Cruz Biotechnology). Equal loading was confirmed with anti-β-tubulin (Cell Signaling, Danvers, MA). To assess OPN expression upon SHH treatment, 105 cells were grown in 6-well plates in the presence of SHH. After 24 h, the cells were lysed in Nonidet P-40 buffer, and 30 μg of each experimental group was assessed by immunoblotting. To assess for the expression of OPN, MMP9, and cathepsin K (CTSK), at the end of day 6 of differentiation, all the different experimental groups were kept in serum-free medium for 24 h, and both the conditioned media and the whole cell lysates were collected and immunoblotted using antibodies to either anti-mouse OPN (Millipore, Bedford, MA) or CTSK (Santa Cruz Biotechnology). Equal loading was confirmed with anti-β-actin (Sigma) antibody. Secreted MMP9 (Santa Cruz Biotechnology) was assessed by loading an equal quantity of protein from the serum-free conditioned medium. Corresponding HRP-conjugated secondary antibodies were used for detection; blots were developed with SuperSignal enhanced chemiluminescence substrate (Pierce) and imaged using a Fuji LAS3000 imager.
Detection and quantification of proteins were done using Fuji LAS 3000 apparatus and MultiGauge version 3.1 software (Fujifilm, Valhalla, NY). Band intensities were measured in arbitrary units. Relative band intensity was obtained as a ratio of individual band intensity to that of the corresponding β-actin band.
Luciferase Assay
Cells were co-transfected with the OPN promoter construct, pGL3-OPNB (16), and pSV-β-galactosidase (Promega). Empty pGL3 vector was used as control. Different concentrations of SHH ligand were added to the well the next day. Cells were lysed in reporter lysis buffer (Promega) 24 h after the addition of SHH, and both β-galactosidase assay and luciferase assays were done following the manufacturer's protocol. Readings were normalized to β-galactosidase. Each parameter was studied in triplicate, and the experiment was repeated at least three times.
Quantitative RT-PCR
cDNA was generated using a High Capacity reverse transcriptase kit (Applied Biosystems, Foster City, CA). Real-time PCR was performed in two cycles using a Bio-Rad iQ5 real-time detection system (Bio-Rad); the first cycle of 95 °C for 10 min was followed by 40 repeats of the second cycle comprising 95 °C for 15 s followed by 60 °C for 1 min. All reactions were done in triplicate. Transcript levels were normalized to GAPDH levels (dCT), which was used to calculate changes in gene expression (2-ddCT). To assess levels of OPN, CTSK, and MMP9, cells (50 × 103) were seeded in each well of 12-well plates. Following the experimental regime as described earlier (for osteoclast differentiation/activity), RNA was isolated using TRIzol (Invitrogen) and assessed as above. The details of the primers used were as follows: OPN (Spp1; Mm00436767_m1), matrix metalloprotease 9 (MMP9; Mm00600163_m1), cathepsin K (Ctsk; Mm00484036_m1), SHH (Hs00179843_m1), GLI1 (Hs01110766_m1), OPN (Hs00959010_m1), hGAPDH (Hs99999905_m1), and GAPDH (Mm99999915_g1).
Statistical Analysis
Statistical differences between groups were assessed using the Mann-Whitney test, Student's t test, or analysis of variance, using GraphPad Prism 4 software. Statistical significance was determined if the analysis reached 95% confidence. The precise p values are listed in the corresponding figure legends. In all figures, the error bars represent S.E.
RESULTS
Hh Signaling Activates OPN Expression in Pre-osteoclasts
We previously published that in melanoma, activated Hh signaling culminates in the transcription of OPN by GLI1, the transcription factor of the Hh pathway (16). We assessed the pre-osteoclastic RAW264.7 cells for their ability to regulate OPN in response to Hh ligands. As seen in Fig. 1A, the RAW264.7 cells show a dose-dependent significant (p < 0.0001) increase in OPN mRNA levels in response to SHH. The increases in OPN mRNA levels are due to an increase in the activity of the OPN promoter in response to SHH (Fig. 1B), indicating that OPN is under regulation of the Hh pathway in this system. This is also reflected in increased protein levels of OPN upon SHH treatment (Fig. 1, C and D).
FIGURE 1.
Hh signaling activates OPN in RAW264.7 cells. A, RAW264.7 cells were treated with the indicated concentrations of recombinant SHH. The levels of OPN were assessed by real-time quantitative RT-PCR. Relative to untreated cells, the cells treated with SHH expressed significantly greater levels of OPN mRNA (p < 0.0001). B, OPN promoter construct (200 ng) and the β-galactosidase plasmid (200 ng) were co-transfected into RAW264.7 cells. Luciferase activity was assayed 24 h after SHH treatment and normalized to β-galactosidase. Each group was assessed in triplicate. The data are depicted as relative luciferase activity and are representative of three independent experiments. The increase in OPN promoter activity is significantly (p < 0.05) higher for the indicated groups relative to the control (untreated) cells. C, SHH treatment of the RAW264.7 cells (at the nm concentrations indicated) stimulates the expression of OPN. Cells were lysed 24 h after SHH treatment, and OPN and β-actin were assessed by immunoblotting. D, densitometric analyses of the immunoblotting results. The results are represented as band intensity relative to respective loading control. Band intensities are represented as arbitrary units.
Breast Cancer Cells Enhance Osteoclast Differentiation and Activity via Hh Signaling
Although reports suggest a role for the Hh pathway in bone development and homeostasis (23), OPN is critical to osteoclast activity, specifically their motility (24, 25). Because both SHH and OPN are secreted molecules, we evaluated the effects of recombinant SHH and OPN on influencing differentiation of RAW264.7 cells. We scored for osteoclast differentiation by staining the cells for TRAP, an indication of differentiated osteoclasts. We scored multinucleate (more than three nuclei per cell) TRAP-positive cells. As seen in Fig. 2A, when compared with DM alone, DM supplemented with SHH or OPN causes a significant (p < 0.005) increase in the numbers of TRAP-stained multinucleate cells, indicating that activation of the Hh pathway enhances osteoclast differentiation. Moreover, OPN-initiated signaling also appears to influence osteoclast differentiation.
FIGURE 2.
Breast cancer cells activate Hh signaling and promote osteoclast differentiation. A, DM supports differentiation of RAW264.7 into osteoclasts. Supplementing DM with recombinant human OPN (100 ng/ml) or SHH (100 nm) significantly (* indicates p < 0.005) increases the numbers of multinucleate (more than three nuclei) TRAP-positive cells. B, conditioned serum-free medium from breast cancer cells, MDA-MB-231, SUM159, and SUM1315, significantly (*, p < 0.01) increases the numbers of multinucleate, TRAP-positive cells. The addition of Hh ligand-neutralizing antibody, 5E1, to differentiation conditions notably (^, p < 0.05) reduces the efficiency of breast cancer cell-conditioned medium to elicit osteoclast differentiation. C, breast cancer cells (MCF10CA clone d, MDA-MB-231, SUM159, and SUM1315) express IHH and SHH ligands. Shown is an immunoblot of the lysate from the breast cancer cells. β-Tubulin serves as a loading control. D, images represent photomicrographs showing the TRAP-stained osteoclasts formed in response to various differentiation conditions. The bar represents 100 μm. Panel a, growth medium (GM); panels b and e, DM; panels c and d, DM supplemented with recombinant OPN (100 ng/ml) (panel c) and SHH (100 nm) (panel d); panel f, DM + 5E1 (2.5 μg/ml); panels g, i, and k, DM supplemented with conditioned medium from MDA-MB-231 cells (panel g), SUM159 cells (panel i), and SUM1315 cells (panel k); panels h, j, and l, 5E1 (2.5 μg/ml) added to DM supplemented with conditioned medium from MDA-MB-231 (panel h), SUM159 (panel j), and SUM1315 cells (panel l). The osteoclasts are marked within circles in panel c. The data were recorded at 10× magnification using the Nikon Eclipse TS 100 microscope and are represented as a percentage of multinucleate TRAP-positive cells relative to the total number of cells in the field. The data were verified by two independent experiments.
Breast cancer cells potentiate osteoclast activity, leading to increased osteolysis. This is brought about, in part, by factors secreted by the breast cancer cells (26, 27). To determine the effects of factors secreted by the breast cancer cells on osteoclast differentiation, we mixed the conditioned medium of breast cancer cells in equal proportion with double strength DM and assessed the effects on differentiation of the RAW264.7 cells. As seen in Fig. 2B, the conditioned medium from three breast cancer cell lines (MDA-MB-231, SUM159, and SUM1315) significantly increases (p < 0.01) the formation of TRAP-positive multinucleate cells.
Breast cancer cells express SHH and IHH (Fig. 2C). The Hh ligands are synthesized in cells and are secreted from the cells and are expressed at the exterior surface of the cell membrane or form a component of the secretome of the cells (28, 29). To determine whether Hh ligands produced by breast cancer cells influence osteoclast differentiation, we added the Hh ligand-neutralizing antibody, 5E1, to the differentiation conditions. The 5E1 antibody is an Hh pathway antagonist that is widely used in Hh-related studies in developmental biology and cancer. The 5E1 antibody blocks binding of all three mammalian Hh ligands to PTCH, thereby inhibiting Hh signaling (30). Thus, using 5E1 provides us with a tool to effectively block Hh signaling. As seen in Fig. 2, B and D, the addition of 5E1 significantly (p < 0.05) diminishes the ability of the breast cancer cell-conditioned medium to influence osteoclast differentiation. This suggests that Hh ligands secreted by breast cancer cells potentiate osteoclast differentiation.
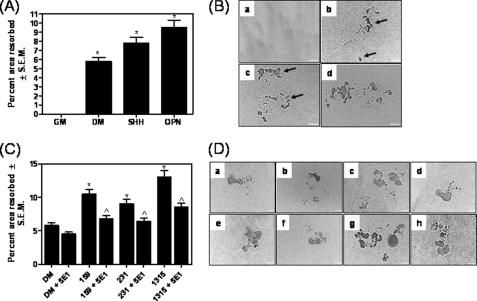
To determine the effects of Hh signaling on osteoclast activity, we cultured the osteoclasts on plates coated with a mineralized bone matrix (carbonated calcium phosphate). The area resorbed was estimated. As seen in Fig. 3, A and B, the DM supplemented with SHH or OPN notably enhances (p < 0.001) osteoclast activity (p < 0.05). Further, the conditioned medium from all three breast cancer cells enhances the ability of osteoclasts to resorb the matrix (Fig. 3, C and D). Although the medium from all three breast cancer cells lines stimulates osteoclasts, (p ≤ 0.01), the medium from SUM1315 cells maximally potentiated osteoclast activity more than the MDA-MB-231 and SUM159 cells. Conversely, depleting the Hh ligands from the secretome of the breast cancer cells using the 5E1 antibody compromises its ability to resorb the matrix. Thus, overall, the results indicate that the Hh ligands secreted by breast cancer cells augment osteoclast differentiation and activity.
FIGURE 3.
Hh signaling initiated by breast cancer cells promotes resorption activity of osteoclasts. A, recombinant human OPN (100 ng/ml) and SHH (100 nm) significantly (* indicates p < 0.001) increase the resorption activity of the differentiated osteoclasts. RAW254.7 cells were induced for differentiation on OAAS plates. At the end of the assay, the area resorbed was quantified. B, images represent photomicrographs showing the areas resorbed by the differentiated osteoclasts in response to various differentiation conditions. The bar represents 100 μm. Panel a, growth medium (GM); panel b, DM; panels c and d, DM supplemented with SHH (100 nm) (panel c) and recombinant OPN (100 ng/ml) (panel d). The arrows point to the area resorbed. C, conditioned serum-free medium from breast cancer cells, SUM159, MDA-MB-231, and SUM1315 significantly (*, p ≤ 0.01) increases the resorption activity of osteoclasts. The addition of the Hh ligand-neutralizing antibody, 5E1, to differentiation conditions notably (^, p < 0.05) decreases the resorption activity of osteoclasts induced by the secretome of breast cancer cells. The difference in the area resorbed by DM and DM + 5E1 is not statistically significant (p = 0.06). Data are represented as a percentage of the area resorbed relative to the total area in the field of view (this corresponds to 568197.12 μm2). The experiment was repeated once. D, images represent photomicrographs showing the areas resorbed by the differentiated osteoclasts in response to various differentiation conditions. The bar represents 100 μm. Panel a, DM; panel b, DM supplemented with 5E1 (2.5 μg/ml); panels c and e, conditioned medium from SUM159 cells (panel c) and MDA-MB-231 (panel e) cells; panel g, SUM1315 cells. Panels d, f, and h, 5E1 (2.5 μg/ml) added to DM supplemented with conditioned medium from SUM159 cells (panel d), MDA-MB-231 cells (panel f), and SUM1315 cells (panel h), respectively.
Activation of Hh Signaling Is Critical to Osteoclast Activity
To assess the role of Hh signaling in osteoclasts in determining their maturation and activity, we supplemented the differentiation medium with the SMOH inhibitor, cyclopamine. As seen in Fig. 4, cyclopamine significantly (p < 0.0001) reduced the ability of DM to elicit differentiation (Fig. 4, A and B) of the pre-osteoclasts into TRAP-positive, multinucleate osteoclasts, without impacting their viability (supplemental Fig. 1B). The ability of breast cancer cell-conditioned medium to potentiate differentiation and resorptive activity of the osteoclasts was also remarkably compromised in the presence of cyclopamine (Fig. 4, A–D). Thus, our results suggest that activation of Hh signaling is an essential event for osteoclast maturation and activity.
FIGURE 4.
Inhibition of Hh signaling interferes with osteoclast differentiation and activity. A and C, inhibiting Hh signaling in the osteoclasts by the Smoothened (SMOH) inhibitor, cyclopamine (Cyclo, 20 μm), significantly (*, p < 0.0001) compromises their ability to differentiate (A) and resorb (C) matrix when stimulated with conditioned medium from breast cancer cells. RAW264.7 cells were cultured under differentiating conditions in the presence of breast cancer cell-conditioned medium/and cyclopamine (20 μm). Images were acquired at 10× magnification using the Nikon Eclipse TS 100 microscope. B and D, images represent differentiation assessed by TRAP staining (B) and resorption activity (D). In both B and D, the differentiation conditions included DM (panel a); DM + cyclopamine (panel b); and conditioned medium from SUM159 (panel c), MDA-MB-231 (panel e), and SUM1315 cells (panel g). Images in panels d, f, and h represent resorption in the presence of conditioned media from SUM159 (panel d), MDA-MB-231 (panel f), and SUM1315 cells (panel h) supplemented with cyclopamine.
Breast Cancer-initiated Hh Signaling Activates OPN, CTSK, and MMP9 Expression by the Osteoclasts
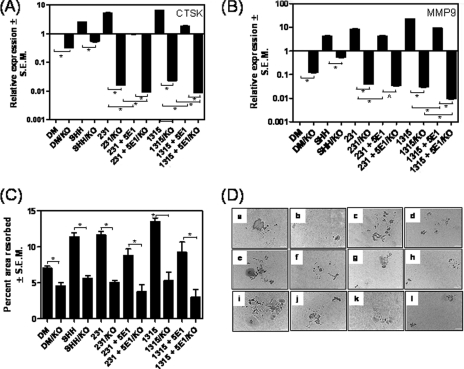
Although OPN enhances osteoclast motility and overall activity, activation of osteoclasts is functionally dictated by the expression of proteases such as MMP9 and CTSK. Thus, to assess whether Hh signaling initiated by breast cancer cells influences the ability of the differentiated osteoclasts to express these key molecules, we assessed the expression of OPN, MMP9, and CTSK by a real-time quantitative PCR. As seen in Fig. 5, A–C, when compared with DM alone, conditioned medium from breast cancer cells increases the expression of OPN, MMP9, and CTSK. Although neutralization of Hh ligands by the 5E1 antibody from the conditioned medium of SUM159 and SUM1315 cells caused a severe reduction (p < 0.001) in the expression of OPN by osteoclasts, the conditioned medium from the MDA-MB-231 cells showed a moderate, but significant (p < 0.01), decrease in OPN expression. The decrease in CTSK and MMP9 expression by the osteoclasts was also statistically significant (p < 0.001 and p < 0.0001, respectively) in the presence of the 5E1 antibody, suggesting that the Hh ligands secreted by the breast cancer cells play a critical role in up-regulating the expression of OPN, CTSK, and MMP9. We also inhibited Hh signaling in the pre-osteoclasts by supplementing DM with cyclopamine. As seen in Fig. 5, A–D, cyclopamine significantly reduces the expression of OPN, MMP9, and CTSK by the osteoclasts. The overall decrease in the expression of OPN, MMP9, and CTSK is likely due to an overall negative impact on differentiation as a result of interfering with Hh signaling in the pre-osteoclasts. The data suggest that inhibiting Hh signaling in the pre-osteoclasts makes them refractory to the stimulative effects provoked by breast cancer cells.
FIGURE 5.
Hh signaling in the osteoclasts regulates expression of OPN and proteases cathepsin K and MMP9. A–C, the levels of OPN (A), MMP9 (B), and cathepsin K (C) were assessed by real-time quantitative RT-PCR and normalized to GAPDH. The levels of gene expression are represented relative to the expression in DM alone. Three breast cancer cell lines, MDA-MB-231, SUM159, and SUM1315, were evaluated. Neutralizing antibody 5E1 significantly decreased levels of OPN (*, p < 0.01), CTSK (*, p < 0.001), and MMP9 (*, p ≤ 0.0001). The effects of cyclopamine (Cyclo) on the expression of OPN, MMP9, and CTSK was assessed by real-time quantitative RT-PCR and normalized to GAPDH. The levels of gene expression are represented relative to the expression in DM alone. Cyclopamine significantly decreased levels of OPN (*, p < 0.0001), CTSK (*, p < 0.0001), and MMP9 (*, p < 0.0001; ^, p < 0.005). D, the expression of proteases cathepsin K and MMP9 is regulated by Hh signaling. The expression of OPN, MMP9 and CTSK was assessed by immunoblotting. The graph represents densitometric analyses of the immunoblotting results. The results are represented as band intensity in arbitrary units relative to respective loading control.
Hh Signaling-initiated OPN Expression in Osteoclasts Is Essential for the Expression of MMP9 and CTSK
Our research (16) (Fig. 1) indicates that Hh signaling transcriptionally promotes the expression of OPN. To determine whether the transcriptional activation of OPN is essential for the expression of MMP9 and CTSK by the osteoclasts in response to breast cancer cell-derived Hh ligands, we abrogated the expression of OPN from the osteoclasts using RNA interference on day 6, after the osteoclast differentiation is complete (supplemental Fig. 2A shows that the differentiation process is not affected by OPN silencing on day 6; supplemental Fig. 2B shows the magnitude of silencing OPN expression). As seen in Fig. 6, silencing OPN in the osteoclasts significantly decreased the expression of CTSK and MMP9 in response to DM alone. The absence of OPN made the osteoclasts refractory to the stimulative effects of SHH. Abrogating OPN from the osteoclasts also made the osteoclasts non-responsive to the effects of the conditioned media from MDA-MB-231 and SUM1315 cells. This was seen as a marked reduction in the expression of both CTSK and MMP9. The most remarkable decrease was seen in the response of osteoclasts that were silenced for OPN expression and exposed to differentiation medium that was depleted of the Hh ligands. Thus, overall, our results implicate a role for Hh ligand-initiated osteopontin expression in osteoclasts in influencing the expression of the proteases CTSK and MMP9. The expression of OPN by the osteoclasts was also critical for their ability to resorb bone matrix. As seen in Fig. 6, C and D, abrogating OPN expression from osteoclasts notably compromised (p < 0.005) their ability to resorb bone in response to DM alone or DM supplemented with SHH. Even in the presence of conditioned medium from the MDA-MB-231 and the SUM1315 cells, the osteoclasts silenced for OPN expression were compromised (p < 0.005) for their resorptive ability. The decrease in resorption in response to depletion of the Hh ligands from the breast cancer cell-conditioned medium was further accentuated when the osteoclasts were unable to express OPN. Cumulatively, our data suggest that the enhanced differentiation and resorptive ability of osteoclasts in response to Hh signaling initiated by breast cancer cells are due to the up-regulated OPN expression by the osteoclasts. OPN is vital to the osteoclast differentiation-associated expression of proteases and resorptive ability (supplemental Fig. 2C).
FIGURE 6.
The ability to up-regulate OPN in response to breast cancer cell-induced Hh signaling is vital to the osteoclasts to express cathepsin K and MMP9 and resorb bone matrix. Silencing the expression of endogenous OPN in osteoclasts decreases their ability to express cathepsin K and MMP9 in response to breast cancer cell-conditioned media. RAW254.7 cells were cultured under differentiating conditions for 6 days to allow for complete differentiation. Differentiation conditions included recombinant SHH (100 nm) or conditioned media from breast cancer cells (MDA-MB-231, SUM159, and SUM1315) with or without the 5E1 antibody (2.5 μg/ml). One set of osteoclasts was silenced on day 6 for OPN expression (KO) using shRNA targeting OPN cloned into pSuper (11). A and B, the expression of cathepsin K (CTSK) (A) and MMP9 (B) was assessed by real-time quantitative RT-PCR on day 7. The levels of gene expression are represented relative to the expression in DM alone. The extent of CTSK expressed by the groups silenced for OPN is significantly lower (*, p < 0.0001) when compared with their respective control (OPN-expressing) for all groups tested. For both breast cancer cell lines tested, the extent of CTSK expression in the 5E1/KO group is significantly lower (*, p < 0.0001) than the 5E1 alone supplemented group. Relative to the respective controls, the levels of MMP9 in the groups silenced for OPN is significantly lower (*, p < 0.0001) for all groups tested. Further, the extent of MMP9 expression in 231 + 5E1/KO is significantly lower (^, p = 0.0002) than the 231 + 5E1 group but not the 231/KO group. Similarly the 1315 + 5E1/KO group expresses significantly (p < 0.0001) lower levels of MMP9 relative to the 1315 + 5E1 group and the 1315/KO group. C, the resorption activity was assessed by conducting the differentiation as described above on OAAS plates followed by quantitation of the resorbed area as described above. The extent of resorption expressed by the groups silenced for OPN is significantly lower (*, p < 0.005) when compared with their respective control (OPN-expressing) for all groups tested. For both breast cancer cell lines tested, the extent of resorption in the 5E1/KO group is significantly lower (*, p < 0.05) than the 5E1 alone supplemented group. The difference between the 5E1/KO group was not statistically different relative to the corresponding KO group. D, images represent photomicrographs showing the areas resorbed by the differentiated osteoclasts in response to various differentiation conditions. The bar represents 100 μm. Panel a, DM; panel b, DM on KO osteoclasts; panel c, recombinant SHH (100 nm); panel d, recombinant SHH on KO osteoclasts; panels e and i, DM supplemented with conditioned medium from MDA-MB-231 cells and SUM1315 cells respectively; panels f and j, represent osteoclasts silenced for OPN expression on day 6 (KO); panels g and k represent differentiation conditions in the presence of the 5E1 antibody (2.5 μg/ml); and panels h and l represent osteoclasts cultured in the presence of 5E1 antibody that were silenced for OPN expression on day 6 (KO). (*, p < 0.05).
Hh Signaling in the Breast Cancer Cells Ameliorates Their Ability to Influence Osteoclast Differentiation and Activity
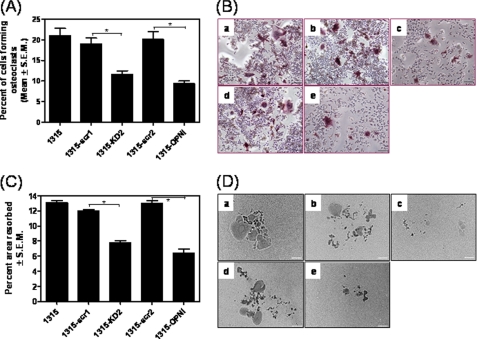
Although Hh ligands expressed by the breast cancer cells clearly play a role in influencing osteoclast differentiation and activity, we assessed the effect of SUM1315 cells that have been abrogated for the expression of the transcription factor, GLI1. As seen in Fig. 7, conditioned media from breast cancer cells silenced for GLI1 expression were deficient in inducing differentiation (p < 0.05) (Fig. 7A) and activity (p = 0.0001) (Fig. 7B) of osteoclasts. Abrogating GLI1 expression in the breast cancer results in a significant decrease (p < 0.05) in the expression of OPN and SHH (supplemental Fig. 2D).
FIGURE 7.
Hh signaling in breast cancer cells ameliorates their ability to induce osteoclast differentiation and activity. RAW264.7 cells were cultured for 6 days in the presence of double strength DM supplemented (1:1) with medium from SUM1315 breast cancer cells. Osteoclast differentiation was assessed by TRAP assay, and the activity was assessed by culturing the osteoclasts as described above, on OAAS plates. A and C, relative to untransfected SUM1315 cells and SUM1315 cells transfected with a scrambled control (scr1 (pSuperior.gfp+neo) and scr2 (pSuper)), the medium from the cells silenced for GLI1 expression (KD2) and OPN (OPNi) was significantly less efficient in inducing osteoclast differentiation (*, p = 0.012 and p = 0.0049, respectively) (A) and resorption activity (p = 0.0001 and p = 0.0005, respectively) (C). Images were acquired at 10× magnification using the Nikon Eclipse TS 100 microscope. B and D, images represent differentiation (B) and resorption (D). Differentiation and resorption conditions included medium from control SUM1315 cells (panel a) or cells transfected with vector control (scr1: pSuperior.egfp.neo) (panel b) or transfected with shRNA targeting GLI1 (panel c), transfected with pSuper vector control (scr2) (panel d), or transfected with shRNA targeting OPN (panel e).
OPN Expression by Breast Cancer Cells Enhances Osteoclast Differentiation and Activity
We have already determined that Hh ligands expressed by the breast cancer cells play a vital role in communicating with the osteoclasts. To directly determine the role of OPN expressed by the SUM1315 cells, we silenced the expression of OPN and assessed the effect of the conditioned media on osteoclast differentiation and activity (supplemental Fig. 2E shows the magnitude of OPN silencing). We see that abrogating OPN expression from the breast cancer cells significantly diminished their ability to influence osteoclast differentiation (p < 0.01) and activity (p = 0.0005) (Fig. 7, A–D).
DISCUSSION
Metastases in the bone occur in 60–80% of advanced breast cancer patients. The bone metastases in breast cancer are predominantly osteolytic, characterized by vigorous bone resorption (bone breakdown). The most common mode of transport of breast cancer cells from the breast to bone is through the vertebral venous system (31) that allows breast cancer cells to come into contact with the axial skeleton, including the ribs, spine, pelvis, and proximal humerus and femur, which is the main distribution of bone metastases in breast cancer patients (32). Once malignant cells have migrated to the bone, their ability to colonize is facilitated by various growth factors that are secreted by the bone. The cross-talk between tumor cells and the microenvironment promotes a vicious cycle of tumor growth and bone loss that perpetuates the formation of bony lesions. When the bone is lysed by osteoclasts, the factors released stimulate malignant tumor growth, which then increases the number of cells available to release the factors that stimulate osteoclastic activity so that more bone is resorbed and the cycle continues (33).
It is believed that factors such as MMPs, chemokine receptor 4 (CXCR4), VEGF, and connective tissue growth factor target metastatic tumor cells to bone and facilitate survival within the bone microenvironment (2, 34). Physical factors within the bone microenvironment, including hypoxia, acidic pH, and extracellular Ca2+, and bone-derived growth factors, such as TGF-β and insulin-like growth factors, activate tumor expression of osteoblast stimulatory factors, such as vascular endothelial growth factor (VEGF), platelet-derived growth factor (PDGF), and endothelin (ET-1) (35). Maturation of osteoblasts is coupled with their release of RANKL that can stimulate osteoclastogenesis (36). Breast cancer cells also express osteoclast stimulatory factors, such as PTHrP, TGF-β, and IL-11. In fact, expression of IL-11 and OPN by breast cancer cells has been found to be critical for the osteolytic activity of breast cancer cells (2).
The Hh signaling pathway involves the binding of an Hh ligand to the receptor PTCH, thereby relieving its inhibitory effect on SMOH, permitting transduction of the Hh signal to intracellular components, culminating in transcriptional activation of downstream genes such as GLI1. The Hh ligands were initially thought to be a transmembrane, non-diffusible signal for neighboring cells. Further research revealed that the Hh ligands are secreted after being post-translationally modified and participate in short and long range signaling (37). Our data show that Hh ligands expressed by breast cancer cells can initiate a cross-talk directly with osteoclasts and promote osteoclast differentiation (assessed by multinucleate cells showing TRAP activity) and resorption activity accompanied by increased expression of OPN, CTSK, and MMP9. TRAP, a glycosylated monomeric metalloenzyme, is highly expressed in osteoclasts and has been implicated in the detachment of cells necessary for initiating cell migration (38). It is up-regulated during osteoclastogenesis along with CTSK and as such used as a histochemical marker for differentiated osteoclasts (39). Multinucleation, an essential step in osteoclast differentiation, is a prerequisite for its efficient bone-resorbing ability. Mononuclear osteoclasts fuse repeatedly to form giant multinucleated osteoclasts, which after the polarization of the membrane and organization of the cytoskeleton result in a mature bone-resorbing osteoclast.
The bone-resorbing ability of mature osteoclast has been attributed to cysteine proteinase CTSK and a host of different matrix metalloproteinases, including MMP9, MMP13, and MMP14. CTSK with its ability to cleave the native helix of collagen at multiple sites has been implicated as the molecule for matrix solubilization (40), whereas collagenolysis-enhancing MMP9 has been found to be critical for osteoclast migration (41). Enhanced osteoclast differentiation and activation elicited by breast cancer cells were concomitant with significantly increased expression of OPN and the proteases CTSK and MMP9. Moreover, Hh ligands expressed by the breast cancer cells play a critical role in inducing changes in the osteoclasts because neutralizing the activity of these ligands from the conditioned medium of the breast cancer cells reduces the efficacy of the breast cancer cells to elicit osteoclast differentiation and resorptive activity. Hh ligands expressed by breast cancer cells are also essential for the production of OPN, CTSK, and MMP9 by the osteoclasts because squelching them with the 5E1 antibody resulted in a significant decrease in expression.
As such, our data show that OPN expression is up-regulated as a downstream event of Hh signaling initiated by the Hh ligands expressed by the breast cancer cells. OPN is particularly abundant at the attachment sites of osteoclasts and is essential for reorganization of the osteoclast cytoskeleton for osteoclast motility. It is not only responsible for activating the bone resorptive ability of osteoclasts but also for their migration (42). Further, OPN enhances the differentiation of pre-osteoclasts to osteoclasts, resulting in up-regulated expression of CTSK and MMP9. Consequently, resorptive activity of the osteoclasts is also negatively impacted by the inability to express OPN. Although Hh signaling in osteoclasts influences their activity, our data reveal that Hh signaling in breast cancer cells is also vital to their ability to elicit osteoclast activation. OPN expressed by breast cancer cells also enhances osteoclast activity. Overall, our study indicates a causal role for Hh signaling in promoting breast cancer cell-mediated osteolytic activity.
Hh signaling has been reported to critically influence osteoblast maturation and the expression of PTHRP leading to RANKL expression, both of which are essential for osteoclastogenesis (23). In osteoblasts, Hh signaling up-regulates expression of BMP2 and Runx2 (43, 44). PTHRP expressed by breast cancer cells also enhances osteolysis and hypercalcemia (45, 46). Hh signaling culminating in activation of the GLI2 transcription factor induces expression of PTHRP and osteolysis by breast cancer cells (47). The effect on osteoclasts has largely been understood to be mediated by osteoblasts. In this study, we show that the Hh ligands expressed by breast cancer cells act as conversational molecules and can directly mediate a paracrine cross-talk with osteoclast precursors leading to osteoclastogenesis and the induction of resorptive activity. Osteoclasts respond to the stimulus provided by breast cancer cells by activating Hh signaling and up-regulating OPN expression that is vital to their differentiation and resorptive activity. Thus, it is likely that the accumulation of OPN and PTHrP in the bone microenvironment in response to Hh signaling can potentially have a cumulative effect on the osteoclasts, resulting in their activation (Fig. 8). We have demonstrated that Hh signaling determines the potency of breast cancer cells to induce osteoclastogenesis and resorption. Further, inhibiting Hh signaling in osteoclasts resulted in significantly reduced osteolytic activity. Hh inhibitors are in clinical trials to test efficacy in combating several malignancies, including breast cancer. Our data demonstrate that these inhibitors can also have an impact on osteoclasts. Specifically, inhibiting the Hh signaling in pre-osteoclasts using cyclopamine hampered the ability of pre-osteoclasts to respond to the stimulatory effects of the breast cancer cells, indicating that Hh signaling is vital to osteoclast activity.
FIGURE 8.
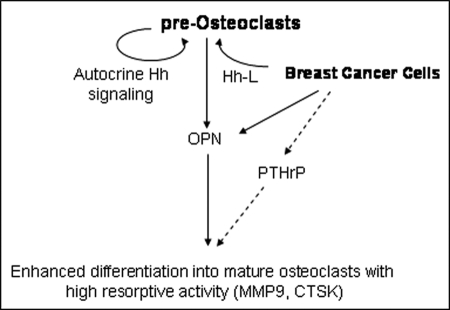
The schematic shows that breast cancer cells express Hh ligands (Hh-L) that activate Hh signaling in pre-osteoclasts. Breast cancer cells also express OPN that can initiate signaling in pre-osteoclasts. The pre-osteoclasts respond to Hh-L secreted by the breast cancer cells as well as autocrine Hh signaling by expressing OPN and differentiating into mature osteoclasts (characterized by expression of MMP9 and CTSK) with increased resorptive activity. Breast cancer cells also express PTHrP in response to Hh signaling (Sterling et al. (47)). Thus, overall, Hh signaling-mediated expression of factors such as OPN and PTHrP cumulatively results in enhanced differentiation and resorptive activity of osteoclasts. The dotted lines denote implications from previously published literature, whereas the solid lines depict data from the present work.
Supplementary Material
This work was supported, in whole or in part, by Department of Defense IDEA Award 07-01-0400 (to L. A. S.). This work was also supported by National Institutes of Health Grant 1R01CA140472-01A1 (to R. S. S.) and support from the Mitchell Cancer Institute, University of South Alabama.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- OPN
- osteopontin
- Hh
- Hedgehog
- IHH
- Indian hedgehog
- SHH
- Sonic hedgehog
- PTCH
- Patched
- SMOH
- Smoothened
- RANKL
- receptor activator of NF-κB ligand
- DM
- differentiation medium
- TRAP
- tartrate-resistant acid phosphatase
- CTSK
- cathepsin K
- PTHrP
- parathyroid hormone-related protein.
REFERENCES
- 1. Siclari V. A., Guise T. A., Chirgwin J. M. (2006) Cancer Metastasis Rev. 25, 621–633 [DOI] [PubMed] [Google Scholar]
- 2. Kang Y., Siegel P. M., Shu W., Drobnjak M., Kakonen S. M., Cordón-Cardo C., Guise T. A., Massagué J. (2003) Cancer Cell 3, 537–549 [DOI] [PubMed] [Google Scholar]
- 3. McKee M. D., Nanci A. (1996) Microsc. Res. Tech. 33, 141–164 [DOI] [PubMed] [Google Scholar]
- 4. Blair H. C., Robinson L. J., Zaidi M. (2005) Biochem. Biophys. Res. Commun. 328, 728–738 [DOI] [PubMed] [Google Scholar]
- 5. Yoneda T., Sasaki A., Mundy G. R. (1994) Breast Cancer Res. Treat 32, 73–84 [DOI] [PubMed] [Google Scholar]
- 6. Chellaiah M. A., Hruska K. A. (2003) Calcif. Tissue Int. 72, 197–205 [DOI] [PubMed] [Google Scholar]
- 7. Flores M. E., Norgård M., Heinegård D., Reinholt F. P., Andersson G. (1992) Exp. Cell Res. 201, 526–530 [DOI] [PubMed] [Google Scholar]
- 8. Carlinfante G., Vassiliou D., Svensson O., Wendel M., Heinegård D., Andersson G. (2003) Clin. Exp. Metastasis 20, 437–444 [DOI] [PubMed] [Google Scholar]
- 9. Bramwell V. H., Doig G. S., Tuck A. B., Wilson S. M., Tonkin K. S., Tomiak A., Perera F., Vandenberg T. A., Chambers A. F. (2006) Clin. Cancer Res. 12, 3337–3343 [DOI] [PubMed] [Google Scholar]
- 10. Standal T., Borset M., Sundan A. (2004) Exp. Oncol. 26, 179–184 [PubMed] [Google Scholar]
- 11. Shevde L. A., Samant R. S., Paik J. C., Metge B. J., Chambers A. F., Casey G., Frost A. R., Welch D. R. (2006) Clin. Exp. Metastasis 23, 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cook A. C., Chambers A. F., Turley E. A., Tuck A. B. (2006) J. Biol. Chem. 281, 24381–24389 [DOI] [PubMed] [Google Scholar]
- 13. Nemoto H., Rittling S. R., Yoshitake H., Furuya K., Amagasa T., Tsuji K., Nifuji A., Denhardt D. T., Noda M. (2001) J. Bone Miner Res. 16, 652–659 [DOI] [PubMed] [Google Scholar]
- 14. Chellaiah M. A., Kizer N., Biswas R., Alvarez U., Strauss-Schoenberger J., Rifas L., Rittling S. R., Denhardt D. T., Hruska K. A. (2003) Mol. Biol. Cell 14, 173–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shijubo N., Uede T., Kon S., Nagata M., Abe S. (2000) Crit. Rev. Oncog. 11, 135–146 [PubMed] [Google Scholar]
- 16. Das S., Harris L. G., Metge B. J., Liu S., Riker A. I., Samant R. S., Shevde L. A. (2009) J. Biol. Chem. 284, 22888–22897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kasper M., Jaks V., Fiaschi M., Toftgård R. (2009) Carcinogenesis 30, 903–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kubo M., Nakamura M., Tasaki A., Yamanaka N., Nakashima H., Nomura M., Kuroki S., Katano M. (2004) Cancer Res. 64, 6071–6074 [DOI] [PubMed] [Google Scholar]
- 19. Stecca B., Mas C., Ruiz i Altaba A. (2005) Trends Mol. Med. 11, 199–203 [DOI] [PubMed] [Google Scholar]
- 20. DiMeo T. A., Anderson K., Phadke P., Fan C., Perou C. M., Naber S., Kuperwasser C. (2009) Cancer Res. 69, 5364–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matsubara T., Ikeda F., Hata K., Nakanishi M., Okada M., Yasuda H., Nishimura R., Yoneda T. (2010) J. Bone Miner Res. 25, 1068–1076 [DOI] [PubMed] [Google Scholar]
- 22. Bailey J. M., Mohr A. M., Hollingsworth M. A. (2009) Oncogene 28, 3513–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mak K. K., Bi Y., Wan C., Chuang P. T., Clemens T., Young M., Yang Y. (2008) Dev. Cell 14, 674–688 [DOI] [PubMed] [Google Scholar]
- 24. Sugatani T., Alvarez U., Hruska K. A. (2003) J. Biol. Chem. 278, 5001–5008 [DOI] [PubMed] [Google Scholar]
- 25. Terai K., Takano-Yamamoto T., Ohba Y., Hiura K., Sugimoto M., Sato M., Kawahata H., Inaguma N., Kitamura Y., Nomura S. (1999) J. Bone Miner Res. 14, 839–849 [DOI] [PubMed] [Google Scholar]
- 26. Kingsley L. A., Fournier P. G., Chirgwin J. M., Guise T. A. (2007) Mol. Cancer Ther. 6, 2609–2617 [DOI] [PubMed] [Google Scholar]
- 27. Bendre M., Gaddy D., Nicholas R. W., Suva L. J. (2003) Clin. Orthop. Relat Res. (suppl.) S39–S45 [DOI] [PubMed] [Google Scholar]
- 28. Dillon R., Gadgil C., Othmer H. G. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10152–10157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shkumatava A., Fischer S., Müller F., Strahle U., Neumann C. J. (2004) Development 131, 3849–3858 [DOI] [PubMed] [Google Scholar]
- 30. Maun H. R., Wen X., Lingel A., de Sauvage F. J., Lazarus R. A., Scales S. J., Hymowitz S. G. (2010) J. Biol. Chem. 285, 26570–26580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harada M., Shimizu A., Nakamura Y., Nemoto R. (1992) Adv. Exp. Med. Biol. 324, 83–92 [DOI] [PubMed] [Google Scholar]
- 32. Abeloff M. D., Armitage J. O., Lichter A. S., Niederhuber J. E. (eds) (1995) Clinical Oncology, Churchill Livingstone, Napeville, IL [Google Scholar]
- 33. Mundy G. R. (2002) Nat. Rev. Cancer 2, 584–593 [DOI] [PubMed] [Google Scholar]
- 34. Guise T. A., Mohammad K. S., Clines G., Stebbins E. G., Wong D. H., Higgins L. S., Vessella R., Corey E., Padalecki S., Suva L., Chirgwin J. M. (2006) Clin. Cancer Res. 12, 6213s-6216s [DOI] [PubMed] [Google Scholar]
- 35. Yin J. J., Mohammad K. S., Käkönen S. M., Harris S., Wu-Wong J. R., Wessale J. L., Padley R. J., Garrett I. R., Chirgwin J. M., Guise T. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10954–10959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thomas R. J., Guise T. A., Yin J. J., Elliott J., Horwood N. J., Martin T. J., Gillespie M. T. (1999) Endocrinology 140, 4451–4458 [DOI] [PubMed] [Google Scholar]
- 37. Goetz J. A., Singh S., Suber L. M., Kull F. J., Robbins D. J. (2006) J. Biol. Chem. 281, 4087–4093 [DOI] [PubMed] [Google Scholar]
- 38. Ek-Rylander B., Andersson G. (2010) Exp. Cell Res. 316, 443–451 [DOI] [PubMed] [Google Scholar]
- 39. Czupalla C., Mansukoski H., Pursche T., Krause E., Hoflack B. (2005) Proteomics 5, 3868–3875 [DOI] [PubMed] [Google Scholar]
- 40. Garnero P., Borel O., Byrjalsen I., Ferreras M., Drake F. H., McQueney M. S., Foged N. T., Delmas P. D., Delaissé J. M. (1998) J. Biol. Chem. 273, 32347–32352 [DOI] [PubMed] [Google Scholar]
- 41. Delaissé J. M., Andersen T. L., Engsig M. T., Henriksen K., Troen T., Blavier L. (2003) Microsc. Res. Tech. 61, 504–513 [DOI] [PubMed] [Google Scholar]
- 42. Rittling S. R., Matsumoto H. N., McKee M. D., Nanci A., An X. R., Novick K. E., Kowalski A. J., Noda M., Denhardt D. T. (1998) J. Bone Miner Res. 13, 1101–1111 [DOI] [PubMed] [Google Scholar]
- 43. Zhao M., Qiao M., Harris S. E., Chen D., Oyajobi B. O., Mundy G. R. (2006) Mol. Cell Biol. 26, 6197–6208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shimoyama A., Wada M., Ikeda F., Hata K., Matsubara T., Nifuji A., Noda M., Amano K., Yamaguchi A., Nishimura R., Yoneda T. (2007) Mol. Biol. Cell 18, 2411–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wysolmerski J. J., Dann P. R., Zelazny E., Dunbar M. E., Insogna K. L., Guise T. A., Perkins A. S. (2002) J. Bone Miner Res. 17, 1164–1170 [DOI] [PubMed] [Google Scholar]
- 46. Guise T. A., Yin J. J., Thomas R. J., Dallas M., Cui Y., Gillespie M. T. (2002) Bone 30, 670–676 [DOI] [PubMed] [Google Scholar]
- 47. Sterling J. A., Oyajobi B. O., Grubbs B., Padalecki S. S., Munoz S. A., Gupta A., Story B., Zhao M., Mundy G. R. (2006) Cancer Res. 66, 7548–7553 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.