Abstract
Glucocorticoids rapidly and robustly induce cell fate decisions in various multipotent cells, although the precise mechanisms of these important cellular events are not understood. Here we showed that glucocorticoids repressed Per3 expression and that this repression was critical for advancing mesenchymal stem cells to the adipocyte fate. Exogenous expression of Per3 inhibited adipogenesis, whereas knocking out Per3 enhanced that fate. Moreover, we found that PER3 formed a complex with PPARγ and inhibited PPARγ-mediated transcriptional activation via Pparγ response elements. Consistent with these findings, Per3 knock-out mice displayed alterations in body composition, with both increased adipose and decreased muscle tissue compared with wild-type mice. Our findings identify Per3 as potent mediator of cell fate that functions by altering the transcriptional activity of PPARγ.
Keywords: Adipocyte, Cell Differentiation, Stem Cell, Steroid Hormone, Steroid Hormone Receptor, GR, Per3, Circadian Rhythm
Introduction
Glucocorticoids play a critical role in inducing the adipocyte cell fate and differentiation both in vivo and ex vivo (1–3). This regulatory process may be relevant to normal physiology and to disease states, as patients exposed chronically to excess glucocorticoids develop increased adiposity (4). Thus, glucocorticoids together with their cognate glucocorticoid receptor protein serve as powerful biological probes of the molecular pathways that govern this subset of cell fate and differentiation decisions. Studies of glucocorticoid receptor action in physiologic processes might eventually advance development of novel therapeutic agents for a broad array of diseases, including those that accompany obesity, such as diabetes and metabolic syndrome (insulin resistance, hypertension, dyslipidemia) (5). Therefore, to discern molecular mechanisms driving adipocyte fate and differentiation, we sought to identify glucocorticoid-regulated target genes whose products participate directly in these decisions and to characterize their activities.
Mesenchymal stem cells, also called mesenchymal stem cells (MSCs),6 are progenitor cells that reside in the bone marrow and vascular wall and are capable of differentiating into cells that form cartilage, muscle, bone, or fat (6–8). MSCs are readily isolated and maintain their multipotency when purified ex vivo in tissue culture (9). Induction of MSC fate determination into distinct lineages can be initiated with defined reagents and monitored at the molecular level. In particular, glucocorticoids potently induce the adipocyte cell fate in these cells, thus, providing a well defined experimental starting point for analyses of mammalian cell fate decisions (10, 11) and for identifying mechanisms by which glucocorticoids control these decisions.
As in other tissues (12–16), glucocorticoids modulate in primary MSCs the expression of various genes that control circadian rhythm (17). Interestingly, clock components affect adipogenesis (18, 19), although the mechanism of this connection is poorly understood. In view of these findings, we were curious to determine whether clock components play functional roles in the glucocorticoid cell fate decision that commits MSCs to adipogenesis.
Here we show that the Period 3 gene (Per3), previously known for a minor role in modulating circadian rhythms, is regulated by glucocorticoids in primary MSCs and is a potent inhibitor of the adipocyte cell fate decision. We demonstrate that this function is achieved through interactions with PPARγ and the formation of a transcriptional regulatory complex. Consistent with these results in isolated cells, our in vivo studies demonstrate that Per3 knock-out mice display altered body composition (increased adipose and decreased muscle) as well as glucose intolerance.
EXPERIMENTAL PROCEDURES
Cell Culture
Mouse MSCs were harvested from the femurs and tibias of mice between 2 and 3 months of age. The ends of the bone were cut off, and the bone marrow was ejected by inserting a 23-gauge needle into the bone marrow cavity and flushing out the marrow with ∼2 ml of Iscove's media with 2% fetal bovine serum per bone. Flushed bone marrow was filtered and placed in culture flasks with mesenchymal stem cell enrichment media (Stem Cell Technologies). Media was changed every day for 3 days to remove the hematopoietic/non-adherent cells and then every other day thereafter. When large circular colonies of MSCs grew in flasks, the cells were trypsinized and passaged to enrich for MSCs. 3T3-L1 cells were purchased from ATCC and cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% calf serum (Invitrogen).
Quantitative RT-PCR
RNA was isolated from cells using RNeasy mini kit (Qiagen). Equal amounts of RNA were reverse-transcribed (∼500 ng of RNA for a 20-μl reverse transcription reaction) using random priming, and the relative transcript level was measured by quantitative PCR using a 7300 Real Time PCR system (Applied Biosystems) or a CFX96 system (Bio-Rad). Primer sequences are available in supplemental Table S1.
Adipogenesis Assay
MSCs were seeded into 12-well plates (Falcon) and allowed to grow to confluence. Cells were maintained at confluence for 2 days and then induced with the glucocorticoid mixture (1 μm dexamethasone, 5 μg/ml insulin, 0.115 mg/ml 3-isobutyl-1-methyl-xanthine) and 25 μm indomethacin) for 3 days followed by 1 day in basal media and then a second induction with glucocorticoid mixture. Thereafter, cells were maintained in basal media, which was changed every other day. Adipogenesis was allowed to continue for a total of 2 weeks. After 2 weeks of culture, cells were photographed under a phase microscope (Nikon), and RNA was extracted as described above.
3T3-L1 cells were passaged fewer than 10 times before experiments. For differentiation, cells were grown to confluence and then switched to DMEM with 10% fetal bovine serum with a glucocorticoid mixture (described above) for 2 days. Cells were then cultured in DMEM with 10% FBS for 6 more days.
Circadian Rhythm Experiments
MSCs were seeded in 12-well plates (Falcon). After cells grew to confluence, media were replaced with mesenchymal stem cell enrichment media (Stem Cell Technologies) containing either vehicle (DMSO) or 1 μm dexamethasone (Sigma) (which has previously been shown to synchronize the clock in confluent cell culture by bringing all cells into the same phase (20)). Cells were harvested at each time point, and total RNA was isolated using RNeasy mini kit (Qiagen).
Chromatin Immunoprecipitation
ChIP from 3T3-L1 cells was performed according to So et al. (17). 12 μg of Per3 antibody (Alpha Diagnostic) or PPARγ antibody (Santa Cruz Biotechnology) or rabbit IgG control (Sigma) were conjugated to 50 μl of Dynabeads (Invitrogen) for each ChIP sample. ChiP-quantitative PCR (qPCR) signals were calculated as percent of input using the formula 100 × 29(adjusted Ct of input − Ct of ChIP)9 to compare with the IgG control and using the ΔΔCt method to compare with other DNA elements. Sequential ChIPs were performed on 3T3-L1 cells grown in 5 × 15-cm diameter dishes and exposed to glucocorticoid mixture for 12 h.
Luciferase Assays
COS-7 cells were grown in 24-well plates (Falcon) to 95% confluence. Cells were transfected with a 3X-PPRE-luc reporter construct (Addgene, submitted by Bruce Spiegelman) as well as pcDNA3.1 expression plasmids with Per3 and PPARγ or an empty pcDNA3.1 control vector. A co-transfected Renilla expression plasmid was used for normalization. 36 h after transfection, cells were lysed, and dual luciferase assays (Promega) were performed.
Immunoprecipitation
COS-7 cells were grown to confluence and transfected with Pparγ (pCMX backbone) or Per3 (pEF6/V5-His backbone) using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Forty-eight hours post-transfection, cells were lysed with modified radioimmune precipitation assay buffer (50 mm Hepes, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 0.1% SDS, 20 mm EDTA). Anti-PPARγ antibody (H-100, Santa Cruz) or control IgG (10 μg each) was pre-bound to Dynabeads (Invitrogen), and proteins were immunoprecipitated according to the manufacturer's instructions. Immunocomplexes were eluted in sample buffer and analyzed by Western blotting. PER3 was detected using an anti-V5 antibody (Invitrogen) followed by goat anti-mouse HRP (Santa Cruz) and PPARγ using the H-100 antibody (Santa Cruz) followed by Clean-Blot IP Detection Reagent-HRP (Thermo Scientific).
For assays using in vitro transcribed and translated protein, PER3 was produced and labeled with [35S]methionine, and PPARγ was generated using unlabeled methionine in vitro using the TNT T7 Quick coupled transcription/translation system (Promega). Reactions were then mixed with binding buffer (20 mm Hepes, pH 8.0, 300 mm KCl, 2.5 mm MgCl2, 0.05% Nonidet P-40, 1 mm DTT, 10% glycerol) overnight at 4 °C under gentle agitation. Protein complexes were then precipitated as described above. Proteins were resolved by SDS-PAGE and transferred to PVDF membranes followed by autoradiography. PPARγ was detected by Western blot.
GST Pulldown Assays
The N-terminal region of Per3 was amplified by PCR and cloned into the EcoRI and XhoI sites of pGEX5 to generate GST fusion proteins. Expression plasmids were grown in Escherichia coli and induced with isopropyl 1-thio-β-d-galactopyranoside. Recombinant 35S-labeled PPARγ, RXRα and luciferase proteins were generated as above. Pulldown assays were carried out using the MagneGST Pull-Down System (Promega) according to the manufacturer's instructions, except that binding/wash buffer was supplemented with 0.05% Nonidet P-40 and 200 mm NaCl.
Animals
The Per3 knock-out allele was made congenic on a C57Bl6/2J background by greater than 10 backcrosses to this strain. Animals were maintained in 12-h light/12-h dark cycles. All animal studies were approved by the Institutional Animal Care and Use Committees.
Glucose Tolerance Testing
Per3−/− (n = 8) and wild-type (n = 8) mice were fasted for 15 h (overnight). Fasting blood sugar was measured using a One Touch Ultra 2 glucometer (Johnson & Johnson) with blood from a nicked tail vein. Two mg/kg of glucose was injected into the peritoneal cavity of each animal. Blood sugar was measured at times 0, 15, 30, 60, and 120 min using the glucometer.
Body Composition
Similar age male C57Bl6/2J mice (wild type (n = 7) and Per3−/− (n = 7)) were fasted for 4 h and anesthetized with isoflurane before scanning. The lean, fat, and total masses were measured using dual energy x-ray absorptiometry (Lunar PIXImus2). The percentage of lean and fat in mice was calculated as the corresponding tissue mass divided by the total animal mass.
RESULTS
Glucocorticoids Regulate Per3 in Primary MSCs
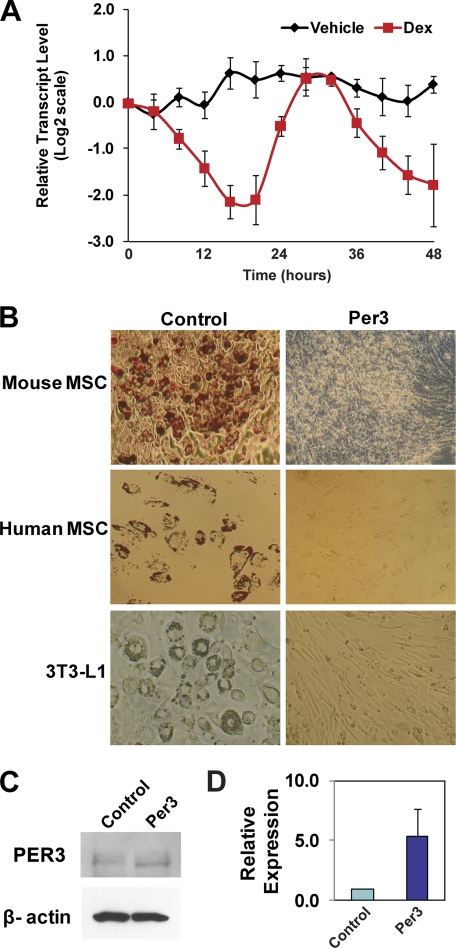
Period 3 (Per3) has been classified as a circadian rhythm gene. However, the function of Per3 is poorly understood, and knock-out mice have been reported to have only subtle alterations in circadian rhythmicity (21). In a previous microarray study performed in the C3H10T1/2 cell line, which possesses some MSC-like properties, we found that Per3 is differentially expressed in cells exposed to glucocorticoids compared with control (22). As glucocorticoids are essential for selecting the adipocyte cell fate in MSCs, we hypothesized that Per3 plays a role in mediating the cell fate actions of glucocorticoids. To begin to assess the significance of the Per3 response to glucocorticoids, we performed time-course qPCR expression studies in primary mouse MSCs. We found that the synthetic glucocorticoid dexamethasone triggers a circadian reduction of Per3 transcript levels in primary mouse MSCs (Fig. 1A).
FIGURE 1.
Per3 is regulated by glucocorticoids and blocks the adipocyte fate. A, dexamethasone (Dex) induces a cycling repression of Per3 transcript levels in MSCs. Real time quantitative PCR of Per3 mRNA levels during a 48-h time course after exposure of mouse MSCs to dexamethasone or vehicle control (DMSO) was performed. Three independent experiments with MSCs isolated from different mice were performed. Error bars represent S.D. between experiments. B, overexpression of Per3 blocks the adipocyte fate. Cells overexpressing Per3 were compared with cells containing control empty plasmid after induction with the glucocorticoid mixture. Adipocytes in human and mouse MSCs were detected by staining the cells with Oil Red O, a dye that stains neutral lipids red. Adipocytes in 3T3-L1 cultures were easily visualized without Oil Red O staining. Overexpression of Per3 blocked adipocyte formation in mouse MSCs, human MSCs, and 3T3-L1 cells. C, cells transfected with the Per3 expression construct overexpress PER3. Western blots, using anti-PER3 and anti-β-actin antibodies, on 3T3-L1 cells transfected with the Per3 expression construct are compared with control. D, shown are real time quantitative PCR data of relative Per3 transcript levels in 3T3-L1 cells transfected with the Per3 expression construct compared with control. Error bars represent S.D. between experiments.
PER3 Regulates the Adipocyte Fate Decision
Knock-out of Per3 in mice has a minimal effect on circadian rhythmicity (21). However, because of previous indications of a link between circadian clock components and adipogenesis and because glucocorticoids are essential for selecting the adipocyte fate in MSCs, we tested whether Per3 might control a cell fate decision in MSCs despite little effect on circadian rhythm. Indeed, we found that constitutive overexpression of PER3 in transfected primary mouse and human MSCs dramatically blocked adipogenesis, whereas control cells maintained competency (Fig. 1B).
Adipocyte formation was visualized under phase microscopy by the appearance of large rounded cells, each containing lipid droplets, which is the classic, unique morphology of an adipocyte. Confirmation of the presence of lipid droplets was obtained by staining the cells with Oil Red O, which stains neutral lipids (Fig. 1B, upper and middle panels). In a process that may be related to their effect on MSCs, glucocorticoids are also an essential component of a mixture of factors that induces adipogenesis in the 3T3-L1 cell line. Therefore, we also tested the effect of constitutive PER3 overexpression on dexamethasone mixture-induced adipogenesis in 3T3-L1. Again, PER3 robustly blocked adipogenesis (Fig. 1B, bottom panels). Thus, enforced PER3 expression inhibits adipocyte formation, suggesting that down-regulation of PER3 expression may be an essential step in the adipocyte cell fate decision.
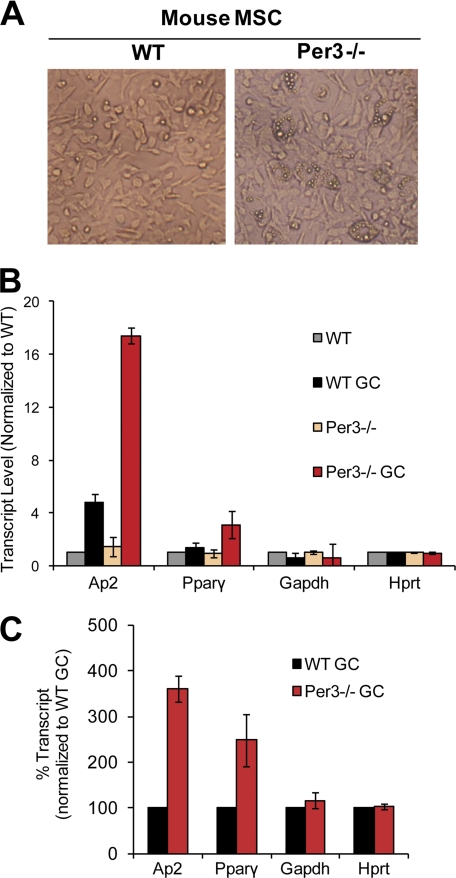
Given the efficacy of PER3 in blocking adipogenesis, we speculated that decreasing PER3 expression might result in an enhancement of the adipocyte fate decision. Indeed, 2 weeks post-induction with glucocorticoid mixture, primary MSCs isolated from Per3 knock-out (Per3−/−) mice had a significantly higher level of adipogenesis than MSCs from wild-type mice (Fig. 2A). As an objective measure of adipogenesis in these assays, we assessed the expression levels of the adipocyte markers aP2 and Pparγ and found that Per3−/− cells had ∼4-fold higher transcript levels of the adipocyte marker aP2 and over 2-fold higher Pparγ levels than wild-type MSCs (Fig. 2, B and C), whereas expression of non-adipocyte control genes was unaffected. These results indicate that endogenous Per3 inhibits the adipocyte fate decision in MSCs and that elimination of Per3 expression enhances adipogenesis.
FIGURE 2.
Disruption of Per3 increases adipogenesis in mesenchymal stem cells. A, adipogenesis was induced in MSCs with the glucocorticoid mixture. Under phase microscopy adipocytes appear as large cells with small intracellular reflective lipid droplets. B, the enhanced adipogenesis of Per3−/− MSCs was confirmed using quantitative real time RT-PCR measuring expression levels of adipocyte markers aP2 and PPARγ. GAPDH and HPRT were used as control genes that do not change during adipogenesis. C, relative transcript levels of marker and control genes were normalized to WT control (vehicle treatment) or WT glucocorticoid mixture-treated (GC). Three independent experiments were performed. Error bars represent S.D. between experiments.
Because certain core clock components other than Per3 are known to impact adipogenesis in murine cell lines (18, 19) and in mice (23–25), perhaps the effect of Per3 on MSC cell fate is secondary to its actions on those other clock components. We, therefore, tested whether Per3 deletion in MSCs produces a global disruption of the greater circadian clock. To this end we synchronized the circadian clock in wild-type and Per3−/− MSCs with glucocorticoids. We then measured the expression levels of the core clock components Cry1 and Rev-erbα at multiple time points over 48 h. We found that the expression levels of these core clock components were indistinguishable between wild-type and Per3−/− MSCs (data not shown). Thus, Per3 deletion increases adipogenesis in MSCs without dramatically disturbing the core clock, implying that the cell fate regulation function of Per3 could be “off pathway” and uncoupled from its role in the circadian clock. In any case, it is apparent that the Per3 effect on cell fate is not merely secondary to an effect on circadian cycling.
PER3 Interacts with PPARγ and Inhibits Transcription
PPARγ is a nuclear receptor that is both necessary and sufficient for adipocyte fate determination (26). Given our finding that PER3 potently blocks this fate decision (Fig. 1B), we speculated that PER3 might directly affect PPARγ activity, perhaps via protein-protein interaction; such an interaction would parallel the finding that the PER2 protein interacts with nuclear receptors PPARα and REV-ERBα and serves as a coregulator (27).
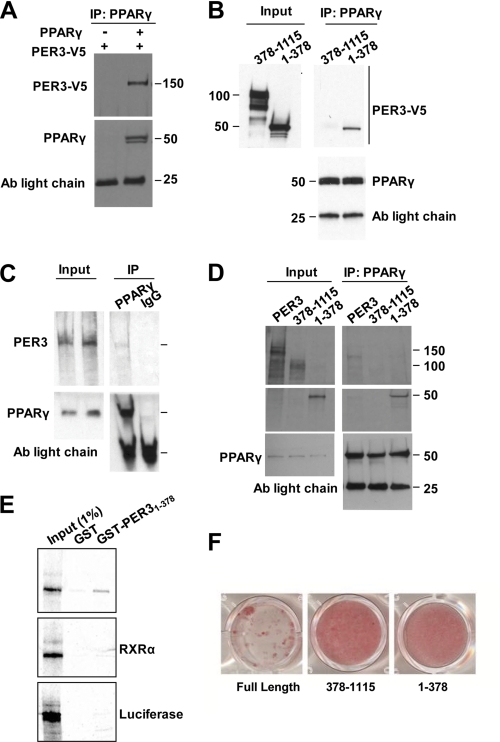
To examine the relationship between PER3 and PPARγ, we first tested whether they could interact in cell extracts. Co-immunoprecipitation (co-IP) assays were performed on cell lysates from COS-7 cells overexpressing PER3 and PPARγ. Indeed, we found that PER3 formed a complex with PPARγ (Fig. 3A). PER3 contains conserved Per-ARNT-SIM (PAS) protein-protein interaction motifs at the N-terminal region (28). Therefore, we co-expressed amino acids 1–378 of PER3 with PPARγ to test if the interaction specifically mapped to this region. We found that this region of PER3 did co-IP with PPARγ, whereas a truncated PER3 protein with this region deleted failed to precipitate (Fig. 3B).
FIGURE 3.
Per3 interacts with PPARγ. A, immunoprecipitation with anti- PPARγ antibody (Ab) from COS-7 cells expressing exogenous PER3-V5 with and without PPARγ followed by Western blots with anti-V5 and anti-PPARγ is shown. B, immunoprecipitation from COS-7 cells co-expressing PPARγ and either the PER3 N-terminal fragment or PER3 with the N terminus deleted with anti-PPARγ antibody followed by Western blots using anti-V5 or anti-PPARγ. C, immunoprecipitation with anti-PPARγ or IgG control antibody of recombinant PPARγ and 35S-labeled recombinant PER3. Input lanes showing PER3 were used to verify that co-precipitated PER3 band is at the correct molecular size. Membranes were probed with anti-PPARγ to confirm the presence or absence of PPARγ. D, immunoprecipitation with anti-PPARγ antibody of recombinant PPARγ and [35S]Met-labeled recombinant full-length PER3 or PER3 deletion mutants. Membranes were probed with anti-PPARγ to confirm the presence or absence of PPARγ. Antibody light chain bands are shown as loading controls. E, a GST fusion with the N-terminal PER3 region was expressed in E. coli, and recombinant [35S]Met-labeled PPARγ was recovered from bacterial lysate in a GST pulldown assay. GST alone did not pull down PPARγ from the lysate. GST-PER3 did not pull down recombinant [35S]Met-labeled RXRα or luciferase. Input controls served as molecular size markers for the recombinant proteins. F, confluent 3T3-L1 cells expressing full-length or fragments of Per3 were induced to undergo adipogenesis with the glucocorticoid mixture and cultured for 6 days. After 6 days, cells were fixed and stained with Oil Red O to identify adipocytes.
We then tested the interaction between PER3 and PPARγ recombinant proteins, generated from in vitro transcription and translation. We confirmed that the recombinant proteins co-IP with cognate but not control IgG antibodies (Fig. 3C). As with the co-IP in cells, the recombinant N-terminal region of PER3 specifically precipitated with PPARγ, whereas PER3 with this region deleted failed to co-IP (Fig. 3D).
To test this interaction with a third approach, we generated a GST fusion with amino acids 1–378 of PER3 and expressed this fusion protein in E. coli. This PER3 fusion protein recovered recombinant PPARγ in a GST pulldown assay, whereas GST alone did not pull down PPARγ (Fig. 3E). As specificity controls, we also tested if the PER3 fusion protein would interact with the nuclear receptor RXRα or the presumably unrelated luciferase protein. PER3 failed to pull down either of these proteins, demonstrating the specificity of the PPARγ-PER3 interaction (Fig. 3E).
To investigate if the domain that interacts with PPARγ is necessary for the adipocyte fate regulatory activity of PER3, we compared the activity of a mutant lacking this domain to full-length PER3 in a 3T3-L1 adipogenesis assay. We found that this domain was necessary for PER3 to block adipogenesis (Fig. 3F). However, expression of the interaction domain alone was not sufficient to block adipogenesis (Fig. 3F), indicating that other parts of the PER3 protein participate in coordinating the activity upon PER3 binding to PPARγ.
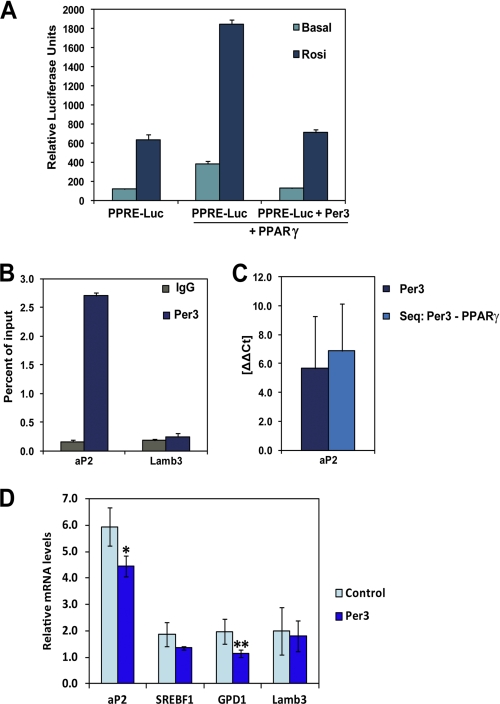
PPARγ is a nuclear receptor that binds to well characterized response elements (PPREs) and modulates transcription (29, 30). To test whether PER3 modulates PPARγ transcriptional regulatory activity, we utilized a reporter system with three tandem PPARγ DNA binding motifs upstream of a minimal promoter. As expected and previously described (31), luciferase reporter activity was induced by PPARγ expression (Fig. 4A). Importantly, co-expression of PER3 with PPARγ blocked the induction of the reporter activity both in the basal state and after stimulation with the PPARγ ligand rosiglitazone (Fig. 4A). Thus, PER3 can inhibit PPARγ activity in this context, consistent with the view that PER3 might function by this mechanism to block adipocyte cell fate and differentiation.
FIGURE 4.
Per3 inhibits PPARγ activity. A, COS-7 cells were transfected with a luciferase reporter gene containing three tandem PPARγ DNA binding motifs (PPRE-luc) with and without 100 nm rosiglitazone. Co-transfection with Pparγ induced reporter gene expression. The induction of reporter activity was significantly inhibited by the addition of Per3. Luciferase activity was internally normalized against the measured activity of a co-transfected Renilla expression vector. Three independent experiments were performed. Error bars represent S.D. between experiments. B, ChIP-qPCR of PER3 revealed enrichment near an endogenous PPRE in the enhancer region located −5.4 kb from the aP2 transcriptional start site compared with a control ChIP-qPCR of this region using a nonspecific IgG antibody. There was no significant enrichment of PER3 at another PPARγ binding site located within the Lamb3 gene (+7.6 kb from the transcriptional start site). The Per3 ChIP was performed three times, and error bars represent S.D. C, single ChIP of Per3 and sequential ChIP of Per3 followed by PPARγ was performed, and relative enrichment at the PPRE in aP2 was compared with the PPARγ binding site in Lamb3. After the sequential ChIP, qPCR of the Lamb3 site approximated the water control, whereas the aP2 site had significant enrichment indicating selective co-occupancy. D, transcript levels of PPARγ target genes in 3T3-L1 cells were measured with qPCR after 24 h of treatment with 100 nm rosiglitazone. Overexpression of Per3 resulted in significant decreased aP2 and Gpd1 transcript levels. Results were internally normalized to β-actin levels within each sample. Between three and five experiments were performed for each condition, and error bars represent S.E. *, p = 0.01; **, p = 0.03.
PER3 Is Enriched at an Endogenous PPARγ Response Element and Modulates Transcription of aP2
We next sought to test whether PER3 could inhibit PPARγ-regulated transcription at an endogenous PPARγ target gene. The adipocyte fatty acid-binding protein gene (aP2, also called Fabp4) was selected for these studies because it is regulated by endogenous levels of PPARγ in 3T3-L1 preadipocytes (30) and is induced by PPARγ during adipogenesis (32). Furthermore, the PPRE regulating aP2 expression has been mapped and characterized (33), and we were able to confirm enrichment of PPARγ at this site in preadipocytes (data not shown). Therefore, we performed chromatin immunoprecipitation of PER3 in 3T3-L1 cells followed by quantitative PCR (ChIP-qPCR) in the region of this PPRE and found a significant enrichment of PER3 around the aP2 PPRE (Fig. 4B). We also assessed PER3 occupancy in the region of a PPARγ binding site identified by Lefterova et al. (29) (PPRE_472) that is located within the third intron of the Lamb3 gene but failed to detect a significant enrichment of PER3 near that site (Fig. 4B), suggesting that PER3 associates with select PPARγ binding sites. To investigate if PER3 and PPARγ co-occupy the aP2 PPRE, we performed a sequential ChIP experiment with PER3 followed by PPARγ. We detected significant co-enrichment of PER3 and PPARγ at the aP2 PPRE compared with trivial levels at the site in Lamb3 (Fig. 4C), indicating that PER3 and PPARγ form a complex at select PPARγ DNA binding sites.
We next tested the functional implications of PER3 enrichment at an endogenous PPRE in the regulation of aP2 expression. Using qRT-PCR, we measured endogenous transcript levels of aP2 in 3T3-L1 cells with overexpression of Per3. We found that overexpression of Per3 inhibited aP2 expression compared with control (Fig. 4D). In addition, we found that shRNA-mediated knockdown of Per3 resulted in the opposite trend, with increased levels of aP2 expression compared with the scramble shRNA control (data not shown). On the other hand, Lamb3 expression levels were not significantly altered by either overexpression or shRNA knockdown of Per3 (Fig. 4D and data not shown). We also measured the expression levels of two other known direct PPARγ target genes and found that Per3 statistically significantly inhibited one (Gpd1) but not the other (Srebf1) (Fig. 4D). Together, these results indicate that Per3 functions as an inhibitor of PPARγ activity at select endogenous response elements.
Per3 Knock-out Mice Have Evidence of Altered Cell Fate in Vivo
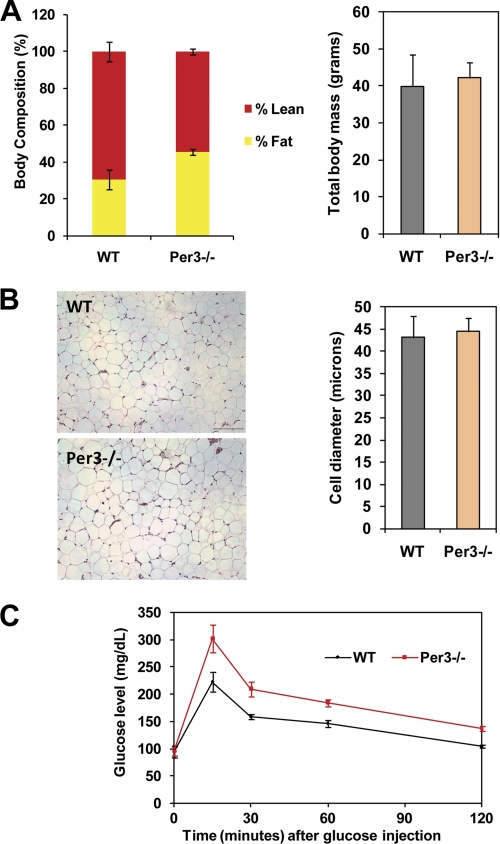
To investigate whether these results have physiological relevance, we then studied the systemic implications of deleting Per3. As mentioned, prior evaluations of Per3−/− mice had identified a slight shortening of the circadian cycle length without change in activity level (21, 34). Our findings, that Per3 functions as a regulator of cell fate prompted us to investigate the impact of deleting Per3 on overall body composition. Therefore, we placed Per3−/− and wild-type littermate mice on a 9% fat diet and monitored total body composition using dual energy x-ray absorptiometry scanning, which quantifies both lean and fat mass (35). Others have shown that Per3−/− mice do not have altered food intake (36). However, although we found that whereas the total body weights of the Per3−/− mice were similar to their wild-type littermates, the body composition of the animals was strikingly distinct (Fig. 5A). Whereas 45% of the total body mass of the Per3−/− animals was adipose tissue, only 30% was present in the wild-type controls. Interestingly, the adipocyte size was similar across the genotypes (Fig. 5B), suggesting that the increased mass was most likely caused by an increased number of adipocytes. Per3−/− mice had only 55% lean mass, whereas wild-type mice were composed of 70% lean mass (Fig. 5A). This demonstrates that not only do Per3−/− mice have increased total adipose tissue, they also have a concomitant decrease in their lean mass. Lean mass is primarily composed of muscle. Therefore, we conclude that this phenotype is consistent with alterations in stem cell fate decisions occurring in vivo.
FIGURE 5.
Per3−/− mice have altered body composition and glucose intolerance. A, total body mass and percent lean and fat mass were quantified using dual energy x-ray absorptiometry scanning (n = 7 wild-type and n = 7 Per3−/−). Per3−/− animals had higher percent body fat (p = 0.02) and lower percent lean mass (p = 0.02) compared with wild-type animals (error bars = S.D). B, histological slides of gonadal fat depots from wild-type and Per3−/− mice were prepared, and the adipocyte size was measured using MetaMorph software (Molecular Devices) from 3 mice of each genotype. Error bars represent S.E. C, after fasting for 15 h, mice (n = 8 wild type and 8 Per3−/−) were injected with 2 mg/kg of glucose. Blood glucose was measured at time = 0, 15, 30, 60, and 120 min after injection. Per3−/− (red) mice had glucose intolerance with higher blood sugars after the bolus compared with wild-type animals (black) (p values: 0 min = 0.9053, 15 min = 0.02, 30 min = 0.0023, 60 min = 0.001, 120 min = 0.0001; error bars represent S.E.).
Adipose and muscle are critical tissues for systemic energy utilization and storage. Muscle is a major site for glucose uptake and increased adipose results in glucose intolerance and diabetes (37, 38). Therefore, we hypothesized that the alterations in body composition in Per3−/− animals would correlate with altered glucose tolerance. In fact, Per3−/− mice were glucose intolerant compared with wild-type controls (Fig. 5C), demonstrating a physiological significance of our findings.
DISCUSSION
Stem cell fate decisions can be influenced and manipulated, opening up the potential for therapeutic interventions. Glucocorticoids have been shown to dramatically influence fate decisions, most notably the induction of adipocyte fate. Studies utilizing this potent effect have revealed critical pathways involved in adipogenesis (39). However, very little is understood about the initial molecular events regulated by glucocorticoids that trigger these differentiation pathways. By focusing on events regulated by glucocorticoids, we discovered a novel function for Per3 as a potent regulator of the adipocyte cell fate decision. It will be interesting in future studies to determine whether Per3 participates in other glucocorticoid-regulated cell fate decisions (40).
It is interesting to speculate whether the regulation of cell fate by Per3 is connected to the circadian clock or if this is an off pathway function. Prior studies have demonstrated that circadian rhythmicity has an important influence on stem cells (41), making the former scenario highly plausible. However, Per3 appears to be excluded from core clock functions, and in vivo deletion of Per3 results in only subtle effects on period (21, 42). In addition, our data indicate that Per3 has some unique properties that may fall outside these pathways. For example, we found that deletion and overexpression of Per3 confer opposite effects on cell fate. If disruption of oscillation were the primary important effect, one would predict both interventions should have similar impacts. Furthermore, disruption of Per3 did not appear to affect oscillation of core clock components. One unifying model is that Per3 is circadian-regulated but is not itself a clock component. This is also consistent with prior reports indicating that Per3 does not play an important role in the circadian clock mechanism regulating locomotor activity (42). In this model, Per3 could serve as a downstream target of the circadian clock in regulating cell fate. This hypothesis is appealing because it provides a further molecular mechanism to the physiological connections between circadian rhythms and cell fate.
Our studies also provide an initial model for the mechanism by which Per3 regulates cell fate; we found that PPARγ can interact with PER3 in cell extracts under conditions of overexpression and that endogenous PER3 and PPARγ co-localize at an endogenous PPRE. We also found that overexpression of Per3 inhibits expression of an endogenous target gene that Per3 and PPARγ co-occupy. We propose that PER3 serves as a co-regulator of PPRE-bound PPARγ, inhibiting its activity and thereby inhibiting the promotion of adipocyte fate by PPARγ. In this model repression is relieved by pro-adipogenic signals that down-regulate Per3 expression, such as glucocorticoids. The significance of the PPARγ-PER3 interaction in extracts remains to be determined. However, it is clear that PER3 and PPARγ co-occupy genomic regions near some PPARγ binding motifs and not others; the selectivity of PER3 occupancy may reflect sequence-specific allosteric effects on DNA-binding protein activity and co-factor association (43).
As predicted from our ex vivo results, Per3−/− mice have systemic alterations consistent with altered cell fate lineage determination including changes in body composition with increased adipose and decreased muscle. In addition, Per3−/− mice have abnormalities in glucose homeostasis. Therefore, in addition to extending our understanding of stem cell biology and the mechanisms that regulate cell fate decisions, these results also have implications for systemic physiology and pathophysiology.
While this manuscript was in post-submission revision, Grimaldi et al. (44) reported that the period paralog protein PER2 regulated lipid metabolism at least in part by inhibiting PPARγ activity. Interestingly, they found that PER2 blocked PPARγ from occupying PPREs as opposed to inhibiting PPARγ activity at response elements as we show occurs with PER3 regulation of adipocyte fate decision. Together, these studies suggest the Per paralogs modulate PPARγ activity in multiple contexts and by using potentially synergistic mechanisms.
Supplementary Material
Acknowledgments
We thank Brian Black, David Feldman, Carlos Pantoja, and Miles Pufall for comments on the manuscript and Teresita Bernal for technical support. We thank Charles Harris, Suneil Koliwad, and Robert Farese, Jr. for guidance and use of the dual energy x-ray absorptiometry scanner. We thank Cristina Alvira for assistance with adipocyte histology. The original Per3−/− mice were kindly provided by David Weaver and Steven Reppert.
This work was supported, in whole or in part, by National Institutes of Health Director's New Innovator Award Program 1-DP2-OD006740 (to B. J. F.) and Grants DK73697 (to B. J. F.), CA20535 (to K. R. Y.), and HL59596 (to Y.-H. F. and L. J. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- MSC
- mesenchymal stem cell
- PPAR
- peroxisome proliferator-activated receptor
- qPCR
- quantitative PCR
- IP
- immunoprecipitation
- PPRE
- peroxisome proliferator-response element.
REFERENCES
- 1. Masuzaki H., Paterson J., Shinyama H., Morton N. M., Mullins J. J., Seckl J. R., Flier J. S. (2001) Science 294, 2166–2170 [DOI] [PubMed] [Google Scholar]
- 2. MacDougald O. A., Mandrup S. (2002) Trends Endocrinol. Metab. 13, 5–11 [DOI] [PubMed] [Google Scholar]
- 3. Roberge C., Carpentier A. C., Langlois M. F., Baillargeon J. P., Ardilouze J. L., Maheux P., Gallo-Payet N. (2007) Am. J. Physiol. Endocrinol. Metab. 293, E1465–E1478 [DOI] [PubMed] [Google Scholar]
- 4. Orth D. N. (1995) N. Engl. J. Med. 332, 791–803 [DOI] [PubMed] [Google Scholar]
- 5. Phinney D. G., Prockop D. J. (2007) Stem Cells 25, 2896–2902 [DOI] [PubMed] [Google Scholar]
- 6. Pittenger M. F., Mackay A. M., Beck S. C., Jaiswal R. K., Douglas R., Mosca J. D., Moorman M. A., Simonetti D. W., Craig S., Marshak D. R. (1999) Science 284, 143–147 [DOI] [PubMed] [Google Scholar]
- 7. Friedenstein A. J., Gorskaja J. F., Kulagina N. N. (1976) Exp. Hematol. 4, 267–274 [PubMed] [Google Scholar]
- 8. Crisan M., Yap S., Casteilla L., Chen C. W., Corselli M., Park T. S., Andriolo G., Sun B., Zheng B., Zhang L., Norotte C., Teng P. N., Traas J., Schugar R., Deasy B. M., Badylak S., Buhring H. J., Giacobino J. P., Lazzari L., Huard J., Péault B. (2008) Cell Stem Cell 3, 301–313 [DOI] [PubMed] [Google Scholar]
- 9. Colter D. C., Class R., DiGirolamo C. M., Prockop D. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 3213–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hong J. H., Hwang E. S., McManus M. T., Amsterdam A., Tian Y., Kalmukova R., Mueller E., Benjamin T., Spiegelman B. M., Sharp P. A., Hopkins N., Yaffe M. B. (2005) Science 309, 1074–1078 [DOI] [PubMed] [Google Scholar]
- 11. Feldman B. J. (2009) Pediatr. Res. 65, 249–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Balsalobre A., Damiola F., Schibler U. (1998) Cell 93, 929–937 [DOI] [PubMed] [Google Scholar]
- 13. Plautz J. D., Kaneko M., Hall J. C., Kay S. A. (1997) Science 278, 1632–1635 [DOI] [PubMed] [Google Scholar]
- 14. Yamazaki S., Numano R., Abe M., Hida A., Takahashi R., Ueda M., Block G. D., Sakaki Y., Menaker M., Tei H. (2000) Science 288, 682–685 [DOI] [PubMed] [Google Scholar]
- 15. Yoo S. H., Yamazaki S., Lowrey P. L., Shimomura K., Ko C. H., Buhr E. D., Siepka S. M., Hong H. K., Oh W. J., Yoo O. J., Menaker M., Takahashi J. S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 5339–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nagoshi E., Saini C., Bauer C., Laroche T., Naef F., Schibler U. (2004) Cell 119, 693–705 [DOI] [PubMed] [Google Scholar]
- 17. So A. Y., Bernal T. U., Pillsbury M. L., Yamamoto K. R., Feldman B. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17582–17587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shimba S., Ishii N., Ohta Y., Ohno T., Watabe Y., Hayashi M., Wada T., Aoyagi T., Tezuka M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12071–12076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang J., Lazar M. A. (2008) Mol. Cell. Biol. 28, 2213–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Balsalobre A., Brown S. A., Marcacci L., Tronche F., Kellendonk C., Reichardt H. M., Schütz G., Schibler U. (2000) Science 289, 2344–2347 [DOI] [PubMed] [Google Scholar]
- 21. Shearman L. P., Jin X., Lee C., Reppert S. M., Weaver D. R. (2000) Mol. Cell. Biol. 20, 6269–6275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. So A. Y., Cooper S. B., Feldman B. J., Manuchehri M., Yamamoto K. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5745–5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu C., Li S., Liu T., Borjigin J., Lin J. D. (2007) Nature 447, 477–481 [DOI] [PubMed] [Google Scholar]
- 24. Turek F. W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E., Laposky A., Losee-Olson S., Easton A., Jensen D. R., Eckel R. H., Takahashi J. S., Bass J. (2005) Science 308, 1043–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rudic R. D., McNamara P., Curtis A. M., Boston R. C., Panda S., Hogenesch J. B., Fitzgerald G. A. (2004) PLoS Biol. 2, e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tontonoz P., Hu E., Spiegelman B. M. (1994) Cell 79, 1147–1156 [DOI] [PubMed] [Google Scholar]
- 27. Schmutz I., Ripperger J. A., Baeriswyl-Aebischer S., Albrecht U. (2010) Genes Dev. 24, 345–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang Z. J., Edery I., Rosbash M. (1993) Nature 364, 259–262 [DOI] [PubMed] [Google Scholar]
- 29. Lefterova M. I., Zhang Y., Steger D. J., Schupp M., Schug J., Cristancho A., Feng D., Zhuo D., Stoeckert C. J., Jr., Liu X. S., Lazar M. A. (2008) Genes Dev. 22, 2941–2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nielsen R., Pedersen T. A., Hagenbeek D., Moulos P., Siersbaek R., Megens E., Denissov S., Børgesen M., Francoijs K. J., Mandrup S., Stunnenberg H. G. (2008) Genes Dev. 22, 2953–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Forman B. M., Tontonoz P., Chen J., Brun R. P., Spiegelman B. M., Evans R. M. (1995) Cell 83, 803–812 [DOI] [PubMed] [Google Scholar]
- 32. Graves R. A., Tontonoz P., Spiegelman B. M. (1992) Mol. Cell. Biol. 12, 1202–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tontonoz P., Hu E., Graves R. A., Budavari A. I., Spiegelman B. M. (1994) Genes Dev. 8, 1224–1234 [DOI] [PubMed] [Google Scholar]
- 34. Bae K., Weaver D. R. (2007) J. Biol. Rhythms 22, 85–88 [DOI] [PubMed] [Google Scholar]
- 35. Brommage R. (2003) Am. J. Physiol. Endocrinol. Metab. 285, E454–E459 [DOI] [PubMed] [Google Scholar]
- 36. Dallmann R., Weaver D. R. (2010) Chronobiol. Int. 27, 1317–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lazar M. A. (2005) Science 307, 373–375 [DOI] [PubMed] [Google Scholar]
- 38. Petersen K. F., Shulman G. I. (2006) Obesity 14, Suppl. 1, 34S–40S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rosen E. D., MacDougald O. A. (2006) Nat. Rev. Mol. Cell Biol. 7, 885–896 [DOI] [PubMed] [Google Scholar]
- 40. Anderson D. J., Axel R. (1986) Cell 47, 1079–1090 [DOI] [PubMed] [Google Scholar]
- 41. Méndez-Ferrer S., Lucas D., Battista M., Frenette P. S. (2008) Nature 452, 442–447 [DOI] [PubMed] [Google Scholar]
- 42. Bae K., Jin X., Maywood E. S., Hastings M. H., Reppert S. M., Weaver D. R. (2001) Neuron 30, 525–536 [DOI] [PubMed] [Google Scholar]
- 43. Lefstin J. A., Yamamoto K. R. (1998) Nature 392, 885–888 [DOI] [PubMed] [Google Scholar]
- 44. Grimaldi B., Bellet M. M., Katada S., Astarita G., Hirayama J., Amin R. H., Granneman J. G., Piomelli D., Leff T., Sassone-Corsi P. (2010) Cell Metab. 12, 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.