Abstract
The expression of a variety of cytoprotective genes is regulated by short cis-acting elements in their promoters, called antioxidant response elements (AREs). A central regulator of ARE-mediated gene expression is the NF-E2-related factor 2 (Nrf2). Nrf2/ARE-regulated genes are crucial for the maintenance of cellular integrity. Hepatitis C virus inhibits the induction of ARE-regulated genes, but neither induction nor inhibition of ARE-regulated gene expression affects HCV replication directly. In HCV-replicating cells the core protein triggers the delocalization of sMaf proteins from the nucleus to the replicon complex. Here sMafs bind to NS3. The extranuclear sMaf proteins prevent Nrf2 from entry in the nucleus and thereby inhibit the induction of Nrf2/ARE-regulated genes. This results in the decreased expression of cytoprotective genes. Consistent with this finding, the elimination of ROI is impaired in HCV-replicating cells as demonstrated by elevated protein oxidation or 8-OH-dG formation, reflecting DNA damage. In conclusion, these data identified a novel mechanism of Nrf2 regulation and suggest that the HCV-dependent inhibition of Nrf2/ARE-regulated genes confers to the HCV-associated pathogenesis by elevation of intracellular ROI that affect integrity of the host genome and regenerative processes.
Keywords: Gene Expression, General Transcription Factors, Hepatitis Virus, Liver, Signal Transduction
Introduction
Hepatitis C virus (HCV)2 infection results in chronic hepatitis in more than 70% of infected individuals. At present more than 170 million people are persistently infected with HCV worldwide. Persistent HCV infection is associated with chronic inflammation of the liver (hepatitis), which can progress to liver fibrosis, cirrhosis, and hepatocellular carcinoma (HCC) (1). HCV is the sole member of the genus hepacivirus that belongs to the flaviridae family. The HCV genome is a single-stranded positive-sense RNA molecule of ∼9600 bases length. The viral RNA codes for a large polyprotein of ∼3100 amino acids, which is post-translationally processed by cellular and viral proteases. The N terminus encompasses the structural proteins core, E1, and E2; the C terminus the p7 protein and the nonstructural (NS) proteins NS2, NS3, NS4A, NS4B, NS5A, and NS5B (2–4).
HCV infection is a major cause of progressive liver disease worldwide. Although the detailed molecular mechanisms by which HCV induces liver injury are not fully understood, there are reports describing that hepatitis C is associated with oxidative stress in human liver cells due to enhanced generation of reactive oxygen intermediates (ROI). The ER-associated viral replication and translation of viral integral membrane proteins (5–8) or ER-associated proteins could cause ER stress. One consequence of the ER stress response is calcium release from the ER, initiating a series of intracellular events, including mitochondrial ROI production induced by Ca2 + signaling (7). Moreover, during the immune response to HCV positive cells, inflammatory cells produce high levels of radicals (8). However, despite the evidence of generation of ROI during the HCV life cylce, little is known about the interference of HCV with cytoprotective mechansims.
Crucial players in the defense against oxidative stress are a variety of cytoprotective genes harboring a short cis acting sequence, the antioxidative response element (ARE), in their promoters(9). The expression of ARE-regulated genes results in the detoxification of electrophiles and radicals. Examples for ARE-regulated genes are NAD(P)H quinione oxidoreductase 1 (NQO1), glutathione peroxidase (pHGPx), the regulatory and catalytic subunits of glutamate-cysteine ligase (GCLM and GCLC) or glutathione S-transferases (GST) ya and π(10–12). An essential factor involved in the defense against oxidative stress is the NF-E2 related factor 2 (Nrf2). Nrf2 is a member of the cap “n” collar family of transcription factors (13–15). In its inactive state Nrf2 is associated with the actin anchored protein Keap1 and localized within the cytoplasm. Keap1-associated Nrf2 is subjected to rapid proteasomal degradation. However, upon its activation initiated by electrophiles or oxidative molecules Nrf2 dissociates from Keap1, escapes proteasomal degration and quickly translocates into the nucleus with the assistance of a nuclear localization signal. In the nucleus Nrf2 binds to ARE sequences and thereby functions in partnership with other nuclear proteins as a strong transcriptional activator of the respective genes (for recent reviews see Refs. 14,15). Here we analyzed the effect of HCV on the expression of ARE-regulated genes, investigated the mechanisms by which HCV interferes, and studied the physiological relevance for the viral life cycle and HCV-associated pathogenesis.
EXPERIMENTAL PROCEDURES
Cell Culture
The Huh7-derived cell clone Huh7.5 (16), which is highly permissive for HCV RNA replication, was used for transfection and infection assays. Cells were grown in Dulbecco's modified Eagle's medium (DMEM; Sigma) supplemented with 2 mm l-glutamine, nonessential amino acids, 100 units of penicillin per ml, 100 μg of streptomycin per ml, and 10% fetal calf serum (DMEM complete). The subgenomic replicon cell lines HuH-7 I377/NS3–3′/wt/9–13(17) (Huh9–13) and HuH-7 7 I389/NS3–3′/LucUbiNeo-ET (18) (Huhet)were kindly provided by Prof. Bartenschlager (University of Heidelberg, Germany). Cultivation and infection of primary human hepatocytes was performed as described (19).
Plasmids
The plasmids pJFH1wt, pJFH1/J6, pJFH1_GND, pJFH1d3C, and pJFH1/DE1E2, were described recently (20–23). Luciferase reporter constructs harboring the AREs from NAD(P)H-dependent quinone oxidoreductase 1 (pNQO1luc), γ-glutamylcysteine ligase (GCL), (pGCLMluc), and gastrointestinal glutathione peroxidase (pGI-GPxluc,) genes were generated using the pGL3-Promoter vector (Promega). Double-stranded oligonucleotides were synthesized containing a part of the rat NQO1 promoter (5′-CTCTAGAGTCACAGTGACTTGGCAAAATCTGAC-3′), the murine Gclm promoter (5′-CCTGGAAGACAATGACTAAGCAGAAAC-3′) or the human GI-GPx promoter (5′-CCTGTTTTGCTAAGTCATCCTGGGGACC-3′), including the antioxidant response elements (underlined). After annealing of the complementary oligonucleotides the double-stranded oligonucleotides were inserted into the pGL3-Promoter vector.
Constitutively active (phcaNrf2) and trans-dominant negative Nrf2 expression constructs (ptdnNrf2) were described recently(24). The PSMB5 reporter constructs (p3.4-luc, p0.5-luc, and p0.2-luc) were kindly provided by Dr. Kwak, Seoul, Korea (25). NQO1 and GCLC expression constructs fused to yellow fluorescence protein (YFP) were purchased from ImaGenes GmbH, Berlin, Germany.
In Vitro Transcription and RNA Transfection
In vitro transcription, electroporation of HCV RNAs, and luciferase assays were performed as described(26,27). The transfection efficiency was up to 65%.
Transient Transfection and Reporter Gene Activity Assay
48 h after electroporation Huh7.5 cells were transfected using linear polyethyleneimine (PEI) (Polysciences, Inc) as described recently (28) and grown for further 48 h. The transfection efficiency was about 70%. Luciferase activity was measured using a luminometer (Berthold Detection Systems, Wildbad, Germany).
Chemicals and Antibodies
Anti-NQO1 (A180), anti-γ-GCL(H-300), anti-sMaf F/G/K (H-100), anti-Nrf2 (C20), anti Lamin-A (H102), and anti-GAPDH (FL-335) antibodies were all purchased from Santa Cruz Biotech.
Anti-PSMB5 was purchased from ABR Affinity BioReagents, and anti-proteasomal β5i-subunit (23–223) from Calbiochem, anti-β-actin from Sigma-Aldrich, anti-GPx (C8C4) from Cell Signaling Technology. Mouse anti-core and anti-NS3 antibodies were purchased from Affinity BioReagents and ViroStat, Marine, respectively. The polyclonal rabbit-derived NS5A-specific serum was described recently (29). Tert-butylhydroquinone (tBHQ) and H2O2 were purchased from Sigma-Aldrich (USA).
Virus Titration
Virus titers were determined as described (30). For detection of HCV-positive cells as NS5A-specific serum (29) was used.
Immunohistochemistry
Consecutive sections of paraffin embedded liver samples derived from HCV-patients or HBV/HCV-negative patients were deparaffinized. The deparaffinized sections were boiled for 4 min in a microwave in citrate buffer pH 6.0. After that the sections were reequilibrated in TBST for 30 min. Endogenous peroxidase activity was blocked by incubaion in 0.3% H2O2 for 20 min. In the next step, the samples were washed twice with TBST and incubated for 20 min in blocking solution (10% sBSA in TBST). The sections were immunostained with anti-sMaf and anti-core antibodies diluted in blocking solution for 90 min. The samples were washed for 30 min in TBST- the buffer was changed five times. Bound antibodies were visualized using a biotinylated secondary antibody and strepatavidin/peroxidase-complex from the Vectastain kit, Vector Laboratories, Inc., according to the instructions of the manufacturer.
Real Time PCR
RNA isolation from liver tissue and from infected primary human hepatocytes was performed using TRIzol (Invitrogen), according to the manufacturer's instructions. For cDNA synthesis, 2–4 mg of total RNA were treated with DNase I. First-strand synthesis was carried out using SupercriptII reverse transcriptase (Invitrogen) according to the Invitrogen protocol.
Real time PCR was performed using the following primers for quantification of GPx-specific transcripts (forward (fw): caaccagttt-gggcatcag; backward(bw): cccaccaggaacttctcaaa) and for GCLC-specific transcripts(fw: cccatgga-ggtgcaattaac; bw: tgcgataaactccctcatcc). The values were referred to GAPDH expression. Three samples from patients with chronic HCV-infection were compared with samples from three HCV-/HBV-negative patients.
Indirect Immunofluorescence Analysis
Fixation and staining were performed as described recently (31). Immunofluorescence staining was analyzed using a confocal laser scanning microscope (CLSM 510 Carl Zeiss).
Subcellular Fractionation
The isolation of the nucleosolic and cytosolic fraction was performed as described (27).
SDS-PAGE and Western Blot Analysis
SDS-PAGE and Western blot analysis were performed according to standard procedures (29). Equal loading was controlled by detection of β-actin. Detection of bound secondary antibodies was performed by ECL using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Freiburg, Germany).
Protein Oxidation and 8-OH-dG Formation Measurement
Protein carbonylation by reactive oxygen intermediates was detected by using OxyBlotTM protein oxidation detection kit (Millipore). 8-OH-dG formation was analyzed using a commercial ELISA system (JalCA).
Analysis of Proteasome Activity
Proteasomes were isolated by differential centrifugation using the protocol of Robek et al. (32). Analysis of constitutive proteasome function was performed using a commercial assay system (20 S Proteasome Activity Assay kit, Millipore) measuring release of the fluorophore 7-amino-4-methylcoumarin (AMC) after cleavage from the labeled substrate peptide LLVY-AMC.
RT-PCR
RNA isolation was performed using Trizol (Invitrogen, Karlsruhe, Germany), according to the instructions of the manufacturer. For cDNA synthesis, 2–4 μg of total RNA were treated with DNase I. First-strand synthesis was carried out using SupercriptII reverse transcriptase (Invitrogen), and semi-quantitative RT-PCR was performed according to the Invitrogen protocol. RT-PCR was performed using the following primers:NQO1fw (5′-GCACTGATCGTACTGGCTCA-3′) NQO1_rev (5′-GAACACTCGCTCAAACCAG-3′), HCVfw (5′-AGTACCACAAGGCCTTTCG-3′) HCVbw (5′-CGGGAGAGCCATAGTGG-3′), and β-actin_fw (5′-GAGCTGCGTGTGGCTCCC-3′) β-actin_rev (5′-ATGTCACGCACGATTTCCCG-3′).
Statistical Analysis
Statistical analysis was performed using the Prism4 statistical program. p values are two-tailed and were calculated using the Mann-Whitney U test. Error bars represent the standard deviation.
RESULTS
HCV Inhibits Induction of ARE-dependent Genes
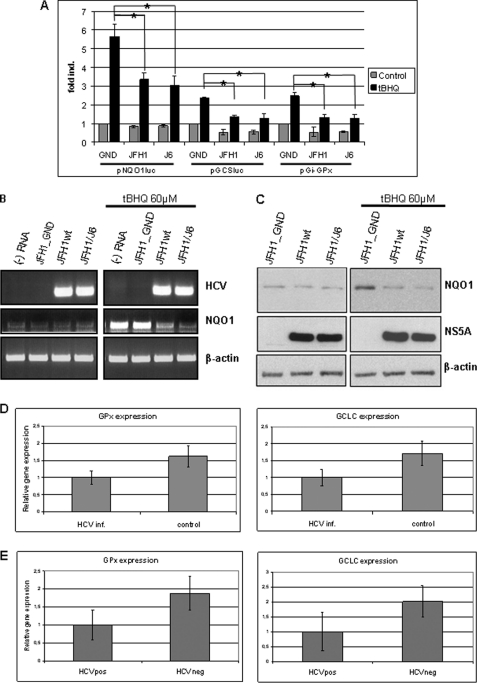
To study the effect of HCV on ARE-dependent gene expression reporter gene assays were performed. HCV-replicating cells were transfected with reporter constructs harboring a luciferase reporter gene under the control of NQO1, GI-GPx, or GCLM-derived ARE sequences. Cells transfected with the replication-deficient GND-mutant served as control. In one set of experiments the effect of HCV on the basal expression of ARE-regulated promoters was analyzed, in the other set the effect of HCV on the electrophil (tBHQ)-dependent induction of ARE-regulated genes was studied. The reporter gene assay shows that both HCV constructs, JFH-1wt and JFH1/J6, caused a slight decrease in the basal activation of the reporter genes as compared with the GND-mutant transfected cells (Fig. 1A). In case of the tBHQ-stimulated cells a significantly smaller induction of the reporter genes was found for the HCV-replicating cells as compared with the GND-transfected cells (Fig. 1A).
FIGURE 1.
Impaired induction of ARE-regulated genes in HCV-replicating cells. A, reporter gene assay in Huh7.5 cells electroporated with HCV replicons JFH1wt or JFH1/J6. Cells electroporated with the replication-deficient construct JFH1_GND served as control. 48 h after electroporation cells were co-transfected with the reporter constructs harboring the NQO1, GI-GPx, or GCLM-derived ARE sequences. Activities are indicated as mean values from three independent experiments. The bars represent the standard deviation. Cells were stimulated with tert-butylhydroquinone (tBHQ) (black bars) or left unstimulated (gray bars). *, p < 0.05). B, analysis of NQO1-expression by reverse transcription polymerase chain reaction (RT-PCR). Electroporated cells were incubated with 60 μm tBHQ for 16 h or treated with the equivalent amount of DMSO as control. Amplification of HCV- and actin-specific sequences served as control. C, Western blot analysis of cellular lysates derived from HuH7.5 cells electroporated with HCV replicons JFH1wt or JFH1/J6 using a NQO1-specific antiserum. Cells electroporated with the replication-deficient construct JFH1_GND served as control. 96 h after electroporation cells were stimulated for 16 h with 60 μm tBHQ (right panel). HCV replication was visualized by a NS5A-specific serum. D, analysis of GPx and GCLC expression normalized to GAPDH in HCV-infected primary human hepatocytes. Uninfected PHH served as control. Expression was analyzed by real time PCR. The data represent mean values from three independent experiments and are shown as relative values. The bars represent the standard deviation. E, analysis of GPx- and GCLC expression normalized to GAPDH in liver samples derived from three patients with chronic hepatitis. Liver samples from three HBV-/HCV-negative patients served as control. Expression was analyzed by real time PCR. The data represent mean values and are shown as relative values. The bars represent the standard deviation.
These data were confirmed by semi-quantitative RT-PCR using NQO1-specific primers (Fig. 1B) and by Western blot analysis (Fig. 1C). Both experiments confirmed that expression of NQO1 was much stronger induced after tBHQ treatment in HCV-negative control cells as compared with the HCV-replicating cells. In the latter, only a weak increase in the expression of NQO1 was observed. Comparable results were obtained for GCLM and GI-GPx (data not shown). To corroborate these data we analyzed the expression of Nrf2/ARE-dependent genes in HCV- infected primary human hepatocytes and in liver samples derived from patients with a chronic HCV infection.
The real time PCR using GCLC- and GPx-specific primers revealed that in the HCV-infected primary human hepatocytes after tBHQ stimulation significant less GPx and GCLC were expressed as compared with the uninfected cells (Fig. 1D).
Real time PCR analyses of liver samples from patients with chronic HCV infection and HCV/HBV-negative samples revealed a decreased expression of GPx and GCLC in the HCV-positive samples as compared with the control (Fig. 1E). Taken together, these data show that HCV exerts an inhibitory effect on the induction of ARE-regulated genes.
Overexpression of ARE-regulated Genes Does Not Affect HCV Replication
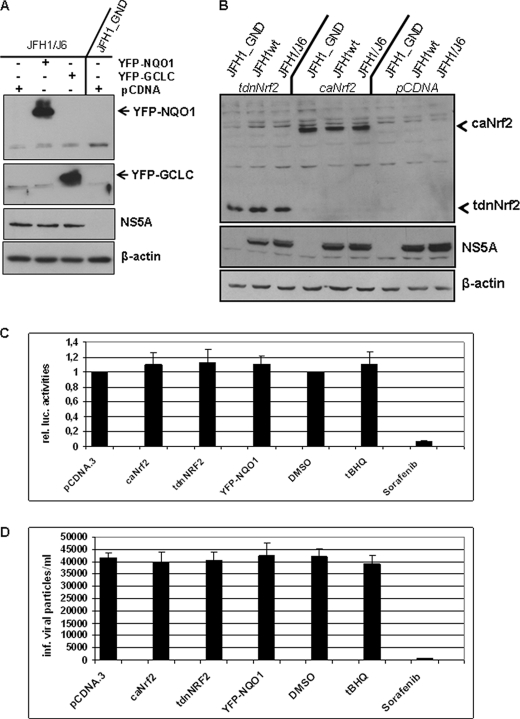
Based on the results described above we analyzed whether overexpression of ARE-regulated genes affects the HCV replication. The Western blot analysis of cellular lysates derived from HCV-replicating cells overexpressing NQO1 or GCLC revealed that neither overexpression of NQO1 nor of GCLC significantly affects the amount of viral proteins (Fig. 2A).
FIGURE 2.
Overexpression of ARE-regulated genes does not affect HCV replication. A, HCV-replicating cells were transfected with expression vectors encoding NQO1 or GCLC as YFP fusion protein. Transfection with the equal amount of pCDNA served as control. Overproduction of YFP-NQO1 or YFP-GCLC was demonstrated by Western blotting. The effect on viral replication was determined using a NS5A-specific antiserum. Lysate from HCV-negative cells (JFH1_GND) served as negative control. B, HCV-replicating cells (JFH1wt, JFH1/J6) were transfected with expression vectors coding for a trans-dominant negative mutant (ptdnNrf2; lanes 1–3) or a constitutively active mutant (pcaNrf2; lanes 4–6) or with the empty expression vector pCDNA (lanes 7–9). Lysate from HCV-negative cells (JFH1_GND) served as negative control. The effect on viral replication was determined by Western blot analysis using a NS5A-specific antiserum. Overproduction of tdnNrf2 or caNrf2 was demonstrated by Western blotting. C, analysis of viral replication by luciferase reporter gene assay. 48 h after electroporation, HCV luc-JFH-1-replicating Huh7.5 cells were co-transfected with pcaNrf2, ptdnNrf2, pYFP-NQO1, or pCDNA.3. Incubation with tBHQ was performed for 16 h. Here, treatment with an equivalent amount of DMSO (0.1%) served as a control treatment with Sorafenib (10 μm) was performed to inhibit HCV replication (26). D, infection of HuH7.5 cells with supernatants derived from pNQO1, ptdnNrf2-, pcaNrf-2, pCDNA3- transfected cells, or tBHQ-stimulated cells. Cells were transfected 48 h after electroporation. Again 48 h later, the supernatant was collected. In case of the tBHQ-treated cells were stimulated 16 h before harvesting with tBHQ. The supernatant from electroporated-untreated cells served as control. The amount of HCV particles was determined by limited dilution. Mean values from three independent experiments are shown. The bars represent the standard deviation.
To determine if modulation of ARE-regulated gene expression affects HCV replication, cells were either treated with tBHQ or co-transfected with an expression construct encoding a constitutively active (ca) mutant of Nrf2 or a transdominant negative (tdn) mutant of Nrf2. Western blot analyses demonstrate that neither the induction of ARE-regulated genes by co-expression of caNrf2 nor the inhibition by co-expression of tdnNrf2 affect HCV replication (Fig. 2B). Comparable results were obtained for tBHQ-stimulated cells (data not shown).
These data were corroborated when cells that replicate a luciferase reporter virus were co-transfected with pcaNrf2, ptdnNrf2, or treated with tBHQ. The reporter gene assays confirm that neither general activation nor inhibition of the Nrf2/ARE-regulated gene expression directly affects HCV replication (Fig. 2C). Moreover dilution assays for determination of the virus titer confirmed that comparable amounts of infectious viral particles were produced from HCV-replicating cells treated with tBHQ, or transfected with pNQO1, pcaNrf2, or ptdnNrf2 as compared with the corresponding controls (Fig. 2D).
Nuclear Nrf2 Levels Are Reduced in HCV-replicating Cells
The transcription factor Nrf2 plays an essential role in the control of ARE-regulated genes. In its inactive state Nrf2 is associated with Keap1, localized in the cytoplasm, and rapidly degraded. Upon activation, Nrf2 enters the nucleus and binds as a heterodimer with small Maf (sMaf) proteins to the ARE sequences.
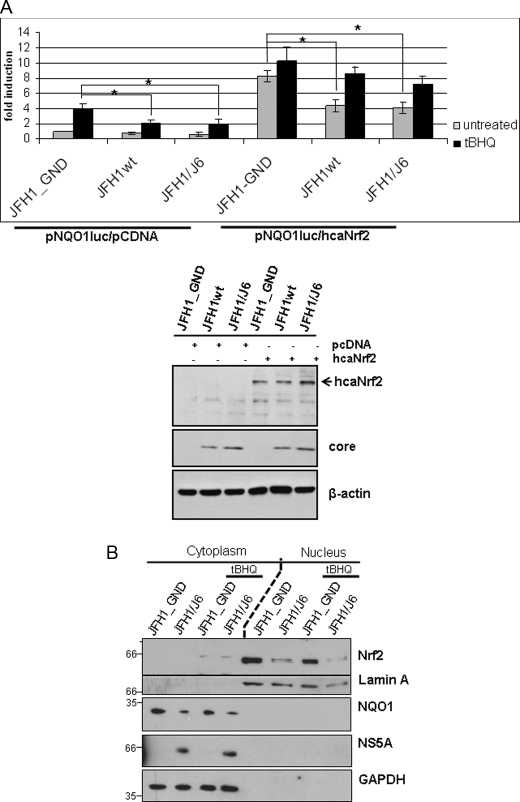
To study the mechanism underlying the HCV-dependent inhibition of Nrf2/ARE-dependent gene expression we analyzed whether co-expression of caNrf2 can rescue the defect in Nrf2/ARE-regulated gene expression seen in HCV-replicating cells. However, reporter gene assays revealed that coexpression of caNrf2 does not fully rescue Nrf2/ARE-dependent gene expression in HCV-replicating cells as compared with the control (Fig. 3A).
FIGURE 3.
Nrf2 accumulation in the nucleus is impaired in HCV-replicating cells. A, reporter gene assay in Huh7.5 electroporated with HCV replicons JFH1wt or JFH1/J6. Cells electroporated with the replication-deficient construct JFH1_GND served as control. 48 h after electroporation cells were cotransfected with the reporter constructs harboring the NQO1-derived ARE sequences and with an expression vector encoding caNrf2 or as control the equal amount of pCDNA.3 vector. Cells were stimulated with tert-butylhydroquinone (tBHQ) (black bars), or left unstimulated (gray bars). Activities presented are mean values from three transfected vials. The bars represent the standard deviation. *, p < 0.05. The Western blot analysis of the lysates used for reporter gene analysis (lower panel) shows the overexpression of caNrf2. For detection of HCV a core-specific serum was used, equal loading was controlled by an actin-specific serum. B, HCV-replicating cells (JFH1/J6) or control cells (JFH1_GND) were stimulated for 2 h with 60 μm tBHQ or left untreated, subjected to subcellular fractionation and analyzed by Western blotting using a Nrf2-specific serum. Purity of the fractions was confirmed by the presence of lamin A and the absence of GAPDH in the nuclear fraction.
Based on this finding we hypothesized that the accumulation of Nrf2 in the nucleus might be impaired in HCV-replicating cells. To study whether the HCV-dependent inhibition of ARE-regulated genes is due to reduced levels of Nrf2 in the nucleus compared with HCV-negative cells we performed subcellular fractionation (27) of HCV-positive and HCV-negative cells and determined the amount of Nrf2 in the nuclear and cytosolic fractions by Western blotting. Indeed, levels of nuclear Nrf2 were much higher in tBHQ-treated HCV-negative cells compared with HCV-positive cells (Fig. 3B). These data demonstrate that less Nrf2 is available for binding to ARE sequences in the nucleus of HCV-replicating cells compared with HCV-negative cells, resulting in a decreased induction of ARE-regulated genes.
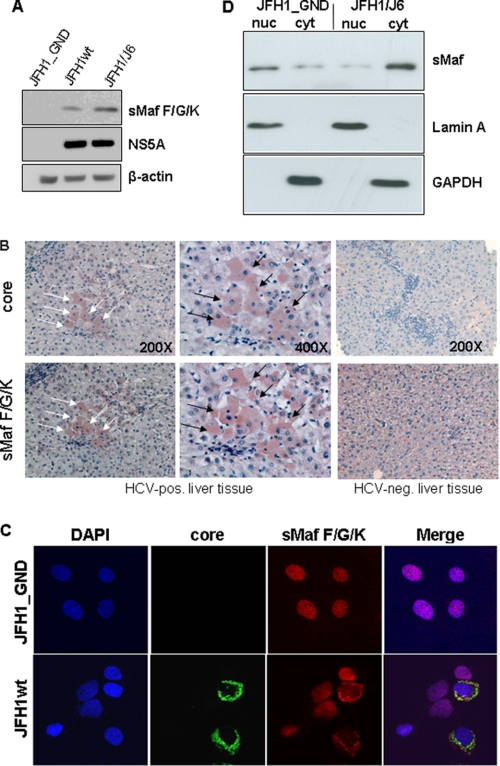
Increased Amounts and Delocalization of sMaf Proteins in HCV-replicating Cells
Small Maf proteins are an essential co-factor regulating Nrf2/ARE-dependent gene expression(14,15). In a recent report it was described that overxpression of the sMaf proteins MafG and MafK impairs ARE-mediated gene expression(33). In light of our data described above we investigated whether the intracellular amounts and/or the distribution of sMaf proteins are affected by HCV. Western blot analyses of cellular lysates derived from HCV-replicating cells revealed significantly higher amounts of sMaf proteins in these cells as compared with the control (Fig. 4A). These data were corroborated by immunohistochemical analysis of consecutive liver sections derived from three HCV-positive patients that gave comparable results. Fig. 4B shows the results obtained for one of these samples at 200- and 400-fold magnification. HCV-positive cells were identified using a core-specific antiserum. The consecutive section was stained using a small Maf-specific antiserum. In the HCV-positive cells we found a much stronger staining of small Maf proteins as compared with the surrounding HCV-negative cells (Fig. 4B). Liver tissue from a HCV-/HBV-negative patient served as control. These data indicate that our in vitro data are indeed relevant for the in vivo situation.
FIGURE 4.
Increased amount and delocalization of sMaf proteins in HCV-replicating cells. A, Western blot analysis of cellular lysates derived from HCV-replicating cells (JFH1; JFH1/J6) or control cells (JFH1_GND) using NS5A and sMaf proteins-specific antisera. B, immunohistochemistry of consecutive liver sections from a patient with chronic HCV infection stained with core- (upper panel) or sMaf-specific antibodies (lower panel). Corresponding cells are labeled with arrows. Liver sections from a HCV-/HBV-negative patient served as negative control. C, confocal immunofluorescence microscopy of HCV-positive cells (JFH1wt) or HCV-negative control cells (JFH1-GND) (630-fold magnification). The immunofluorescence staining was performed using the polyclonal sMaf-specific serum (red) and a core-specific antibody (red). Nuclei were stained with DAPI. D, subcellular fractionation of HCV-positive (JFH1/J6)-replicating Huh7.5 cells and HCV-negative (JFH1_GND) cells. The nucleosolic (nuc) and the cytoplasmatic (cyt) fraction were analyzed by Western blotting analyzed by Western blotting using a sMaf-specific serum. Purity of the fractions was confirmed by the presence of lamin A and the absence of GAPDH in the nuclear fraction.
For a more detailed analysis, confocal double immunofluorescence microscopy of HCV-positive and HCV-negative control cells was performed. The double immunofluorescence microscopies show that in HCV-negative cells almost all of the small Maf-proteins are found within the nucleus. However, in case of the HCV-positive cells a strong sMaf-specific signal was observed in the perinuclear region, while the nucleus remained unstained or weakly stained (Fig. 4C).
This could be confirmed by cell fractionation experiments of HCV-positive cells and HCV-negative cells and subsequent Western blot analyses of the extranuclear (cyt) and nucleosolic fraction. In case of the HCV-negative cells almost all of the SMafs was found in the nucleosolic fraction, while in the extranuclear fraction significant less was found. In case of the subcellular fractions derived from HCV-positive cells comparable amounts of sMafs were found in the extranuclear and nucleosolic fraction (Fig. 4D). These results demonstrate that the levels of sMaf proteins are increased in HCV-positive cells. In addition, sMafs are predominantly localized in the extranuclear compartment of infected cells, whereas their localization is mainly nuclear in control cells.
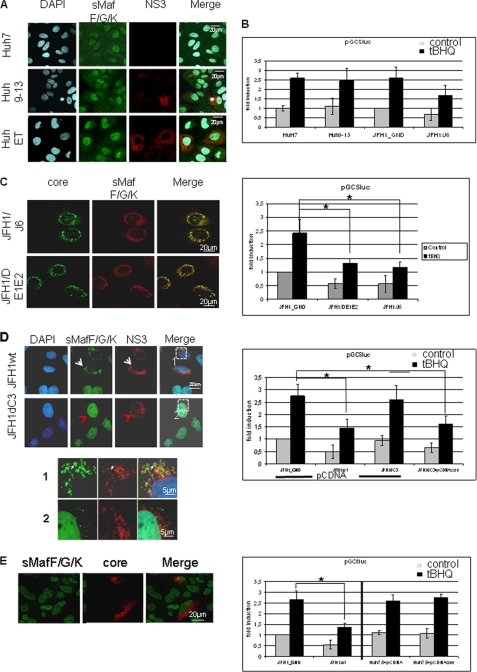
Core Is Required but Not Sufficient for the Delocalization of sMaf Proteins
In the next set of experiments we aimed to perform a more detailed analysis of the HCV-dependent delocalization of sMafs. Huh7.5 cells were transfected with various HCV replicon constructs and analyzed with regard to their effect on the subcellular distribution of sMafs and their interference with the Nrf2/ARE-regulated gene expression.
First, the effect of the subgenomic replicons Huh 9–13 and HuhEt that lack the structural proteins and encompass the NS3-NS5B proteins were analyzed. The confocal double immunofluorescence microscopy shows that in cells replicating these subgenomic replicons no delocalization of sMafs can be observed (Fig. 5A) indicating that the nonstructural proteins are not sufficient to delocalize sMafs. In accordance with this finding, co-transfection of Huh9–13 cells with an ARE-dependent luc reporter construct revealed that this subgenomic replicon does not exert an inhibitory effect on Nrf2/ARE-dependent gene expression (Fig. 5B).
FIGURE 5.
HCV-dependent inhibition of Nrf2/ARE-regulated genes depends on core. A, confocal immunofluorescence microscopy (×630 magnification) of the cell lines Huh9–13 or HuhEtneo carrying the subgenomic replicons. HCV-negative cells (Huh7.5) served as control. Immunofluorescence staining was performed using the polyclonal sMaf-specific serum (green fluorescence) and an NS3-specific antibody (red). B, reporter gene assay of HuH9–13, Huh7.5, Huh7.5JFH1/J6, and Huh7.5/JFH1_GND cells transfected with the luciferase reporter construct harboring the GCS-derived ARE. The bars represent the standard deviation. *, p < 0.05. C, confocal immunofluorescence microscopy of Huh7.5 cells replicating the complete HCV genome (JFH1/J6) or the E1/E2 deletion construct (JFH1DE1/E2). Immunofluorescence staining was performed using the polyclonal sMaf-specific serum (red) and a core-specific antibody (green). The corresponding reporter gene assay was performed using the luciferase reporter construct harboring the GCS-derived ARE sequences. The bars represent the standard deviation. *, p < 0.05. D, confocal immunofluorescence microscopy of Huh7.5 cells replicating the complete HCV genome (JFH1wt) or the core deletion construct (JFH1dC3) Immunofluorescence staining was performed using the polyclonal sMaf-specific serum (green) and a NS3-specific antibody (red). Higher magnification of the indicated fields is shown in 1 and 2. The corresponding reporter gene assay of JFH1wt- or JFH1dC3-replicating HuH7.5 cells is shown below. 48 h after electroporation cells were co-transfected with the reporter constructs harboring the GCS-derived ARE sequences and pCDNA3 vector as control. Co-transfection with pCDNAcore that encodes HCV core rescues the inhibitory effect of the core-deficient replicon construct. The bars represent the standard deviation. *, p < 0.05. E, confocal immunofluorescence microscopy of Huh7.5 cells that selectively overexpress HCV core after transient transfection with a core expression vector. Immunofluorescence staining was performed using the polyclonal small Maf-specific serum (green) and a core-specific antibody (red). Corresponding reporter gene assay of Huh7.5 cells that were co-transfected with pCDNAcore and the reporter constructs harboring the GCS-derived ARE sequences is shown below. Transfection with pCDNA3 vector or cotransfection of JFH1wt or JFH1-GND cells served as controls. The bars represent the standard deviation. *, p < 0.05. A–E, cells were stimulated with tert-butylhydroquinone (tBHQ) (black bars) or left untreated (gray bars).
These data suggested that the structural proteins might be causative for sMaf delocalization and the resulting inhibition of Nrf2/ARE-dependent gene expression. In cells replicating the E1/E2-deficient replicon JFH1DE1/E2 the delocalization of sMaf proteins was found indistinguishable from cells replicating the wild-type replicon JFH1 (Fig. 5C). In accordance with this finding, reporter gene experiments showed a comparable inhibition of ARE-dependent gene expression in cells replicating JFH1DE1/E2 or JFH1/J6 (Fig. 5C).
Next, the effect of the core-deficient replicon construct JFH1d3C on sMaf delocalization and Nrf2/ARE-dependent gene expression was analyzed. Immunofluorescence microscopy shows no delocalization of sMaf for the core-deficient replicon (Figs. 5D and 6A). Furthermore, no inhibitory effect on the Nrf2/ARE-dependent gene expression was observed using reporter assays (Fig. 5D). However, co-transfection of Huh7.5 cells replicating the core-deficient JFH1d3C construct with a core expression construct restored the inhibitory potential on the Nrf2/ARE-dependent driven reporter gene (Fig. 5D). Surprisingly, the selective overexpression of core in transiently transfected cells neither causes delocalization of sMaf proteins (Fig. 5E) nor impairs Nrf2/ARE-dependent gene expression (Fig. 5E).
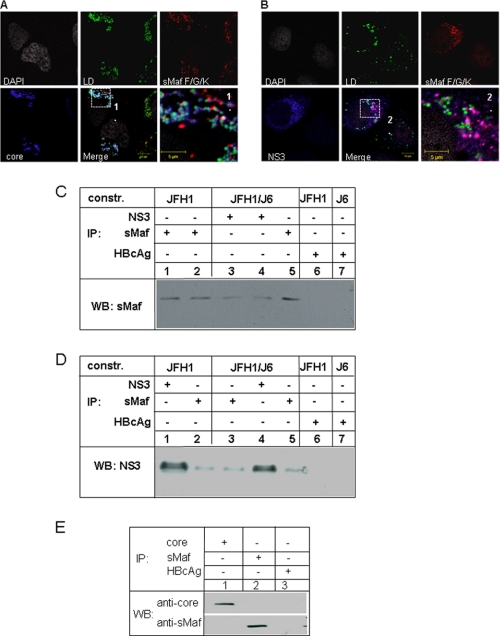
FIGURE 6.
In HCV-replicating cells sMafs co-localize with NS3 but not with core. A and B, confocal immunofluorescence microscopy of Huh7.5 cells replicating the complete HCV genome (JFH1wt). Immunofluorescence staining was performed using the polyclonal sMaf-specific serum (red), a core-specific antibody (blue) (A) or a NS3-specific antibody (blue) (B). Lipid droplets are stained in green, nuclei in white. Higher magnification of the indicated fields is shown in 1 and 2. The co-localization of sMaf with NS3 is shown in magenta. C, HCV-replicating cells (JFH1, lanes 1, 2, and 6 and JFH1/J6 lanes 3, 4, and 7) were lysed 48 h (lanes 1, 3, 5, 6, and 7) or 72 h (lanes 2 and 4) after electroporation. The lysates were precipitated using using NS3- (lanes 3 and 4) and sMaf-specific antisera (lanes 1, 2, and 5). HBcAg-specific antiserum (lanes 6 and 7) served as control. The precipitates were analyzed by Western blotting using sMaf-specific antiserum for detection. D, HCV-replicating cells (JFH1, lanes 1, 2, and 6 and JFH1/J6 lanes 3, 4, and 7) were lysed 48 h (lanes 1, 3, 5, 6, and 7) or 72 h (lanes 2 and 4) after electroporation. The lysates were precipitated using NS3- (lanes 1 and 4) and sMaf-specific antisera (lanes 2, 3, and 5). HBcAg-specific antiserum (lanes 6 and 7) served as control. The precipitates were analyzed by Western blotting using NS3-specific antiserum for detection. E, HCV-replicating cells (JFH1/J6) were lysed 48 h after electroporation. The lysates were precipitated using core (lane 1) or sMaf-specific antisera (lane 2). HBcAg-specific antiserum (lane 3) served as control. The precipitates were analyzed by Western blotting using core- or sMaf-specific antiserum for detection.
Taken together, these data indicate that HCV triggers delocalization of small Maf proteins by core that requires the presence of the NS proteins. The resulting question was whether there is a direct interaction of the NS-proteins or of core with sMaf. To address this question, HCV-replicating cells were stained with NS3, core and sMaf-specific antisera and analyzed by confocal immunofluorescence microscopy. The detailed analysis reveals for HCV positive cells a colocalization of NS3 with sMaf that is absent in cells replicating the core deficient mutant (Fig. 5D). Further analysis including staining of lipid droplets revealed that there is no co-localization of sMafs with core (Fig. 6A) but with NS3 (Fig. 6B). The interaction of sMaf with NS3 in HCV-replicating cells could be confirmed by coimmunoprecipitation experiments using cellular lysates derived from HCV-replicating cells obtained 48 or 72 h after electroporation (Fig. 6, C and D). However, coimmunoprecipitation experiments using a core-specific antiserum failed to co-precipitate sMafs and vice versa (Fig. 6E). These experiments revealed that sMafs could be specifically coprecipitated by a NS3-specific antiserum and vice versa (Fig. 6D) but not by a core-specific serum (Fig. 6E).
These data suggest that core triggers by an indirect mechanism i.e. by affecting intracellular signal transduction cascades but not by a direct interaction the delocalization of sMafs. In accordance with this, sMafs do not colocalize with core in the cytoplasm but with NS3. This suggests that core results in modification of sMafs or NS3 that are required for their interaction.
Reduced Proteasomal Activity and Increased Oxidative Damage in HCV-replicating Cells
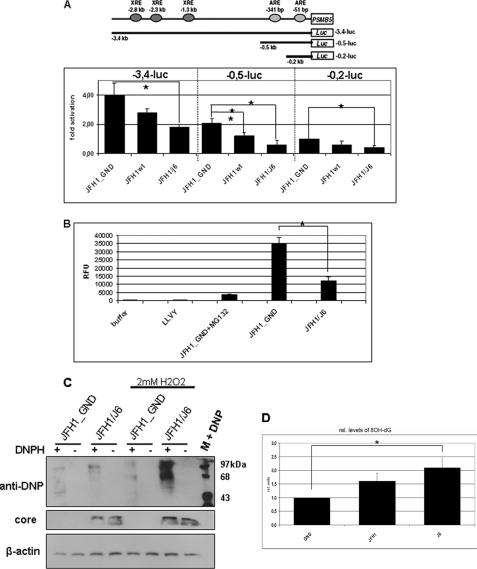
The expression of some proteasomal subunits is affected by the Nrf2/ARE-system: i.e. the expression of the catalytically active PSMB5 subunit depends on three XRE (xenobiotic response element) and two ARE sequences(25). Reporter gene experiments using a luciferase reporter gene driven by the complete promoter of PSMB5 (p3,4luc) showed a decreased induction of the reporter gene in HCV-replicating cells as compared with the GND control. To demonstrate the relevance of the ARE sequences for the observed effect, the XRE sequences were deleted. Transfection of HCV-replicating cells with this truncated reporter construct that still harbors the two ARE sequences demonstrated an impaired induction in HCV-replicating cells as compared with the GND control (Fig. 7A). In accordance with this finding, the analysis of the constitutive proteasomal activity revealed a significantly decreased activity for proteasomes isolated from HCV-replicating cells as compared with the control cells (Fig. 7B).
FIGURE 7.
Impaired proteasomal activity and reduced elimination of ROI in HCV-replicating cells. A, reporter gene assay of HCV-replicating cells co-transfected with luciferase reporter constructs harboring different fragments of the promoter of the gene encoding the proteasomal subunit PSMB5: the complete promoter in case of p3.4 kb luc (encompassing 3 XRE- and 2 ARE sequences), or the 0.2 kb fragment that still harbors 1 ARE sequence were used. The bars represent the standard deviation. *, p < 0.05). B, analysis of constitutive proteasomal activity by measuring the cleavage of the substrate peptides LLVY-AMC. Proteasomes were isolated from HCV-replicating cells or from JFH1_GND cells (control). MG132 was used to inhibit proteasomal activity. The bars represent the standard deviation. *, p < 0.05. C, oxyblot analysis of cellular lysates derived from HCV positive (JFH1/J6) or negative cells (JFH1_GND). In the case of lanes 5–8 cells were treated for 30 min with 2 mm H2O2. Protein oxidation was analyzed using 2,4,-dinitrophenylhydrazine (DNPH) (lanes 1, 3, 5, 7) for covalent modification of oxidized proteins. 2,4 DNPH was omitted in lanes 2, 4, 6, and 8 to demonstrate the specificity of the observed signals. Probing with a core-specific serum was performed to detect HCV replication. D, analysis of 8-OH-dG formation by ELISA. Cells were treated for 4 h with 150 μm H2O2 before chromosomal DNA was analyzed. 8-OH-dG levels are referred to the control that was arbitrarily set as 1. The values are mean values from three independent experiments. The bars represent the standard deviation. *, p < 0.05.
Nrf2/ARE-regulated genes play an essential role in the protection of cells against damage by electrophiles and ROI (13–15). The observed inhibitory effect of HCV on the induction of Nrf2/ARE-regulated genes suggests an impaired elimination of ROI. To test this hypothesis, we analyzed the formation of oxidized proteins by oxyblots and the formation of DNA adducts by measuring 8-OHdG formation. While in case of untreated cells only a slight increase in the amount of oxidized proteins was observed, a significant difference was found when the intracellular levels of ROI were increased by H2O2 or glucose oxidase. In the case of the lysates derived from the HCV-positive cells a significantly elevated level of oxidized proteins was observed as compared with the lysates derived from the GND control (Fig. 7C).
In accordance with this finding, a significantly elevated level of 8-OHdG formation was observed in H2O2-treated HCV-replicating cells as compared with the control (Fig. 7D). These data indicate that in HCV-replicating cells the protective mechanisms preventing oxidative damage are impaired.
DISCUSSION
In this study we demonstrate the capacity of HCV to impair the induction of Nrf2/ARE-regulated genes. In a recent report it was shown that an increased amount of small Mafs negatively regulates the expression of Nrf2-ARE-regulated genes(33). The preferential formation of sMaf homodimers under these conditions and their binding to the ARE despite the presence of sMaf/Nrf2 heterodimers, were considered as causative for the inhibitory effect. Indeed, in case of HCV-replicating cells in vitro and in vivo an increased amount of sMafs is observed. Interestingly, a detailed analysis revealed that in HCV-replicating cells the sMafs are preferentially localized in the perinuclear region. The observed extranuclear localization of sMafs in HCV-replicating cells suggests that the inhibitory effect on ARE-dependent gene expression results from abnormal localization of sMafs and not from their increased expression with subsequent formation of homodimers. In this case sMafs would be localized in the nucleus(13). The observed delocalization of small Mafs suggests a different mechanism. The subcellular fractionation experiment indicates that in HCV-replicating cells the accumulation of Nrf2 in the nucleus is impaired. Moreover, reporter gene assays demonstrated that co-expression of caNrf2 does not restore ARE-regulated gene expression to the level of the control cell. These data strongly suggest that the extranuclear sMafs might bind Nrf2 and thereby prevent the entry of Nrf2 into the nucleus. Furthermore, due to their extranuclear localization, the small Maf proteins are not available for the formation of Nrf2/sMaf heterodimers in the nucleus, thereby preventing ARE-regulated gene expression (15).
The observed co-localization of sMafs with NS3 in HCV-replicating cells suggests an interplay between core, sMafs and NS3. There is no evidence for a direct interaction of core with sMafs. Core as regulatory protein (34, 35) might trigger intracellular signaling, resulting in posttranslational modification of sMaf and/or NS3 that enables their interaction, resulting in the delocalization of sMaf to the replicon complex.
Our data demonstrated that HCV replication is not affected directly by induction or inhibition of Nrf2/ARE-regulated gene expression. Recent reports based on Con1-based or subgenomic replicons, however, demonstrated that strong overexpression of heme oxygenase 1 (HO-1), which resulted in increased biliverdin levels, suppressed HCV replication. The HO-1 product biliverdin triggers the antiviral interferon response and thereby reduces HCV replication (36, 37). In light of this finding, the HCV-dependent inhibition of Nrf2/ARE-regulated genes, which include the HO-1 gene, could result in decreased biliverdin levels and subsequent stimulation of HCV replication.
The HCV-dependent inhibition of Nrf2/ARE-regulated genes could be relevant for the HCV-associated pathogenesis. During the viral life cycle the virus producing cells encounter an increased level of ROI (8). This can be caused i.e. by the immune response or by ER-overload due to the overproduction of viral membrane proteins. Furthermore, the decreased activity of the constitutive proteasome can lead to an impaired elimination of misfolded proteins and thereby induce cellular stress that confers to increased ROI levels (38–40). Therefore, it may well be that the HCV-dependent inhibition of cytoprotective gene expression results in an impaired detoxification of ROIs. Indeed, oxyblots and analysis of 8-OH-dG formation demonstrated that the elimination of ROIs in HCV-replicating cells is impaired as compared with the control. The elevated ROI level in HCV-replicating cells can affect the genetic integrity of the host cell and thereby contribute to the development of HCC. Apart from this, increased ROI levels could increase the mutation rate of the viral genome and thereby enhance to the genetic variability of the virus. In a recent report it was demonstrated that in Nrf2-deficient mice the process of liver regeneration is impaired due to increased ROI levels that negatively affect insulin/IGF-1 signaling(41). Therefore, the HCV-dependent inhibition of Nrf2/ARE-regulated gene expression could be relevant for the HCV-associated formation of liver fibrosis and cirrhosis due to an inhibitory effect on liver regeneration (42).
Interestingly, in contrast to HCV, hepatitis B virus (HBV) activates the Nrf2/ARE-dependent gene expression (43). In accordance to this a better elimination of ROI can be observed in HBV-positive cells as compared with the control. Moreover, the HBV-dependent induction of Nrf2/ARE-regulated genes results in an increased activity of the constitutive proteasome that is associated with a decreased activity of the immunoproteasome This might aggravate the elimination of HBV-infected cells. In HCV-replicating cells the decreased activity of the constitutive proteasome is not compensated by an increased activity of the immunoproteasome (data not shown).
Taken together, our study revealed a novel mechanism of Nrf2 regulation in virus-infected cells that depends on de-localization of sMaf proteins. The resulting inhibition of Nrf2/ARE-regulated genes might be relevant for the HCV-associated pathogenesis for several reasons: (i) increased ROI levels due to the impaired induction of cytoprotective genes cause DNA-damage in the host cell and could enhance the genetic variability of the viral genome; (ii) elevated ROI levels impair insulin/IGF-1 signaling and thereby negatively affect liver regeneration. In the future it will be interesting to determine if rescue of Nrf2/ARE activity can be therapeutically explored for prevention of HCV-associated pathogenesis.
Acknowledgments
We thank Prof. Sabine Werner and Franziska Lieder, ETH Zurich, for many helpful comments and for the generous gift of the ARE-reporter constructs, and of the Nrf2 mutants. We thank Dr. M. K. Kwak, Yeungnam University, South Korea for the generous gift of the PSMB5-reporter constructs.
Footnotes
- HCV
- hepatitis C virus
- ROI
- reactive oxygen intermediates
- ARE
- antioxidant response elements
- NS
- nonstructural
- DNPH
- 2,4,-dinitrophenylhydrazine.
REFERENCES
- 1. Levrero M. (2006) Oncogene 25, 3834–3847 [DOI] [PubMed] [Google Scholar]
- 2. Choo Q. L., Kuo G., Weiner A. J., Overby L. R., Bradley D. W., Houghton M. (1989) Science 244, 359–362 [DOI] [PubMed] [Google Scholar]
- 3. Hijikata M., Mizushima H., Tanji Y., Komoda Y., Hirowatari Y., Akagi T., Kato N., Kimura K., Shimotohno K. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 10773–10777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Penin F., Dubuisson J., Rey F. A., Moradpour D., Pawlotsky J. M. (2004) Hepatology 39, 5–19 [DOI] [PubMed] [Google Scholar]
- 5. Tellinghuisen T. L., Marcotrigiano J., Gorbalenya A. E., Rice C. M. (2004) J. Biol. Chem. 279, 48576–48587 [DOI] [PubMed] [Google Scholar]
- 6. Boudreau H. E., Emerson S. U., Korzeniowska A., Jendrysik M. A., Leto T. L. (2009) J. Virol. 83, 12934–12946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dionisio N., Garcia-Mediavilla M. V., Sanchez-Campos S., Majano P. L., Benedicto I., Rosado J. A., Salido G. M., Gonzalez-Gallego J. (2009) J. Hepatol. 50, 872–882 [DOI] [PubMed] [Google Scholar]
- 8. Bartosch B., Thimme R., Blum H. E., Zoulim F. (2009) J. Hepatol. 51, 810–820 [DOI] [PubMed] [Google Scholar]
- 9. Wasserman W. W., Fahl W. E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5361–5366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vasiliou V., Ross D., Nebert D. W. (2006) Hum. Genomics 2, 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jaiswal A. K. (2004) Free Radic. Biol. Med. 36, 1199–1207 [DOI] [PubMed] [Google Scholar]
- 12. Aleksunes L. M., Manautou J. E. (2007) Toxicol Pathol. 35, 459–473 [DOI] [PubMed] [Google Scholar]
- 13. Li W., Kong A. N. (2009) Mol. Carcinog. 48, 91–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kensler T. W., Wakabayashi N. (2010) Carcinogenesis 31, 90–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kensler T. W., Wakabayashi N., Biswal S. (2007) Annu. Rev. Pharmacol. Toxicol. 47, 89–116 [DOI] [PubMed] [Google Scholar]
- 16. Blight K. J., McKeating J. A., Rice C. M. (2002) J. Virol. 76, 13001–13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lohmann V., Körner F., Koch J., Herian U., Theilmann L., Bartenschlager R. (1999) Science 285, 110–113 [DOI] [PubMed] [Google Scholar]
- 18. Frese M., Barth K., Kaul A., Lohmann V., Schwärzle V., Bartenschlager R. (2003) J. Gen. Virol. 84, 1253–1259 [DOI] [PubMed] [Google Scholar]
- 19. Lan L., Gorke S., Rau S. J., Zeisel M. B., Hildt E., Himmelsbach K., Carvajal-Yepes M., Huber R., Wakita T., Schmitt-Graeff A., Royer C., Blum H. E., Fischer R., Baumert T. F. (2008) J. Immunol. 181, 4926–4935 [DOI] [PubMed] [Google Scholar]
- 20. Wakita T., Pietschmann T., Kato T., Date T., Miyamoto M., Zhao Z., Murthy K., Habermann A., Kräusslich H. G., Mizokami M., Bartenschlager R., Liang T. J. (2005) Nat. Med. 11, 791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koutsoudakis G., Kaul A., Steinmann E., Kallis S., Lohmann V., Pietschmann T., Bartenschlager R. (2006) J. Virol. 80, 5308–5320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miyanari Y., Atsuzawa K., Usuda N., Watashi K., Hishiki T., Zayas M., Bartenschlager R., Wakita T., Hijikata M., Shimotohno K. (2007) Nat. Cell Biol. 9, 1089–1097 [DOI] [PubMed] [Google Scholar]
- 23. Pietschmann T., Kaul A., Koutsoudakis G., Shavinskaya A., Kallis S., Steinmann E., Abid K., Negro F., Dreux M., Cosset F. L., Bartenschlager R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7408–7413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. auf dem Keller U., Huber M., Beyer T. A., Kümin A., Siemes C., Braun S., Bugnon P., Mitropoulos V., Johnson D. A., Johnson J. A., Hohl D., Werner S. (2006) Mol. Cell. Biol. 26, 3773–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kwak M. K., Kensler T. W. (2006) Biochem. Biophys. Res. Commun. 345, 1350–1357 [DOI] [PubMed] [Google Scholar]
- 26. Himmelsbach K., Sauter D., Baumert T. F., Ludwig L., Blum H. E., Hildt E. (2009) Gut. 58, 1644–1653 [DOI] [PubMed] [Google Scholar]
- 27. Sauter D., Himmelsbach K., Kriegs M., Carvajal Yepes M., Hildt E. (2009) J. Hepatol. 50, 861–871 [DOI] [PubMed] [Google Scholar]
- 28. Ehrhardt C. S., Matzke M., Knoblauch A., Will A., Wixler V., Ludwig S. (2006) Signal Transduction 6, 179–184 [Google Scholar]
- 29. Bürckstümmer T., Kriegs M., Lupberger J., Pauli E. K., Schmittel S., Hildt E. (2006) FEBS Lett. 580, 575–580 [DOI] [PubMed] [Google Scholar]
- 30. Steinmann E., Brohm C., Kallis S., Bartenschlager R., Pietschmann T. (2008) J. Virol. 82, 7034–7046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brandenburg B., Stockl L., Gutzeit C., Roos M., Lupberger J., Schwartlander R., Gelderblom H., Sauer I. M., Hofschneider P. H., Hildt E. (2005) Hepatology 42, 1300–1309 [DOI] [PubMed] [Google Scholar]
- 32. Robek M. D., Garcia M. L., Boyd B. S., Chisari F. V. (2007) J. Virol. 81, 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dhakshinamoorthy S., Jaiswal A. K. (2000) J. Biol. Chem. 275, 40134–40141 [DOI] [PubMed] [Google Scholar]
- 34. Spaziani A., Alisi A., Sanna D., Balsano C. (2006) J. Biol. Chem. 281, 10983–10989 [DOI] [PubMed] [Google Scholar]
- 35. Chou A. H., Tsai H. F., Wu Y. Y., Hu C. Y., Hwang L. H., Hsu P. I., Hsu P. N. (2005) J. Immunol. 174, 2160–2166 [DOI] [PubMed] [Google Scholar]
- 36. Zhu Z., Wilson A. T., Mathahs M. M., Wen F., Brown K. E., Luxon B. A., Schmidt W. N. (2008) Hepatology 48, 1430–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lehmann E., El-Tantawy W. H., Ocker M., Bartenschlager R., Lohmann V., Hashemolhosseini S., Tiegs G., Sass G. (2010) Hepatology 51, 398–404 [DOI] [PubMed] [Google Scholar]
- 38. Oyadomari S., Yun C., Fisher E. A., Kreglinger N., Kreibich G., Oyadomari M., Harding H. P., Goodman A. G., Harant H., Garrison J. L., Taunton J., Katze M. G., Ron D. (2006) Cell 126, 727–739 [DOI] [PubMed] [Google Scholar]
- 39. Jung T., Bader N., Grune T. (2007) Arch. Biochem. Biophys. 462, 231–237 [DOI] [PubMed] [Google Scholar]
- 40. Breusing N., Grune T. (2008) Biol. Chem. 389, 203–209 [DOI] [PubMed] [Google Scholar]
- 41. Beyer T. A., Xu W., Teupser D., auf dem Keller U., Bugnon P., Hildt E., Thiery J., Kan Y. W., Werner S. (2008) EMBO J. 27, 212–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu W., Hellerbrand C., Köhler U. A., Bugnon P., Kan Y. W., Werner S., Beyer T. A. (2008) Lab. Invest. 88, 1068–1078 [DOI] [PubMed] [Google Scholar]
- 43. Schaedler S., Krause J., Himmelsbach K., Carvajal-Yepes M., Lieder F., Klingel K., Nassal M., Weiss T. S., Werner S., Hildt E. (2010) J. Biol. Chem. 285, 41074–41086 [DOI] [PMC free article] [PubMed] [Google Scholar]