Abstract
The oxaloacetate decarboxylase primary Na+ pump (OAD) is an essential membrane protein complex that functions in the citrate fermentation pathway of some pathogenic bacteria under anaerobic conditions. OAD contains three different subunits: Oad-α, a biotinylated extrinsic protein that catalyzes the α-ketodecarboxylation of oxaloacetate; Oad-γ, a structural bitopic membrane protein whose cytosolic tail (named as Oad-γ′) binds tightly to Oad-α; and Oad-β, a multispan transmembrane α-helical protein that constitutes the Na+ channel. How OAD is organized structurally at the membrane and what the molecular determinants are that lead to an efficient energy coupling mechanism remain elusive. In the present work, we elucidate the stoichiometry of the native complex as well as the low resolution structure of the peripheral components of OAD (Oad-α and Oad-γ′) by small angle x-ray scattering. Our results point to a quaternary assembly similar to the pyruvate carboxylase complex organization. Herein, we propose a model in which the association in pairs of Oad-α dimers, mediated by Oad-γ, results in the acquisition of a functional oligomeric state at the bacterial membrane. New structural insights for the conformational rearrangements associated with the carboxylbiotin transfer reaction within OAD are provided.
Keywords: Bioenergetics, Biotin, Computer Modeling, Decarboxylase, Sodium Transport, Molecular Machineries, Energy Coupling, Oxaloacetate Decarboxylase Primary Na+ Pump, Small Angle X-ray Scattering, Sodium Motif Force
Introduction
The metabolism of a variety of bacteria completely relies on the production of a sodium motive force as a source of energy (1). Similarly to the proton cycle, sodium motive force generators (primary Na+ pumps) and sodium motive force consumers are functionally coordinated. The first primary Na+ pump described in bacteria was the oxaloacetate decarboxylase (OAD)5 Na+ primary pump in 1980 (2). OAD is a multifunctional enzyme that converts the free energy of oxaloacetate decarboxylation reaction into an electrochemical gradient of Na+ ions across the membrane (for a review see Ref. 3). This membrane protein complex is functional and essential in the anaerobic citrate fermentation pathway of some bacteria and archaea. It contains three different subunits: Oad-α, a biotinylated extrinsic protein that catalyzes the α-ketodecarboxylation of oxaloacetate; Oad-γ, a structural bitopic membrane protein whose cytosolic tail (named as Oad-γ′) binds tightly to Oad-α; and Oad-β, a multispan transmembrane α-helical protein that constitutes the Na+ channel. The functional complex has generally been assumed to be present in a 1:1:1 stoichiometry (4).
Biochemical studies during the last decades have contributed important information to the mechanism of action of OAD (4–6). As part of an essential metabolic step in the citrate fermentation pathway, the carboxyl transferase (CT) domain of Oad-α catalyzes the decarboxylation of oxaloacetate to pyruvate. The subtracted carboxyl group remains covalently attached to Oad-α via a biotinylated lysine on its biotin carboxyl carrier (BCC) domain. It has been proposed that the carboxyl transfer from the CT domain to the BCC domain provokes a conformational rearrangement in Oad-α that triggers a chemical attack on the reactive carboxylbiotin bond enabled by a Na+-dependent proton transfer from Oad-β (7, 8). CO2 is then released to the cytosol, and Oad-α is ready again for a new catalytic cycle. This reaction is coupled to a molecular reorganization within Oad-β which switches the channel into an open conformation and promotes the translocation of two Na+ ions out of the cytosol. Subsequently, Oad-β consumes a periplasmic-derived proton, and the channel switches back to its original conformation.
Despite the abundance of biochemical data on this particular system, the only structural information currently available on the OAD complex corresponds to the CT domain of Oad-α (9). The high resolution structure of this domain revealed an α8β8 TIM barrel fold in a homodimer organization similar to other members of the biotin carboxylase (BC) family. Because the isolated CT and BCC domains interact in solution (9), an intramolecular exchange of the carboxyl group between the two Oad-α domains was invoked. Besides, Oad-γ has been demonstrated to play an essential role in the catalytic activity of Oad-α (10). Although it was initially proposed that Oad-γ contains a Zn(II)-bound metal and, therefore, an active role of in the catalytic reaction on OAD, the crystal structure of the CT domain demonstrated that indeed Zn(II) is coordinated to Oad-α (9), which resembles the metal requirements for the catalytic activity of other members of the BC family (11). The specific function of Oad-γ in OAD thus remains to be explored. Many structural and functional aspects of Oad-β remain also unknown. Topological studies suggested the presence of nine transmembrane helices in Oad-β (12), and mutational and functional analyses in reconstituted liposomes identified a number of amino acid residues that likely form part of the two Na+ binding sites or participate in the decarboxylation of carboxylbiotin on Oad-α (8, 13–15). The clarification of the interaction mechanism between these two subunits still requires further investigation.
As an essential point in understanding the mechanisms that govern the coordination of events within the OAD complex, we have explored the structural organization of Oad-α and its interaction with the soluble domain of Oad-γ (Oad-γ′) by means of biochemical analysis in combination with homology modeling and structural analyses by small angle x-ray solution scattering (SAXS). Our results strongly suggest that, within its native complex, Oad-α shares similarities with the tetrameric pyruvate carboxylase (PC) protein with respect to the intermolecular transfer of the BCC domain between active sites during the reaction cycle (for a review, see Ref. 16). A model is proposed in which the pair association of Oad-α dimers, mediated by Oad-γ, at the bacterial membrane results in the acquisition of an oligomeric state crucial for enzymatic activity and essential for energy coupling. New mechanistic insights into the conformational rearrangements associated with the carboxylbiotin transfer within the OAD complex are discussed.
EXPERIMENTAL PROCEDURES
Protein Purification
Different Oad-α and Oad-α/γ′ constructs (Fig. 1), corresponding to the protein sequences of the OAD complex isoform 2 from Vibrio cholerae (17), were produced in Escherichia coli Rosetta (C43) strain and purified by standard methodologies. Purification of the OAD complex from E. coli membranes was done essentially as described before (18). Further details can be found in supplemental Experimental Procedures.
FIGURE 1.
Protein constructs used in this study. Different versions of Oad-α from V. cholerae (first column) were co-expressed and purified with or without the predicted soluble domain of Oad-γ (Oad-γ′; second column): a, full-length Oad-α; b, full-length Oad-α and Oad-γ′; c, N-terminal CT domain of Oad-α; d, C-terminal BCC domain of Oad-α together with Oad-γ′; e, BCC domain-depleted Oad-α mutant simultaneously with Oad-γ′; f, the same as b but with a Oad-γ′ double mutant in which Leu72-Leu73 residues were substituted with serines (L72S/L73S). The molecular mass was theoretically calculated for a single polypeptide chain from the amino acid sequence (Mw theor.) and experimentally analyzed by SEC-SLS experiments (Mw SLS), except for construct f, in which the molecular mass estimation was performed from HPLC experiments (Fig. 5). The last column summarizes the estimated oligomerization state in solution derived from this study. The orange rectangle represents a His tag. Note that comparable results were obtained in the absence of the His tag. Values were obtained from Ref. 9; n/d, not determined.
Oligomerization State
The oligomerization states of the different Oad-α and Oad-α/γ′ proteins were assessed by SLS, using a miniDAWN TriStar equipped with an Optilab rEX refractometer (Wyatt Technology Corp.), coupled to size-exclusion chromatography (SEC). Blue native-PAGE (BN-PAGE) was performed as described in Ref. 19.
Stoichiometry Determination of the Oad-α/γ′ Subcomplex by in Vitro Assembly
A peptide corresponding to the predicted soluble α-helical secondary structural element of the Oad-γ subunit (see below) fused to fluorescein isothiocyanate at the N terminus (FITC-KANPNQNQGELLAVLTAAVHHHKTQQKLS) was synthesized using solid phase (University of Lausanne). The in vitro assembly reactions were performed incubating 20 μm Oad-α with a range of concentrations (0–100 μm) of the synthetic peptide at 4 °C in 20 mm Tris-HCl, pH 7.6, 150 mm NaCl, and 2 mm β-mercaptoethanol. After 1 h, the mixture was applied to a Superdex 200 PC 3.2/30 column, and the values of absorbance at 280 nm (protein and peptide) and 494 nm (peptide) were monitored. The amount of bound peptide after incubation with the protein was obtained from the difference between the total peptide concentration and the amount of free peptide obtained from the peak elution area. The concentration of the protein was determined spectrophotometrically at 280 nm.
Estimation of the Apparent Affinity Constant for the Oad-α/γ′ Complex
100 nm FITC-Oad-γ′ was incubated with increasing amounts of Oad-α ranging from 0 to 24 μm in a buffer containing 20 mm Tris-HCl, pH 7.6, 150 mm NaCl, 0.1% BSA, and 0.05% Tween 20. Fluorescence anisotropy was monitored using 485 and 535 nm polarization filters (20 and 25 nm cut-off, respectively) at room temperature in a Tecan UltraEvolution system. Assuming a 2:1 stoichiometry, i.e. one Oad-α dimer per one FITC-Oad-γ′ peptide, the experimental anisotropy data were iteratively fitted to a four-parameter sigmoidal dose-response model using SigmaPlot (Jandel Scientific) that results in estimations of apparent EC50 and the Hill coefficients.
Three-dimensional Modeling
A homology model was built for the Oad-α/γ′ subcomplex by Swiss model server in project mode (20) using the high resolution structure of PC as template.
SAXS
X-ray scattering data were collected at cSAXS (x12sa) beamline at SLS (Paul Scherrer Institute, Villigen) using a Pilatus 2M image plate detector, with a sample to detector distance of 2.15 m at 12.4 keV (1-Å wavelength). SAXS measurements were performed on capillaries of 1-mm diameter at 10 °C. The scattering intensities of 10 different points (0.5-mm distance) in the capillary were acquired for 0.25 or 0.5 s each, and the 10-points scan was repeated 20 times in array. Protein sample solutions were adjusted to concentrations between 2.5 and 7.5 mg/ml in 20 mm Tris-HCl, pH 7.6, 150 mm NaCl, and 2 mm β-mercaptoethanol. After checking for radiation damage, the scans were radially averaged using home-written software.6 Standard analysis and parameters determination were performed using ATSAS software (21), according to standard procedures. Ab initio low resolution models were built by DAMMIN (22) and DAMMIF (23), and model alignment and averaging were performed using DAMAVER (24). The theoretical scattering intensities of models and homologous structures were calculated using CRYSOL (25).
RESULTS
Oligomerization State of Oad-α
Previous results from our group demonstrated that Oad-α CT domain (amino acid residues 1–455) dimerizes in solution (Fig. 1c), and the crystal structure of CTD revealed the detailed structure of the extended dimer interface (9). Here, we have analyzed the oligomerization state of full-length Oad-α in solution using SLS-SEC. With a calculated molecular mass of 69 kDa for monomeric Oad-α, the molecular mass of 140 kDa derived from SLS-SEC suggests a dimeric full-length Oad-α (Fig. 1a). The BCC domain construct (amino acid residues 527–599) was found to behave as a monomer (Fig. 1d). We conclude that Oad-α dimerization occurs through its CT domain. The Stokes radius of Oad-α calculated from the comparison of the retention time on the size-exclusion column with globular standard proteins suggests that the protein has an elongated shape in solution (Fig. 2A, red curve).
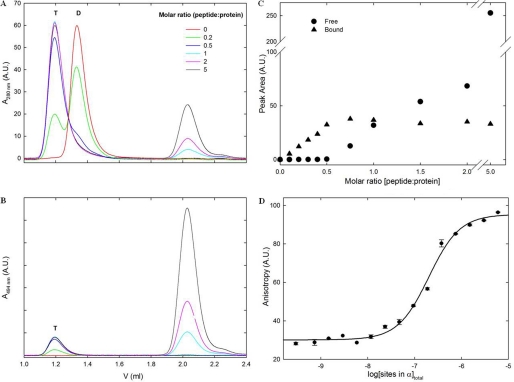
FIGURE 2.
Stoichiometry determination of the Oad-α/γ′ complex. The apparent affinity constant for the Oad-γ′ binding on Oad-α was determined by analytical SEC and steady-state fluorescence anisotropy measurements. During SEC, dimer (D) and tetramer (T) species of Oad-α can be distinguished. A, titration experiment of Oad-α in the presence of different concentrations of a synthetic Oad-γ′ peptide fused at its N terminus to a fluorescence tag (FITC-γ′). B, monitoring the absorbance at 495 nm showed that the FITC-γ′ peptide was either found as free peptide (retention volume of 2.05 ml) or associated with Oad-α tetramers (1.19 ml) but never bound to Oad-α dimers (1.33 ml). C, determination of the apparent affinity constant of Oad-α with FITC-γ′. A fixed amount of FITC-γ′ (100 nm) was titrated with increasing amounts of Oad-α (0–12 μm, dimer concentration), and the change in fluorescence anisotropy of the FITC probe was monitored. D, curve fitting of the fluorescence anisotropy binding assay. A.U., arbitrary units.
Effect of Oad-γ′ on Oad-α Oligomerization
A stable complex between Oad-α and Oad-γ′ (Oad-α/γ′) was generated by co-expressing the two genes encoding each protein in E. coli cells (17). After purification of the Oad-α/γ′ complex to homogeneity via a His tag fused to the N terminus of Oad-γ′, the Oad-α/γ′ molecular mass was estimated by SLS-SEC (Figs. 1b and 2A). Compared with Oad-α, the SEC retention time of the Oad-α/γ′ complex shifted to higher molecular mass (almost double), albeit one Oad-γ′ contributes only 6 kDa to the molecular mass of the complex, and a stoichiometry of 1:1 was expected (4). SLS analysis yielded a value of 260 kDa for the molecular mass of the complex, suggesting dimerization of two Oad-α dimers in the presence of Oad-γ′.
Stoichoimetry and Binding Affinity of the Oad-α/γ′ Complex
The stoichiometry of the Oad-α/γ′ complex was further investigated by in vitro assembly using purified Oad-α and a synthetic Oad-γ′ peptide fused at its N terminus to a fluorescence tag (FITC). The Oad-α/FITC-γ′ complex formation after incubation of a fixed amount of Oad-α with increasing FITC-γ′ peptide concentration was assessed by analyzing the relative concentration of free and bound peptide at 495 nm after separation by analytical HPLC gel filtration (Fig. 2, B and C). Control experiments with purified Oad-α and Oad-α/γ′ and free FITC-γ′ peptide yielded the retention volume of each of these species as 1.19, 1.33, and 2.05 ml, respectively (Fig. 2A). With increasing concentrations of free FITC-γ′, the retention volume of Oad-α shifted to the value observed for the Oad-α/γ′ complex, indicative of a complex containing four Oad-α subunits. Monitoring the absorbance at 495 nm showed that the FITC-γ′ peptide was either found as free peptide (retention volume of 2.05 ml) or associated with Oad-α tetramers (1.19 ml) but never bound to Oad-α dimers (1.33 ml) (Fig. 2B). Apparently, association of Oad-α with FITC-γ′ leads to the formation of higher order oligomers, a process that is complete at a FITC-γ′ concentration half that of Oad-α (molar ratio peptide:monomeric Oad-α equals 0.5), and further addition of FITC-γ′ resulted in increasing concentration of free peptide (Fig. 2, A and B). The most likely stoichiometry for the Oad-α/γ′ complex in solution is therefore 4:2. The calculated molecular mass of such a Oad-α/γ′ complex is 288 kDa, in very good agreement with the molecular mass estimated from SLS-SEC (see above).
To determine the apparent affinity constant, that is, the binding of the FITC-γ′ peptide to an Oad-α dimer (coupled to the formation of higher order oligomers), a fixed amount of FITC-γ′ (100 nm) was titrated with increasing amounts of Oad-α (0–12 μm, dimer concentration), and the change in fluorescence anisotropy of the FITC probe was monitored (Fig. 2D). The fitting of the curve to a sigmoidal dose-response model allowed the determination of the apparent EC50 as 0.2 μm. The derived Hill coefficient is 1.3, indicating that Oad-γ′ binding to the Oad-α dimer and Oad-α/γ′ higher order complex formation are weakly cooperative. In conclusion, these experiments show that Oad-γ′ has a profound effect on the quaternary organization of the Oad-α subunit (Fig. 2A) and that binding and oligomerization are tightly coupled.
Homology Modeling of Oad-α
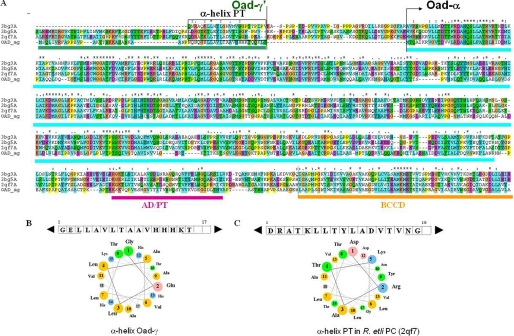
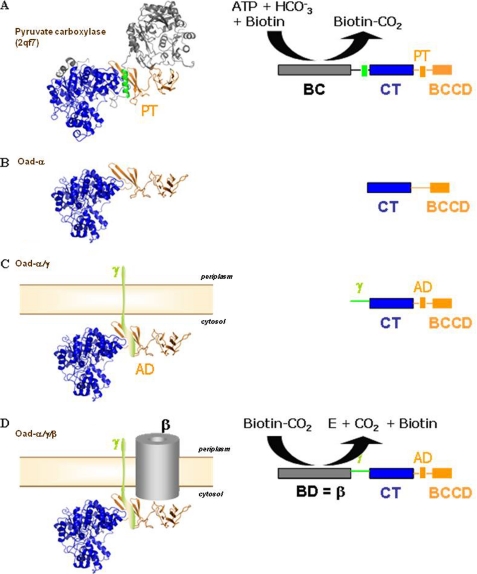
Sequence similarity searches in the protein databases showed that Oad-α is sufficiently similar to PC, without its N-terminal part comprising the ATP-dependent BC domain, to use it as a template for homology modeling of full-length Oad-α (34 and 53% sequence identity and similarity, respectively; Fig. 3). Several structures of different PCs have only recently become known (26–28). The C-terminal part of PC is composed of three domains, the CT domain followed by a small domain responsible for tetramerization (PT domain), and the BCC domain (Fig. 4A). These domains are easily recognized in Oad-α (Figs. 3A and 4B), and homology modeling was performed using as a template the structure of Rhizobium etli PC (Protein Data Bank code 2qf7) that shares the highest sequence identity with Oad-α. The in silico combination of the x-ray structure of the Oad-α CT domain with the computationally modeled C-terminal Oad-α structure, comprising the association and the BCC domains, provided a structural model of full-length Oad-α (Fig. 4B). A significant difference between model and template is the presence of an α-helix packing against the highly twisted β-sheet of the PT domain of PC (Fig. 4A, green). This helical segment is part of the polypeptide chain linking the BC and CT domains of PCs and is absent in Oad-α (compare Fig. 4, A and B). Interestingly, a previous mutational analysis concluded that the equivalent of the PT domain in Oad-α (residues 480–520) is interacting with Oad-γ′(17), the so-called association domain (AD). Besides, Oad-γ′ is predicted to form a soluble amphipathic α-helix of length similar to the α-helix stabilizing the PT domain of PC (Fig. 3C). Indeed, the sequence alignment between PC and Oad-α can be extended by adding the Oad-γ′ sequence upstream of the Oad-α N terminus (Fig. 3A). In this way, the predicted α-helix of Oad-γ′ would substitute for the intrachain α-helix of PC that interacts with its PT domain (Fig. 4, A and C, green color).
FIGURE 3.
Sequence comparison between Oad subunits and PC (without its BC domain). A, multiple sequence alignment between an Oad-α/γ′ chimera and different PC proteins of known structure represented by the codes in the Protein Data Bank: 3bg3, human PC; 3bg5, Staphylococcus aureus PC; 2qf7, R. etli PC; OAD_αγ, Oad-α, and the soluble domain of Oad-γ (Oad-γ′) sequence of V. cholerae oxaloacetate decarboxylase 2. The amino acid sequence of the γ subunit of OAD is marked in green; the CT domain, the association domain/tetramerization domain (AD/TD), and the BCC domain of Oad-α are marked in cyan, purple, and orange, respectively. B, α-helix in PC equivalent to Oad-γ that is involved in PT stabilization is depicted in a rectangle. C, properties of the α-helix in Oad-γ′ related to an equivalent α-helix in PT domain of R. etli are PC illustrated by the amino acid sequence and helical wheel presentation.
FIGURE 4.
Equivalence between PC and OAD systems. Stepwise comparison of different domains in the multimodular proteins is shown. A, high resolution structure and schematic single-chain monomer representation of R. etli PC (PDB code: 2qf7) are shown. PC monomers are composed of four domains: the N-terminal BC, the central CT domain, the tetramerization or allosteric domain (TD), and the BCC domain. B, homology modeling of Oad-α from V. cholera is based on R. etli PC. Oad-α is composed of the N-terminal CT and the C-terminal BCC domains (BCCD) that are homologs to those in 2qf7. C, schematic compares a putative chimera composed of Oad-α/Oad-γ subunits and PC. A similar module organization is noticeable between the two systems: the AD in Oad-α is equivalent to the tetramerization domain in PC, and the soluble α-helical element in Oad-γ likely mimics the α-helix in PC that stabilizes the PT domain (green color in A; see also Fig. 3B). D, although performing the opposite chemical reaction, the Oad-β subunit, acting as a biotin decarboxylase (BD), sharing functional similarities with BC in PC.
Molecular Determinants of the Oad-α/γ′ Complex Formation
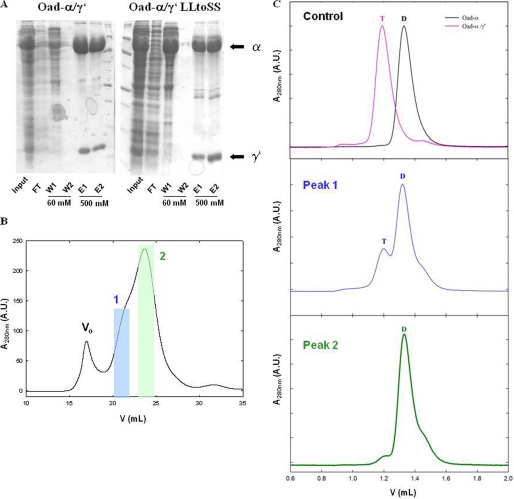
Analysis of the helix-sheet interaction interface in different PCs reveals conserved hydrophobic interactions that appear important for stabilization of the PT domain structure. To verify experimentally whether similar interactions apply to the postulated Oad-γ′/Oad-α interaction, we designed two independent double mutants in which candidate hydrophobic residues of Oad-γ′ were disrupted (Fig. 3A). More specifically, Leu72-Leu73 and Val75-Leu76 in Oad-γ′ were substituted with serines (L72S/L73S Oad-γ′ and V75S/L76S Oad-γ′, respectively), and each double mutant was co-expressed with Oad-α in E. coli cells. Oad-α and Oad-γ′ association was assessed in a pulldown assay analyzing the occurrence of Oad-α in the eluate upon Oad-γ′ purification via a fused His tag at its N terminus (Fig. 1f). Whereas the His-tagged L72S/L73S Oad-γ′ mutant co-elutes with Oad-α during affinity purification (Fig. 5A), co-expression with the V75S/L76S Oad-γ′ mutant does not yield a soluble complex, and both gene products are present in inclusion bodies (data not shown). Interestingly, the binding affinity of the L72S/L73S Oad-γ′ mutant to Oad-α was seriously compromised and resulted in the dissociation of the Oad-α/γ′ complexes into Oad-α dimers during SEC (Fig. 5, B and C). Based on these results, we propose that Oad-γ′ binds in an α-helical conformation to one face of the β-sheet of the Oad-α association domain (Fig. 4C) as predicted from homology modeling (Fig. 4A).
FIGURE 5.
Disruption of Oad-α and Oad-γ′ interaction by point mutations in Oad-γ′. A, SDS-PAGE protein separation of Oad-α/γ′and Oad-α/γ′L72S/L73S complexes (constructs b and f, respectively, Fig. 1) during protein affinity purification. Input, flow-through (FT), first (W1) and last (W2) washes with 60 mm imidazole and elution with 500 mm imidazole (E1 and E2) are shown. B, quaternary structure analysis of the mutant complex by SEC. Two peaks containing Oad species are clearly distinguished (blue and green, respectively). C: first panel on the right, control experiment in which the elution volume of Oad-α and Oad-α/γ′ allows distinguishing between dimer (D) and tetramer (T) species. Second and third panels on the right, second run over a SEC column of peaks 1 (blue) and 2 (green), respectively. Dimer or tetramer species are indicated with a D and T, respectively. Note that Oad-γ′ does not absorb at 280 nm due to the absence of aromatic amino acids in its sequence.
PC and OAD Structural Comparisons
Structurally, PCs are homotetrameric assemblies held together by distinct symmetry interfaces (26–28). Essential structural interactions in PC are mediated by CT and PT domains; the BC interface is, however, not mandatory for a proper assembly (27). Interestingly, the CT-CT dimer interface found in PC structures is conserved in the structure of the CT domain of OAD (9). Based on the sequence and structural similarities between the two systems, it is tempting to speculate that the Oad-α/γ′ subcomplex is the result of the AD-mediated interaction of two Oad-α dimers, which is dependent on the Oad-γ′-mediated stabilization of the association domain in Oad-α in much the same way as observed with the PT domain of PCs. To demonstrate that the PT domain is truly involved in the higher order oligomer formation of Oad-α, we prepared an Oad-α mutant devoid of the BCC domain (Oad-αΔBCCD), and this mutant was co-expressed with Oad-γ′ in E. coli cells. As expected, the Oad-αΔBCCD/γ′ complex was highly stable, and the formation of the 4(Oad-α)/2(Oad-γ′) complex was not impaired (Fig. 1e). Altogether, our results indicate a similar oligomer organization. between PC (without the BC domain) and Oad-α/γ′.
Solution Structure Analysis of Oad-α Complexes by SAXS
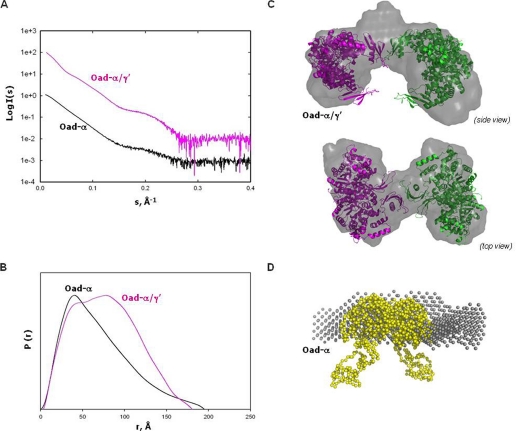
To validate our hypothesis of a similar quaternary organization between OAD and PC, SAXS experiments of the Oad-α/γ′ complex and the Oad-α subunit were performed. The experimental scattering curves showed no dependence on protein concentration. Kratky plots corresponded to folded proteins, and the Guinier plots showed a linear correlation between the scattered intensity and the q distance at values of qRg lower than 1.3 (data not shown). The SAXS profiles extrapolated to zero concentration are shown in Fig. 6A, and the basic hydrodynamic parameters are summarized in Table 1. Estimated excluded Porod volumes were consistent with a 4(α)/2(γ′) assembly for Oad-α/γ′ and a dimer for Oad-α (Table 1). The pair-distance distribution functions calculated from the SAXS data (Fig. 6B) revealed the presence of two maxima (about 43 and 72 Å; Table 1) for the Oad-α/γ′ complex, i.e. two characteristic distances appear with a higher probability, indicating a dumbbell-like structure. On the other hand, the pair-distance distribution function derived from Oad-α SAXS data in the absence of Oad-γ′ is unimodal and has the characteristic end shape of an elongated particle. Thus, the first peak likely represents pair-distances within an Oad-α dimer whereas the second peak is related to distances between two Oad-α dimers formed in the presence of Oad-γ′.
FIGURE 6.
SAXS analysis of Oad-α/γ′ and Oad-α. A, extrapolation to zero concentration of the SAXS experimental scattering profiles. B, pair-distribution functions of the curves in A. C, rigid-body modeling and ab initio shape reconstruction (gray) of Oad-α/γ′. Two Oad-α dimers (pink and green, respectively) are fitted into the ab initio models. The two dimers interact by two association domains coupled with two Oad-γ′ subunits. D, overlapping of a Oad-α dimer model (yellow) constructed by fusion of the 2nx9 and PT-BCC domain from 2qf7 with the low resolution envelope obtained from ab initio shape reconstitution (gray) from SAXS-derived data of purified Oad-α protein.
TABLE 1.
Biophysical properties of the Oad-α/γ′ constructs derived from SAXS
| Construct | Rga | Dmaxb | First peakc | Second peakc | Vporod (× 103) | Molecular massd |
|---|---|---|---|---|---|---|
| Å | Å | kDa | ||||
| Oad-α/γ′ | 59.2 ± 0.8 | 180 | 43.2 | 72 | 443.7 | 266 |
| Oad-α | 55 ± 2 | 200 | 38.4 | 252.7 | 151 |
a Radius of gyration determined from Guinier analysis.
b Maximum particle distance determined by GNOM.
c Maximum peak position in the pair-distribution function.
d Molecular mass determination of the scatter species determined from the Porod volume.
Low Resolution Shape Restoration of the Oad-α Complexes by SAXS
Different crystallographic models for the tetramer organization of PC are known, in which substantial differences in dimer-to-dimer orientations and compactness of the domains are observed (26–28). When the theoretical scattering profiles for these crystallographic models depleted of the BC domain are calculated (supplemental Fig. 1), a systematic deviation is in the 0.12–0.13 Å−1 region of the scattering curve, suggesting some dissimilarities between PC crystal structures and the Oad-α/γ′ protein in solution (29). Several studies have already pointed out an intrinsic built-in flexibility for the BCC domain that is essential for complex functionality in terms of domain coordination and communication mechanisms (16, 30). According to the detected similarities in sequence and structure in the OAD and PC systems, some degree of flexibility is also expected in OAD, which might preclude flexible BCC domains from reliable modeling. A SAXS-derived ab initio model is shown in Fig. 6C. It contains a central dense region with two nonparallel arms oriented oppositely (Fig. 6C). Each arm has the dimensions of one Oad-α dimer, and the central region would represent the interaction between two association domains together with the flexible BCC domains interacting in a fashion similar to the PT domains in PCs (Fig. 6C). Despite the low resolution, the SAXS-derived model pointed out an overall oligomeric organization similar to that in PC but, unlike PC, the AD-mediated interaction between monomers in one of the PC layers would be broken in Oad-α/γ′. These observations are in accordance with the finding that only one Oad-γ′ subunit is found per Oad-α dimer (see above), and therefore just one AD interface would become available for interaction per Oad-α dimer in the presence of Oad-γ′. Our data do not directly show which of the four putative binding sites in Oad-α are bound to Oad-γ′, but it appears reasonable to suggest that the two interacting association domains have Oad-γ′ bound. This would explain the differences in the SAXS scattering profiles between PC and OAD discussed above.
We have also analyzed the SAXS data obtained from the Oad-α dimer. In the absence of Oad-γ′, the Oad-α dimer has a strongly elongated shape according to the SAXS measurements (Fig. 6, B and D). The known dimeric structure of the CT domain of Oad-α (9) can be fitted well into the central part of the envelope, and the considerable extra density remaining at both ends could be attributed to the additional residues that composed the AD and BCC domains and are likely floppy in solution (Fig. 6D), probably because the association domain is not fully structured in the absence of Oad-γ′. We proposed that Oad-γ′ binding is necessary to structure the AD region, similar to what it is observed in PCs, creating a valid interaction interface that results in the association of two Oad-α dimers. Further, this would explain why the Oad-α subunit has only basal catalytic activity in the absence of the Oad-γ subunit (31).
Oligomerization State of the OAD Membrane Complex
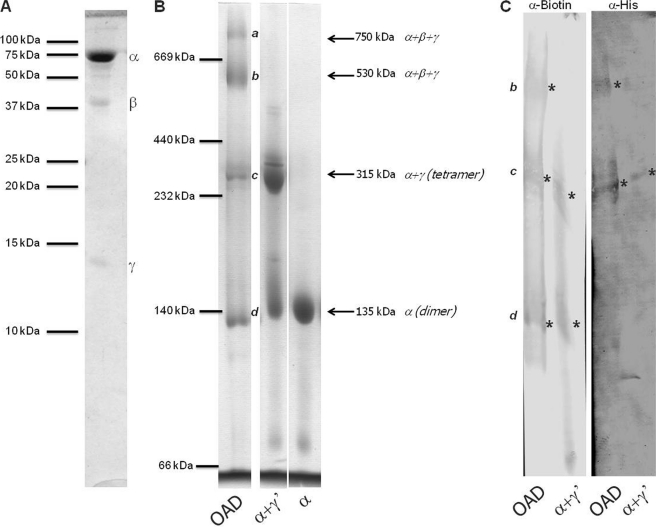
An obvious question thus was whether this tetramerization is also relevant for the structure of the native membrane complex. To this end, we have done BN-PAGE to analyze the OAD membrane protein complex in Triton X-100-solubilized membranes of E. coli, a detergent already known to maintain the integrity of the complex (18). As shown in Fig. 7, the OAD complex does not migrate as a discrete band in BN-PAGE but rather with four distinct molecular masses of about 135, 315, 530, and 750 kDa (estimated using soluble protein as molecular markers). When the running behavior of OAD in BN-PAGE is compared with purified Oad-α and Oad-α/γ′ proteins, it is easily recognized that the bands at 135 kDa and 315 correspond to Oad-α and Oad-α/γ proteins, respectively (Fig. 7B). This suggests that the higher molecular mass bands are a consequence of the presence of Oad-β. Western blotting analysis using anti-biotin (for Oad-α) confirmed the presence of these subunits in the 135, 315, and 530 kDa bands, whereas anti-His tag antibodies (for Oad-γ′) reacted with the bands at 315 and 530 kDa (Fig. 7C). The reactivity with the 750 kDa was very weak, and no further conclusions could be made. Besides, a second dimension SDS-PAGE demonstrates that only the bands at 530 and 750 kDa contain Oad-β. The nature of each band at the BN-PAGE was additionally analyzed by mass spectrometry (data not shown). It was found that both the 750 and 530 kDa bands are constituted of the three subunits and may represent different protein oligomerization state of the OAD complex or different lipid/detergent composition. The 315 kDa band is composed just of Oad-α, and Oad-γ and may represent the tetrameric complex (theoretical molecular mass of 288 kDa). The band at 135 kDa is solely composed of Oad-α (theoretical molecular mass of 140 kDa). Assuming that the running behavior of hydrophobic membrane proteins in BN-PAGE is perturbed by the (unknown) amount of lipids, detergent molecules and the Coomassie dye, the most prominent band at 530 kDa is likely composed of a tetrameric Oad-α/γ plus a dimeric (or tetrameric) Oad-β. Further studies are necessary to assess the stoichiometry of Oad-β in the OAD complex. Nevertheless, these results confirm the hypothesis that Oad-α in the presence of Oad-γ is organized as a tetramer in a native complex and, consequently, at the membrane.
FIGURE 7.
Stoichiometry analysis of the OAD complex in the membrane. A, 16% SDS-PAGE of the purified recombinant OAD membrane complex. B, 6.5–12% BN-PAGE of the purified OAD membrane complex, Oad-α/γ′ complex, and Oad-α dimer. Coomassie-detected bands are marked from a to d. C, immunoblot analysis of the BN-PAGE bands from OAD using antibodies against Oad-α (α-Biotin) and Oad-γ (α-His).
DISCUSSION
This work provides the first structural information for the full-length α-subunit of OAD in complex with the soluble domain of its Oad-γ′ interaction partner. The main conclusions derived from our study are: (i) binding of Oad-γ seems to reduce the intrinsic flexibility of Oad-α around residues 480–520 by stabilizing the 4-stranded β sandwich with Oad-γ likely acting as a hinge; (ii) Oad-γ promotes the dimerization of Oad-α dimers in such a way that Oad-α/γ′ association and Oad-α tetramerization have a reciprocal dependence; (iii) Oad-α/γ′ organization in solution resembles that of the PC. Interestingly, PC has an intrinsic OAD activity because of its ability to stabilize a pyruvate enolate intermediate (32). Similarly, OAD is able to catalyze pyruvate carboxylation in vitro under certain conditions (33). The common features of Oad-α/γ′ and PC and the reversibility of the catalyzed reactions are indicative of a similar quaternary structure organization, and, therefore, PC might serve as a useful system to get insights into OAD structure and mechanism.
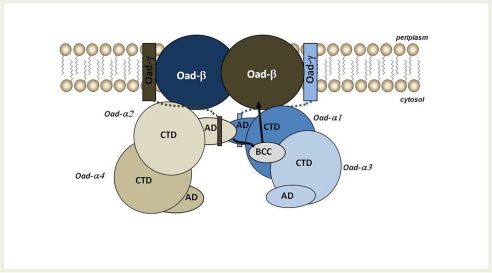
A key feature of pyruvate carboxylase functionality is the way in which the carboxylbiotin, attached to a conserved lysine of the BCC domain, is transferred between the BC of its own monomer and the active site of the CT domain of a monomer which shares no interface with it in the homotetrameric assembly (16). Previously, a swing mechanism has been considered for OAD in which the carboxyl group removed from oxaloacetate at the active site of the CT domain of Oad-α is transferred via the biotin prosthetic group of its own BCC domain to the decarboxylation site of Oad-β at the membrane (17). However, how this large movement occurs was unclear. Based on our recent experimental results, sequence comparisons and structural homology considerations as described above, we propose a new model of carboxylbiotin transfer from the CT domain to the decarboxylation site at the membrane for the OAD complex (Fig. 8). This new model implies a dimeric organization of two Oad-α dimers, whose functional conformation is only achieved upon the interaction with the soluble C-terminal α-helix of the Oad-γ subunit at the membrane. Once at the membrane, Oad-α would function following a swing mechanism in which the biotin at the BCC domain of one Oad-α monomer is carboxylated by the CT domain of another Oad-α monomer in the opposite dimer, itself carboxylated by OAA in the cytosol (Fig. 8). In the proposed mechanism, the four monomers of Oad-α are involved in the coupled reaction but with different roles: the decarboxylation reaction mediated by the CT domains of two Oad-α monomers in opposite dimers would perform the carboxylation reaction of the BCC domains of the other two monomers that are stabilized by interactions of their association domains mediated by Oad-γ, close to the membrane. The transfer reaction within the Oad-α/γ complex would promote a motion of the carboxylated BCC domain to position the carboxylbiotin close to the catalytic site in Oad-β. Concomitant with biotin decarboxylation, a conformational rearrangement of Oad-β would result in Na+ translocation through the membrane and, eventually, OAD would be ready for a new catalytic cycle. This model explains the observed stoichiometry of the Oad-α/γ complex: due to the presence of the bacterial lipidic membrane, only two Oad-γ subunits could interact with a tetrameric Oad-α. Thus a half-site reaction mechanism is envisaged, similar to that proposed for PC (16). The model suggests that Oad-β is a dimer in the membrane, occupying the position of two-interacting BC domain in PC. Indeed, albeit catalyzing reverse reactions, the BC domains in PC is conceptually equivalent to the membrane-embedded Oad-β subunit that must interact with the BCC domains during the catalytic cycle to perform the (de)carboxylation of biotin (Fig. 4D).
FIGURE 8.
Working model of OAD complex organization in the membrane. According to this model, two Oad-α dimers form a functional unit upon interaction with Oad-γ at the membrane. The soluble α-helices of two intrinsic Oad-γ subunits (shown as rectangles connected to the membrane by dotted lines) would promote the interaction between AD of two Oad-α monomers of opposite dimers. As a consequence, the BCC domain of one AD-mediated interacting Oad-α subunit would be located near the CT domain of an opposite monomer. The most likely mechanism supports a conformational flexibility of the BCC domain and would involve the carboxyl transfer to biotin in the BCC domain from the CT domain of an opposite monomer. Upon a conformational reorganization, the BCC domain would move toward one Oad-β subunit at the membrane, favoring its decarboxylation that results in the translocation of sodium ions across the bacterial membrane.
In summary, herein we present a new working model for OAD in bacteria, based on its homology to the PC system. According to this model the decarboxylation of oxaloacetate to pyruvate catalyzed by the Oad-α subunit would occur at the bacterial membrane when Oad-α acquires an active conformation via its interaction with the intrinsic Oad-γ. The interaction with the intrinsic subunit would promote the dimerization of Oad-α dimers in much the same way as PC tetramers are formed in eukaryotes and bacteria. Besides, Oad-α interaction with Oad-γ would position the biotin cofactor close to the membrane channel, Oad-β, favoring the energy transduction across the bacterial membrane. Again, the ternary complex in OAD shows mechanistic similarities with PC. The working model presented here represents a useful system to design new experiments and further characterize the long range communication and the coordination mechanism functioning in OAD.
Supplementary Material
Acknowledgments
We thank John Missimer for support in SAXS data collection and processing and Andreas Menzel at cSAXS-PSI for technical support.
This work was supported by Swiss National Science Foundation Grant 1122.0036 NF (to X.-D. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1, Experimental Procedures, and additional references.
J. Missimer, unpublished results.
- OAD
- oxaloacetate decarboxylase
- AD
- association domain
- BC
- biotin carboxylase
- BCC
- biotin carboxy carrier
- BN-PAGE
- blue native-PAGE
- CT
- carboxyl transferase
- PC
- pyruvate carboxylase
- PT
- pyruvate tetramerization
- SAXS
- small angle x-ray scattering
- SEC
- size exclusion chromatography
- SLS
- static light scattering.
REFERENCES
- 1. Mulkidjanian A. Y., Dibrov P., Galperin M. Y. (2008) Biochim. Biophys. Acta 1777, 985–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dimroth P. (1980) FEBS Lett. 122, 234–236 [DOI] [PubMed] [Google Scholar]
- 3. Dimroth P., Jockel P., Schmid M. (2001) Biochim. Biophys. Acta 1505, 1–14 [DOI] [PubMed] [Google Scholar]
- 4. Dimroth P., Thomer A. (1983) Eur. J. Biochem. 137, 107–112 [DOI] [PubMed] [Google Scholar]
- 5. Dimroth P. (1982) Eur. J. Biochem. 121, 435–441 [DOI] [PubMed] [Google Scholar]
- 6. Dimroth P., Thomer A. (1993) Biochemistry 32, 1734–1739 [DOI] [PubMed] [Google Scholar]
- 7. Di Berardino M., Dimroth P. (1996) EMBO J. 15, 1842–1849 [PMC free article] [PubMed] [Google Scholar]
- 8. Jockel P., Schmid M., Choinowski T., Dimroth P. (2000) Biochemistry 39, 4320–4326 [DOI] [PubMed] [Google Scholar]
- 9. Studer R., Dahinden P., Wang W. W., Auchli Y., Li X. D., Dimroth P. (2007) J. Mol. Biol. 367, 547–557 [DOI] [PubMed] [Google Scholar]
- 10. Schmid M., Wild M. R., Dahinden P., Dimroth P. (2002) Biochemistry 41, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 11. Jitrapakdee S., Wallace J. C. (2003) Curr. Protein Peptide Sci. 4, 217–229 [DOI] [PubMed] [Google Scholar]
- 12. Jockel P., Di Berardino M., Dimroth P. (1999) Biochemistry 38, 13461–13472 [DOI] [PubMed] [Google Scholar]
- 13. Jockel P., Schmid M., Steuber J., Dimroth P. (2000) Biochemistry 39, 2307–2315 [DOI] [PubMed] [Google Scholar]
- 14. Schmid M., Vorburger T., Pos K. M., Dimroth P. (2002) Eur. J. Biochem. 269, 2997–3004 [DOI] [PubMed] [Google Scholar]
- 15. Wild M. R., Pos K. M., Dimroth P. (2003) Biochemistry 42, 11615–11624 [DOI] [PubMed] [Google Scholar]
- 16. Jitrapakdee S., St. Maurice M., Rayment I., Cleland W. W., Wallace J. C., Attwood P. V. (2008) Biochem. J. 413, 369–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dahinden P., Pos K. M., Dimroth P. (2005) FEBS J. 272, 846–855 [DOI] [PubMed] [Google Scholar]
- 18. Dahinden P., Auchli Y., Granjon T., Taralczak M., Wild M., Dimroth P. (2005) Arch. Microbiol. 183, 121–129 [DOI] [PubMed] [Google Scholar]
- 19. Wittig I., Braun H. P., Schägger H. (2006) Nat. Protocols 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 20. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 21. Konarev P. V., Petoukhov M. V., Volkov V. V., Svergun D. I. (2006) J. Appl. Crystallogr. 39, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Svergun D. I. (1999) Biophys. J. 76, 2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Franke D., Svergun D. I. (2009) J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Volkov V. V., Svergun D. I. (2003) J. Appl. Crystallogr. 36, 860–864 [Google Scholar]
- 25. Svergun D., Barberato C., Koch M. H. J. (1995) J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 26. St. Maurice M., Reinhardt L., Surinya K. H., Attwood P. V., Wallace J. C., Cleland W. W., Rayment I. (2007) Science 317, 1076–1079 [DOI] [PubMed] [Google Scholar]
- 27. Xiang S., Tong L. (2008) Nat. Struct. Mol. Biol. 15, 295–302 [DOI] [PubMed] [Google Scholar]
- 28. Yu L. P., Xiang S., Lasso G., Gil D., Valle M., Tong L. (2009) Structure 17, 823–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Putnam C. D., Hammel M., Hura G. L., Tainer J. A. (2007) Q. Rev. Biophys. 40, 191–285 [DOI] [PubMed] [Google Scholar]
- 30. Lasso G., Yu L. P., Gil D., Xiang S., Tong L., Valle M. (2010) Structure 18, 1300–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Di Berardino M., Dimroth P. (1995) Eur. J. Biochem. 231, 790–801 [DOI] [PubMed] [Google Scholar]
- 32. Attwood P. V., Cleland W. W. (1986) Biochemistry 25, 8191–8196 [DOI] [PubMed] [Google Scholar]
- 33. Dimroth P., Hilpert W. (1984) Biochemistry 23, 5360–5366 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.