Abstract
In the presence of oligomeric chondroitin 4-sulfate (C4-S), cathepsin K (catK) forms a specific complex that was shown to be the source of the major collagenolytic activity in bone osteoclasts. C4-S forms multiple contacts with amino acid residues on the backside of the catK molecule that help to facilitate complex formation. As cathepsin L does not exhibit a significant collagenase activity in the presence or in the absence of C4-S, we substituted the C4-S interacting residues in catK with those of cathepsin L. Variants revealed altered collagenolytic activities with the largest inhibitory effect shown by the hexavariant M5. None of the variants showed a reduction in their gelatinolytic and peptidolytic activities when compared with wild-type catK, indicating no structural alteration within their active sites. However, the crystal structure of the M5 variant in the presence of oligomeric C4-S revealed a different binding of chondroitin 4-sulfate. C4-S is not continuously ordered as it is in the wild-type catK·C4-S complex. The orientation and the direction of the hexasaccharide on the catK surface have changed, so that the hexasaccharide is positioned between two symmetry-related molecules. Only one M5 variant molecule of the dimer that is present in the asymmetric unit interacts with C4-S. These substitutions have changed the mode of catK binding to C4-S and, as a result, have likely affected the collagenolytic potential of the variant. The data presented here support our hypothesis that distinct catK/C4-S interactions are necessary for the collagenolytic activity of the enzyme.
Keywords: Chondroitin Sulfate, Collagen, Cysteine Protease, Protein Domains, Protein Structure, Protein-Protein Interactions, Proteoglycan Structure, Cathepsin, Collagenase
Introduction
Collagen degradation is a natural process observed in bone remodeling, wound healing, and organ development. However, excessive collagen degradation is implicated in many serious diseases such as osteoporosis, different forms of arthritis, and some vascular disorders. Triple-helical collagens are the major organic components of bone matrix (type I collagen) and of cartilage (type II collagen). These collagens are highly resistant to general proteolysis, and it requires specific peptidases for their degradation. Cathepsin K (catK),7 a member of the papain family of lysosomal cysteine peptidases, is the predominant peptidase of bone-degrading osteoclasts (1, 2). It has the ability, unique among mammalian proteinases, of cleaving triple-helical collagen at multiple sites (3, 4). Analysis of catK activities revealed that the collagen-degrading activity is not proportional to the general peptidase activity suggesting some other factors affect the specificity of catK. It was shown (5, 6) that the ability to cleave collagen is highly dependent on the formation of an oligomeric complex of catK with glycosaminoglycans, in particular, with chondroitin 4-sulfate (C4-S). Glycosaminoglycans are naturally present in bone and cartilage in sufficient quantities to form complexes with catK. Complex formation with glycosaminoglycans is unique for catK among other papain-like proteases (6, 7). It is remarkable that monomeric catK has no significant collagenase activity (6). Therefore, any disruption of the complex formation between catK and glycosaminoglycans might provide a therapeutic effect in collagen degradation-related diseases. On the other hand, the inhibition of catK by excess concentrations of glycosaminoglycans might be a contributing factor to bone anomalies as seen in mucopolysaccharidoses (8). Earlier we had solved the structure of the WT catK:C4-S complex that showed that each hexasaccharide segment of the C4-S binds one catK molecule (9). As a result, an oligomer of 17-kDa C4-S binds 12 catK molecules. The C4-S oligosaccharide runs continuously through the crystal lattice and spans six unit cell lengths of the a axis (42.0 Å). The interpretation of WT catK·C4-S complex structure did not fully explain the effect that C4-S binding had on the catK collagenolytic activity. The oligosaccharide chain binding site was far from the active site Cys25, and the latter was completely exposed to substrates. There were no changes in the catK structure compared with the many structures of catK determined earlier in the absence of C4-S. At least 12 amino acid residues on the catK surface were identified as being involved in the C4-S binding. As cathepsin L (catL) does not show a significant collagenase activity neither in the presence nor absence of C4-S (7), we have substituted six C4-S interacting residues that are specific for catK binding by the appropriate residues present in catL. The aim of this study has been to determine whether these residues contribute to the formation of the collagenolytically active C4-S·catK complex. Several variants addressing the individual interactions were generated, and their activities toward triple-helical collagen and noncollageneous substrates were determined. The hexavariant (M5) revealed that it has the strongest effect on the collagenolytic activity (∼60% inhibition). Interestingly, this variant is still capable of binding C4-S, though in an altered manner. This strongly suggests that the collagenase activity of cathepsin K requires a highly specific interaction with C4-S. Here, we report the structure of the M5 catK variant in complex with the 17-kDa C4-S.
EXPERIMENTAL PROCEDURES
Materials
Z-Gly-Pro-Arg-MCA and Z-Leu-Arg-MCA were purchased from Bachem Feinchemikalien, Inc. (Bubendorf, Switzerland). C4-S, DTT, and EDTA were obtained from Sigma. Recombinant WT human catK was expressed in Pichia pastoris as described previously (10).
Construction of CatK Variants Containing Substitutions at Chondroitin 4-Sulfate Binding Sites and Their Expression in P. pastoris
Human pro-catK cDNA (11) was used as a template to construct catK variants at C4-S binding sites by site-directed mutagenesis. For the creation of multiple site variants, appropriate variant cDNAs were used as the PCR template. Each point substitution was introduced into the cDNA using the PCR ligation method with Pfu polymerase (New England Biolabs, Beverly, MA). The catK variants K9E (M1), K9E/I171E/Q172S (M2), N190M/K191G/L195K (M3), K9E/N190M/K191G/L195K (M4), and K9E/I171E/Q172S/N190M/K191G/L195K (M5) were designed to contain the corresponding cathepsin L residues (Table 1). Oligonucleotides used for mutagenesis were as follows: K9E, 5′-GAC TCT GTC GAC TAT CGA GAG AAA GGA TAT GTT ACT CCT-3′ (primer 1); I171E/Q172S, 5′-GCA GTG GGA TAT GGA GAG TCG AAG GGA AAC AAG CAC TGG-3′ (primer 2), and N190M/K191G/L195K, 5′-GGA GAA AAC TGG GGA ATG GGA GGA TAT ATC AAA ATG GCT CGA AAT AAG AAC AAC GC-3′ (primer 3). The substituted nucleotides at protein positions 9, 171, 172, 190, 191, and 195 are underlined. All three primers were synthesized in the forward and reverse orientations. The flanking primers were 5′-CGG TCG TAA CGA CGA TTT CTT CTT CCC CAT A-3′ and 5′-GGC AAA TGG CAT TCT GAC ATC CTC TTG ATT-3′. To generate the appropriate cDNAs for the individual variants, primer set 1 was used to generate variant 1 using WT catK cDNA as template. Variant 2 was generated with primer set 2 using variant 1 cDNA as template, variant 3 with primer set 3 with WT catK cDNA template, variant 4 with primer set 3 and variant 1 cDNA as template, and variant 5 with primer set 2 and variant 4 cDNA as template. Amplified PCR products were cleaved by EcoRI and NotI, at the 5′ and 3′ ends and were ligated into the pPIC9K expression vector that had its counterpart excised by the same restriction enzymes (Invitrogen). The substitutions were subsequently confirmed by DNA sequencing using a series of primers derived from either the vector or the internal cDNA sequences. Sequence analyses were performed using an Applied Biosystems model 3777 automated sequencer. The pPIC9K vectors each containing the appropriate variant cDNA were linearized with BglII and then electroporated into P. pastoris GS115 cells (Invitrogen). Several catK variant protein expressing clones were obtained after phenotype screening according to the manufacturer's instructions.
TABLE 1.
Cathepsin K variants
| catK variants | Residues mutated |
|---|---|
| Variant 1 (M1) | K9E |
| Variant 2 (M2) | K9E, I171E, Q172S |
| Variant 3 (M3) | N190M, K191G, L195K |
| Variant 4 (M4) | K9E, N190M, K191G, L195K |
| Variant 5 (M5) | K9E, I171E, Q172S, N190M, K191G, L195K |
Clones were grown in shaker flasks for 4 days, and the culture supernatant was concentrated using an YM10 ultrafiltration membrane with a cut-off size of 10 kDa (Amicon Inc., Beverly, MA). The expression of the variant protein was detected by Western blot analysis using a rabbit polyclonal antibody raised against human catK (MS2) as described previously (12).
Activation of Precursor Protein and Purification of Active CatK Variants
The conversion of the precursor form into the active enzyme was accomplished by treatment with pepsin as described previously (13). Briefly, the concentrated yeast culture supernatant containing the precursor forms of catK variants was diluted with 2.5 volumes to a final solution containing 0.1 m sodium acetate, pH 4.0, 2.5 mm DTT, and 2.5 mm EDTA. Porcine pepsin (Sigma) at a final concentration of 0.4 mg/ml was added, and the mixture was incubated for several minutes at 37 °C. The activation was monitored using the fluorogenic substrate Z-Leu-Arg-MCA (2 μm) and the activation was stopped at reaching maximal activity by increasing the pH to 5.5. The activated form of each variant protein was purified by ion-exchange chromatography using N-butyl-Sepharose 4 Fast Flow (Amersham Biosciences) as described (14) and proved homogeneous in SDS electrophoresis. The active site concentration of each variant peptidase was determined by titration with E64 (15).
Substrate Assays using Z-Peptidyl-MCA Peptides
Steady state kinetics were performed with the fluorogenic substrates Z-Leu-Arg-MCA and Z-Gly-Pro-Arg-MCA as described previously (11). The excitation and emission wavelengths used for the assays were set at 380 and 460 nm, respectively, using the PerkinElmer spectrofluorometer LB50. Michaelis-Menten constants (Km) and specificity constants (kcat/Km) were determined using nonlinear regression analysis. All enzymes were assayed at room temperature at fixed enzyme concentrations (1–5 nm) and variable substrate concentrations (1–200 μm) in 100 mm sodium acetate buffer, pH 5.5, containing 2.5 mm DTT, and 2.5 mm EDTA. Stability measurements of cathepsin activities in the presence or absence of C4-S were conducted by incubating 600 nm WT or variant proteins for 8 h in 100 mm sodium acetate buffer, pH 5.5, containing 2.5 mm DTT, 2.5 mm EDTA, and 0.4 mg/ml of soluble calf skin type I collagen at 28 °C. At indicated time points, residual activities toward the synthetic substrate Z-Leu-Arg-MCA (2 μm) were determined.
Collagen and Gelatin Digests
Active WT catK (600 nm) and the five catK variant proteins (each 600 nm) were incubated with 0.4 mg/ml of soluble calf skin type I collagen (U.S. Biochemical Corp., Cleveland, OH) in 100 mm sodium acetate buffer, pH 5.5, containing 2.5 mm DTT, and 2.5 mm EDTA. Collagen digestion was performed at 28 °C for 8 h in the presence of 0.1% (w/v) C4-S. The digestion reaction was stopped by the addition of 10 μm of E64. To measure the gelatinase activity of each cysteine protease, type I collagen was heated for 30 min at 70 °C prior to incubation with the proteases. In the presence of WT catK (20 nm) or each catK variant (20 nm), the reaction mix was incubated for 30 min at 28 °C. Before adding E64, residual activity of each enzyme was monitored spectrofluorometrically using the fluorogenic substrate Z-Leu-Arg-MCA (2 μm). Collagen and gelatin digests were subjected to SDS-polyacrylamide electrophoresis using 4–20% Tris/glycine gels (Novex, San Diego, CA), and degradation products were visualized by Coomassie Blue staining and then analyzed by using the software NIH ImageJ (version 1.62). Signal densities in a window containing the α1- and α2-chains of collagen in presence or absence of C4-S were measured. Data obtained for the digest reactions in the absence of C4-S for each variant protein (no or only minimal intrahelical degradation) were set as 100% and compared with the amounts of α1 and α2-chains in the presence of C4-S. Densitometric measurements were taken from at least three independent experiments, averaged and expressed in percentage of α-chain cleavage. For statistical analysis the Student's t test was performed. Significance was concluded when p < 0.05.
CD Spectroscopy
The conformations of the catK variants were estimated by CD spectroscopy and compared with that of the WT catK. CD spectra were measured in the far ultraviolet regions (250 to 200 nm) by a Jasco J-810 spectropolarimeter at 23 °C. The recombinant catK and the corresponding variants were diluted in 0.1 m sodium acetate, pH 5.5, and the concentration of each protein was determined by UV spectroscopy at 280 nm and confirmed by protein determination using the Bio-Rad Protein Assay (Bio-Rad) and then adjusted to 0.2 mg/ml before each measurement. The cell length was 1 mm. Ten spectra were averaged for each derivative and corrected by background subtraction for the spectrum obtained with the buffer alone or the buffer containing C-4S. The observed elipticity was normalized to units of degrees cm2·dmol−1.
Gel Shift Assay and Kd Determination
Gel shift assays demonstrating the formation of a complex between the M5 variant and C4-S were performed as described for the WT protein (6). The Kd value for the binding of C4-S (37-kDa fraction) to the M5 variant was determined as described previously (9).
Crystallization and Data Collection
The M5 variant of catK (E64 inhibited) and the 17-kDa fraction of C4-S were mixed prior to crystallization in a 1:1 molar ratio to a final concentration of 20 mg/ml. The complex was co-crystallized by the hanging-drop vapor diffusion method by mixing the protein with an equal volume of reservoir solution that contained 30% (v/v) 2-methyl-2,4-pentanediol, 0.1 m sodium acetate buffer, pH 4.5, and 20 mm CaCl2. Macroseeding was used to improve the size and quality of the crystals. Crystals grew to a size of 0.25 × 0.05 × 0.02 mm over a period of 2 weeks. Crystals were flash-cooled in a nitrogen stream at 100 K. The data were collected at beam line 8.3.1 at the Advanced Light Source (Berkeley, CA). The data were processed using HKL2000 (16).
Structure Determination
The structure was determined by molecular replacement using the coordinates of the previously determined structure of catK in complex with C4-S (PDB entry 3C9E) (9). M5/E64 was localized using molecular replacement methods with the CNS package (17). The C4-S (PDB 1C4S) was then fit into the difference density using Coot (18). The subsequent refinement was carried out with the PHENIX package and a maximum likelihood target (19). The secondary structure and stereochemistry of the protein were analyzed by PROCHECK (20). Calculations of the buried surface area and ΔG were carried out with the EMBL-EBI PISA server (21, 22). The program ALIGN (23) was used to calculate root mean square deviation values between analogous structures. Figures were made with PyMOL (24). The coordinates and structure factors have been deposited in the PDB under accession code 3H7D.
RESULTS
Expression and Characterization of CatK Variants
To investigate whether surface amino acid residues of catK that were identified to interact with C4-S are critical for the collagenase activity of catK, we generated five catK variants by replacing the C4-S interacting residues Lys9, Ile171, Gln172, Asn190, Lys191, and Leu195 with the equivalent residues present in human catL (Table 1). CatL does not exhibit collagenase activity in the presence or absence of C4-S (7). The variants were designed to evaluate the distinct interaction sites of the C4-S chain to WT catK and to determine their individual contributions to the collagenase activity of the protease. Briefly, we used the Lys9/BDP5 interaction as an “anchor points” (M1) and subsequently added appropriate interaction sites with sugars BDP3 and ASG4 (M2) and sugars BDP1 and ASG2 (M4). As the interactions with sugars BDP1 and ASG2 were realized via three interaction sites, we also generated this variant separately. Finally, in the M5 variant, all catK-specific residues were replaced with the appropriate catL residues. The corresponding variant full-length cDNAs were expressed in P. pastoris and activated into mature proteins and purified as described previously (11). All variants revealed SDS-PAGE homogeneity and were completely processed into their catalytically active mature forms. Variant and WT proteins were titrated with E64 resulting in the following stock concentrations: 3 μm (variant M1), 39 μm (variant M2), 41 μm (variant M3), 4 μm (variant M4), and 25 μm (variant M5), respectively. Enzyme stocks were stable for months when stored at −80 °C.
CatK Variant Activities toward Synthetic Fluorogenic Substrates, CD Spectral Analysis, Peptidase Stability, and Gel Shift Analysis
To determine whether the introduction of substitutions would impair the activity of the variant proteases, the kinetic parameters Km and kcat were determined. Proteolytic activity measurements of each variant and the WT catK were performed using the general cathepsin substrate (Z-Leu-Arg-MCA) and a catK specific substrate (Z-Gly-Pro-Arg-MCA). Comparable Michaelis-Menten constants (Km) and second order rate constants (kcat/Km) were obtained for the WT protein and the variants for each of the substrates (Table 2). As for the WT enzyme, Z-Gly-Pro-Arg-MCA was hydrolyzed ∼40× less efficiently than Z-Leu-Arg-MCA by all five variant proteins. The Km values for both substrates varied by less than a factor of 1.9, the kcat values by <1.9 and the kcat/Km values by less than a factor of 2.2 among all catK variants, including WT. Variant M5 and the WT enzyme varied by factors <1.3 in their kinetic constants. This suggested that the introduced substitutions neither significantly altered the integrity of the catalytic site nor was the substrate binding region of the variant enzymes altered or disrupted when compared with WT catK.
TABLE 2.
Kinetic parameters for the hydrolysis of Z-Leu-Arg-MCA and Z-Gly-Pro-Arg-MCA by recombinant cathepsin K variants
| Variant | kcat | Km | kcat/Km |
|---|---|---|---|
| s−1 | μm | × 106m−1s−1 | |
| Z-Leu-Arg-MCA | |||
| Wild-type | 6.2 ± 2.3 | 3.3 ± 0.5 | 1.9 |
| Variant 1 | 5.7 ± 2.9 | 3.5 ± 1.0 | 1.6 |
| Variant 2 | 6.4 ± 0.4 | 2.4 ± 0.1 | 2.7 |
| Variant 3 | 9.1 ± 1.4 | 4.6 ± 0.6 | 2.0 |
| Variant 4 | 7.6 ± 1.1 | 3.7 ± 0.4 | 2.1 |
| Variant 5 | 5.6 ± 1.4 | 3.7 ± 0.2 | 1.5 |
| Z-Gly-Pro-Arg-MCA | |||
| Wild-type | 2.9 ± 1.4 | 48 ± 9 | 6.0 |
| Variant 1 | 2.0 ± 0.4 | 34 ± 2 | 5.9 |
| Variant 2 | 2.1 ± 0.1 | 37 ± 1 | 5.7 |
| Variant 3 | 3.7 ± 0.3 | 38 ± 3 | 9.7 |
| Variant 4 | 1.9 ± 0.6 | 43 ± 9 | 4.4 |
| Variant 5 | 3.6 ± 0.8 | 58 ± 2 | 6.2 |
Comparison of the CD spectra of the variant proteins with that of WT catK did not reveal any significant differences, thus indicating that the cumulative introduction of up to six substitutions did not change the secondary structure of the variant proteins (data not shown). Moreover, all catK proteins (WT and variant proteins) displayed residual activities of ∼40–50% in the presence of type I collagen and C4-S compared with 2–14% of activity without C4-S at 28 °C after 8 h of incubation, indicating comparable enzyme stabilities. The stability time courses for WT and variants were highly similar (Fig. 1A).
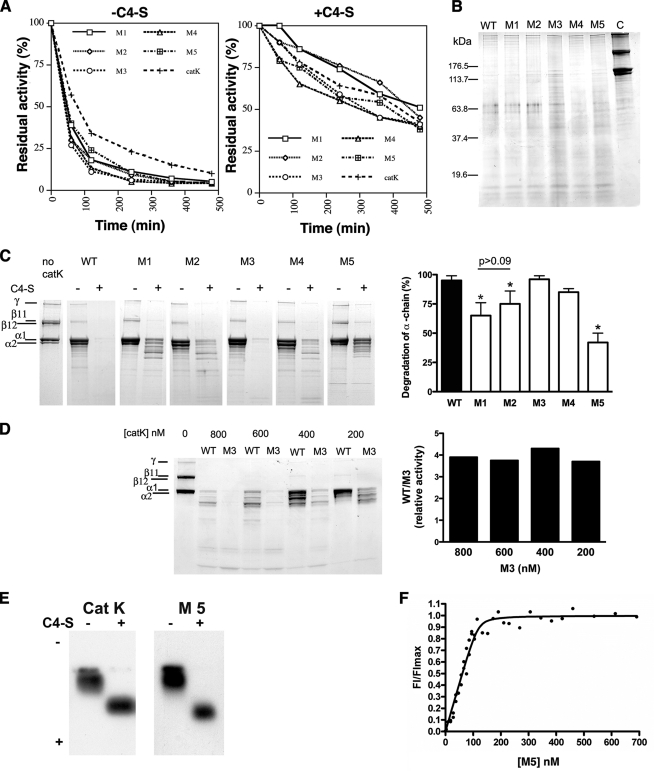
FIGURE 1.
Stability, substrate specificity, and complex formation of catK. A, cathepsin stability. Active WT catK (600 nm) and the five catK variant proteins (600 nm each) were incubated at 28 °C with 0.4 mg/ml of soluble calf skin type I collagen in 100 mm sodium acetate buffer, pH 5.5, containing 2.5 mm DTT and 2.5 mm EDTA. Residual activity of each enzyme in absence (right panel) or in presence of 0.1% C4-S (w/v) (left panel) was monitored spectrofluorometrically using the fluorogenic substrate, Z-Leu-Arg-MCA (2 μm) at different time points. B, hydrolysis of gelatin by WT catK and variants. SDS-PAGE of degradation products of gelatin (heat-denatured type I collagen) after incubation with WT catK or C4-S interaction site variants of catK at pH 5.5 and 28 °C for 30 min (enzyme concentration, 20 nm). C, left panel, SDS-PAGE of degradation products of triple-helical type I collagen after incubation with WT catK or the C4-S interaction site variants of catK at pH 5.5 and 28 °C for 8 h (enzyme concentration, 800 nm); right panel, densitometric quantification of remaining α-chains after 8 h of incubation with WT catK or variants. D, left panel, SDS-PAGE of degradation products of triple-helical type I collagen after incubation with M3 at pH 5.5 and 28 °C for 8 h at different enzyme concentrations; right panel, densitometric quantification of remaining α-chains after 8 h of incubation with M3. E, separation of [129I]DCG-04-labeled M5 and WT catK complexes and monomeric variant and WT forms of catK by 0.5% agarose gel electrophoresis. F, Plot of FI/FImax versus increasing M5 concentrations in the presence of 37-kDa C4-S fractions.
Gelatinase Activity
Heat-denatured collagen (gelatin) was used as a protein substrate to compare the general proteolytic activities of the catK variant proteins with the WT enzyme. All peptidase variants were highly active at low enzyme concentrations (0.5 nm) and hydrolyzed gelatin similarly to the WT catK gelatin at pH 5.5 into a ladder of peptides within 30 min (Fig. 1B). The potent and comparable gelatinase activities of catK and its variant proteins suggest that the introduced substitutions on the surface of the R-domain of the protease do not impair the active site and thus the catalytic activity of the variant proteins.
Degradation of Triple-helical Type I Collagen by CatK Variants
In the presence of C4-S, WT catK degrades triple-helical type I collagen efficiently into small peptides that are not detectable by SDS-PAGE. In the absence of C4-S, catK activity is limited to a cleavage in the nonhelical telopeptide regions leading to the disappearance or reduction of the high molecular weight β- and γ-bands of collagen and an enrichment of the α-chains (Fig. 1C). WT and all variant proteins lacked significant collagenase activities in the absence of C4-S, and the generation of collagen α-chains was comparable among all catK variants. In the presence of C4-S, however, significant differences in the ability of the variants to cleave collagen were observed. The replacement of Lys9 by the appropriate catL residue Glu increased the presence of remaining α-chains by 35% when compared with the WT enzyme (Fig. 1C). Lys9 forms strong salt bridges with two sugar residues of C4-S (GCU5 and ASG6) in the WT complex (9), and thus the effect of its replacement with a negatively charged catL residue on the collagenase activity suggests a critical involvement of Lys9 in the formation of a collagenolytically active complex. Ile171 and Asn172 interact directly with ASG4 and GCU3 and via water molecules with ASG2 and GCU3 (9). Substituting these two residues together with Lys9 in variant 2 led to a slightly less inhibition of the degradation of the α-chains (25% reduction compared with 35% in the K9E single variant: Fig. 1C), although the difference was not significant (p > 0.09). This implies that the major effect can be attributed to Lys9. Residues Asn190, Lys191, and Leu195 are further from binding site of C4-S. Their replacement with catL residues in variant 3 had no inhibitory effect on the collagenase activity of catK. On the contrary, this variant appears to be more potent as a collagenase than the WT enzyme. Comparing the collagenolytic activity of variant M3 with that of WT catK over a concentration range from 200 to 800 nm enzyme in the collagen digest reaction displayed a 3.5–4-fold higher collagenase activity for the variant peptidase (measured by the decrease in the amount of α-chains for the variant-mediated collagen digest compared with that of the WT enzyme) (Fig. 1D). This may also explain, why variant M4 that contains substitutions in Lys9, Asn190, Lys191, and Leu195 is significantly more efficient than variant M1 (K9E). The activating effect of the N190M/K191G/L195K substitutions appears to compensate for the inhibitory effect of the K9E substitution. Interestingly, the combination of all six amino acid replacements in the M5 variant (K9E/I171E/Q172S/N190M/K191G/L195K) displayed a cumulative effect and inhibited the collagenolytic activity by ∼60%. As shown in Fig. 1A, this inhibition cannot be attributed to an increased instability of this variant when compared with the other catK variants.
To determine whether M5 forms a stable complex with C4-S, we performed a protein gel shift assay using radiolabeled inhibitor marked M5 in the presence or absence of C4-S. Similar to WT, the M5 variant showed a gel shift in the presence of C4-S, suggesting a complex formation (Fig. 1E). The Kd value for C4-S binding to the M5 variant (1.9 ± 1.4 nm) was smaller as the value determined previously for the WT protein (10.4 ± 7.8 nm) (9). Interestingly, the lower Kd value corresponded to a higher number of disaccharide units interacting with the M5 variant when compared with WT catK (5.5 ± 03 versus 3.8 ± 0.5 for the WT protease (9)). To understand the structural basis of M5 variant complex with C4-S and its 60% loss in collagenolytic activity, this variant in complex with C4-S was crystallized and its structure determined.
Crystallization and Structure Determination
The active site Cys25 of catK was inhibited by E64, l-3-carboxy-trans-2,3 epoxypropionylleucylamido-(4-guanidino)butane, prior to the complexation with C4-S. The oligomeric complex of M5/E64 with C4-S (M5·C4-S) was produced by mixing the protein with the 17 kDa C4-S in a 1:1 ratio. The complex was crystallized in the same conditions as described earlier for the WT catK·C4-S (9). The structure of the M5·C4-S complex was determined by the molecular replacement method using the structure of WT catK·C4-S (PDB code 3C9E) (9) as the search model. The M5·C4-S complex crystals belong to the centered monoclinic space group C2; the asymmetric unit of the cell contains two E64-inhibited catK molecules, one hexasaccharide segment of C4-S and two Ca2+ ions lying on a crystallographic 2-fold axis (Fig. 2A). The E64 inhibitor forms a covalent bond with the sulfur atom of Cys25, and it is positioned very similarly to the other catK·E64 complexes. The crystallographic and structure refinement data are summarized in Table 3. The structures of the two M5 molecules in the asymmetric unit are very similar to the previously determined structures of E64 inhibited catK complexes with and without C4-S (for example, PDB codes 3C9E and 1ATK). The root mean square difference is 0.20 Å (>210 Cα atom pairs) and 0.29 Å (>213 Cα atom pairs), respectively.
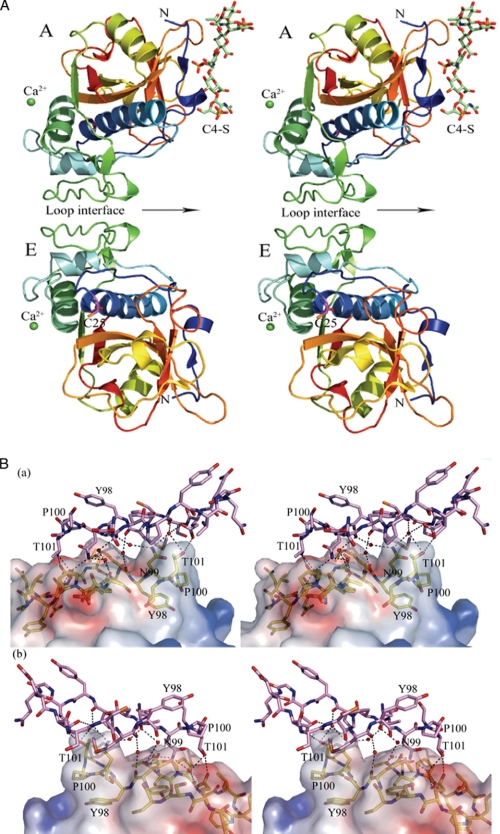
FIGURE 2.
Tyr98 loop dimer of the catK·C4-S complex. A, the structure of the M5 variant of catK·C4-S complex in the space group C2. The asymmetric unit contains two M5 molecules (A and E) interacting through a loop interface, one hexasaccharide segment (shown in sticks; green carbon atoms, blue nitrogen atoms, and red oxygen atoms) of a 17-kDa C4-S and two Ca2+ ions (green spheres) in special positions (on the crystallographic 2-fold axes). The M5 variant molecules are represented in rainbow colors (blue for the N terminus, red for the C terminus). A noncrystallographic 2-fold axis relating monomers of the dimer is shown by an arrow. The active site Cys25 (sticks, with magenta carbon atoms) has a covalently bound E64 inhibitor (not shown). B, the details of the loop dimer interface. The two views are of the opposite sides of the interface. The surface relief provides a view of the complementary fit of the two molecules. The loop segment (residues 85–102) of molecule A (yellow carbon atoms) is shown inside the electrostatic surface. The identical loop segment of molecule E (violet carbon atoms) is represented by sticks. This interface is largely hydrophobic.
TABLE 3.
Crystallographic data for the M5 variant of cathepsin K·E64·C4-S complex
| Crystallographic data | |
| Space group | C2 |
| Unit cell constants | |
| a, b, c (Å) | 140.56, 42.01, 87.51 |
| α, β, γ | 90.0°, 94.19°, 90.0° |
| Data collection | |
| Temperature (K) | 100 |
| Detector | ADSC Q210 (ALS)a |
| Wavelength (Å) | 1.11587 |
| Resolution (Å) | 50-2.25 |
| High resolution shell (Å) | 2.33-2.25 |
| Total observations | 88,259 |
| Unique reflections | 24,692 (2411)b |
| Redundancy | 3.6 (3.4)b |
| I/σ(I) | 10 (2.5)b |
| Completeness (%) | 99.6 (98.3)b |
| Rsym (%) | 13.3 (53.5)b |
| Structure Refinement | |
| Refinement resolution (Å) | 40-2.25 |
| Unique reflections (all/test) | 24,685/1256 |
| Rwork/Rfreec(%) | 17.5/23.6 |
| No. of atomsd | 3286/50/90/260 |
| r.m.s.d. from idealitye | |
| Bond lengths (Å) | 0.010 |
| Bond angles | 1.295° |
| Ramachandran plot for the M5 variant of cathepsin K | |
| Favored (%) | 96.5 |
| Allowed (%) | 3.5 |
| Outliers (%) | 0.0 |
| Mean B factors (Å2)f | 24.1/30.0/58.3/31.5 |
a ADSC, Area Detector System Corporation; ALS, Advanced Light Source at Lawrence Berkeley Laboratory, the beamline 8.3.1.
b Values within parentheses refer to the highest resolution shell.
c Rfree was calculated using 5% of all unique data.
d Number of non-hydrogen atoms of M5/non-hydrogen of E64 inhibitor atoms/non-hydrogen atoms of C4-S/water molecules.
e r.m.s.d. indicates root mean square deviation.
f Mean B factors for protein, inhibitor, oligosaccharide, and water, respectively.
Tyr98 Loop Dimer Interface
To determine how the catK molecules are assembled for collagen degradation, we looked at several catK dimer interfaces. The two largest molecular interfaces that have been found in the present structure and that are also present in the WT catK·C4-S complex structure bury ∼364 Å2 per molecule (or 730 Å2 total per interface). This is not large enough to make a stable dimer in solution, and as a result, WT catK does not form dimers. However, in the presence of C4-S, WT catK forms oligomers containing several such dimers bound to the C4-S chains. The combined interface would multiply several times and would make the oligomeric complex stable. Size exclusion chromatography of the oligomeric complex revealed molecular masses between 180 and 310 kDa (6), indicating that there can be up to four dimers in the complex bound to C4-S in solution (data not shown). One of these dimers represents the asymmetric unit in which two M5 molecules, A and E, are related by a noncrystallographic 2-fold axis (Fig. 2A). The root mean square deviation between the protomers is very low: 0.127 Å (overall 215 Cα atom pairs). This dimer interface is formed by the loop segments (residues 86–101) from both molecules (Fig. 2B). A mutation of Tyr98 into a cysteine residue has been identified previously to cause pycnodysostosis, an autosomal recessive bone disorder lacking collagenolytic activity (25). The majority of interactions across the interface are van der Waals (a total of 13 contacts) with −6.0 kCal/mol of ΔGi (solvation free energy change upon interface formation), indicating its overall hydrophobic nature. Only two hydrogen bonds between Glu94 and Thr101 of each molecule are present. Several interactions occur through water molecules. Six water molecules connect the interfacing residues of the dimer: Ala86, Pro88, Glu94, Met97, Asn99, and Thr101.
Calcium Dimer Interface
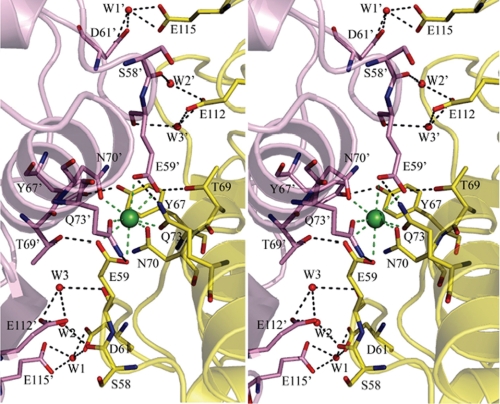
The calcium dimer interface buries an equivalent surface area as does the Tyr98 loop interface; however, the majority of the contacts between the interfacing molecules are electrostatic and hydrogen bonding. These dimers are related by crystallographic 2-fold axes. There are very small structural differences between the dimers of the A molecules and of the E molecules (Fig. 2A). The most significant set of interactions comes from calcium ion coordination. Calcium ions lie on crystallographic 2-fold axis in a similar manner as they do in the WT catK·C4-S complex. Each calcium ion interacts with four oxygen ligands from each molecule of the dimer, totaling eight oxygen atoms. The Ca2+–O distances lie in the range from 2.47 to 2.74 Å, slightly longer that the average distance observed for the most common heptacoordinated calcium ion, 2.4 Å (26). The calcium ions play a pivotal role in the formation of this interface, as well as in the intermolecular packing of this crystal form (Fig. 3). However, several additional hydrogen-bonding contacts, mainly through water molecules, also contribute to this dimer formation. Water molecules, W1 and W1′, form bifurcated hydrogen bonds with Asp61 and Glu115. Similarly, pairs of water molecules W2 and W2′, and W3 and W3′ make bifurcated hydrogen-bonding interactions between the carboxylate oxygens of Glu112 and the carbonyl oxygens of Ser58 and Glu59, respectively, in the A dimer (Fig. 3). The hydroxyl groups of Tyr110 Oη and Ser58 Oγ interact through water molecules in the E dimer (data not shown). It was found that the presence or absence of Ca2+ ions does not influence the collagenolytic activity (9). That makes this interface a less likely candidate for a molecular assembly formation of the active collagenase complex. However, this dimer interface was found to form not only in the presence of C4-S and Ca2+, but also in their absence (PDB code 2FTD) (27), although without Ca2+ the dimer would likely be much less stable.
FIGURE 3.
The calcium-binding interface. The Ca2+ ion is shown as a green sphere. It is positioned on a 2-fold axis and coordinates eight oxygen ligands, four from each M5 variant molecule. Residues involved in hydrogen-bonding interactions are shown in sticks and colored yellow for carbon atoms of the asymmetric unit and violet for carbon atoms of the 2-fold-related molecule. Other atoms are shown in elemental colors. W1, W2, and W3 represent water molecules that are shown as red spheres.
Binding of Chondroitin 4-Sulfate to M5
C4-S is a copolymer of the disaccharide of β-d-galactopyranuronic acid (BDP) and 2′-deoxy-2′-acetamido-β-d-galactose-4-sulfate (ASG). A hexasaccharide segment of the C4-S has been found to bind to one (molecule A) of the two molecules in the asymmetric unit (Fig. 2A). The C4-S molecule runs approximately parallel to the b axis (42.0 Å in length) of the unit cell. It forms a broken cosine-like wave configuration in which the ends of the hexasaccharides are separated by several disordered sugar residues (Fig. 4A). Interestingly, the C4-S in the present structure runs in the opposite direction to that in the WT catK·C4-S complex (Fig. 4B). The molecular mass of the oligosaccharide used for crystallization was 17 kDa; this means that the C4-S chain must run through several unit cells in the b axis direction. The b axis length in this space group is equal in length to the a axis dimension in the WT catK·C4-S unit cell (42.0 Å) and would therefore form a similar period of the cosine wave that the hexasaccharide forms in the WT complex (9).
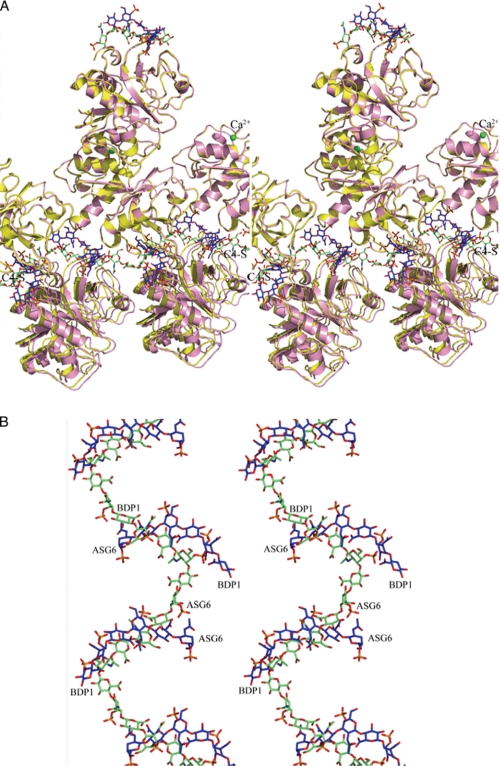
FIGURE 4.
Orientation of catK and C4-S molecules. A, superposition of several unit cells of the present structure (pink ribbons and blue sticks) onto several unit cells of the WT catK·C4 complex (yellow ribbons, green sticks). The C4-S is represented by sticks as a continuous cosine wave (green carbon atoms) for the WT complex and as a broken wave (blue carbon atoms) for the variant complex. The arrangement of the enzyme molecules in both complexes did not change along the b axis. B, a close-up of the superimposed C4-S structures. The two oligosaccharide chains run in opposite directions.
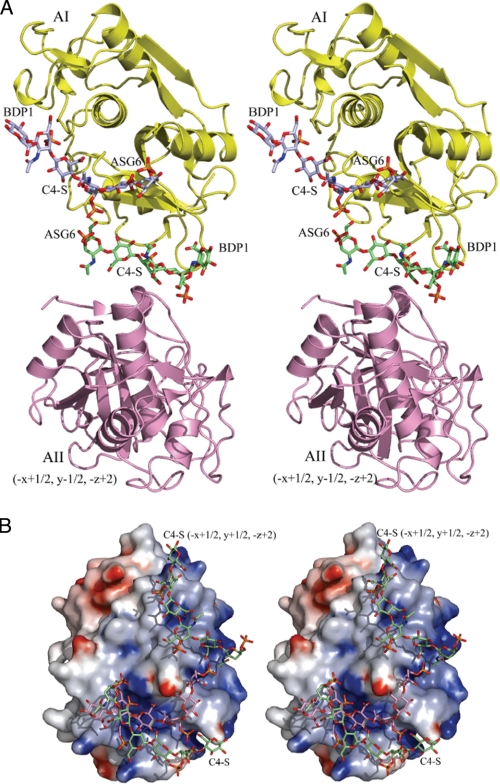
The C4-S hexasaccharide in the present structure binds in the space between two A molecules (AI from the same asymmetric unit and AII from the symmetry related molecule operation −x+1/2, y−1/2, −z+2); it makes contacts with both of them (Fig. 5A). In turn, each molecule A of the asymmetric unit interacts with two segments of the C4-S (Fig. 5A). One segment belongs in the same asymmetric unit (green carbon atoms) and the other segment (violet) belongs in the symmetry-related asymmetric unit (−x+1/2, y+1/2, −z+2). The surface of the M5 variant (molecule A) with two C4-S segments (green carbon atoms) is shown in Fig. 5B. The position of the C4-S (pink carbon atoms) in the complex with WT catK is shown for comparison.
FIGURE 5.
Interaction of M5 variant with C4-S. A, each catK molecule A binds two hexasaccharides: one in the same asymmetric unit (green carbon atoms) and another (violet carbon atoms) from the symmetry operation −x+1/2, y+1/2, −z+2. In turn, each hexasaccharide binds between two A molecules (in the asymmetric unit (yellow) and from the symmetry operation −x+1/2, y−1/2, −z+2, pink). As a result, a broken wave of the saccharide oligomer runs through many unit cells (see also Fig. 4A). B, the electrostatic surface of the M5 variant with two C4-S hexasaccharides shown in sticks (green carbon atoms). The M5 variant surface is largely divided into the negatively charged (red) and positively charged (blue) sides. The C4-S oligosaccharide binds to the positively charged side of the surface. The C4-S path on the surface of the WT catK is also shown (sticks, pink carbon atoms).
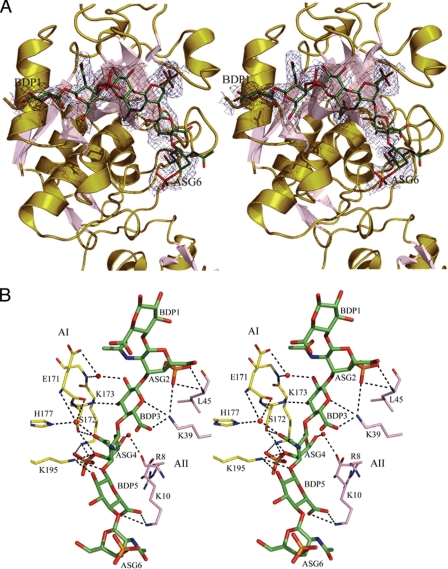
Only four saccharide residues make direct contacts with the protein and have well defined electron density (Fig. 6A). The two terminal sugar residues (BDP1 and ASG6) have no contacts with the protein and have the least well defined electron density. Several residues of the molecule AI (Fig. 6B, yellow carbon atoms) make direct hydrogen bonding and electrostatic interactions with the C4-S hexasaccharide: Lys195, Lys173, and Ser172 contact C4-S residues from three to five. Residues of the symmetry-related molecule AII (pink carbon atoms), in particular, residues Lys39, Lys10, and Leu45 make several hydrogen bonding and electrostatic interactions with residues two, three, and five of the hexasaccharide. In total, four sugar residues make ∼15 hydrogen bonds (at distances <3.5 Å) with variant M5 (Table 4). Several interactions between the protein and the carbohydrate residues also occur through water molecules. Calculated torsion angles of chondroitin 4-sulfates in their unbound form or bound to WT catK and the M5 variant are summarized in Table 5.
FIGURE 6.
Electron density map of C4-S binding on the catK molecule and contact sites of C4-S with catK. A, the |Fo| − |Fc|, αc, electron density map contoured at 1.2σ over the atoms of the C4-S hexasaccharide. The first sugar residue is BDP1, and the last sugar residue is ASG6. Loops and helices of M5 are colored olive, and the strands of β-sheets are in pink. B, binding site of the C4-S hexasaccharide (green carbon atoms). The hydrogen-bonding interactions are represented by broken lines. Water molecules are shown as red spheres. Molecules AI of the asymmetric unit (yellow carbon atoms) and the symmetry-related AII (pink carbon atoms, symmetry operation −x+1/2, y−1/2, −z+2) interact with four sugar residues (from 2 to 5). The majority of interactions are hydrogen-bonding and electrostatic.
TABLE 4.
Contacts between C4-S and the M5 variant of cathepsin K
| C4-S residue atom | Protein atom | Distance |
|---|---|---|
| Å | ||
| Direct C4-S/M5 (molecule A) interactions in one asymmetric unit | ||
| BDP3 O3 | Lys173 N | 2.75 |
| ASG4 O4 | Lys195 Nζ | 3.22 |
| ASG4 O4 | Ser172 Oγ | 3.45 |
| ASG4 O7 | Ser172 Oγ | 3.0 |
| ASG4 OSA | Lys195 Nζ | 3.46 |
| ASG4 OSB | Lys173 Nζ | 2.69 |
| ASG4 OSC | Lys173 Nζ | 2.95 |
| BDP5 O1 | Lys195 Nζ | 3.07 |
| BDP5 O2 | Lys195 Nζ | 3.45 |
| Direct C4-S interactions with the symmetry-related M5 (molecule A) | ||
| ASG2 OSA | Lys39 Nζ (−x+1/2, y-1/2, z+2) | 3.41 |
| ASG2 OSA | Leu45 N (−x+1/2, y−1/2, z+2) | 3.25 |
| ASG2 OSB | Leu45 N (−x+1/2, y−1/2, z+2) | 2.88 |
| BDP3 O5 | Lys39 Nζ (−x+1/2, y−1/2, z+2) | 3.26 |
| BDP3 O6B | Lys39 Nζ (−x+1/2, y−1/2, z+2) | 2.68 |
| BDP5 O6A | Lys10 Nζ (−x+1/2, y−1/2, z+2) | 3.14 |
| BDP5 O6B | Lys10 Nζ (−x+1/2, y−1/2, z+2) | 3.07 |
| C-4S residue atom | Sugar-to-water distance | Protein atoms | Protein-to-water distance |
|---|---|---|---|
| Å | Å | ||
| C-4S/M5 interactions through water | |||
| BDP3 O2 | 2.76 | Glu171 Oϵ1 | 2.93 |
| Gly174 N | 2.79 | ||
| ASG4 O7 | 2.66 | Glu171 N | 3.49 |
| His177 Nϵ2 | 2.90 | ||
| ASG6 O6 | 2.90 | Arg8 Nη2 | 2.85 |
| Met190 O | 2.63 |
TABLE 5.
Selected torsion angles of chondroitin 4-sulfate structures
Torsion angles were calculated using COOT according to IUPAC rules for naming of polysaccharide conformations. β(1–4), Φ = O5(i)-C1(i)-O1(i)-C4(i−1) and Ψ = C1(i)-O1(i)-C4(i−1)-C3(i−1); β(1–3) Φ = O5(i)−C1(i)-O1(i)-C3(i−1) and Ψ = C1(i)-O1(i)-C3(i−1)-C2(i−1).
| C4-S structure | Torsion angle Φ | Torsion angle Ψ | Torsion angle Φ | Torsion angle Ψ |
|---|---|---|---|---|
| ASG4-BDP3 (β(1–4)) | ASG4-BDP3 (β(1–4)) | BDP5-ASG4 (β(1–3)) | BDP5-ASG4 (β(1–3)) | |
| Fiber (sodium salt) | −79.9 | 110.5 | −86.4 | −110.7 |
| Fiber (calcium salt) | −97.8 | 65.5 | −79.6 | −132.0 |
| WT catK·C4-S | −70.6 | 126.8 | 56.3 | −116.6 |
| M5 variant catK·C4-S | −72.3 | 126.1 | −85.3 | −99.3 |
The second M5 molecule (E) in the asymmetric unit does not bind a C4-S segment (Fig. 2A), as the environment surrounding molecule E is different. Modeling of the C4-S segment to molecule E (by superposition of molecule A on molecule E) shows that the C4-S oligosaccharide could not form interactions with any of the symmetry-related molecules E; they were too far apart.
DISCUSSION
We have previously demonstrated that the collagenase activity of catK requires specific interactions with bone- and cartilage-resident glycosaminoglycans such as chondroitin sulfate (5, 6). The crystal structure of the complex between WT catK and C4-S revealed 12 amino acid residues interacting directly or indirectly via water molecules with the glycosaminoglycan (9). From these residues, six amino acid residues are specific for catK when compared with the amino acid sequence of the related catL. As catL does not exhibit any significant collagenase activity (14) and in contrast to WT, catK does not show an electrophoretic mobility shift in the presence of C4-S (6), we replaced the equivalent C4-S interacting residues in catK with the appropriate amino acid residues present in catL. Five variant proteins reflecting different binding sites of the WT·C4-S complex were generated and analyzed for the effect of their substitutions on the proteolytic and specifically on the collagenolytic activities. As the interaction sites with C4-S are remote from the active site of the peptidase and complex formation between catK and C4-S did not alter the three-dimensional structure of the protein (9), it was not surprising that the specificity constants kcat/Km for all variants were comparable to that of the WT enzyme (Table 2). Moreover, the hydrolytic efficacy toward gelatin was not altered by any of the substitutions and was again comparable with that of the WT enzyme. However, these substitutions affected the collagenase activity of catK to various degrees and thus confirmed that the specific binding of catK to C4-S at the sites identified in the x-ray structure of the complex is indeed one critical parameter for the degradation of triple-helical collagens. The M5 variant comprising all six substitutions showed the lowest collagenase activity with a reduction of 60% when compared with the WT enzyme. Interestingly, these mutations did not prevent the binding of C4-S to the variant form of catK as initially hypothesized. Two of the substitutions (Q172S and L195K) actually contributed to new contacts. However, the binding mode of C4-S to the variant is different from that of the WT catK; two neighboring protein molecules are needed to hold the hexasaccharide segment. Only four sugar residues have interactions with the protein; the terminal residues on the both ends of the hexasaccharide have no interactions with the M5 variant and have much higher B factors and significantly weaker electron density. The C4-S chain is not continuously ordered; there are gaps in the electron density between the hexasaccharide terminal residues. In the WT catK complex, each sugar residue in the hexasaccharide has interactions with the protein, and the C4-S is continuously ordered (9). The C4-S hexasaccharide runs in a deep groove on the WT catK surface that fits its shape very well. In the M5 variant, the C4-S does not run in a groove; it is situated on a bulging surface. As a result, it does not make enough contacts with one molecule; a second molecule is needed to provide more interactions for a stable complex to form. When the second molecule is not close enough (as in the case of molecule E in the asymmetric unit) C4-S does not bind to it. Although the individual binding sites appear weaker in the M5 variant, additional interaction sites were revealed on the M5 variant (Fig. 5B). This may explain the surprisingly lower Kd value for the binding of C4-S to the M5 variant. This may partially also explain why no difference in the electrophoretic mobility shift assay between the WT and M5 variant was observed. Thus, it is tempting to speculate that the collagenase activity of the catK·C4-S complex requires a highly specific binding of C4-S to the surface of catK, which when disturbed by alternative binding modes and affinities, leads to the observed loss of this activity. Alternative binding modes may interfere with the docking of triple-helical collagen to the complex and thus reduce or prevent the unfolding and or cleavage of collagen.
It is of interest that WT catK as well as its M5 variant form significant protein-protein interfaces that may play a role in the formation of the active collagenase complex of cathepsin K with C4-S. It has been concluded from a number of studies that a buried surface area larger than 600–850 Å2 is biologically relevant (22). In the catK·C4-S complex, the Tyr98 loop interface buried 360 Å2 per one catK molecule, which would be insignificant. However, we have polymeric strings with protein molecules (beads) interfacing each other, the combined buried area of several interfaces could be significant. As a result, these particular multimeric assemblies are likely to exist in solution. Interestingly, one of the interfaces contains Tyr98, a residue that has been shown to influence complex formation with C4-S, but has no effect on the general proteolytic activity (6, 25). The effect of the Y98C mutation on the collagen-degrading activity might be explained by the breakup of Tyr98 loop interface as a result of the mutation. This is corroborated by the finding that the Y98C variant does not show an electrophoretic mobility shift in the presence of C4-S (6). As the protein-protein interactions were not affected in the M5 variant, and the electromobility shift was maintained, it is likely that protein-protein interactions significantly contribute to the stability of the complex. It further implies that for a collagenolytically active catK·C4-S complex, both the protein/C4S and protein/protein interactions are critical.
As it was mentioned above, the M5 variant retains a portion of the collagenolytic activity. It is of interest that the introduction of two new interaction sites by the substitutions Q172S and L195K had some enhancing effect on the collagenase activity of these variants. Whereas the single variant M1, K9E, lost ∼35% of its collagenolytic activity, the triple variant K9E/I171E/Q172S (M2) lost only ∼25% of its collagenase activity when compared with the WT enzyme. Moreover, M3 (N190M/K191G/L195K) potentially making additional interactions of the Lys195 with C4-S was an even better collagenase than the WT peptidase. Using lower enzyme concentrations that allowed for a more accurate quantification of α-chain degradation, M3 left ∼4× fewer α-chains intact than did WT catK. This may also explain why the composite M4 variant that combines the inhibitory activity of the K9E substitution with those of the activating activity of M3 was almost as active as the WT enzyme. It should be noted that alterations in the collagenase activity are not due to altered stabilities of the variants as all enzymes including the WT protein displayed comparable residual activities toward the synthetic substrate Z-Leu-Arg-MCA after 8 h of incubation with type I collagen. Moreover, identical CD spectra and the identical folds of M5 and WT catK exclude major structural alterations due to the substitutions.
This allows us to speculate that the similarities and differences between the structures of the two catK·C4-S complexes (M5 variant and WT) can provide insight into the collagenolytic molecular assembly. 1) The specific binding of C4-S to catK is required to allow the formation of an oligomeric complex of catK and C4-S molecules capable to digest triple-helical collagen. Alterations of this binding mode may effect the formation of the collagenolytically productive CatK·C4-S complex. 2) Two neighboring catK molecules are required to be positioned in a very similar way on the C4-S “string” relative to one another. This dimer (attached to a C4-S string) with a docked collagen triple helix was suggested previously as an element of the multimeric collagen-degrading molecular complex. 3) Another important element of this multimer could be the loop interface containing the Tyr98 residue. The importance of Tyr98 is underlined by the fact that its mutation leads to pycnodysostosis in humans, which is characterized by a specific loss of its collagenase activity while retaining its gelatinase activity (25). It is tempting to speculate that the main function of this complex is the partial unfolding of triple-helical collagen and thus to increase the accessibility of the scissile bond to the active site of catK. All three interactions might be essential for the formation of the productive collagenase complex. Ongoing studies will analyze these and additional structural requirements for a productive collagenolytic assembly.
Acknowledgments
We thank Ernst Bergmann and the staff at the beamline 8.3.1 of the Advanced Light Source at Lawrence Berkeley Laboratory for help in data collection.
This work was supported in part by a Canada Research Chair Award (Protease in Diseases to D. B.), National Institutes of Health Grant AR 48669 (to D. B.), and Canadian Institutes of Health Research Grant FRN 12831 (to M. N. G. J.) and from the Alberta Heritage Foundation for Medical Research (to M. N. G. J.).
The atomic coordinates and structure factors (code 3H7D) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- catK
- cathepsin K
- catL
- cathepsin L
- Z
- benzyloxycarbonyl
- MCA
- 4-methyl-7-coumarylamide
- BDP
- β-d-galactopyranuronic acid
- ASG
- 2′-deoxy-2′-acetamido-β-d-galactose-4-sulfate
- E64
- L-3-carboxy-trans-2,3-epoxypropionyl-leucylamido-(4-guanidino)butane
- C4-S
- chondroitin 4-sulfate
- PDB
- Protein Data Bank.
REFERENCES
- 1. Brömme D., Okamoto K. (1995) Biol. Chem. Hoppe Seyler 376, 379–384 [DOI] [PubMed] [Google Scholar]
- 2. Drake F. H., Dodds R. A., James I. E., Connor J. R., Debouck C., Richardson S., Lee-Rykaczewski E., Coleman L., Rieman D., Barthlow R., Hastings G., Gowen M. (1996) J. Biol. Chem. 271, 12511–12516 [DOI] [PubMed] [Google Scholar]
- 3. Kafienah W., Brömme D., Buttle D. J., Croucher L. J., Hollander A. P. (1998) Biochem. J. 331, 727–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garnero P., Borel O., Byrjalsen I., Ferreras M., Drake F. H., McQueney M. S., Foged N. T., Delmas P. D., Delaissé J. M. (1998) J. Biol. Chem. 273, 32347–32352 [DOI] [PubMed] [Google Scholar]
- 5. Li Z., Hou W. S., Brömme D. (2000) Biochemistry 39, 529–536 [DOI] [PubMed] [Google Scholar]
- 6. Li Z., Hou W. S., Escalante-Torres C. R., Gelb B. D., Bromme D. (2002) J. Biol. Chem. 277, 28669–28676 [DOI] [PubMed] [Google Scholar]
- 7. Li Z., Yasuda Y., Li W., Bogyo M., Katz N., Gordon R. E., Fields G. B., Brömme D. (2004) J. Biol. Chem. 279, 5470–5479 [DOI] [PubMed] [Google Scholar]
- 8. Wilson S., Hashamiyan S., Clarke L., Saftig P., Mort J., Dejica V. M., Brömme D. (2009) Am. J. Pathol. 175, 2053–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Z., Kienetz M., Cherney M. M., James M. N., Brömme D. (2008) J. Mol. Biol. 383, 78–91 [DOI] [PubMed] [Google Scholar]
- 10. Linnevers C. J., McGrath M. E., Armstrong R., Mistry F. R., Barnes M. G., Klaus J. L., Palmer J. T., Katz B. A., Brömme D. (1997) Protein Science 6, 919–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lecaille F., Choe Y., Brandt W., Li Z., Craik C. S., Brömme D. (2002) Biochemistry 41, 8447–8454 [DOI] [PubMed] [Google Scholar]
- 12. Hou W. S., Li Z., Gordon R. E., Chan K., Klein M. J., Levy R., Keysser M., Keyszer G., Brömme D. (2001) Am. J. Pathol. 159, 2167–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brömme D., Nallaseth F. S., Turk B. (2004) Methods 32, 199–206 [DOI] [PubMed] [Google Scholar]
- 14. Brömme D., Okamoto K., Wang B. B., Biroc S. (1996) J. Biol. Chem. 271, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 15. Barrett A. J., Kembhavi A. A., Hanada K. (1981) Acta Biol. Med. Ger. 40, 1513–1517 [PubMed] [Google Scholar]
- 16. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 17. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D. Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 18. Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 19. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Cryst. 26, 283–291 [Google Scholar]
- 21. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 22. Krissinel E. (2010) J. Comput. Chem. 31, 133–143 [DOI] [PubMed] [Google Scholar]
- 23. Cohen G. E. (1997) J. Appl. Cryst. 30, 1160–1161 [Google Scholar]
- 24. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 25. Hou W. S., Brömme D., Zhao Y., Mehler E., Dushey C., Weinstein H., Miranda C. S., Fraga C., Greig F., Carey J., Rimoin D. L., Desnick R. J., Gelb B. D. (1999) J. Clin. Invest. 103, 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Strynadka N. C., James M. N. (1989) Annu. Rev. Biochem. 58, 951–998 [DOI] [PubMed] [Google Scholar]
- 27. Yamashita D. S., Marquis R. W., Xie R., Nidamarthy S. D., Oh H. J., Jeong J. U., Erhard K. F., Ward K. W., Roethke T. J., Smith B. R., Cheng H. Y., Geng X., Lin F., Offen P. H., Wang B., Nevins N., Head M. S., Haltiwanger R. C., Narducci Sarjeant A. A., Liable-Sands L. M., Zhao B., Smith W. W., Janson C. A., Gao E., Tomaszek T., McQueney M., James I. E., Gress C. J., Zembryki D. L., Lark M. W., Veber D. F. (2006) J. Med. Chem. 49, 1597–1612 [DOI] [PubMed] [Google Scholar]