Abstract
The synthesis of deoxy-thymidine di-phosphate (dTDP)–l-rhamnose, an important component of the cell wall of many microorganisms, is a target for therapeutic intervention. The first enzyme in the dTDP–l-rhamnose biosynthetic pathway is glucose-1-phosphate thymidylyltransferase (RmlA). RmlA is inhibited by dTDP–l-rhamnose thereby regulating l-rhamnose production in bacteria. The structure of Pseudomonas aeruginosa RmlA has been solved to 1.66 Å resolution. RmlA is a homotetramer, with the monomer consisting of three functional subdomains. The sugar binding and dimerization subdomains are unique to RmlA-like enzymes. The sequence of the core subdomain is found not only in sugar nucleotidyltransferases but also in other nucleotidyltransferases. The structures of five distinct enzyme substrate– product complexes reveal the enzyme mechanism that involves precise positioning of the nucleophile and activation of the electrophile. All the key residues are within the core subdomain, suggesting that the basic mechanism is found in many nucleotidyltransferases. The dTDP–l-rhamnose complex identifies how the protein is controlled by its natural inhibitor. This work provides a platform for the design of novel drugs against pathogenic bacteria.
Keywords: drug design/nucleotidyltransferase/Pseudomonas aeruginosa/pyrophosphorylase/rhamnose
Introduction
The development of resistance towards antibiotics poses a permanent threat in the fight against pathogenic microorganisms. Consequently, there is a constant need for new drugs to treat infectious diseases. The bacterial cell wall is a target for many current antibiotics. This is because it is not only vital for cell integrity, providing the microorganism with a natural shield against a hostile environment, but it consists of peptidoglycan and lipopolysaccharide instead of phospholipids; thus it is radically different from the eukaryotic cell membrane. Moreover, many of the chemical building blocks that make up the bacterial cell wall are not found in humans.
l-rhamnose, a 6-deoxyhexose, is located in the cell wall of both Gram-negative and Gram-positive bacteria. In the Gram-negatives it is one of the important residues of the O-antigen of lipopolysaccharide, a key determinant factor for the virulence of these species. Gram-positive organisms such as streptococci utilize l-rhamnose in their cell wall or capsule. Mycobacteria, on the other hand, use l-rhamnose in the arabinogalactan that attaches the lipid mycolic acid layer to the peptidoglycan layer (McNeil et al., 1990). It has been demonstrated that this attachment is of vital importance to mycobacteria: inhibitors of the formation of the arabinan portion of arabinogalactan, e.g. ethambutol, can stop cell growth and are effective drugs (e.g. see Deng et al., 1995).
l-rhamnose is derived from a glucose scaffold in four steps, starting with glucose-1-phosphate (G-1-P) and deoxy-thymidine tri-phosphate (dTTP), yielding deoxy-thymidine di-phospho (dTDP)–l-rhamnose. The enzymes catalysing the conversion are glucose-1-phosphate thymidylyltransferase (RmlA, E.C. 2.7.7.24), dTDP–d-glucose 4,6-dehydratase (RmlB), dTDP–6-deoxy-d-xylo-4-hexulose 3,5-epimerase (RmlC) and dTDP–6-deoxy-l-lyxo-4-hexulose reductase (RmlD). Significantly, these proteins are highly conserved amongst microorganisms (e.g. see Ma et al., 1997; Graninger et al., 1999) and therefore conclusions drawn from the structure of a protein from one species will have strong implications for the corresponding enzyme structure of another origin. RmlB (Allard et al., 2000) and RmlD (Giraud et al., 1999), as NAD(P)-dependent enzymes, possess the famous Rossman fold and are members of the short-chain dehydrogenase superfamily. RmlC has a novel structure that was described first this year in the enzyme from Salmonella typhimurium (Giraud et al., 2000) and more recently from Methanobacterium thermoautotrophicum (Christendat et al., 2000). However, the Protein Data Bank (PDB) does not contain any structures with an identifiable sequence relationship to RmlA, the first enzyme in the l-rhamnose biosynthetic pathway. The enzyme combines dTTP with G-1-P to yield dTDP–d-glucose and pyrophosphate (Figure 1). The reaction is effectively the transfer of deoxy-thymidine mono-phosphate (dTMP) to G-1-P. It has been demonstrated that RmlA is inhibited by dTDP–l-rhamnose in a competitive and non-competitive manner (Melo and Glaser, 1965). This feedback makes RmlA the point of control for the whole pathway, marking RmlA as a highly attractive candidate in the search of inhibitors against l-rhamnose biosynthesis.
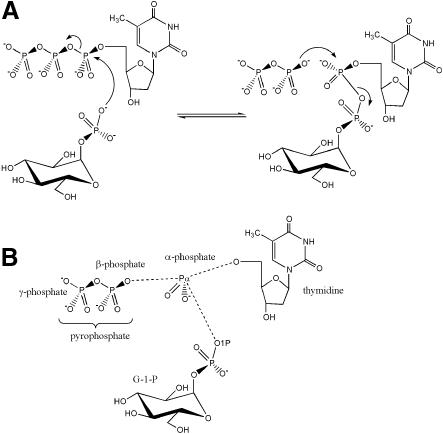
Fig. 1. (A) The mechanism of the reaction catalysed by RmlA. (B) The distinct chemical groups that form the ternary complex with the protein.
Pseudomonas aeruginosa is a Gram-negative bacterium that colonizes many children with cystic fibrosis, where it is a significant cause of morbidity and mortality. In addition it is an opportunistic pathogen that can cause a wide variety of infections, particularly in victims of severe burns and in patients who are for any reason immunosuppressed. As a consequence, P.aeruginosa is one of the most prevalent pathogens in hospital-acquired infections. Due to its high resistance to antibiotics, it is a particularly dangerous pathogen and any approach towards its control is highly desirable. l-rhamnose is a key component of the lipopolysaccharide core oligosaccharide (Rahim et al., 2000) and is also found in the secreted rhamnolipids, an important factor of virulence, in P.aeruginosa.
We describe here the structure of RmlA from P.aeruginosa and the following complexes: G-1-P (substrate), dTTP (substrate), dTDP–d-glucose (product), dTMP, thymidine plus G-1-P, and dTDP–l-rhamnose (competitive and non-competitive inhibitors). Based on these component crystal structures, we describe the structural basis for the catalytic mechanism of the enzyme and identify the key residues involved in catalysis. Our results explain the basis of competitive inhibition of RmlA by dTDP–l-rhamnose and discuss models for the non-competitive control of RmlA by dTDP–l-rhamnose. These insights have obvious implications for many other systems, including those involving distantly related nucleotidyltransferases. It is our hope that these results will prove helpful in the development of novel antibiotics.
Results
Overall structure
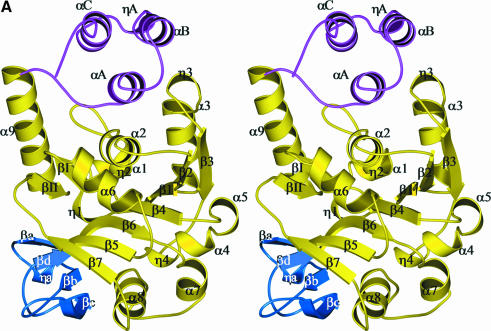
Monomer. The structure of the monomer is shown in Figure 2. An automatic domain identification procedure using the DOMAK algorithm (Siddiqui and Barton, 1995) recognizes the monomer’s fold as a single domain. It is dominated by a mixed seven-stranded β-sheet that is covered mainly by α-helical layers. The monomer would thus be classified as an αβα sandwich in the CATH (Class, Architecture, Topology, Homology) nomenclature (Orengo et al., 1997). For functional reasons, we have divided the monomer further into three subdomains (Figure 2), although there is no large movement of them relative to each other in the various data sets. The core subdomain (residues 1–135 and 173–246) contains the central β-sheet. This seven-stranded sheet has six parallel strands and one strand (β6) that is anti-parallel (Figure 2). One face of the central sheet packs exclusively against helices that almost completely occlude the face of the sheet. The other face, which binds the nucleotide, stacks against a large α-helix (α8) typical of nucleotide binding proteins. Other helices sit against this face but leave an exposed region. The fold of this subdomain has obvious similarities to the Rossman fold of NAD(P)-dependent dehydrogenases and contains all residues that are in contact with the active centre nucleotide in RmlA. The second subdomain sits against the exposed face of the central β-sheet. It is a small, four-stranded, antiparallel β-sheet structure (residues 136–172). Important residues for the recognition of the sugar-phosphate moiety are found here and it will therefore be termed the sugar-binding subdomain. The α8 helix and the sugar-binding subdomain act as the sides of a channel with the exposed face of the central β-sheet at the bottom. The third subdomain is composed of three α-helices at the C-terminus (amino acids 247–293). Helices αB and αC are amphipathic, packing their hydrophobic faces against each other. Several leucine residues are found at the interface. As this subdomain interacts extensively with other monomers, it is designated the dimerization subdomain. In cooperation with the core subdomain of the same monomer and the core subdomain of an adjacent monomer, the dimerization subdomain forms a second binding site for thymidine-containing compounds.
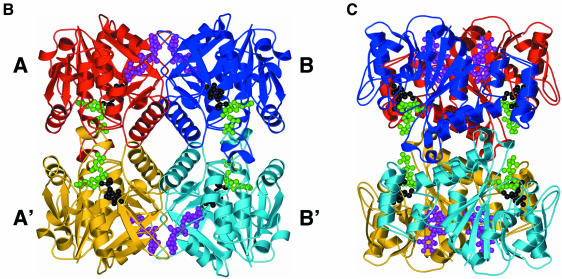
Fig. 2. (A) Stereo ribbon diagram of the RmlA monomer with location of secondary structure elements. The different colours denote the three subdomains. Yellow is the core binding subdomain, light blue is the sugar-binding subdomain and magenta the dimerization subdomain. The η character represents a 310 helix, and α and β have their normal meaning. Secondary structure was assigned with DSSP (Kabsch and Sander, 1983). (B) A ribbon representation of the RmlA tetramer. The monomers are coloured red, monomer A; blue, monomer B; yellow, monomer A′; and light blue, monomer B′. G-1-P (black) and dTTP (green) are shown at the active sites in ball-and-stick format. dTDP–l-rhamnose (magenta) is shown in the secondary binding sites, again as a ball-and-stick diagram. (C) Same as (B), rotated by 90° around the y-axis. All molecular representations are prepared with BOBSCRIPT (Esnouf, 1997) through the GL_RENDER interface (L.Esser and J.Deisenhofer, unpublished data) and were rendered with POV-RayTM.
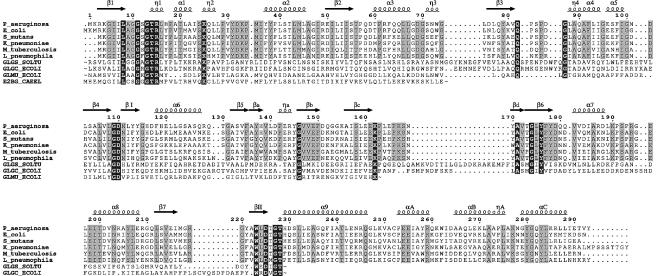
A search for similar structures in the PDB (Sussman et al., 1998) using DALI (Holm and Sander, 1993) reveals a clear relationship between the monomer and SpsA, a nucleotide-diphospho-sugar transferase from Bacillus subtilis that is possibly involved in the production of a mature spore coat (Charnock and Davies, 1999; Z-score of 8.3 compared with a Z-score of 53.2 when RmlA is overlaid with itself, and a ‘nonsense’ score of 2 for unrelated proteins). This finding is pertinent as sugar nucleotides themselves are delivered to the cell wall, where they are transferred to the growing carbohydrate chain by the release of the dinucleotide. Although the reactions differ in detail, there are chemical similarities. In addition to SpsA, more distant relationships of RmlA with other nucleotide binding proteins exist, but the similarities are restricted to the central β-sheet of the core subdomain. The amino acid sequence of the core subdomain of RmlA is related to the N-terminal domain of N-acetylglucosamine-1-phosphate uridyltransferase (GlmU) (21% identity) from Escherichia coli (Figure 3). Although this structure has recently been described in the literature, the coordinates are not yet available from the PDB (Brown et al., 1999). Inspection of the published figures indicates structural similarity between the N-terminal domain of GlmU and the core subdomain of RmlA with mechanistically important and conserved residues residing at similar structural locations. The mutagenesis data reported for GlmU is therefore relevant to RmlA.
Fig. 3. Sequence alignment, prepared with CLUSTAL_X (Thompson et al., 1997), and manually edited to avoid deletions and insertions in secondary structure elements. Non-RmlA sequences were truncated as indicated by ‘∼’ in clearly unrelated regions. White letters on a black background represent conservation throughout all sequences while conservation within RmlA proteins only is shown by black letters on grey background. The RmlA sequences are as follows: P_aeruginosa, P.aeruginosa; E_coli, E.coli; S_mutans, Streptococcus mutans; K_pneumoniae, Klebsiella pneumoniae; M_tuberculosis, Mycobacterium tuberculosis; L_pneumophila, Legionella pneumophilia. Other sequences are: GLGS_SOLTU, Solanum tuberosum G-1-P adenylyltransferase small subunit (precursor); GLGC_ECOLI, E.coli G-1-P adenylyltransferase; GLMU_ECOLI, E.coli N-acetylglucosamine 1-phosphate uridyltransferase; E2BG_CAEEL, Caenorhabditis elegans putative translation initiation factor eIF2B γ-subunit. The figure was prepared with EsPript (Gouet et al., 1999).
Tetramer. RmlA is a 222 tetrameric molecule with approximate dimensions of 81 × 73 × 67 Å (Figure 2). In its centre, an α-helix from each monomer contributes to a striking four-helix motif (Figure 2). Although a tetramer, RmlA can be viewed as a ‘dimer of dimers’ for simplicity of discussion. Figure 2 shows the tetramer with subunits labelled A, B, A′ and B′. The AB dimer involves both the dimerization and core subdomains and buries a total of 3700 Å, 56% of which is hydrophobic. The AA′ interface is less substantial than the AB, withdrawing from solvent a total of ∼2500 Å of surface area, 66% of which is hydrophobic. In total the tetramer occludes ∼23% of each monomer’s surface area.
The active centre
The catalytically relevant complexes dTTP, G-1-P, dTDP–d-glucose, dTMP and thymidine/G-1-P locate the active site. If we split dTDP–d-glucose into three chemophores, G-1-P, thymidine and α-phosphate (Figure 1), each part superimposes very closely upon its equivalent in the other complexes, i.e. the same residues always bind thymidine, G-1-P and the α-phosphate in the same manner.
The active centre of RmlA lies in a deep pocket formed by core and sugar-binding domains (Figures 2 and 4). It has approximate dimensions of 17 × 17 × 13 Å, and is lined by a number of polar residues that are conserved in related nucleotidyltransferases and in more distantly related proteins, including glycosyltransferases. We assign these residues to one of three groups, depending on their function in RmlA. Group one contains amino acids that are involved in the catalytic reaction, while groups two and three recognize the nucleotide and carbohydrate components of dTDP–d-glucose, respectively.
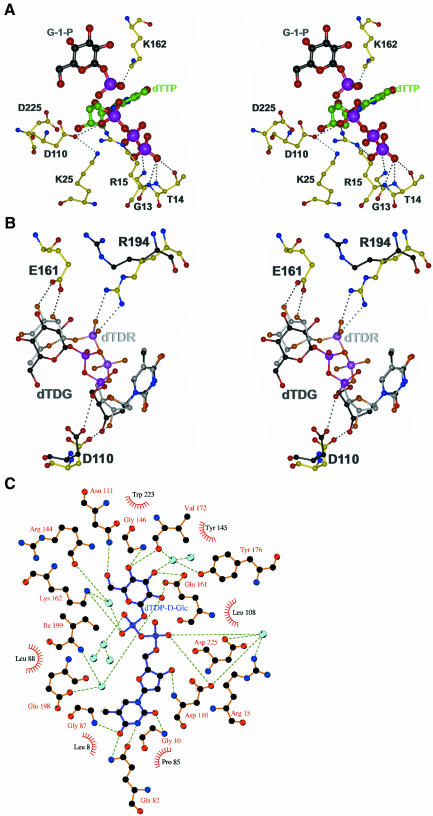
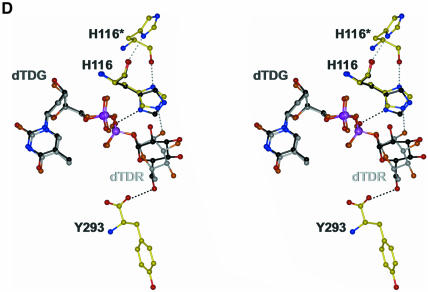
Fig. 4. (A) Model of a ternary RmlA–dTTP–G-1-P complex with important protein residues shown as a ball-and-stick representation. The protein and dTTP coordinates are taken from the dTTP data set. The G-1-P position was derived from the thymidine/G-1-P structure. Oxygen atoms are coloured red, nitrogen atoms blue, phosphorus atoms pink, protein carbon atoms yellow, G-1-P carbon atoms black and dTTP carbon atoms green. The different carbon colours were chosen to emphasis the three components of the complex. (B) Stereoview of dTDP–d-glucose and dTDP–l-rhamnose in the active site; their positions are derived from superposition of the corresponding protein structures. The colour scheme is the same as in (A) except darker shades are used for dTDP–d-glucose and lighter shades for dTDP–l-rhamnose. R194, which differs between the complexes, is shown with black carbon atoms for dTDP–d-glucose complex. dTDG, dTDP–d-glucose; dTDR, dTDP–l-rhamnose. (C) LIGPLOT representation (Wallace et al., 1995) of the dTDP–d-glucose protein interactions in the active centre. Water molecules are shown as light blue spheres, hydrophobic interactions are indicated by part circles. (D) The second binding site showing both dTDP–d-glucose and dTDP–l-rhamnose bound. The colour scheme is the same as (B). The asterisk at H116 denotes that this residue is from the neighbouring monomer. The protein atoms are taken from the dTDP–l-rhamnose complex. The imidazole ring from the dTDP–d-glucose is shown and is rotated relative to the dTDP–l-rhamnose complex. Hydrogen bonds for both orientations of H166 are shown.
Based on their contacts with substrates and their absolute sequence conservation amongst sugar nucleotidyltransferases, we have identified the residues R15, K25, D110, K162 and D225 as catalytically important (Figure 4). Their exact role in catalysis is discussed in the section on mechanism below. R15 belongs to a highly conserved sequence motif (G)GXGXR(L) that resides in a flexible region consisting of residues G11 to I23. R15 sits on the N-terminal end of this loop, which adopts a tight helical conformation. The only cis-peptide bond in RmlA is found in this loop at the conserved P18. The loop is located at the AA′ dimer interface within the immediate vicinity of the same loop from the A′ monomer. Part of the loop is disordered in the apo enzyme but becomes traceable in the thymidine-containing complexes. In the dTDP–d-glucose complex, the loop adopts a different orientation in the AB dimer than in the A′B′ dimer. This conformational ‘pairing’ is notable because the loops touch each other at I23 in the AA′ dimer, suggesting a possible route for cross talk between active sites. R15 binds the triphosphate group of dTTP. The flexibility of the loop is possibly important in allowing binding of dTTP and departure of pyrophosphate (the β- and γ-phosphates of dTTP). R194 is involved in binding of dTDP–l-rhamnose but in other complexes the side chain faces away from the active site. Although conserved in RmlA proteins, this residue is not conserved more widely. This is consistent with a role only in regulation of RmlA and therefore tailored to the rhamnose pathway rather than a catalytic role, which would almost certainly be conserved in related sugar nucleotide transferases.
Specificity for thymidine is provided by G10, Q82 and G87, all of which form hydrogen bonds with the pyrimidine ring (Figure 4). A tight loop in the main chain between Q82 and G87 selects for pyrimidine bases over purine bases. A hydrogen bond to OE1 of Q26 anchors the O3 of the deoxyribose ring. As there is no specific interaction of the methyl group of the pyrimidine ring and a pocket for the 2-OH of ribose exists, it is not surprising that RmlA accepts UTP and dUTP as substrates (Melo and Glaser, 1965).
Glucose is bound to the protein by a mixture of hydrogen bonds and van der Waals interactions (Figure 4). The bidentate interaction between the side chain of E161, and O2′ and O3′ of glucose is a classic motif in protein carbohydrate interactions (e.g. see Moothoo and Naismith, 1998). O4′ makes hydrogen bonds to the main chain of G146 (amide nitrogen) and L172 (carbonyl). O6′ is bound to ND2 of N111. In addition, O6′, O5′, O3′ and O2′ all hydrogen bond to bridging water molecules. W223, L108 and G174 provide a hydrophobic surface for the apolar patch of glucose (C5′). The side chains of L108 and L8 limit the size of the active site by providing a hydrophobic cap.
Second binding site
A second binding site for compounds that contain thymidine is found in the interface between two neighbouring monomers (Figure 2). The organization of the tetramer is such that two of these binding sites lie in close proximity to each other at the AB dimer interface. Residues from the core and dimerization subdomains plus the core subdomain of the adjacent monomer form a cleft of dimensions 21 × 12 × 13 Å. Residues L45, Y114, G115, H119, V250, A251, I256 and R259, with G218 and R219 from the neighbouring monomer, line the site into which thymidine and the phosphate linker bind (Figure 4). There are few hydrogen bonds in this pocket, indicating that this site is rather non-specific. In the dTDP–d-glucose complex, O2′ makes three hydrogen bonds to the protein (D117 and D120 bidentate) and O6′ a single hydrogen bond to Y293. In the dTDP–l-rhamnose complex, the β-glycosidic linkage places the plane of the sugar ring in a different orientation, but the position of two phosphate groups is conserved. Only the axial O2′ of rhamnose appears to be recognized by a single hydrogen bond to H116 (Figure 4). The side chain of H116 of the second binding site is within hydrogen bonding distance of the main chain carbonyl oxygen of H116 in the neighbouring monomer. This hydrogen bond is only realized in the dTDP–l-rhamnose complex; in all other structures, H116 interacts with the phosphate of the nucleotide or an adventitious sulfate ion (G-1-P and apo), preventing the imidazole ring from rotating. Amino acids in this region of the structure show a lesser degree of conservation than the ones in the active centre, even within RmlA sequences from other organisms. Interestingly, A251 adopts an unusual (+/+)-φ/ψ-backbone conformation; both it and G115 are conserved, suggesting that the conformation of backbone structure around the second site is preserved.
Discussion
Insights into the reaction mechanism
RmlA catalyses the condensation of dTTP and G-1-P to form dTDP–d-glucose. The reverse reaction, the pyrophosphorolysis of dTDP–d-glucose, gives the enzyme its other name, dTDP–d-glucose pyrophosphorylase. The reaction mechanism that RmlA and related enzymes follow has not yet been described in detail. An early study (Melo and Glaser, 1965) showed that the enzyme normally follows a sequential ordered bi bi mechanism in which the substrates bind in an obligate order, react and then depart, again in a specific sequence. Only in the case of dTDP–d-glucose pyrophosphorolysis were ‘ping pong’ kinetics observed, implying the existence of a covalent dTMP–enzyme intermediate. The possibility of such a species, however, could be eliminated by isotope exchange experiments, as RmlA does not incorporate 14C-glucose-1-phosphate into dTDP–d-glucose. More recent data (Lindquist et al., 1993) has again suggested a ping pong type mechanism, in contradiction to detailed kinetic studies on the closely related ADP–glucose pyrophosphorylase, for which only a sequential ordered bi bi mechanism fits the data (Paule and Preiss, 1971). Subtle ambiguity remains as to whether the second substrate binds and reacts simultaneously [the so called ‘hit and run’ mechanism (Theorell and Chance, 1951)], or binds and then reacts. These experiments also showed that the nucleotide triphosphate binds to the protein first. Our work, in which a stable dTTP enzyme complex has been obtained, argues further against a covalent intermediate. Taken together, these data are only consistent if the reaction is of the SN2 type, with dTTP binding to the protein, followed by G-1-P, which acts as the nucleophile attacking the α-phosphate of dTTP. The β- and γ-phosphates of dTTP are displaced and leave as pyrophosphate. In the reverse direction, the α-phosphate of dTDP–d-glucose is subject to nucleophilic attack by pyrophosphate and G-1-P is displaced. For simplicity we restrict discussion to the forward reaction, dTDP–d-glucose synthesis, since in the light of reversibility it is trivial to use the data to interpret the reverse direction (dTDP–d-glucose decomposition).
In SN2 type reactions, the leaving group and the attacking nucleophile reside at opposite faces of the central atom, in this case the phosphorus atom (Pα) of the α-phosphate. This agrees with the crystallographic data presented here. The β- and γ-phosphates of dTTP bind to the conserved loop around R15, folding back over the nucleotide and leaving the space opposite the pyrophosphate entity free for G-1-P binding. This strained conformation of dTTP activates it for nucleophilic attack at the α-phosphate. Superposition of Cα traces from the dTTP, the thymidine/G-1-P and the G-1-P complexes shows that the O1P atom of G-1-P is correctly positioned for attack at the Pα atom of dTTP. The bond between pyrophosphate and Pα, which is broken during the reaction (Figure 1), is, as required, opposite to the trajectory of the nucleophilic attack. The distance between O1P and Pα in this model is 3.1 Å, almost the sum of their van der Waals radii. The orientation of the phosphate group in G-1-P and the orientation α-phosphate of dTTP with respect to the protein and each other are identical to those seen in the dTDP–d-glucose complex. This orientation of the O1P atom of G-1-P is accomplished not only by binding the glucose ring but also by a very strong electrostatic interaction bond (∼2.5 Å) of the O4P atom of G-1-P to the side chain of K162 (Figure 3). This lysine is an absolutely conserved amino acid in sequences of other sugar nucleotidyl transferases. Mutagenesis and biochemical studies of the equivalent residue in ADP– glucose pyrophosphorylase had shown it to interact with G-1-P (Parsons and Preiss, 1978; Hill et al., 1991). In addition to this locking function, the interaction stabilizes the negative charge on the phosphate of G-1-P, increasing the nucleophilicity of the O1P atom.
Electrostatic repulsions between the negative charges at the phosphate of G-1-P and the phosphates of dTTP would presumably hinder nucleophilic attack and G-1-P binding. A number of conserved basic side chains surround the phosphates at the active site (R15, K25, K162). Nearby are R194 (conserved) and R144 (not conserved). Additional counterbalancing charges come from the N-terminal dipole of the helical turn at R15 and the main chain amide nitrogen pocket formed by residues G12 to T14. In the study of GlmU mutated residues (equivalent to 11, 15 and 25 in RmlA) reduce, but do not completely abolish, activity (Brown et al., 1999). Mutation of R15 (RmlA numbering) had the most dramatic effect, reducing kcat almost 6000-fold, while KM was doubled, confirming that this residue has an important role in orientating and/or charge compensation of the pyrophosphate.
The activity of RmlA-related enzymes is known to require Mg2+ ions (e.g. Bernstein and Robbins, 1965), which could further neutralize negative charges. However, the Mg2+ binding mode of RmlA is not clear as co-crystallization experiments with either Mg2+ or Mn2+ in the presence or absence of dTDP–d-glucose revealed no extra electron density for a metal ion, and co-crystallization with dTTP and Mg2+ or Mn2+ has not been successful. It is possible that the cation coordinates to the β- and γ-phosphate groups of dTTP in a chelated manner, as observed in kanamycin nucleotidyltransferase (Pedersen et al., 1995), and leaves the active centre as a complex with pyrophosphate. In the dTTP complex we have obtained, there is no metal ion at this position. K25, D110 and D225 are reminiscent of a metal ion binding site in inorganic pyrophosphatase (Heikinheimo et al., 1996). However, the catalytic metal ion in RmlA cannot be bound here unless the positioning of the substrates and several side chains in all our complexes changes in the presence of a metal ion.
The OD2 atom of the conserved residue D110 is ∼3.0 Å in distance from an oxygen of the α-phosphate group of dTDP–d-glucose. Since it is inconceivable that two negatively charged atoms would be this close, there must be a hydrogen atom bound between the atoms. We propose that this hydrogen bond is a key factor in activating the α-phosphate for nucleophilic attack. Examination of the OD2 atom of D110 shows it makes an electrostatic bond (3.0 Å) with the absolutely conserved K25. Our hypothesis is that this interaction is crucial for the formation of a hydrogen bond between D110 and the α-phosphate. The Asp interaction will substantially reduce the charge on the α-phosphate, making it more electrophilic, and at the same time minimize electrostatic repulsion with the negatively charged nucleophile. In the dTTP complex, the distance between the α-phosphate and OD2 of D110 varies between 3.4 and 4.1 Å, but only a very small movement is required during enzyme turnover to shorten this distance. A similar arrangement of α-phosphate and glutamic acid is observed in the structurally unrelated kanamycin nucleotidyltransferase (Pedersen et al., 1995). The side chain of E52 is 2.2 Å away from the α-phosphate in this structure. The site-directed mutants D110A and D110N are very weakly active, showing a decrease in kcat of at least 1000-fold. However, circular dichroism (CD) spectroscopy shows that both mutants do not exhibit an identical spectrum to native protein.
Inhibition of enzymatic activity by dTDP–l-rhamnose
The feedback control of P.aeruginosa RmlA by dTDP– l-rhamnose has been studied extensively by Melo and Glaser (1965). dTDP–l-rhamnose was found to be a competitive and a non-competitive inhibitor in the direction of both synthesis and pyrophosphorolysis, with a Ki value of 22 µM (Melo and Glaser, 1965). They observed that inhibition was reversed by dTDP–d-glucose and with very high concentrations of pyrophosphate.
The competitive inhibition of dTDP–l-rhamnose is easily explained, as it binds at the same sites in roughly the same manner as the substrates in both the forward and reverse direction. The structure of the complex shows why dTDP–l-rhamnose is not a substrate for the enzyme and provides further evidence for our mechanistic proposal. With its β-linkage, dTDP–l-rhamnose adopts a different orientation in the protein than dTDP–d-glucose. Unlike the second site, where this change is manifested in a different orientation to the planes of the carbohydrate rings, here the carbohydrate rings are much closer to co-planarity. E161 recognizes the rhamnose ring with a bidentate hydrogen bond to the O2′ and O3′ carbohydrate, similar to the situation in the dTDP–d-glucose complex. It is the two phosphate groups that adopt different conformations (Figure 4). As a consequence, the ribose O3 oxygen becomes the hydrogen bonding partner of D110 instead of the α-phosphate. We propose that this lack of a hydrogen bond between the α-phosphate and the side chain of D110 stabilizes dTDP–l-rhamnose to decomposition by pyrophosphorolysis, making it a competitive inhibitor. The positions of the two phosphates are stabilized by strong salt contacts with R194, which moves into the active site. Harnessing this interaction could provide a potential basis for inhibitor design.
The basis for the non-competitive inhibition of RmlA by dTDP–l-rhamnose is less clear. There are two conditions that could lead to the observed non-competitive inhibition and deviation from Michaelis–Menten behaviour: (i) if by dTDP–l-rhamnose binding at one active site in the tetramer it disrupted the catalytic mechanism in another monomer; and (ii) if dTDP–l-rhamnose was bound, other than at the active site, and as a result affected the catalytic activity within that monomer or within another monomer. In either case, the cause of non-competitive inhibition has to lie in the differences between dTDP–l-rhamnose and dTDP–d-glucose (a competitive inhibitor) (Melo and Glaser, 1965).
Superposition of the dTDP–d-glucose or G-1-P or dTTP complexes on the dTDP–l-rhamnose complex shows there are no gross changes in main chain position. The active site at the R15 loop is subtly different. The change manifests itself in the separation of D110 and R15. In the closed form, the distance is 12.6 Å, and in the open form it is 13.9 Å. If this change in loop conformation in monomer A is transmitted via I23 to the same loop in the neighbouring monomer A′, it could non-competitively inhibit the activity of A′. However, as all four active sites are filled with dTDP–l-rhamnose in our structure, we have no direct structural evidence for this proposal. Interestingly, in both the two closed chains of the dTDP–d-glucose complex, R15 binds a sulfate ion at the identical position to γ-phosphate from the dTTP complex. Only the closed form is found in the dTTP complex. This suggests that the closed form occurs only when R15 makes a salt contact, which would indicate movement of this loop is important for, and occurs during, the catalytic cycle.
At the second site, superposition of Cα atoms from all the complexes shows that the positions of the protein main chain and the atomic positions of the thymidyl-diphospho groups are almost indistinguishable. Consequently, if non-competitive inhibition results from dTDP–l-rhamnose binding to this site it has to be achieved by differences between the carbohydrate portions of dTDP–d-glucose and dTDP–l-rhamnose. Since dTTP is not a non-competitive inhibitor, the rhamnose ring would have to make a unique interaction. The only specific interaction observed is with H116. However, H116 is not conserved (found as proline). Clearly, more studies will be needed to elucidate the role, if any, of residues of this second binding site in RmlA.
Implications for sugar nucleotidyltransferases
Hexose-1-phosphate nucleotidyltransferases are not only involved in transformation reactions, but are also important enzymes in energy storage processes such as glycogen or starch generation. In these pathways, ADP–glucose pyrophosphorylase, the most thoroughly studied RmlA-related protein, is the point of control for starch synthesis in plants and for glycogen synthesis in bacteria (Preiss, 1982, 1984). The crystallization of both the α4-homotetrameric bacterial and the α2β2-heterotetrameric plant protein has been described, but a structure has not yet been published (Mulichak et al., 1988; Binderup et al., 2000). ADP–glucose pyrophosphorylase from plants (Preiss, 1991), cyanobacteria (Iglesias et al., 1991) and green algae (Iglesias et al., 1994) is allosterically activated by 3-phosphoglycerate and inhibited by phosphate, whereas the protein from enteric bacteria is activated by fructose-1,6-bisphosphate and inhibited by AMP (Preiss and Romeo, 1989). In the plant enzyme, the smaller 50 kDa monomer is believed to catalyse the reaction, while the larger 51 kDa monomer is mainly responsible for controlling activity (Ballicora et al., 1995). Residues that determine the allosteric behaviour of ADP–glucose pyrophosphorylase are located at its C-terminus (Ball and Preiss, 1994; Greene et al., 1996; Ballicora et al., 1998), a region that has no homology with RmlA.
A PSI-BLAST databank search (Altschul et al., 1997) identifies several structurally uncharacterized proteins with a sequence relationship to RmlA. In addition to sugar-nucleotidyltransferases, the ε- and γ-subunits of eukaryotic translation initiation factor 2B (eIF2B) appear to be distant relatives of RmlA, as has already been reported by others (Price et al., 1996). eIF2Bγ and eIF2Bε are part of a heteropentameric complex that acts as a nucleotide-exchange protein for eIF2, reactivating this factor by exchanging GDP for GTP. The profiles of eIF2Bγ, eIF2Bε and RmlA are clearly similar for approximately the first 80 residues, and the sequence signature around D225 is preserved, allowing prediction of the nucleotide binding regions of eIF2Bγ and eIF2Bε. Large differences exist in the region of the sugar-binding subdomain of RmlA and in the C-termini of these proteins. The RmlA structure shows that the crucial catalytic residues for nucleotide transfer reside solely in the core subdomain. This observation, taken together with the appearance of the conserved catalytic residues in proteins that are not sugar nucleotidyltransferases, indicates that the core subdomain can be viewed as a distinct module with nucleotidyltransferase activity. We predict that this module can be added to other modules to produce enzymes with tailored activities centered around nucleotide transfer.
Conclusion
The crystal structures described here present a comprehensive view of the RmlA enzyme. From these results it has been possible to identify the structural basis of the mechanism of this class of nucleotidyltransferases. The work highlights the importance of several residues, in particular an aspartic acid (D110) that activates the α-phosphate for nucleophilic attack by G-1-P. The nucleotidyltransferase activity resides entirely in a core subdomain, whose sequence is found in many other structurally uncharacterized proteins. This strongly suggests that the proposed mechanism is found in many other nucleotidyltransferases, including those only distantly related to RmlA. The results show how dTDP–l-rhamnose regulates the activity of RmlA without itself becoming a substrate. This is an important finding as this inhibition is the feedback control of the l-rhamnose biosynthetic pathway, a new and important therapeutic target in pathogenic bacteria. These data provide a basis upon which rational design of novel antibiotics can proceed.
Materials and methods
Protein expression and crystallization
RmlA of P.aeruginosa was purified and crystallized as described previously (Blankenfeldt et al., 2000). Briefly, crystals were obtained from 9–11% (w/v) PEG 6000, 0.5 M Li2SO4 and 0.1 M Na-citrate pH 4.6. Se-methionine-labelled protein was obtained by inhibition of the methionine biosynthesis pathway in BL21(λDE3) cells (Doublie, 1997). RmlA ligand complexes were generated by co-crystallization with 1 µl of 10–100 mM ligand throughout. Diffraction data was measured from shock frozen crystals that were cryoprotected in mother liquor substituted with 16–20% (w/v) PEG 600. Crystals generally belonged to spacegroup P1 with cell parameters of a = 71.5 Å, b = 73.1 Å, c = 134.7 Å, α = 89.9°, β = 80.9° and γ = 81.1°, containing two RmlA tetramers in the asymmetric unit. For crystals grown in the presence of dTTP or dTDP–l-rhamnose, it was necessary to re-optimize the original crystallization conditions. The protein was pre-incubated with 10 mM ligand in these cases. The crystals grown in the presence of dTDP–l-rhamnose [precipitant 0.2 M Li2SO4, 10% (w/v) PEG 6000, 0.1 M Na-citrate pH 4.0] possessed a P212121 cell with unit cell parameters a = 71.1 Å, b = 138.6 Å, c = 139.7 Å; α = β = γ = 90.0°, corresponding to one RmlA tetramer per asymmetric unit. Two tetramers were found for protein crystallized in the presence of dTTP. These crystals belonged to space group P21 with cell parameters a = 73.0 Å, b = 134.4 Å, c = 140.9 Å; α = γ = 90.0°, β = 98.2°. The crystals were obtained with a precipitant solution of 5% (w/v) PEG 6000 and 0.1 M Na-citrate pH 4.0. PEG 600 [25% (v/v)] in mother liquor was necessary to achieve cryoprotection. All data were indexed and reduced with MOSFLM (Leslie, 1992) and CCP4 (CCP4, 1994). Full details of all data collection are given in Tables I and II.
Table I. Data collection and refinement statistics for data sets used in the phasing procedure.
| MAD (G-1-P) |
dTMP | |||
|---|---|---|---|---|
| Peak | Inflection | Remote | ||
| Data collection | ||||
| wavelength (Å) | 0.979 | 0.9791 | 0.885 | 0.978 |
| total measurements | 221 219 | 220 585 | 224 654 | 1 141 819 |
| unique reflections | 121 843 | 121 982 | 121 112 | 297 160 |
| average redundancy | 1.8 (1.0) | 1.8 (1.0) | 1.9 (1.7) | 3.8 (2.0) |
| I/σ | 31.7 (9.0) | 31.6 (8.8) | 29.7 (9.8) | 10.9 (2.0) |
| completeness (%) | 93.0 (63.2) | 93.0 (63.2) | 92.7 (69.4) | 94.2 (81.2) |
| anomalous completeness (%)a | 85.7 (51.4) | 86.0 (52.0) | 85.4 (60.6) | – |
| Rmergeb | 2.2 (8.9) | 2.3 (9.5) | 2.4 (8.9) | 5.3 (33.2) |
| f′/f′′ | –7.9/6.6 | –8.8/2.8 | –2.0/3.1 | – |
| resolution (highest shell, Å) | 30.0–2.8 (2.87–2.80) | 32.12–1.66 (1.74–1.66) | ||
| space group | P1 | P1 | ||
| cell constants (Å; °) | a = 71.6, b = 73.9, c = 133.8 | a = 71.5, b = 73.1, c = 134.7 | ||
| α = 89.8, β = 80.3, γ = 80.2 | α = 89.9, β = 80.9, γ = 81.1 | |||
| VM | 2.54 | 2.54 | ||
| Refinement | ||||
| resolution (highest shell, Å) | 72.55–2.80 (2.87–2.80) | 72.55–1.66 (1.70–1.66) | ||
| R-factorc | 19.0 (23.2) | 14.2 (22.5) | ||
| Rfreed | 22.7 (28.7) | 19.6 (30.3) | ||
| r.m.s.d. bonds (Å)/angles (°) | 0.033/2.27 | 0.016/2.32 | ||
| B-factor deviation (bonds/angles, Å2) | ||||
| main chain | 1.0/1.9 | 2.7/2.8 | ||
| side chain | 2.1/3.7 | 2.9/3.7 | ||
| residues in Ramachandran core (%)e | 90.0 | 92.9 | ||
| protein atoms | 18 280 | 18 858 | ||
| water atoms | – | 3365 | ||
| ligand atoms | 193 | 548 | ||
| average B-factor (Å2) | 28 | 14 | ||
| PDB accession code | 1g23 | 1fxo | ||
aCompleteness calculations treat F+ and F– as separate observations and are based on data used in the MAD calculation.
bRmerge = ΣΣ I(h)j – <I(h)> / ΣΣ I(h)j, where I(h)j is the measured diffraction intensity and the summation includes all observations.
cR-factor = (Σ | Fo | – Σ | Fc |) / Σ | Fo |
dRfree is the R-factor calculated using 5% of the data that were excluded from the refinement.
eRamachandran core refers to the most favoured regions in the φ/ψ-Ramachandran plot as defined by Laskowski et al. (1993).
Table II. Data collection and refinement statistics for all other data sets.
| Apo | dTTPa | Thymidine/G-1-P | dTDP–d-glucose | dTDP–l-rhamnosea | |
|---|---|---|---|---|---|
| Data collection | |||||
| wavelength (Å) | 0.934 | 1.542 | 0.934 | 0.934 | 1.542 |
| resolution (highest shell, Å) | 40.49–1.90 (1.99–1.90) | 50.00–2.60 (2.74–2.60) | 36.27–1.87 (1.97–1.87) | 40.49–1.77 (1.86–1.77) | 49.90–2.70 (2.84–2.70) |
| space group | P1 | P21 | P1 | P1 | P212121 |
| cell constants (Å; °) | a = 71.7, b = 73.7, c = 134.5 | a = 73.0, b = 134.4, c = 140.9 | a = 71.3, b = 73.1, c = 133.7 | a = 71.6, b = 73.4, c = 134.3 | a = 71.1, b = 138.6, c = 139.7 |
| α = 90.0, β = 80.9, γ = 80.9 | α = 90.0, β = 98.2, γ = 90.0 | α = 90.0, β = 81.4, γ = 81.6 | α = 89.9, β = 80.6, γ = 80.9 | α = 90.0, β = 90.0, γ = 90.0 | |
| VM | 2.56 | 2.53 | 2.52 | 2.54 | 2.55 |
| total measurements | 377 690 | 310 827 | 333 697 | 483 036 | 260 832 |
| unique reflections | 202 988 | 75 557 | 183 207 | 202 988 | 36 311 |
| average redundancy | 1.9 (1.9) | 4.1 (3.0) | 1.8 (1.3) | 2.0 (1.3) | 7.2 (5.7) |
| I/σ | 9.2 (1.9) | 7.4 (4.6) | 6.9 (2.4) | 7.1 (2.4) | 6.8 (2.9) |
| completeness (%) | 96.1 (95.6) | 91.5 (60.1) | 84.7 (66.8) | 93.9 (77.4) | 94.2 (70.8) |
| Rmerge | 5.4 (28.6) | 7.0 (15.4) | 6.7 (29.1) | 7.2 (30.2) | 10.2 (26.6) |
| Refinement | |||||
| resolution (highest shell, Å) | 72.55–1.90 (1.95–1.90) | 100.00–2.60 (2.67–2.60) | 72.55–1.87 (1.92–1.87) | 72.55–1.77 (1.83–1.77) | 100.00–2.70 (2.77–2.70) |
| R | 17.6 (30.6) | 21.5 (28.3) | 14.7 (23.2) | 15.2 (24.7) | 20.5 (25.5) |
| Rfree | 25.1 (45.4) | 24.2 (34.4) | 22.1 (39.5) | 21.7 (35.0) | 23.0 (33.3) |
| r.m.s.d. bonds (Å)/angles (°) | 0.025/1.87 | 0.028/2.27 | 0.022/2.52 | 0.018/2.38 | 0.014/2.39 |
| B-factor deviation bonds/angles (Å2) | |||||
| main chain | 2.6/3.5 | 0.8/1.6 | 2.1/2.7 | 4.9/4.8 | 0.5/0.8 |
| side chains | 5.6/6.1 | 3.0/5.0 | 5.3/6.1 | 3.7/4.3 | 1.1/1.8 |
| residues in Ramachandran core (%) | 92.0 | 91.6 | 91.6 | 92.6 | 90.3 |
| protein atoms | 18 516 | 18 260 | 18 626 | 18 688 | 9148 |
| water atoms | 2478 | – | 2236 | 2971 | – |
| ligand atoms | 100 | 464 | 499 | 717 | 305 |
| average B-factor (Å2) | 21 | 13 | 18 | 12 | 13 |
| PDB accession code | 1fzw | 1g2v | 1g0r | 1g1l | 1g3l |
Symbols and abbreviations are the same as in Table I.
aThese data sets were measured inhouse on an R-axis VI++ rotating anode.
All chemicals were purchased from Sigma. dTDP–l-rhamnose was prepared following the protocol of Graninger et al. (1999).
Phase determination and structure refinement
A three-wavelength MAD-data set was collected on BM14 at the ESRF-Grenoble in France (Blankenfeldt et al., 2000). Care was taken to obtain accurate rather than high resolution data. Seven-hundred-and-twenty non-overlapping 0.5° oscillations, to a resolution of 2.8 Å, were collected at a wavelength corresponding to the maximum value of f′′ (peak), the minimum value of f′ (inflection) and a remote point with minimal f′ contribution. SOLVE 1.17 (Terwilliger and Berentzen, 1999) was used to identify the position of selenium atoms and to calculate a first phase set. A total of 24 Se sites out of a possible 48 were unequivocally identified. These sites could easily be separated into groups of three, allowing the determination of non-crystallographic symmetry operators. Drawing a 30 Å sphere around the midpoint of an Se triplet generated a simple monomer mask. Overlapping regions in this mask were removed with the program NCSMASK of the CCP4 package (CCP4, 1994). The optimized mask was used to improve SOLVE phases by non-crystallographic symmetry (NCS) averaging and solvent flattening in DM (Cowtan, 1994). This phase set was transferred to the highest resolution data (dTMP) and slowly extended to higher resolution with DM. The electron density map was of high quality and allowed the further application of automated structure refinement procedures. Warp/Arp in the warpNtrace mode (Perrakis et al., 1999) was used to obtain a first protein chain trace in a mixed free-atom/protein model. Using this procedure, a total of 2126 out of 2344 amino acids in 39 protein chain fragments were traced resulting in an electron density map of superb quality. New NCS operators were derived from this trace. These NCS operators were inverted to superimpose all protein chain fragments onto a single location. By combining the fragments, the complete monomer backbone structure was obtained. The flexible loop around R15 and a smaller flexible surface loop around residue P193 were missing, but could be traced manually in the residual electron density. Unambiguous building of the monomer’s side chains was possible at this stage. The asymmetric unit contents were then reconstructed from the traced monomer and the NCS operators. Only the N-terminal His6-tag and the N-terminal methionine residue of most chains could not be located. The model was refined with CNS (Brünger et al., 1998). Warp/Arp in the solvent mode was used to improve the model’s quality further by adding water molecules, leading to an R-factor of 20.6% and an Rfree of 26.4%, with excellent protein geometry. Water molecules were then removed from the dTMP binding sites and difference electron density maps calculated after refinement in CNS to allow addition of ligand molecules into unbiased electron density. The refinement in other high resolution data sets followed a protocol of rigid body, positional and B-factor refinement using CNS. Water molecules were added in Warp/Arp and removed from the active and second site by hand. Addition of ligand molecules and manual adjustment of the model were made through program O (Jones et al., 1991). No NCS restraints were applied to the high resolution data sets as lower free R-factors could be achieved without them. REFMAC5 (Murshudov et al., 1997) became available at later stages and was used to complete the refinement. The addition of H atoms in their riding positions and the application of TLS (translation/libration/screw-rotation) and anisotropic B-factor refinement further improved the R-factor to 14.2% and Rfree to 19.6% in the case of the highest resolution structure (dTMP complex).
In the lower resolution rotating anode data sets, tight NCS restraints were employed throughout the REFMAC5 refinement. Water molecules were not included in these structures. In the case of the dTDP–l-rhamnose and dTTP complex data, the position of the tetramers was determined by molecular replacement using MOLREP (Vagin and Teplyakov, 1997). Clear difference electron density for these ligands was observed. Refinement statistics are given in Tables I and II for all structures.
Coordinates and structural factors for all structures described in this paper have been deposited in the PDB; accession codes are given in Tables I and II.
Site-directed mutagenesis
The plasmid encoding RmlA from P.aeruginosa (Blankenfeldt et al., 2000) was used to introduce a site-specific mutation at position 110 (D110A or D110N), using Pfu DNA polymerase and the Stratagene Quick Change mutagenesis protocol (Stratagene, La Jolla, CA) with the following oligonucleotides: GCGCTGGTCCTGGGCGCCAACCTCTATTACGGC with GCCGTAATAGAGGTTGGCGCCCCAGGACCAGCGC for D110A; and GCGCTGGTCCTGGGCAACAACCTCTATTACGGC with GCCGTAATAGAGGTTGTTGCCCCAGGACCAGCGC for D110N. Mutations were confirmed by nucleotide sequencing using the Sequenase 2.0 protocol (Amersham Pharmacia Biotech) and an ABI 377 automated DNA sequencer (Applied Biosystems Inc.).
The mutants were expressed in E.coli BL21(λDE3) pLysS cells (Novagen Inc.) and purified as described previously (Blankenfeldt et al., 2000). As the protocol involves a nickel chelation, contamination with E.coli RmlA is unlikely. Both mutants are folded and have the same CD spectrum (BBSRC CD Centre, Glasgow). However, this spectrum is not identical to the native protein (see supplementary data).
Enzymatic activity was measured for both wild-type and mutated RmlA following the generation of pyrophosphate with a commercially available coupled enzyme essay (O’Brien, 1976; Sigma, product number P7275) according to the manufacturer’s instructions. Normally the assay uses 50 ng of wild-type protein. The coupled assay is not suitable to determine accurate kinetic values, but is a guide to rates of reaction. Both mutants exhibit enzyme turnover at high protein concentrations (2000 ng). The ratio of the rates indicates the mutants are >1000-fold slower. Controls of boiled protein and 220 000 ng of E.coli (the same strain but lacking the expression plasmid) soluble cell extract do not show any detectable activity. This would suggest there is no endogenous RmlA activity in the mutant preparations.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful for beam time at the ESRF. We thank Uwe Dengler of EBI for help with domain identification and Garib Murshudov for assistance with dictionaries and REFMAC. We thank Sharon Kelly and Nicholas Price for their assistance in performing CD spectra at the BBSRC-funded Scottish Circular Dichroism Facility. This work is supported by a grant to J.H.N. from The Wellcome Trust and by a grant to J.S.L. from the Canadian Bacterial Disease Network.
References
- Allard S.T.M., Giraud,M.F., Messner,P., Whitfield,C. and Naismith,J.H. (2000) Overexpression, purification, crystallisation and preliminary structural study of dTDP–4,6-glucose dehydratase (RmlB) the second enzyme of the dTDP–rhamnose synthesis pathway, from Salmonella entrica serovar Typhimurim. Acta Crystallogr. D, 56, 222–225. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball K. and Preiss,J. (1994) Allosteric sites of the large subunit of the spinach leaf ADPglucose pyrophosphorylase. J. Biol. Chem., 269, 24706–24711. [PubMed] [Google Scholar]
- Ballicora M.A., Laughlin,M.J., Fu,Y., Okita,T.W., Barry,G.F. and Preiss,J. (1995) Adenosine 5′-diphosphate-glucose pyrophosphorylase from potato tuber. Significance of the N terminus of the small subunit for catalytic properties and heat stability. Plant Physiol., 109, 245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballicora M.A., Fu,Y., Nesbitt,N.M. and Preiss,J. (1998) ADP-glucose pyrophosphorylase from potato tubers. Site-directed mutagenesis studies of the regulatory sites. Plant Physiol., 118, 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein R.L. and Robbins,P.W. (1965) Control aspects of uridine 5′-diphosphate glucose and thymidine 5′-diphosphate glucose synthesis by microbial enzymes. J. Biol. Chem., 240, 391–397. [PubMed] [Google Scholar]
- Binderup K., Watanabe,L., Polikarpov,I., Preiss,J. and Arni,R.K. (2000) Crystallization and preliminary X-ray diffraction analysis of the catalytic subunit of ADP-glucose pyrophosphorylase from potato tuber. Acta Crystallogr. D, 56, 192–194. [DOI] [PubMed] [Google Scholar]
- Blankenfeldt W., Giraud,M.F., Leonard,G., Rahim,R., Creuzenet,C., Lam,J.S. and Naismith,J.H. (2000) The purification, crystallisation and preliminary structural characterisation of glucose-1-phosphate thymidylyltransferase (RmlA), the first enzyme of the dTDP- l-rhamnose synthesis pathway from Pseudomonas aeruginosa. Acta Crystallogr. D, 56, 1501–1504. [DOI] [PubMed] [Google Scholar]
- Brown K., Pompeo,F., Dixon,S., Mengin-Lecreulx,D., Cambillau,C. and Bourne,Y. (1999) Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily. EMBO J., 18, 4096–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Charnock S.J. and Davies,G.J. (1999) Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry, 38, 6380–6385. [DOI] [PubMed] [Google Scholar]
- Christendat D.S., Saridakis,V., Dharamsi,A., Bochkarev,A., Pai,E.F., Arrowsmith,C.H. and Edwards,A.M. (2000) Crystal structure of dTDP-4-keto-6-deoxy-d-hexulose 3,5-epimerase from Methano bacterium thermoautotrophicum complexed with dTDP. J. Biol. Chem., 275, 24608–24612. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cowtan K. (1994). Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography, 31, 34–38. [Google Scholar]
- Deng L., Mikusova,K., Robuck,K.G., Scherman,M., Brennan,P.J. and McNeil,M.R. (1995) Recognition of multiple effects of ethambutol on metabolism of mycobacterial cell envelope. Antimicrob. Agents Chemother., 39, 694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublie S. (1997) Preparation of selenomethionyl proteins for phase determination. Methods Enzymol., 276, 523–530. [PubMed] [Google Scholar]
- Esnouf R.M. (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model., 15, 132–134, 112–113. [DOI] [PubMed] [Google Scholar]
- Giraud M.F., McMiken H.J., Leonard,G.A., Messner,P., Whitfield,C. and Naismith,J.H. (1999) Overexpression, purification, crystallisation and preliminary structural study of dTDP-6-deoxy-lyxo-4-reductase (RmlD) the fourth enzyme of the dTDP-rhanose synthesis pathway, from Salmonella entrica serovar Typhimurim. Acta Crystallogr. D, 55, 2043–2046. [DOI] [PubMed] [Google Scholar]
- Giraud M.F., Leonard,G.A., Field,R.A., Berlind,C. and Naismith,J.H. (2000) RmlC, the third enzyme of dTDP-l-rhamnose pathway, is a new class of epimerase. Nature Struct. Biol., 7, 398–402. [DOI] [PubMed] [Google Scholar]
- Gouet P., Courcelle,E., Stuart,D.I. and Metoz,F. (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics, 15, 305–308. [DOI] [PubMed] [Google Scholar]
- Graninger M., Nidetzky,B., Heinrichs,D.E., Whitfield,C. and Messner,P.J. (1999) Characterization of dTDP-4-dehydrorhamnose 3,5-epimerase and dTDP-4-dehydrorhamnose reductase, required for dTDP-l-rhamnose biosynthesis in Salmonella enterica serovar Typhimurium LT2. J. Biol. Chem., 274, 25069–25077. [DOI] [PubMed] [Google Scholar]
- Greene T.W., Woodbury,R.L. and Okita,T.W. (1996) Aspartic acid 413 is important for the normal allosteric functioning of ADP-glucose pyrophosphorylase. Plant Physiol., 112, 1315–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikinheimo P., Lehtonen,J., Baykov,A., Lahti,R., Cooperman,B.S. and Goldman,A. (1996) The structural basis for pyrophophatase catalysis. Structure, 4, 1491–1508. [DOI] [PubMed] [Google Scholar]
- Hill M.A., Kaufmann,K., Otero,J. and Preiss,J. (1991) Mutagenesis of a catalytic residue of ADP-glucose pyrophosphorylase from Escherichia coli. J. Biol. Chem., 266, 12455–12460. [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Iglesias A.A., Kakefuda,G. and Preiss,J. (1991) Regulatory and structural properties of the cyanobacterial ADP-glucose pyrophos phorylase. Plant Physiol., 97, 1187–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias A.A., Charng,Y.Y., Ball,S. and Preiss,J. (1994) Characterization of the kinetic, regulatory, and structural properties of ADP-glucose pyrophosphorylase from Chlamydomonas reinhardtii. Plant Physiol., 104, 1287–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.H., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993). PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Leslie A.G.W. (1992). CCP4 and ESF-EACMB Newsletter on Protein Crystallography, 26. [Google Scholar]
- Lindquist L., Kaiser,R., Reeves,P.R. and Lindberg,A.A. (1993) Purification, characterization and HPLC assay of Salmonella glucose-1-phosphate thymidylyl-transferase from the cloned rfbA gene. Eur. J. Biochem., 211, 763–770. [DOI] [PubMed] [Google Scholar]
- Ma Y., Mills,J.A., Belisle,J.T., Vissa,V., Howell,M., Bowlin,K., Scherman,M.S. and McNeil,M. (1997) Determination of the pathway for rhamnose biosynthesis in mycobacteria: cloning, sequencing and expression of the Mycobacterium tuberculosis gene encoding α-d-glucose-1-phosphate thymidylyltransferase Microbiology, 143, 937–945. [DOI] [PubMed] [Google Scholar]
- McNeil M., Daffé,M. and Brennan,P.J. (1990) Evidence for the nature of the link between the arabinogalactan and peptidoglycan of mycobacterial cell walls. J. Biol. Chem., 265, 18200–18206. [PubMed] [Google Scholar]
- Melo A. and Glaser,L. (1965) The nucleotide specificity and feedback control of thymidine diphosphate d-glucose pyrophosphorylase. J. Biol. Chem., 240, 398–405. [PubMed] [Google Scholar]
- Moothoo D.N. and Naismith,J.H. (1998) Concanavalin A distorts the GlcNAc b1-2 Man linkage of the pentasaccharide core upon binding. Glycobiology, 8, 173–181. [DOI] [PubMed] [Google Scholar]
- Mulichak A.M., Skrzypczak-Jankun,E., Rydel,T.J., Tulinsky,A. and Preiss,J. (1988) Crystallization and preliminary diffraction data of Escherichia coli ADP glucose pyrophosphorylase. J. Biol. Chem., 263, 17237–17238. [PubMed] [Google Scholar]
- Murshudov G.N., Vagin,A.A. and Dodson,E.J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D, 53, 240–255. [DOI] [PubMed] [Google Scholar]
- O’Brien W.E. (1976) A continuous spectrophotometric assay for argininosuccinate synthetase based on pyrophosphate formation. Anal. Biochem., 76, 423–430. [DOI] [PubMed] [Google Scholar]
- Orengo C.A., Michie,A.D., Jones,S., Jones,D.T., Swindells,M.B. and Thornton,J.M. (1997) CATH–a hierarchic classification of protein domain structures. Structure, 5, 1093–1108. [DOI] [PubMed] [Google Scholar]
- Parsons T.F. and Preiss,J. (1978) Isolation and characterization of the pyridoxal-P allosteric activator site and the ADP-glucose protected pyridoxal-P binding site of Escherichia coli B ADP–glucose synthase. J. Biol. Chem., 253, 7638–7645. [PubMed] [Google Scholar]
- Paule M.R. and Preiss,J. (1971) The kinetic mechanism of adenosine diphosphoglucose pyrophosphorylase from Rhodospirillum rubrum. J. Biol. Chem., 246, 4602–4609. [DOI] [PubMed] [Google Scholar]
- Pedersen L.C., Benning,M.M. and Holden,H.M. (1995) Structural investigation of the antibiotic and ATP-binding sites in kanamycin nucleotidyltransferase. Biochemistry, 34, 13305–13311. [DOI] [PubMed] [Google Scholar]
- Perrakis A., Morris,R. and Lamzin,V.S. (1999) Automated protein model building combined with iterative structure refinement. Nature Struct. Biol., 6, 458–463. [DOI] [PubMed] [Google Scholar]
- Preiss J. (1982) Regulation of the biosynthesis and the degradation of starch. Annu. Rev. Plant. Physiol., 33, 432–451. [Google Scholar]
- Preiss J. (1984) Bacterial glycogen synthesis and its regulation. Annu. Rev. Plant. Physiol., 38, 419–458. [DOI] [PubMed] [Google Scholar]
- Preiss J. (1991) Biology and molecular biology of starch synthesis and its regulation. In Miflin,B.J. (ed.), Surveys of Plant Molecular and Cell Biology, Vol. 7. Oxford University Press, Oxford, UK, pp. 59–114. [Google Scholar]
- Preiss J. and Romeo,T. (1989) Physiology, biochemistry and genetics of bacterial glycogen synthesis. Adv. Microb. Physiol., 30, 183–238. [DOI] [PubMed] [Google Scholar]
- Price N.T., Kimball,S.R., Jefferson,L.S. and Proud,C.G. (1996) Cloning of cDNA for the γ-subunit of mammalian translation initiation factor 2B, the guanine nucleotide-exchange factor for eukaryotic initiation factor 2. Biochem. J., 318, 631–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahim R., Burrows,L.L., Monteiro,M.A., Perry,M.B. and Lam,J.S. (2000) Involvement of the rml locus in core oligosaccharide and O polysaccharide assembly in Pseudomonas aeruginosa.Microbiology, 146, 2803–2814. [DOI] [PubMed] [Google Scholar]
- Siddiqui A.S. and Barton,G.J. (1995) Continuous and discontinuous domains: An algorithm for the automatic generation of reliable protein domain definitions. Protein Sci., 4, 872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman J.L., Lin,D., Jiang,J., Manning,N.O., Prilusky,J., Ritter,O. and Abola,E.E. (1998) Protein Data Bank (PDB): database of three-dimensional structural information of biological macromolecules. Acta Crystallogr. D, 54, 1078–1084. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theorell H. and Chance,B. (1951) Liver alcohol dehydrogenase. II. Kinetics of the compound of horse-liver alcohol dehydrogenase and reduced diphospho pyridene nucleotide. Acta Chem. Scand., 5, 1127–1144. [Google Scholar]
- Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 24, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace A.C., Laskowski,R.A. and Thornton,J.M. (1995) LIGPLOT: a program to generate schematic diagrams of protein–ligand interactions. Protein Eng., 8, 127–134. [DOI] [PubMed] [Google Scholar]
- Vagin A.A. and Teplyakov,A. (1997) MOLREP: an Automated Program for Molecular Replacement. J. Appl. Crystallogr., 30, 1022–1025. [Google Scholar]