Abstract
Precise trafficking, localization, and activity of inward rectifier potassium Kir2 channels are important for shaping the electrical response of skeletal muscle. However, how coordinated trafficking occurs to target sites remains unclear. Kir2 channels are tetrameric assemblies of Kir2.x subunits. By immunocytochemistry we show that endogenous Kir2.1 and Kir2.2 are localized at the plasma membrane and T-tubules in rodent skeletal muscle. Recently, a new subunit, Kir2.6, present in human skeletal muscle, was identified as a gene in which mutations confer susceptibility to thyrotoxic hypokalemic periodic paralysis. Here we characterize the trafficking and interaction of wild type Kir2.6 with other Kir2.x in COS-1 cells and skeletal muscle in vivo. Immunocytochemical and electrophysiological data demonstrate that Kir2.6 is largely retained in the endoplasmic reticulum, despite high sequence identity with Kir2.2 and conserved endoplasmic reticulum and Golgi trafficking motifs shared with Kir2.1 and Kir2.2. We identify amino acids responsible for the trafficking differences of Kir2.6. Significantly, we show that Kir2.6 subunits can coassemble with Kir2.1 and Kir2.2 in vitro and in vivo. Notably, this interaction limits the surface expression of both Kir2.1 and Kir2.2. We provide evidence that Kir2.6 functions as a dominant negative, in which incorporation of Kir2.6 as a subunit in a Kir2 channel heterotetramer reduces the abundance of Kir2 channels on the plasma membrane.
Keywords: Intracellular Trafficking, Ion Channels, Potassium Channels, Skeletal Muscle, Trafficking
Introduction
Inward rectifier potassium (Kir2) channels are key skeletal muscle components involved in determination of muscle resting potential, regulation of electrical excitability, repolarization of the action potential, and clearance of K+ from the T-tubule2 system (1–4). Electrophysiological studies in skeletal muscle and immunocytochemical studies in cardiac muscle indicate that Kir2 channels are localized at the plasma membrane and T-tubules (Refs. 1 and 5–15; reviewed in Refs. 3 and 16). The abundance of Kir2 channels is critical to muscle function, and their importance is highlighted when the channels are mutated or absent. Andersen-Tawil syndrome is characterized by periodic paralysis of skeletal muscle, cardiac arrhythmia, and developmental dysgenesis, resulting from loss-of-function mutations in KCNJ2, the gene that encodes Kir2.1 potassium channels (17, 18); gain-of-function mutations cause some forms of short QT syndrome (19). Targeted deletion of Kir2.1 in mice also causes defective cerebral vasodilation, and deletion of either Kir2.1 or Kir2.2 causes a loss or decrease in cardiac IK1 currents (20, 21).
Recently, a search for genes involved in thyrotoxic periodic paralysis (TPP) revealed KCNJ18, which encodes a novel subtype of inward rectifier potassium channel subunit, Kir2.6 (22). Kir2.6 is expressed primarily in skeletal muscle and shares >98% identity with Kir2.2 (22). Mutations in human Kir2.6 confer susceptibility to TPP, and it has been shown that these disease-associated mutations contribute to atypical current signatures and altered cell excitability (22). Expression studies in heterologous cells demonstrate that Kir2.6 subunits are able to form inwardly rectifying channels and that disease-associated mutations include truncated subunits that do not form functional channels, as well as gain-of-function mutations that increase electrical activity because of misregulation by phosphorylation (22).
In addition to their electrophysiological properties, ion channels contribute to electrical events by their abundance on the plasma membrane. Targeting ion channels to subcellular sites and the plasma membrane is important for shaping the electrical response of muscle. Surface expression levels and channel activity are crucial for determining muscle excitability. However, trafficking of Kir2.6 to the plasma membrane has not yet been explored. Moreover, the role of wild type Kir2.6 in normal muscle physiology has not been established.
Several trafficking motifs in Kir2 subunits have been identified and are well conserved among the inward rectifier subunits that are found in human skeletal muscle (Kir2.1, Kir2.2, Kir2.3, and Kir2.6). Anterograde trafficking signals in Kir2.1 channels promote surface expression by enhancing ER and Golgi export early in the secretory pathway (23–26). In particular, a di-acidic sequence motif in the C terminus of Kir2.1 facilitates traffic from the ER to the Golgi (23, 24). Two additional regions within Kir2.1, a positively charged motif in the N terminus and a conserved YXXΦ sequence on the membrane-proximal region of the C terminus, have been associated with export from the Golgi to the plasma membrane (25, 26). The Kir2.1, Kir2.2, and Kir2.3 C termini also contain a PDZ-binding motif that permits binding to scaffolding and clustering MAGUK proteins that may direct channel localization and abundance (7, 10, 11, 27–32).
Additionally, Kir channel subunits may influence the properties of muscle by coassembly with other Kir2 subunits. Inward rectifier channels are formed by the homotetrameric or heterotetrameric assembly of subunits. Previous reports demonstrate that Kir2.1, Kir2.2, and Kir2.3 subunits can coassemble to form heterotetrameric channels (33–36). Notably, many Kir2.1 mutants involved with Andersen-Tawil syndrome dominantly suppress inward rectifier current through tetramerization with Kir2.1, Kir2.2, and Kir2.3 (33).
Here we describe the interaction and trafficking of wild type Kir2.1, Kir2.2, and Kir2.6 in mouse skeletal muscle and in COS-1 cells. Although Kir2.6 shares conserved ER and Golgi trafficking motifs with Kir2.1 and Kir2.2, we show that Kir2.1 and Kir2.2 traffic to the plasma membrane and T-tubules, whereas Kir2.6 is largely retained in the ER. We demonstrate that Kir2.6 can associate with Kir2.1 and Kir2.2 in cultured cells and in vivo and causes partial ER retention of Kir2.1 or Kir2.2. Thus, Kir2.6 has a dominant negative effect on the forward trafficking of Kir2.1 and Kir2.2. Our results suggest that through heterotetramerization of subunits, Kir2.6 may control Kir2.1 and Kir2.2 abundance on the muscle plasma membrane, thus providing a mechanism to fine tune electrical responses.
EXPERIMENTAL PROCEDURES
DNA Constructs
cDNAs encoding human Kir2.2, human Kir2.6, and mouse Kir2.1 were subcloned into the mammalian expression vector pGW1 with a Myc or HA tag in the extracellular M1-to-P linker region. GFP-Kir2.1, GFP-Kir2.2, and GFP-Kir2.6 were cloned into pEGFP-C1 to produce GFP fused to the N terminus of the Kir channels. pECFP-Golgi (Clontech) is a trans-medial Golgi marker that encodes a ECFP-β1,4-galactosyltransferase fusion protein (37).
Site-directed Mutagenesis
Site-directed mutagenesis was performed using Platinum Pfx DNA polymerase (Invitrogen) and Pfu Turbo® DNA polymerase (Stratagene) according to the manufacturer's instructions. Kir2.6 and Kir2.2 share ∼98% amino acid identity, and the Kir2.6 cDNA in this study was a typical sequence found in human populations (GenBankTM accession number FJ434338.1) that does not include rare nucleotide polymorphisms (22). Site-directed mutagenesis was used to reverse the amino acids that differ between Kir2.6 and Kir2.2. Constructs include Kir2.6 single point mutations (Kir2.6-L15S, Kir2.6-Q39R, Kir2.6-H40R, Kir2.6-H118R, Kir2.6-L156P, Kir2.6-H192Q, and Kir2.6-G430E), Kir2.6 double point mutations (Kir2.6-Q39R/H40R, Kir2.6-L15S/G430E, Kir2.6-H118R/L156P, Kir2.6-L156P/H192Q, and Kir2.6-L156P/G430E), Kir2.6 triple point mutations (Kir2.6-L15S/Q39R/H40R, Kir2.6-Q39R/H40R/G430E, Kir2.6-H118R/L156P/G430E, and Kir2.6-L156P/H192Q/G430E), Kir2.6 quadruple point mutation (Kir2.6-Q39R/H40R/V249I/G430E), Kir2.6 quintuple point mutations (Kir2.6-L15S/Q39R/H40R/V249I/G430E), Kir2.2 single point mutations (Kir2.2-S15L, Kir2.2-I100V, Kir2.2-R118H, Kir2.2-P156L, Kir2.2-Q192H, and Kir2.2-E430G), and Kir2.2 double point mutations (Kir2.2-R39Q/R40H, Kir2.2-R118H/P156L, and Kir2.2-I249V/E430G). The mutations were confirmed by sequencing the entire cDNA.
Antibodies
The primary antibodies used include rat anti-Myc and rabbit anti-GFP (Abcam), mouse anti-Myc (Santa Cruz Biotechnology), rat anti-HA (Roche Applied Science), mouse anti-HA (Covance), rabbit anti-protein disulfide isomerase and mouse anti-α-actinin (Sigma), mouse anti-GM130 (BD Biosciences), mouse anti-dihydropyridine receptor α2 (Affinity Bioreagents), mouse anti-ryanodine receptor (34C; Developmental Studies Hybridoma Bank), and rabbit anti-Kir2.1 and rabbit anti-Kir2.2 (38). The secondary antibodies used include anti-rat Cy3, anti-mouse Cy3, and anti-mouse Cy2 (Jackson ImmunoResearch Laboratories), anti-rat Alexa Fluor 647, anti-mouse Alexa Fluor 647, rabbit anti-GFP-Alexa Fluor 488, α-bungarotoxin conjugated Alexa Fluor 647, and anti-rabbit Alexa Fluor 680 (Invitrogen), and anti-rat IRDye® 800 (Rockland).
Transfection and Expression in COS-1
COS-1 cells were grown and maintained (10% CO2, 37 °C) in DMEM-supplemented with 10% fetal calf serum. The cells were cotransfected with cDNAs encoding tagged Kir2 subunits and pECFP-Golgi constructs utilizing FuGENETM 6 (Roche Applied Science), using 0.4 μg of total DNA/15-mm well (0.2 μg each when cotransfected with two DNAs; empty vector was not used). One day post-transfection, the cells were fixed at room temperature with 4% paraformaldehyde, 4% sucrose in PBS for 8 min and then blocked with 3% BSA, 1% goat serum, 1% donkey serum in PBS. The cells were incubated with rat anti-Myc (1:1000) or mouse anti-HA (1:500) primary antibodies for 1 h at room temperature, then washed three times with PBS, and incubated with anti-rat or anti-mouse Cy3 (1:300) secondary antibodies for detection of surface expression. To label intracellular epitopes, the cells then were washed, permeabilized with blocking buffer containing 0.15% Triton X-100, and incubated with rat anti-HA (1:500) or mouse anti-Myc (1:500) primary antibodies, washed, and incubated with anti-rat or anti-mouse Alexa Fluor 647 (1:300) secondary antibodies. In experiments that used pECFP-Golgi, the cells also were labeled with rabbit anti-GFP-conjugated Alexa Fluor 488 (1:1000).
COS-1 Cell Surface and Internal Measurements
The images were captured using widefield fluorescence microscopy with a Zeiss Axiovert 200 microscope with either Plan Apochromat 100×/1.4 or Plan Apochromat 20×/0.8 objectives. The images were taken with identical exposure times for all measurements of surface and internal labeling within an experiment. For each transfection, surface and internal fluorescence intensity and cell area were measured from 100 cells using AxioVision 4.6 analysis software, and the data were expressed as the average fluorescent intensity. Statistically significant differences were assessed using the unpaired Student's t test. The data are given as the averages ± S.E. expressed as relative surface/internal expression level.
Electrophysiology
COS-1 cells were cotransfected with cDNAs encoding 0.5 μg of Myc-tagged Kir2.x and 0.1 μg of pEGFP-C1 utilizing FuGENETM 6 (Roche Applied Science). One day post-transfection, the cell currents were recorded using standard methods for whole cell voltage clamp studies. The currents were acquired using a Dagan 3900/3911A patch clamp, low pass-filtered at 5 kHz, and sampled at 20 kHz with an ITC-16 digitizer (Instrutech). Data acquisition was controlled with Pulse (HEKA) software. Pipettes had resistances of 1–3 megohms. After establishing the whole cell configuration, the cell capacitance was cancelled, and series resistance was compensated (>50%). The currents were elicited by 100-ms pulses applied in 20-mV increments to potentials ranging from −100 to 60 mV from a holding potential of 0 mV. The pipette solution contained 110 mm potassium aspartate, 20 mm KCl, 10 mm EGTA, 1 mm MgCl2, 5 mm glucose, 10 mm Hepes, pH 7.3 (adjusted with KOH). The extracellular bath solution contained 115 mm NaCl, 30 mm KCl, 2 mm CaCl2, 1 mm MgCl2, 5 mm glucose, 10 mm Hepes, pH 7.4 (adjusted with NaOH). Recordings were made at room temperature (22–25 °C) within 23–32 h of transfection. For comparison of current magnitudes for different Kir2 channels, current amplitude at −100 mV was averaged in the interval from 5–15 ms after the onset of the voltage pulse, and leakage current was subtracted using the scaled current for pulses to 60 mV. The data were analyzed and displayed with Pulse and SigmaPlot software.
Immunoprecipitation
Thirty-six hours post-transfection, COS-1 cells (six-well dish) were washed twice with PBS and then lysed in 0.6 ml of immunoprecipitation buffer (20 mm Hepes, 1 mm EDTA, 150 mm NaCl, 1% Nonidet P-40, 1× Complete protease inhibitor (Roche Applied Science), pH 7.4). Solubilized protein was precleared with protein G-agarose (1 h), followed by the addition of mouse anti-HA (2 μg) (1 h), and protein G-agarose overnight at 4 °C. Then resins were washed once with the immunoprecipitation buffer, three times with high salt immunoprecipitation buffer (500 mm NaCl), and once with immunoprecipitation buffer and eluted in SDS sample buffer. The eluted proteins were separated by 10% SDS-PAGE. Proteins were transblotted to Hybond ECLTM nitrocellulose membrane (GE Healthcare) and incubated with rabbit anti-GFP (1:2000) and rat anti-HA (1:200) followed by anti-rabbit Alexa Fluor 680 (1:5000) and anti-rat IRDye® 800 (1:5000). The membranes were imaged with an Odyssey infrared imaging system (LI-COR Biosciences).
In Vivo Mouse Muscle Experiments
Tibialis cranialis skeletal muscles of 8–10-week-old Swiss Webster mice were injected with hyaluronidase (25 μl/muscle, 4 unit/μl (Sigma)) under isoflurane anesthesia. Two hours later, cDNAs encoding tagged Kir2.1, Kir2.2, and Kir2.6 and site-directed mutants were electroporated under anesthesia into the muscle using a BTX ECM830 with fine needle electrodes (25 μg each DNA construct, 0.5-cm electrode gap; 5 × 150 V/cm pulses, each 20 ms in duration) (39–42).
Confocal Microscopy
Seven days following electroporation, the mice were euthanized with CO2. The muscles were dissected, fixed with 4% paraformaldehyde, 4% sucrose in PBS for 20 min, then washed four times for 20 min with PBS, and blocked (3% BSA, 2% goat serum, 2% donkey serum, 0.2% Triton X-100, 0.04% saponin in PBS). The fibers were incubated overnight at 4 °C with primary antibodies (rat anti-HA (1:200), rat anti-Myc (1:1000), rabbit anti-PDI (1:50), and/or mouse GM130 (1:50)), washed four times for 40 min with PBS and incubated overnight with secondary antibodies (anti-rat Cy3 (1:200), rabbit anti-GFP-Alexa Fluor 488 (1:1000), anti-mouse Cy2 (1:200), and α-bungarotoxin-Alexa Fluor 647 (1:500)) and then washed again. The subcellular localization of channel subunits was determined by colabeling with α-bungarotoxin to identify neuromuscular junctions, with anti-PDI for ER and with anti-GM130 for Golgi (43–45). Single muscle fibers were dissected and mounted on slides using DAPI ProLong® Gold (Invitrogen). Endogenous channels in rat skeletal muscle were labeled similarly using primary rabbit antibodies specific for rat Kir2.1 or Kir2.2 (38). The images were captured using an Olympus Fluoview 500 laser-scanning confocal microscope with a UPlanFL 40×/1.3 or PlanApo 60×/1.4 objective and were merged and displayed with Adobe Photoshop.
Skeletal Muscle Surface and Internal Labeling
Skeletal muscles electroporated with HA-Kir2.1 alone or HA-Kir2.2 alone or coelectroporated with GFP-Kir2.6 were carefully dissected and fixed as above, then washed, and blocked (3% BSA, 2% goat serum, 2% donkey serum in PBS). Surface labeling was performed overnight at 4 °C with rat anti-HA (1:200), then washed four times with PBS, and incubated for 4 h with anti-rat Cy3 (1:200). The fibers were permeabilized with block containing 0.2% Triton X-100, 0.04% saponin, and then internal labeling was performed with rat anti-HA (1:200) overnight, followed by washing and internal labeling with anti-rat Alexa Fluor 647 (1:200) and rabbit anti-GFP-Alexa Fluor 488 (1:1000) overnight.
RESULTS
Kir2.6 Is Retained in the ER when Expressed in COS-1 Cells
To investigate the trafficking of Kir2.1, Kir2.2, and Kir2.6 to the plasma membrane, tagged channel constructs were transfected into COS-1 cells, and their surface and internal expression were evaluated using immunocytochemistry. Surface expression was evaluated by labeling fixed cells 24 h after transfection; internal expression was assessed in the same cells by labeling subsequent to a permeabilization step. Expressing data as a surface-to-internal ratio for each cell controlled for variations in expression levels in individual cells or between different constructs. A 24-h post-transfection time point was chosen to allow adequate time for channel biosynthesis and processing without overexpression effects that can saturate the trafficking machinery.
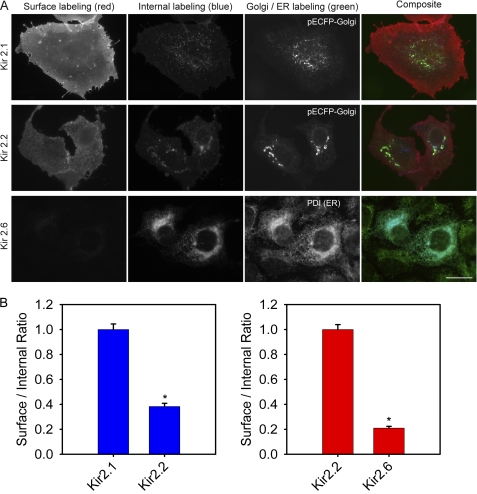
Robust plasma membrane fluorescence was consistently observed for Kir2.1 and Kir2.2, together with a low level of internal fluorescence that colocalized with the Golgi markers pECFP-Golgi or GM130, suggesting that Kir2.1 and Kir2.2 are trafficked through the Golgi, then highly expressed on the plasma membrane of COS-1 cells (Fig. 1A, top and middle rows). Conversely, Kir2.6 channels displayed very low surface fluorescence and were predominantly intracellular in the perinuclear region colocalized with the ER marker PDI (Fig. 1A, bottom row). Note that the ER localization of Kir2.6 was not affected by the epitope tag on the subunit, and identical results were obtained for channels tagged with Myc on the extracellular M1-to-P linker (Fig. 1), with GFP on the intracellular N terminus (see Fig. 9), or with V5 on the N terminus (not shown). Quantitative analysis of channel expression for a large number of cells revealed that the relative ratio of surface/internal fluorescence was highest for Kir2.1, was decreased to ∼40% for Kir2.2 compared with Kir2.1, and was only ∼20% for Kir2.6 compared with Kir2.2. These data suggest that in COS-1 cells, Kir2.6 channels are mainly retained in the ER (Fig. 1B).
FIGURE 1.
Kir2.1 and Kir2. 2 traffic to plasma membrane, but Kir2.6 is retained in ER in COS-1 cells. A, cells were transfected with HA-Kir2.1, Myc-Kir2.2, or Myc-Kir2.6. The cells were fixed 24 h later, labeled with mouse anti-HA (or rat anti-Myc) and anti-mouse Cy3 (or anti-rat Cy3, red) for surface expression, then permeabilized, and labeled with rat anti-HA (or mouse anti-Myc) and anti-rat Alexa 647 (or anti-mouse Alexa 647, blue) for internal expression. Cotransfection with pECFP-Golgi or anti-PDI (green) identified Golgi and ER, respectively. Bar, 20 μm. B, cells expressing HA-tagged Kir2.1 or Kir2.2 (left) or Myc-tagged Kir2.2 or Kir2.6 (right) were analyzed by measuring the fluorescence intensity for surface and internal labeling; the normalized value of the surface/internal ratio is displayed (n = 100). The error bars indicate ± S.E. *, statistically significant (p < 0.0001).
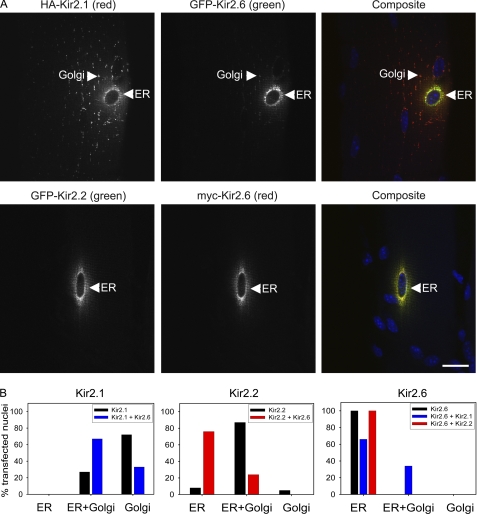
FIGURE 9.
Kir2.2 and Kir2. 1 are partially retained in the ER following coelectroporation with Kir2.6 in mouse skeletal muscle. A, mouse tibialis cranialis skeletal muscle fibers were coelectroporated in vivo with HA-Kir2.1 and GFP-Kir2.6 or with GFP-Kir2.2 and Myc-Kir2.6. Seven days later, tissue was fixed, permeabilized, and labeled with rat anti-Myc (or rat anti-HA, red), rabbit anti-GFP (green), and DAPI (blue). Both HA-Kir2.1 and GFP-Kir2.2 are partially retained in the ER colocalized with Kir2.6. Bar, 20 μm. B, localization of Kir2.1, Kir2.2, and Kir2.6 when expressed alone or coexpressed with another Kir2.x subunit in skeletal muscle. Channel subunit distribution in skeletal muscle was visually scored for presence or absence in ER and/or Golgi from widefield fluorescence images (n = 41–102 electroporated nuclei for each condition).
To ascertain whether the ER retention of Kir2.6 was dependent on the cellular context, we compared its trafficking in the HL-1 mouse cardiac muscle cell line. Kir2.6 was similarly retained in the ER and colocalized with PDI, whereas Kir2.1 and Kir2.2 were trafficked to the plasma membrane as in COS-1 cells, a monkey kidney cell line (data not shown).
Kir2.6 Forms Functional Homotetrameric Channels in COS-1 Cells
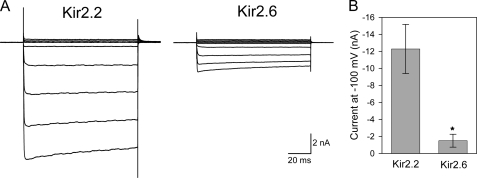
Although Kir2.6 traffics poorly to the cell surface, immunofluorescence data suggested that a small fraction of the protein was present on the plasma membrane (Fig. 1B). We used whole cell voltage clamp recording in COS-1 cells to assess the ability of Kir2.6 subunits to form functional homotetrameric channels. The hallmark of Kir2 channels is their strong inward rectification, which in muscle allows them to carry currents near the resting potential but not at positive potentials. Both Kir2.2 and Kir2.6 exhibit strong inward rectification, displaying larger inward currents at potentials negative to the potassium equilibrium potential and small outward currents at more positive voltages, in agreement with previous findings (Fig. 2A) (22, 46). Because the Kir2.6 surface/internal ratio by immunofluorescence was significantly lower than Kir2.1 and Kir2.2 ratios, we expected to find differences in current magnitude between Kir2.2 and Kir2.6, and indeed the whole cell current magnitude for Kir2.6 was ∼15% compared with Kir2.2 (Fig. 2B). Because single channel conductance and open probability are similar for Kir2.6 and Kir2.2 (22), this reduced Kir2.6 current most likely corresponds to a decrease in the number of channels on the plasma membrane.
FIGURE 2.
Kir2.6 produces small inwardly rectifying potassium currents. A, whole cell currents in COS-1 cells transfected with Kir2.2 or Kir2.6. The currents were elicited by 100-ms pulses applied in 20-mV increments to potentials ranging from −100 to 60 mV from a holding potential of 0 mV. Both Kir2.2 and Kir2.6 form functional channels with characteristic inward rectification. B, whole cell current magnitudes for Kir2.2 and Kir2.6 channels were measured at −100 mV. *, Kir2.6 current amplitude is significantly different compared with Kir2.2 (p < 0.01).
Kir2.2 Proline 156 Is a Critical Determinant of Channel Trafficking to the Plasma Membrane
Because of the large differences in surface expression observed between Kir2.6 and Kir2.2, despite the conservation of known forward trafficking signals, we compared channel sequences to identify potentially new amino acid regions that might be important for trafficking. Site-directed mutagenesis was used to alter each of the nine amino acids that varies between Kir2.2 and Kir2.6 by replacing sites in Kir2.6 with the corresponding Kir2.2 counterparts. These mutations targeted sites in the N-terminal region (three sites: Kir2.6-L15S, Kir2.6-Q39R, and Kir2.6-H40R), the M1 helix (Kir2.6-V100I), between the M1 and the P loop (Kir2.6-H118R), between the P-loop and the M2 helix (Kir2.6-L156P), and three sites in the C terminus (Kir2.6-H192Q located next to the PIP2 binding domain, Kir2.6-V249I, and Kir2.6-G430E positioned adjacent to the PDZ binding domain). The constructs were transfected into COS-1 cells and labeled for internal and surface expression as described above.
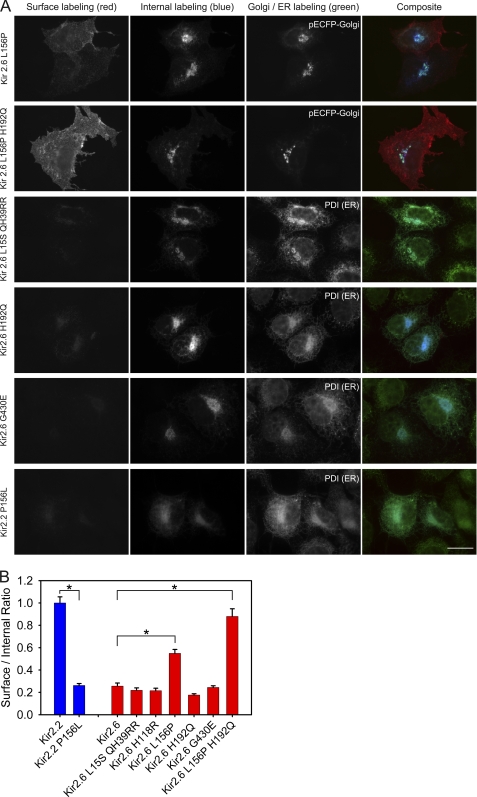
Immunocytochemical analysis revealed that Pro-156 is essential for efficient channel trafficking to the plasma membrane (Fig. 3A, top row). Kir2.6-L156P displays a surface-to-internal ratio significantly higher than Kir2.6 and more than half that of Kir2.2. The reverse Kir2.2 mutation, Kir2.2-P156L, is sufficient to retain the channel in the ER (Fig. 3). Further, we found that a combination of Pro-156 and Gln-192 is able to confer full plasma membrane expression, and the high expression ratio of Kir2.6-L156P/H192Q was not significantly different from Kir2.2 (Fig. 3). In contrast, mutations at other sites were unable to overcome the ER retention of Kir2.6 (Fig. 3 and data not shown).
FIGURE 3.
Proline/leucine difference between Kir2.2 and Kir2.6 at position 156 plays an essential role in trafficking to the plasma membrane. A, COS-1 cells were transfected with cDNAs encoding Myc-Kir2.6 mutants or Myc-Kir2.2 mutants and labeled for surface and internal channel expression. Cotransfection with pECFP-Golgi or labeling with PDI antibody identified Golgi and ER, respectively. Bar, 20 μm. B, quantitative analysis of the normalized surface/internal ratio shows that proline 156 was required for high channel surface expression. The surface expression of Kir2.2 was dramatically reduced in the Kir2.2-P156L mutant; high surface expression was conferred to Kir2.6 by the complementary mutation Kir2.6-L156P (n = 100). The error bar indicates ± S.E. *, statistically significant (p < 0.0001).
Kir2.6 Assembles with Kir2.1 and Kir2.2 to Form Heterotetrameric Channels
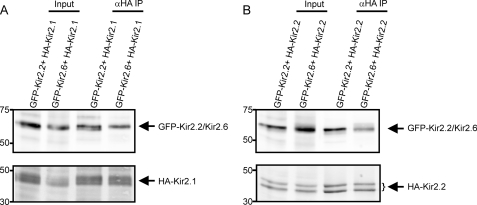
Kir2.1 and Kir2.2 subunits have the ability to coassemble with other Kir2.x subunits to form heterotetrameric channels (33, 34). To determine whether Kir2.1 or Kir2.2 can coassemble with Kir2.6, we cotransfected COS-1 cells with GFP-Kir2.6 and either HA-Kir2.1 or HA-Kir2.2. HA-tagged Kir2.1 or Kir2.2 channels were immunoprecipitated from detergent-solubilized proteins and then probed with anti-GFP to detect coassembled GFP-Kir2.6. Western blot analysis reveals prominent coprecipitating bands, demonstrating that both Kir2.1 (Fig. 4A) and Kir2.2 (Fig. 4B) can coassemble with Kir2.6 in cells.
FIGURE 4.
Kir2.6 associates with Kir2. 1 and Kir2.2 subunits. Detergent-solubilized proteins from COS-1 cells transiently transfected with the constructs indicated were immunoprecipitated with mouse anti-HA antibodies to immunoprecipitate HA-Kir2.1 (A) or HA-Kir2.2 (B). Then the immunoblot was probed with rat anti-HA to detect immunoprecipitated HA-Kir2.1 (A, lower panel) or HA-Kir2.2 (B, lower panel) and with anti-GFP to detect coimmunoprecipitated GFP-Kir2.2 or GFP-Kir2.6 (upper panels). Inputs (left lanes) represent 2.5% of the protein used in the corresponding immunoprecipitations.
Kir2.1 and Kir2.2 Expression on the Plasma Membrane Is Decreased by Coexpression with Kir2.6 in COS-1 Cells
Because some Kir2.1 mutations responsible for Andersen-Tawil syndrome have a dominant negative affect on currents carried by channels formed by heterotetrameric assembly with other Kir2.x subunits (33), we sought to examine the trafficking of heterotetrameric Kir2.6-Kir2.x channels. Following cotransfection of tagged Kir2.x and Kir2.6 in COS-1 cells, we evaluated surface and internal localization using immunocytochemistry.
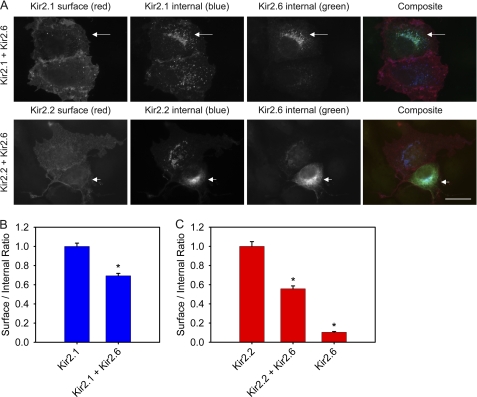
Direct measurement of surface and internal expression revealed that Kir2.6 subunits associate with Kir2.1 and Kir2.2 subunits and result in their partial retention in the ER (Fig. 5A). To illustrate the effect of Kir2.6 on trafficking of other Kir2.x, each of these images shows a pair of cells, one of which expresses Kir2.6 (arrow), and both of which express either Kir2.1 or Kir2.2. The cells expressing Kir2.6 (arrow) show an increase in the intracellular abundance of Kir2.1 or Kir2.2 that colocalizes with Kir2.6 and a corresponding decrease in surface expression of Kir2.1 or Kir2.2 compared with the neighboring cell without Kir2.6. Quantitative analysis showed that coexpression of Kir2.6 with Kir2.1 reduced Kir2.1 surface/internal expression ratio by ∼30% (Fig. 5B). Similarly, coexpression of Kir2.6 with Kir2.2 reduced Kir2.2 surface/internal ratio by ∼50% (Fig. 5C). Total protein levels of the subunits, evaluated by immunoblot, were comparable (supplemental Fig. S1). Hence, Kir2.6 impedes surface expression of Kir2.1 and Kir2.2 when coexpressed in COS-1 cells, suggesting that Kir2.6 subunits are dominant in their ability to retain heterotetrameric channels in the ER.
FIGURE 5.
Kir2.1 and Kir2. 2 are partially retained in the ER when coexpressed with Kir2.6 in COS-1 cells. A, cells were cotransfected with HA-Kir2.1 and GFP-Kir2.6 (upper panels) or Myc-Kir2.2 and GFP-Kir2.6 (lower panels). The cells were fixed 24 h later and labeled for surface expression of Kir2.1 or Kir2.2 with mouse anti-HA or rat anti-Myc, respectively. Then cells were permeabilized and labeled for internal expression of Kir2.1 or Kir2.2 with rat anti-HA or mouse anti-Myc, respectively. Total Kir2.6 in permeabilized cells also was labeled with rabbit anti-GFP. Pairs of cells with high or low Kir2.6 expression are shown in each image. Note that the cell of each pair with high Kir2.6 expression (arrow) displays correspondingly high internal expression of Kir2.1 or Kir2.2 that colocalizes with Kir2.6 and low surface expression of Kir2.1 or Kir2.2. Bar, 20 μm. B, the ratios of surface to internal expression were measured using anti-HA antibodies for HA-Kir2.1 expressed alone or HA-Kir2.1 when cotransfected with GFP-Kir2.6. The surface/internal ratios were normalized to the values for HA-Kir2.1 alone. C, the ratios of surface to internal expression were measured using anti-Myc antibodies for Myc-Kir2.2 expressed alone, Myc-Kir2.2 when cotransfected with GFP-Kir2.6, or Myc-Kir2.6 expressed alone. The surface/internal ratios were normalized to the values for Myc-Kir2.2 alone. For each condition, n = 100. The error bar indicates ± S.E. *, statistically significant when compared with Kir2.1 or Kir2.2 (p < 0.0001).
Endogenous Kir2.1 and Kir2.2 Localize at T-tubules and the Plasma Membrane in Skeletal Muscle
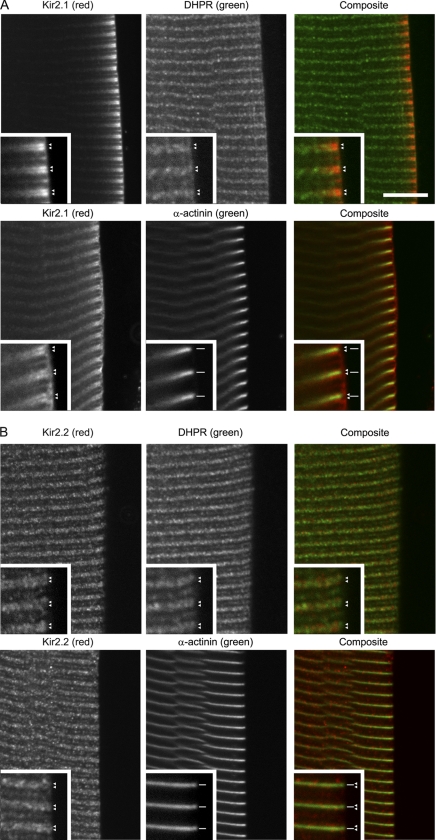
Targeting of ion channels to subcellular sites is important for shaping the response of muscle. We evaluated the localization of endogenous Kir2.1 and Kir2.2 channels in rat tibialis cranialis skeletal muscle using channel-specific antibodies. Kir2.6 is present in humans and other primates, but the gene encoding Kir2.6 has not been identified in the data bank for rodents or other nonprimate species (22), so endogenous Kir2.6 was not assessed. Kir2.1 labeling was abundant on the cell surface and as transverse striations regularly spaced at intervals of ∼2.5 μm in moderately stretched fibers (Fig. 6A). Optical confocal sectioning showed that Kir2.1 labeling was high on striations near the surface membrane and was lower toward the center of muscle cells. At high magnification in moderately stretched fibers, the transverse labeling could be resolved as double rows that colocalized with rows of dihydropyridine receptor α2 subunit (DHPR), a marker of T-tubules (Fig. 6A, insets, arrowheads). The double rows of Kir2.1 labeling bracketing the Z line (identified with anti-α-actinin; Fig. 6A, insets) were continuous with DHPR and with the same spacing and were slightly narrower than double rows of ryanodine receptor that mark the sarcoplasmic reticulum at the triad junction (data not shown). In mammalian skeletal muscle, there are two closely spaced transverse tubules per sarcomere, and the striated Kir2.1 labeling pattern is consistent with localization at T-tubules (47). Altogether the data suggest that endogenous Kir2.1 is abundant on the plasma membrane and on T-tubules near the periphery of muscle fibers and with lower abundance on T-tubules in the center of fibers.
FIGURE 6.
Endogenous Kir2.1 and Kir2.2 are localized at T-tubules and plasma membrane in skeletal muscle. Rat tibialis cranialis skeletal muscle fibers were colabeled with rabbit anti-Kir2.1 (A) or rabbit anti-Kir2.2 (B) and anti-rabbit Cy3 (red) together with either mouse anti-DHPR or anti-α-actinin and anti-mouse-Alexa 647 (green). At higher magnification (inset), Kir2.1 and Kir2.2 labeling could be resolved as double rows (arrowheads) continuous with DHPR labeling that identifies T-tubules (arrowheads) and bracketing α-actinin labeling that defines the Z line (line segments). The bar represents 10 μm for main panels; 5 μm for insets.
Endogenous Kir2.2 showed a distribution that is distinct but overlapping with Kir2.1. Kir2.2 was present in transverse striations with a distribution that extended into the center of muscle fibers (Fig. 6B). Colabeling with antibodies to markers indicated that these striations corresponded to T-tubules (47); Kir2.2 labeling resolved as double transverse rows that colocalize with rows of DHPR (Fig. 6B, insets, arrowheads), that bracket the Z-line identified with anti-α-actinin (Fig. 6B), and that are slightly narrower than double rows of ryanodine receptor (not shown). Interestingly, the distribution of both Kir2.1 and Kir2.2 was punctate along the T-tubules and appeared adjacent to and with some overlap with DHPR, suggesting that the Kir2 channels and DHPR are spatially segregated in discrete clusters on the T-tubules. In addition, Kir2.2 was abundant at the neuromuscular junction and was present in a punctate Golgi distribution both adjacent to the neuromuscular junction and throughout the muscle fibers (data not shown; see also Ref. 7).
Kir2.6 Is Predominantly Retained in the ER in Mouse Skeletal Muscle
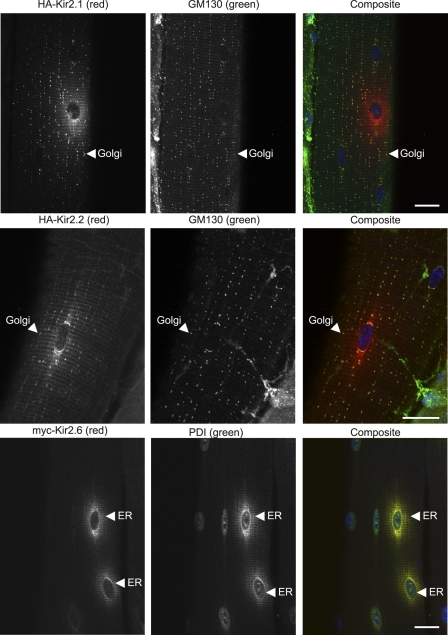
To study Kir trafficking in muscle in vivo, we electroporated tagged channel cDNAs into mouse tibialis cranialis skeletal muscle, and 7 days later evaluated channel expression and localization by immunocytochemistry. We observed that channel expression was spatially restricted in muscle fibers to regions of the fiber near electroporated nuclei. Kir2.1 was particularly abundant on the plasma membrane and peripheral T-tubules (supplemental Fig. S2) and showed a clear punctate intracellular Golgi distribution that colabeled with the Golgi marker GM130 (45) (Fig. 7). Kir2.1 was in low abundance in the ER, which is positioned as a perinuclear region closely associated with each of the nuclei in skeletal muscle (43–45). Thus, the trafficking of tagged Kir2.1 (Fig. 7 and supplemental Fig. S2) closely matched the localization of endogenous Kir2.1 (Fig. 6), and its relative abundance suggests that Kir2.1 is rapidly trafficked out of the ER to the Golgi, peripheral T-tubules, and plasma membrane.
FIGURE 7.
Kir2.6 is retained in the ER, whereas Kir2. 1 and Kir2.2 are trafficked out of the ER to the Golgi, following electroporation in mouse skeletal muscle. Mouse tibialis cranialis skeletal muscle fibers were electroporated in vivo with HA-Kir2.1, HA-Kir2.2, or Myc-Kir2.6. Seven days later, tissue was fixed, permeabilized, and labeled with rat anti-HA (or rat anti-Myc, red), mouse anti-GM130 (Golgi marker), or rabbit anti-PDI (ER marker, green) and DAPI (blue). HA-Kir2.1 and HA-Kir2.2 are trafficked to the Golgi and T-tubules, whereas Myc-Kir2.6 is retained in the ER. Bar, 20 μm.
The expression pattern of electroporated Kir2.2 (Fig. 7) gave high expression in Golgi and low expression in ER, which corresponds with labeling of endogenous Kir2.2 channels (Fig. 6B), indicating that Kir2.2 traffics efficiently from the ER through the Golgi (Fig. 7, middle row). Kir2.2 also was present in a striated pattern consistent with localization in T-tubules (Fig. 7).
In contrast, we observed a strikingly different localization for Kir2.6, which was restricted to the ER, with no detectable expression in Golgi or plasma membrane. Colocalization with the ER marker PDI confirmed retention of Kir2.6 to the ER (43, 44) (Fig. 7, bottom row).
Kir2.6-L156P Is Trafficked to the Golgi in Mouse Skeletal Muscle
In vivo electroporation was utilized to investigate trafficking and localization of mutant Kir2 channels in skeletal muscle. We found that forward trafficking of the channels could be most clearly assessed by comparison of the abundance of Kir2.x subunits in Golgi relative to the ER because of the concentrated and discrete nature of those cellular compartments and because of their close proximity in individual confocal sections. Of all of the Kir2.6 single amino acid mutants evaluated, only Kir2.6-L156P resulted in partial release from the ER and trafficking to the Golgi, indicating that among the amino acids that differ between Kir2.2 and Kir2.6, Pro-156 is the most critical determinant for forward trafficking (Fig. 8, top row). As in COS-1 cells, forward trafficking of Kir2.6-L156P was less complete than Kir2.2, and the double mutation Kir2.6-L156P/H192Q was required to promote maximal transfer from the ER to the Golgi (Fig. 8, bottom row).
FIGURE 8.
Kir2.6-L156P is trafficked to Golgi following electroporation in mouse skeletal muscle. Mouse tibialis cranialis skeletal muscle fibers were electroporated in vivo with Myc-Kir2.6-L156P or Myc-Kir2.6-L156P/H192Q. Seven days later, tissue was fixed and labeled with anti-Myc (red), anti-GM130 (green), and DAPI (blue). The Kir2.6-L156P mutant is trafficked out of the ER to the Golgi (arrowhead), and trafficking is enhanced in the double mutant Kir2.6-L156P/H192Q. Bar, 20 μm.
Kir2.1 and Kir2.2 Are Partially Retained in the ER when Coexpressed with Kir2.6 in Mouse Skeletal Muscle
The affect of Kir2.6 on the subcellular localization of Kir2.1 and Kir2.2 trafficking was further studied in vivo by coelectroporation of pairs of cDNAs into mouse skeletal muscle. By itself Kir2.1 was rarely observed in the ER. However, when Kir2.1 and Kir2.6 were coexpressed, Kir2.1 was present in both the ER and Golgi in most coelectroporated nuclei, and the fraction of Kir2.1 that was retained in the ER colocalized with Kir2.6 (Fig. 9). Affects of Kir2.6 on Kir2.2 trafficking were even more severe. Coexpression of Kir2.2 and Kir2.6 resulted in complete retention of Kir2.2 in the perinuclear ER region in the vast majority of the coelectroporated nuclei examined (Fig. 9). In contrast to the strong effect of Kir2.6 on the localization of Kir2.1 and Kir2.2, these subunits had only a weak influence on Kir2.6 localization. The ER localization of Kir2.6 was not affected by coexpression with Kir2.2; Kir2.6 displayed only minor trafficking to the Golgi in some nuclei when coexpressed with Kir2.1 (Fig. 9).
These results were extended by measuring the effect of coelectroporated Kir2.6 on the surface/internal ratio of expressed Kir2.1 or Kir2.2 in muscle (Fig. 10, A–D). Nuclei expressing Kir2.6 (Fig. 10, A and C, arrows) showed increased internal expression and/or decreased surface expression of coexpressed Kir2.1 or Kir2.2 compared with nearby nuclei in which Kir2.6 expression was low or absent (Fig. 10, A and C, arrowheads). Quantitative analysis demonstrated that surface/internal ratios were reduced from muscle coelectroporated with Kir2.6 and Kir2.1 or Kir2.2 compared with muscle electroporated with Kir2.1 or Kir2.2 alone (Fig. 10, B and D).
FIGURE 10.
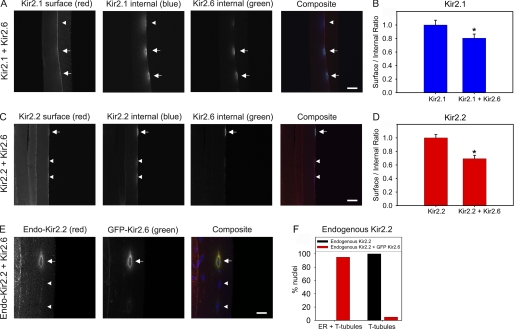
Surface-to-internal ratios of HA-Kir2.1 and HA-Kir2.2 are reduced when coexpressed with GFP-Kir2.6, and endogenous Kir2.2 is partially retained in the ER with GFP-Kir2.6 in skeletal muscle. A and C, mouse tibialis cranialis skeletal muscle fibers were electroporated in vivo with HA-Kir2.1 and GFP-Kir2.6 (A) or HA-Kir2.2 and GFP-Kir2.6 (C). Seven days later, tissue was fixed, and surface HA-Kir2.1 or HA-Kir2.2 was labeled with rat anti-HA and anti-rat Cy3 (red). Tissue was permeabilized and labeled for internal HA-Kir2.1 or HA-Kir2.2 with rat anti-HA and anti-rat Alexa 647 (pseudo blue) and for GFP-Kir2.6 with rabbit anti-GFP-conjugated Alexa 488 (green). Three nuclei are shown in each image; expression of GFP-Kir2.6 (arrows) is correlated with high internal expression and/or low surface expression of HA-Kir2.1 or HA-Kir2.2 compared with nearby nuclei in which GFP-Kir2.6 expression is low or absent (arrowheads). Bar, 40 μm. B and D, the ratios of surface to internal expression of HA-Kir2.1 or HA-Kir2.2 were measured from muscle electroporated with these subunits alone or together with GFP-Kir2.6. The surface/internal ratios were normalized to the values for HA-Kir2.1 or HA-Kir2.2 alone. For B, n = 40; *, statistically significant when compared with Kir2.1 (p < 0.05). For D, n = 50; *, statistically significant when compared with Kir2.2 (p < 0.001). The error bars indicate ± S.E. E, mouse tibialis cranialis skeletal muscle fibers were electroporated in vivo with GFP-Kir2.6. Seven days later, tissue was fixed, permeabilized, and labeled with rabbit anti-Kir2.2 and anti-rabbit Cy3 (red) to label endogenous Kir2.2, mouse anti-GFP, and anti-mouse Cy2 (green) to label GFP-Kir2.6 and DAPI (blue). Endogenous Kir2.2 is retained in the ER only surrounding nuclei that express GFP-Kir2.6 (arrow) and not for untransfected nuclei (arrowheads). Bar, 20 μm. F, localization of endogenous Kir2.2 from control skeletal muscle fibers or from muscle electroporated with GFP-Kir2.6. Endogenous Kir2.2 subunit distribution in skeletal muscle was visually scored for presence in the ER and T-tubules or only in the T-tubules from widefield fluorescence images from control fibers or for nuclei expressing GFP-Kir2.6 from electroporated fibers (n = 100 nuclei for each condition).
Endogenous Kir2.2 Is Partially Retained in the ER by Kir2.6 Expression
Strong support for a regulatory role of Kir2.6 in Kir2.x channel trafficking was further obtained by examining the effect of Kir2.6 expression on the localization of endogenous channels in mouse muscle. In control skeletal muscle fibers, endogenous Kir2.2 was not observed in the ER, but after in vivo electroporation of Kir2.6, nearly all of the nuclei expressing Kir2.6 showed retention of endogenous Kir2.2 in the ER surrounding those nuclei, which colocalized with Kir2.6 (Fig. 10, E and F). Nearby nuclei not expressing Kir2.6 were unaffected (Fig. 10E, arrowheads), and muscle fibers electroporated with unrelated constructs (myristoylated-GFP or GFP-Ca2+-ATPase) were unaffected (data not shown). In contrast to the effects on endogenous Kir2.2, we did not observe ER retention of endogenous Kir2.1 (data not shown), perhaps because of a smaller effect on Kir2.1 (Figs. 5, 9, and 10) or the lower sensitivitiy of the Kir2.1 antibody to endogenous mouse Kir2.1. Taken together, these data indicate that Kir2.6 has a dominant negative affect on the trafficking of other Kir2.x channel subunits in skeletal muscle, with the strongest effect on Kir2.2 and a more moderate effect on Kir2.1.
DISCUSSION
In this study we have investigated the trafficking, cellular localization, and subunit interactions of a recently identified human inward rectifier potassium channel Kir2.6. This subunit was discovered as a gene that when mutated confers susceptibility to TPP, a condition characterized by muscle weakness or paralysis accompanied by hypokalemia and thyrotoxicosis (22). Here we characterize wild type Kir2.6 to understand how the typical native Kir2.6 contributes to muscle cell biology. Surprisingly, we find that despite its high sequence similarity to other Kir2.x subunits (>98% identity with Kir2.2), Kir2.6 is very poorly trafficked to the surface membrane and instead primarily resides in the endoplasmic reticulum. Whole cell currents of Kir2.6 by itself are very small relative to other inward rectifier subunits because of its low surface expression. However, inward rectifier channels are composed of a heterotetrameric assembly of subunits, and we show that Kir2.6 readily coassembles with other Kir2.x subunits. Because of its dominant trafficking phenotype, Kir2.6 exerts an important regulatory control on the trafficking of inward rectifier channels through dominant negative retention in the ER. To our knowledge, this is the first demonstration of regulation of the trafficking of strong inward rectifier Kir2 potassium channels by dominant negative interaction with wild type channel subunits.
Localization of potassium channels in skeletal muscle is important to their function; Kir2 channels play a major role in setting the cell resting potential, in controlling muscle cell excitability by determining the extent of sodium channel inactivation, and in clearing activity-dependent accumulation of K+ from T-tubules. We have shown by immunocytochemistry that the major skeletal muscle Kir channels Kir2.1 and Kir2.2 are well positioned for these functions, with Kir2.1 channels located on the plasma membrane and the peripheral regions of T-tubules, and Kir2.2 located in central regions of T-tubules and neuromuscular junction (Fig. 6). These results agree well with the proposed localization of inward rectifier channels based on electrophysiological studies in frog skeletal muscle (8, 12–14) and on biochemical membrane fractionation studies in rat skeletal muscle (1). Indeed a compelling case for Kir channel localization on T-tubules and plasma membrane was made by Wallinga et al. (15), who showed by modeling that maintenance of skeletal muscle excitability and excitation-contraction coupling during repetitive action potential firing requires the presence of Kir channels in these sites for clearance of K+ and prevention of positive shifts in the resting membrane potential. Our results also are in agreement with studies in cardiac muscle in which T-tubule and plasma membrane localization, and their roles in K+ accumulation have been demonstrated (5, 6, 10, 11).
Although Kir2.2 and Kir2.6 share more than 98% identity and are thought to have arisen from gene duplication (22) with several diversifications acquired throughout evolution, their trafficking and localization are widely different. Exogenous expression of Kir2.6 in both COS-1 cells and mouse skeletal muscle showed colocalization with the ER marker PDI, indicating that Kir2.6 is retained in the ER and trafficked poorly to the plasma membrane (Figs. 1, 7, and 9). In contrast Kir2.1 and Kir2.2 trafficked out of the ER through the Golgi, shown by colocalization with Golgi markers, and finally expressed on T-tubules and the plasma membrane (Figs. 1, 7, and 10 and supplemental Fig. S2).
Even though Kir2.6 primarily is retained in the ER, a small proportion of the channel is trafficked to the cell surface in COS-1 cells. We speculated that this surface Kir2.6 formed functional homotetrameric channels. Indeed, electrophysiological investigation confirmed that Kir2.6 can form functional channels on the plasma membrane, in agreement with findings from Ryan et al. (22). Kir2.6 currents display characteristic inward rectification, with more pronounced inward currents at potentials negative to EK than at potentials positive to EK. However, when compared with Kir2.2, whole cell currents reveal a marked reduction in magnitude, implying that fewer channels are present on the plasma membrane.
It is possible that high expression levels and longer expression times in some cultured cells may allow Kir2.6 channels to escape cellular trafficking and quality control mechanisms and may account for larger Kir2.6 current magnitudes observed by Ryan et al. (22) in 293T cells. In mouse skeletal muscle, however, even after 7–14 days of expression in vivo, we found no evidence for Kir2.6 trafficking beyond the ER when Kir2.6 was expressed alone, suggesting that it is effectively retained in the ER (Fig. 7). When expressed with Kir2.1 in mouse skeletal muscle, a very low level of Kir2.6 trafficked to the Golgi (Fig. 9). In primate skeletal muscle, it is possible that a tissue-specific and species-specific accessory subunit or chaperone may aid the forward trafficking of Kir2.6; thus, a greater fraction of Kir2.6 might traffic to surface membranes in human skeletal muscle.
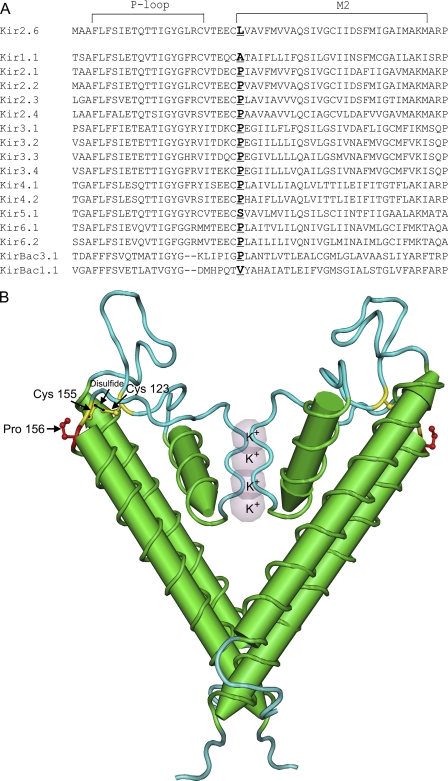
Although the Kir2.6 sequence includes all known anterograde trafficking signals that are present in other Kir2.x channels, it is still retained in the ER (Figs. 1, 5, 7, 9, and 10). Our studies show that a previously unidentified site, proline 156, is the most essential amino acid for surface trafficking of Kir2.2, and notably, the leucine at position 156 of Kir2.6 caused its retention in the ER (Figs. 3 and 8). Alignment of all known human Kir family sequences to identify conserved amino acids shows that the amino acid pair cysteine 155 and proline 156 is conserved among nearly all eukaryotic Kir subunits, but a leucine is present in place of the proline in Kir2.6 (Fig. 11A). Prolines have an established role in α-helical turn structure and are commonly found as the first residue of an α-helix (48). Indeed, in the three-dimensional structure of Kir2.2 (Protein Data Bank file 3JYC), proline 156 is located at a turn that forms the beginning of the M2 inner helix (Fig. 11B) (49). Significantly, the adjacent cysteine 155 is part of an intrasubunit disulfide with conserved cysteine 123 that is essential for channel function and that bridges the two extracellular loops in eukaryotic Kir2 channels (49–52). It has been shown that mutation of either of these conserved cysteines of Kir2.1 in one subunit of a channel tetramer is sufficient to eliminate wild type Kir2.1 currents in a dominant negative manner (50). We assume that proline 156 in Kir2.2 induces bending that initiates the M2 helix. Leucine at this position in Kir2.6 likely impacts channel conformation and may alter folding efficiency or disulfide bond formation that could result in retention of Kir2.6 in the ER.
FIGURE 11.
Alignment of Kir2 family reveals a conserved proline located between the P-loop and the M2 helix. A, multisequence alignment of human Kir family members was performed with Clustal W (1.81) with Kir sequences from IUPHAR. Leucine 156 in Kir2.6 is instead a conserved proline that initiates the M2 transmembrane helix in nearly all other Kir channels (site is shown in bold text). B, two subunits of chicken Kir2.2 (Protein Data Bank code 3JYC) are shown, with conserved proline 156 and an adjacent disulfide bond between cysteine 155 and cysteine 123 (49).
Interestingly, the absence of a conserved proline has been implicated in a loss of function of two voltage-gated potassium channel subunits. Kv6.3 and Kv8.1 are electrically silent homotetrameric channels because of retention in the ER, a phenomenon likely resulting from divergence of a proline-containing motif. Study of chimeric subunits between Kv6.3 and Kv2.1 and point mutation analysis in Kv8.1 revealed that a PXP motif, located in the S6 transmembrane domain in the gating hinge, is altered to PXT or PXA in the silent channels, respectively (53, 54) (Protein Data Bank file 2A79).
We have shown that Kir2.6 associates with Kir2.1 and Kir2.2 by immunoprecipitation (Fig. 4), with important consequences on channel trafficking, plasma membrane abundance, and localization (Figs. 3, 5, 9, and 10). Kir2 channels are known to form by homotetrameric and heterotetrameric assembly of Kir2.x subunits, with the resulting channels reflecting the physiological properties of their subunits (3). However, the subunit composition, the extent of heteromultimerization, and the roles of individual subunits in different cell types are not known. Gene knock-out studies in mice showed that Kir2.1 and Kir2.2 contribute to K+ currents in heart in a nonadditive manner, suggesting that these subunits can coassemble in vivo (21). In cardiac myocytes, the diversity of channel conductances, Ba2+ block properties, and pH sensitivity also support a model in which channels are composed of homotetrameric and heterotetrameric subunits (33–36). Our data showing the overlapping distribution of endogenous Kir2.1 and Kir2.2 channels in skeletal muscle T-tubules are consistent with the idea that a fraction of these subunits coassemble in vivo in skeletal muscle (Fig. 6).
Our trafficking studies, in which Kir2.6 causes ER retention of other Kir2.x subunits and colocalization of subunits, provide strong support for heteromultimeric formation of Kir2 channels in skeletal muscle, cardiac muscle cells, and COS-1 cells (Figs. 3, 5, 9, and 10). Kir2.6 is the dominant subunit when it forms a heteromultimer with Kir2.1 or Kir2.2 in vivo and has a stronger dominant negative trafficking effect on Kir2.2 compared with Kir2.1, suggesting that those subunits that are most similar in sequence may have a greater propensity to coassemble. A dominant negative trafficking phenomenon also is a well known explanation for disease-associated mutations in Andersen-Tawil syndrome where Kir2.1 Δ95–98 and Kir2.1 Δ314–315 do not traffic to the plasma membrane. Instead, they are distributed inside the cell, although they retain their ability to coassemble with wild type channels (18). Bendahhou et al. (18) speculate that deletion of amino acids 95–98, located in the outer M1 helix, results in protein misfolding and consequent ER retention. Coexpression of wild type Kir2.1 with Kir2.1 Δ95–98 or Kir2.1 Δ314–315 trapped the wild type subunit at intracellular sites. Reduction in Kir2.1 expression on the plasma membrane can interfere with regulation of electrical excitability and muscle resting potential (18).
It is interesting to note that another gene encoding Kir2.5/Kir2.2v, also closely related to Kir2.2, may act as a negative regulator of Kir2.2 channel activity as well, but in that case the Kir2.5 subunit is electrically silent possibly because of alterations of the conserved GYG selectivity filter or C terminus (55).
Although our studies do not address the function of Kir2.6 mutations that are associated with TPP, we speculate that some of those mutations may alter channel trafficking, leading to changes in the magnitude of Kir2 currents on surface membrane and T-tubules. Two of the TPP-associated mutations, Kir2.6 R399X and Kir2.6 Q407X, cause truncation of the Kir2.6 C terminus and result in the absence of a putative PDZ-binding motif (22), a region known to be important for subcellular localization of Kir2 channels (7, 10, 11, 28, 30, 31).
The Kir2.6 gene contains a thyroid-responsive element that may regulate gene transcription (22) and consequently lead to changes in its protein abundance, ER retention of Kir2.x subunits, and reduction in inward rectifier currents. Thus, thyrotoxicity may alter a delicate balance of electrical activity in muscle. Modeling studies and human disease-associated mutations in the genes encoding Kir2.1 and Kir2.6 suggest that too little or too much inward rectifier current can alter the electrical excitability of muscle and lead to paralysis (15, 17, 22, 56). Because TPP patients normalize when treated with antithyroid agents (reviewed in Ref. 57), the consequences of high T3 on muscle conductance is of interest.
In summary, we suggest that Kir2.6 is a regulatory subunit that is mainly retained in the ER as a consequence of leucine 156. Association between Kir2.6 and Kir2.1 or Kir2.2 promotes ER retention of the heterotetramer. The amount of inward rectifier channels on the plasma membrane may depend on the ratio between the Kir2.1, Kir2.2, and Kir2.6 subunits. We speculate that homeostasis is maintained in healthy individuals, enabling a stable amount of cell surface channel expression. Together, our data support the concept that ER retention is an important mechanism in controlling skeletal muscle excitability.
Supplementary Material
This work was supported by grants from American Heart Association Western Affiliate Grant 0655234Y, the Muscular Dystrophy Association Grant 4137, and the Cottage Hospital Research Program (to C. A. V.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- T-tubule
- transverse tubule
- DHPR
- dihydropyridine receptor α2
- ER
- endoplasmic reticulum
- TPP
- thyrotoxic hypokalemic periodic paralysis
- ECFP
- enhanced cyan fluorescent protein
- PDI
- protein disulfide isomerase.
REFERENCES
- 1. Kristensen M., Hansen T., Juel C. (2006) Am. J. Physiol. Regul. Integr. Comp. Physiol. 290, R766–R772 [DOI] [PubMed] [Google Scholar]
- 2. Bichet D., Haass F. A., Jan L. Y. (2003) Nat. Rev. Neurosci. 4, 957–967 [DOI] [PubMed] [Google Scholar]
- 3. Hibino H., Inanobe A., Furutani K., Murakami S., Findlay I., Kurachi Y. (2010) Physiol. Rev. 90, 291–366 [DOI] [PubMed] [Google Scholar]
- 4. Sacconi S., Simkin D., Arrighi N., Chapon F., Larroque M. M., Vicart S., Sternberg D., Fontaine B., Barhanin J., Desnuelle C., Bendahhou S. (2009) Am. J. Physiol. Cell Physiol. 297, C876–C885 [DOI] [PubMed] [Google Scholar]
- 5. Clark R. B., Tremblay A., Melnyk P., Allen B. G., Giles W. R., Fiset C. (2001) J. Physiol. 537, 979–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Melnyk P., Zhang L., Shrier A., Nattel S. (2002) Am. J. Physiol. Heart Circ. Physiol. 283, H1123–H1133 [DOI] [PubMed] [Google Scholar]
- 7. Leonoudakis D., Conti L. R., Anderson S., Radeke C. M., McGuire L. M., Adams M. E., Froehner S. C., Yates J. R., 3rd, Vandenberg C. A. (2004) J. Biol. Chem. 279, 22331–22346 [DOI] [PubMed] [Google Scholar]
- 8. Ashcroft F. M., Heiny J. A., Vergara J. (1985) J. Physiol. 359, 269–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Christé G. (1999) J. Mol. Cell Cardiol. 31, 2207–2213 [DOI] [PubMed] [Google Scholar]
- 10. Leonoudakis D., Mailliard W., Wingerd K., Clegg D., Vandenberg C. (2001) J. Cell Sci. 114, 987–998 [DOI] [PubMed] [Google Scholar]
- 11. Vaidyanathan R., Taffet S. M., Vikstrom K. L., Anumonwo J. M. (2010) J. Biol. Chem. 285, 28000–28009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Almers W. (1972) J. Physiol. 225, 57–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schneider M. F., Chandler W. K. (1976) J. Gen. Physiol. 67, 125–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Standen N. B., Stanfield P. R. (1979) J. Physiol. 294, 497–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wallinga W., Meijer S. L., Alberink M. J., Vliek M., Wienk E. D., Ypey D. L. (1999) Eur. Biophys. J. 28, 317–329 [DOI] [PubMed] [Google Scholar]
- 16. Anumonwo J. M., Lopatin A. N. (2010) J. Mol. Cell Cardiol. 48, 45–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Plaster N. M., Tawil R., Tristani-Firouzi M., Canún S., Bendahhou S., Tsunoda A., Donaldson M. R., Iannaccone S. T., Brunt E., Barohn R., Clark J., Deymeer F., George A. L., Jr., Fish F. A., Hahn A., Nitu A., Ozdemir C., Serdaroglu P., Subramony S. H., Wolfe G., Fu Y. H., Ptácek L. J. (2001) Cell 105, 511–519 [DOI] [PubMed] [Google Scholar]
- 18. Bendahhou S., Donaldson M. R., Plaster N. M., Tristani-Firouzi M., Fu Y. H., Ptácek L. J. (2003) J. Biol. Chem. 278, 51779–51785 [DOI] [PubMed] [Google Scholar]
- 19. Priori S. G., Pandit S. V., Rivolta I., Berenfeld O., Ronchetti E., Dhamoon A., Napolitano C., Anumonwo J., di Barletta M. R., Gudapakkam S., Bosi G., Stramba-Badiale M., Jalife J. (2005) Circ. Res. 96, 800–807 [DOI] [PubMed] [Google Scholar]
- 20. Zaritsky J. J., Eckman D. M., Wellman G. C., Nelson M. T., Schwarz T. L. (2000) Circ. Res. 87, 160–166 [DOI] [PubMed] [Google Scholar]
- 21. Zaritsky J. J., Redell J. B., Tempel B. L., Schwarz T. L. (2001) J. Physiol. 533, 697–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ryan D. P., da Silva M. R., Soong T. W., Fontaine B., Donaldson M. R., Kung A. W., Jongjaroenprasert W., Liang M. C., Khoo D. H., Cheah J. S., Ho S. C., Bernstein H. S., Maciel R. M., Brown R. H., Jr., Ptácek L. J. (2010) Cell 140, 88–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ma D., Zerangue N., Lin Y. F., Collins A., Yu M., Jan Y. N., Jan L. Y. (2001) Science 291, 316–319 [DOI] [PubMed] [Google Scholar]
- 24. Stockklausner C., Ludwig J., Ruppersberg J. P., Klöcker N. (2001) FEBS Lett. 493, 129–133 [DOI] [PubMed] [Google Scholar]
- 25. Stockklausner C., Klocker N. (2003) J. Biol. Chem. 278, 17000–17005 [DOI] [PubMed] [Google Scholar]
- 26. Hofherr A., Fakler B., Klöcker N. (2005) J. Cell Sci. 118, 1935–1943 [DOI] [PubMed] [Google Scholar]
- 27. Cohen N. A., Brenman J. E., Snyder S. H., Bredt D. S. (1996) Neuron 17, 759–767 [DOI] [PubMed] [Google Scholar]
- 28. Nehring R. B., Wischmeyer E., Döring F., Veh R. W., Sheng M., Karschin A. (2000) J. Neurosci. 20, 156–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Inanobe A., Fujita A., Ito M., Tomoike H., Inageda K., Kurachi Y. (2002) Am. J. Physiol. Cell Physiol. 282, C1396–C1403 [DOI] [PubMed] [Google Scholar]
- 30. Olsen O., Liu H., Wade J. B., Merot J., Welling P. A. (2002) Am. J. Physiol. Cell Physiol. 282, C183–C195 [DOI] [PubMed] [Google Scholar]
- 31. Leonoudakis D., Conti L. R., Radeke C. M., McGuire L. M., Vandenberg C. A. (2004) J. Biol. Chem. 279, 19051–19063 [DOI] [PubMed] [Google Scholar]
- 32. Leyland M. L., Dart C. (2004) J. Biol. Chem. 279, 43427–43436 [DOI] [PubMed] [Google Scholar]
- 33. Preisig-Müller R., Schlichthörl G., Goerge T., Heinen S., Brüggemann A., Rajan S., Derst C., Veh R. W., Daut J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7774–7779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zobel C., Cho H. C., Nguyen T. T., Pekhletski R., Diaz R. J., Wilson G. J., Backx P. H. (2003) J. Physiol. 550, 365–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schram G., Pourrier M., Wang Z., White M., Nattel S. (2003) Cardiovasc. Res. 59, 328–338 [DOI] [PubMed] [Google Scholar]
- 36. Panama B. K., McLerie M., Lopatin A. N. (2010) Pflugers Arch. 460, 839–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kung A. W. (2006) J. Clin. Endocrinol. Metab. 91, 2490–2495 [DOI] [PubMed] [Google Scholar]
- 38. Fang Y., Schram G., Romanenko V. G., Shi C., Conti L., Vandenberg C. A., Davies P. F., Nattel S., Levitan I. (2005) Am. J. Physiol. Cell Physiol. 289, C1134–C1144 [DOI] [PubMed] [Google Scholar]
- 39. McMahon J. M., Signori E., Wells K. E., Fazio V. M., Wells D. J. (2001) Gene Ther. 8, 1264–1270 [DOI] [PubMed] [Google Scholar]
- 40. Bloquel C., Fabre E., Bureau M. F., Scherman D. (2004) J. Gene Med. 6, (Suppl. 1) S11–S23 [DOI] [PubMed] [Google Scholar]
- 41. Wong S. H., Lowes K. N., Quigley A. F., Marotta R., Kita M., Byrne E., Kornberg A. J., Cook M. J., Kapsa R. M. (2005) Neuromuscul. Disord. 15, 630–641 [DOI] [PubMed] [Google Scholar]
- 42. Grady R. M., Akaaboune M., Cohen A. L., Maimone M. M., Lichtman J. W., Sanes J. R. (2003) J. Cell Biol. 160, 741–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rahkila P., Väänänen K., Saraste J., Metsikkö K. (1997) Exp. Cell Res. 234, 452–464 [DOI] [PubMed] [Google Scholar]
- 44. Kaisto T., Metsikkö K. (2003) Exp. Cell Res. 289, 47–57 [DOI] [PubMed] [Google Scholar]
- 45. Percival J. M., Froehner S. C. (2007) Traffic 8, 184–194 [DOI] [PubMed] [Google Scholar]
- 46. Wible B. A., De Biasi M., Majumder K., Taglialatela M., Brown A. M. (1995) Circ. Res. 76, 343–350 [DOI] [PubMed] [Google Scholar]
- 47. Flucher B. E., Morton M. E., Froehner S. C., Daniels M. P. (1990) Neuron 5, 339–351 [DOI] [PubMed] [Google Scholar]
- 48. Richardson J. S., Richardson D. C. (1988) Science 240, 1648–1652 [DOI] [PubMed] [Google Scholar]
- 49. Tao X., Avalos J. L., Chen J., MacKinnon R. (2009) Science 326, 1668–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cho H. C., Tsushima R. G., Nguyen T. T., Guy H. R., Backx P. H. (2000) Biochemistry 39, 4649–4657 [DOI] [PubMed] [Google Scholar]
- 51. Bendahhou S., Fournier E., Sternberg D., Bassez G., Furby A., Sereni C., Donaldson M. R., Larroque M. M., Fontaine B., Barhanin J. (2005) J. Physiol. 565, 731–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Leyland M. L., Dart C., Spencer P. J., Sutcliffe M. J., Stanfield P. R. (1999) Pflugers Arch. 438, 778–781 [DOI] [PubMed] [Google Scholar]
- 53. Salinas M., de Weille J., Guillemare E., Lazdunski M., Hugnot J. P. (1997) J. Biol. Chem. 272, 8774–8780 [DOI] [PubMed] [Google Scholar]
- 54. Ottschytsch N., Raes A. L., Timmermans J. P., Snyders D. J. (2005) J. Physiol. 568, 737–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Namba N., Inagaki N., Gonoi T., Seino Y., Seino S. (1996) FEBS Lett. 386, 211–214 [DOI] [PubMed] [Google Scholar]
- 56. Struyk A. F., Cannon S. C. (2008) Muscle Nerve 37, 326–337 [DOI] [PubMed] [Google Scholar]
- 57. Fontaine B. (2008) Adv. Genet. 63, 3–23 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.