Abstract
Individuals with Down syndrome (DS) will inevitably develop Alzheimer disease (AD) neuropathology sometime after middle age, which may be attributable to genes triplicated in individuals with DS. The characteristics of AD neuropathology include neuritic plaques, neurofibrillary tangles, and neuronal loss in various brain regions. The mechanism underlying neurodegeneration in AD and DS remains elusive. Regulator of calcineurin 1 (RCAN1) has been implicated in the pathogenesis of DS. Our data show that RCAN1 expression is elevated in the cortex of DS and AD patients. RCAN1 expression can be activated by the stress hormone dexamethasone. A functional glucocorticoid response element was identified in the RCAN1 isoform 1 (RCAN1-1) promoter region, which is able to mediate the up-regulation of RCAN1 expression. Here we show that overexpression of RCAN1-1 in primary neurons activates caspase-9 and caspase-3 and subsequently induces neuronal apoptosis. Furthermore, we found that the neurotoxicity of RCAN1-1 is inhibited by knock-out of caspase-3 in caspase-3−/− neurons. Our study provides a novel mechanism by which RCAN1 functions as a mediator of stress- and Aβ-induced neuronal death, and overexpression of RCAN1 due to an extra copy of the RCAN1 gene on chromosome 21 contributes to AD pathogenesis in DS.
Keywords: Alzheimer Disease, Apoptosis, Caspase, Neurodegeneration, Transcription, Down Syndrome, RCAN1
Introduction
Down syndrome (DS)4 is the most common genetic cause of mental retardation, affecting approximately one in 800–1,000 babies (1). Individuals with DS inevitably develop characteristic Alzheimer disease (AD) neuropathology, which includes neuritic plaques, neurofibrillary tangles, and neuronal cell death following middle age (2–5). DS is a valuable model system for understanding AD pathogenesis. Both DS and AD feature prominent neuronal losses in the hippocampus and cortex, resulting in progressive cognitive deficits in AD patients (5). Apoptosis has been implicated in playing a major role in neuronal cell death in both AD and DS (6–9). Increased immunoreactivity of activated caspase-3 is observed in neurons of AD and DS (7); however, the underlying mechanism leading to neuronal apoptosis in AD and DS remains elusive.
The development of DS is caused by the presence of an extra copy of human chromosome 21 (10, 11). The DSCR1 (Down syndrome critical region 1) gene was identified and located on chromosome 21 (12, 13). Dysregulation of a regulatory circuit involving DSCR1-calcineurin-nuclear factor of activated T cells (NFAT) plays an important role in DS development (14). DSCR1 proteins physically interact with calcineurin subunit A and inhibit calcineurin activity in vitro and in vivo (15–19). DSCR1 was accordingly renamed as RCAN1 (regulator of calcineurin 1) (20). RCAN1 is phosphorylated at Ser112 by BMK1 (big MAP kinase 1), which is the priming site for the subsequent phosphorylation at Ser108 by GSK-3 (21–24). The phosphorylation form of RCAN1 can release the inhibition effect on calcineurin. Furthermore, phosphorylated RCAN1 is a substrate for calcineurin (79). RCAN1 FLISPP motif phosphorylation could increase its ability to inhibit the calcineurin activity (25) and accelerate its degradation rate (26). However, it was reported that GSK-3 kinase can enhance the calcineurin activity through the RCAN1 phosphorylation in the yeast system (23). RCAN1 could also be phosphorylated by NF-κB-inducing kinase (NIK), which increases the RCAN1 stability (27). Furthermore, RCAN1 interacts with TAB2 (TAK1 (TGF-β-activated kinase 1)-binding protein 2) to be phosphorylated by TAK1 (28). The phosphorylated form of the RCAN1 could serve as a calcineurin facilitator. NFAT is a major substrate for calcineurin, and dephosphorylation of NFAT facilitates NFAT nuclear translocation and activation of its target genes' transcription. RCAN1 can repress the NFAT signaling pathway by inhibition of calcineurin (14). Also, as a downstream gene of the NFAT signaling pathway, RCAN1 can be activated by a subset of molecules, including VEGF, angiotensin II, G protein-coupled receptor 54, TNF-α, thrombin, and other activators of the calcineurin-NFAT pathway, such as calcium ionophore (29, 30). RCAN1 can also be activated by dephosphorylation in neural cells via calcium current increase through the L-type calcium channel (31).
RCAN1 has been shown to be involved in cardiac valve development, cardiac hypertrophy, inflammation, angiogenesis, and cancer. RCAN1 has also been implicated in learning and memory. However, the role of RCAN1 in neurodegeneration in AD and DS is unknown. In this study, we show that RCAN1 is overexpressed in cortical tissues from AD and DS patients. To investigate the mechanism of RCAN1 overexpression in AD and DS, we characterized human RCAN1 gene promoters and identified a functional glucocorticoid response element (GRE), through which RCAN1 expression is up-regulated by stress hormone dexamethasone. Here we show that RCAN1 mediates glucocorticoid-induced neuronal apoptosis. We found that overexpression of RCAN1-1 in primary neurons activates the caspase-9 and caspase-3 apoptotic pathway, thereby rendering neurons more vulnerable to apoptosis induced by dexamethasone and Aβ. Our data suggest that RCAN1 overexpression may contribute to AD pathogenesis by mediating neuronal death in the brains of DS and AD patients.
EXPERIMENTAL PROCEDURES
Cloning of the RCAN1 Gene Promoter and Construction of Chimeric Luciferase Reporter Plasmids
A forward primer corresponding to −684 bp (5′-ccgctcgaggtcctcttatttttccgctatttc-3′) of the transcriptional start site and a reverse primer corresponding to the coding sequence (5′-cacaagcttgtcctgcaggtccacctc-3′) were used to PCR-amplify the 5′-UTR region of the RCAN1 gene exon 1 from the genome of human neuroblastoma cells SH-SY5Y. The DNA fragment was cloned into pGL3-Basic upstream of the luciferase reporter gene to construct pRCANluc-G. The fragment from −684 to +46 bp was amplified and cloned using primers 5′-ccgctcgaggtcctcttatttttccgctatttc and 5′-cacaagctttgtcagcagtctcccagc, corresponding to +46 bp of the transcriptional start site on RCAN1 gene exon 1. Primers 5′-gctagctagcaatatattgtgaacc and 5′-cacctcgagtgagcagacatgtctc were used to amplify the region between −1650 and −685 bp in the 5′-UTR of RCAN1 gene exon 1. The amplified fragment was inserted in front of the −684 to 46 bp fragment in pRCANluc-B to obtain the pRCANluc-A. Primers were designed to include restriction enzyme sites so that the resulting PCR-amplified fragment could be easily cloned into the multicloning sites of vector pGL3-Basic (Promega). Deletion plasmids pRCANluc-C, -D, and -E were created from pRCANluc-A by utilization of restriction enzyme sites ApaI, PvuII, and EcoRI, respectively.
Cell Culture
Neuronal tissues for primary cultures originating from Wistar rat embryos at 17–18 days of gestation were dissected and gently digested with trypsin (0.025% EDTA; Invitrogen). The cells were suspended in neurobasal medium supplemented with B27 (Invitrogen) and plated at a density of 1–2 × 105 cells/well onto poly-d-lysine (0.01 mg/ml; Sigma)-coated 24-well plates (Nunc). The cultures were maintained at 37 °C in a humidified atmosphere containing 5% CO2 and used for experiments after 7–14 days. Primary neuronal cultures derived from caspase-3 knock-out (caspase-3−/−) (32) and wild type newborn mice were isolated and cultured as described (33). SH-SY5Y cells and human embryonic kidney 293 cells (HEK293) were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% FBS, 1 mm sodium pyruvate, 2 mm l-glutamine, 50 units/ml penicillin G sodium, and 50 μg/ml streptomycin sulfate (Invitrogen). All cells were maintained in a 37 °C incubator containing 5% CO2.
Preparation of β-Amyloid Fibrils
Synthesized Aβ(1–40) was dissolved in sterile double-distilled H2O to 1 mm. The dissolved Aβ was incubated at 37 °C for 1 h and diluted again with the same volume of PBS (1:1 dilution). Aβ was aged at 37 °C for 4 days, allowing the stable growth of Aβ fibrils.
Transfection and Luciferase Assay
Cells were grown to ∼70% confluence and transfected with 2 μg of plasmid DNA/35-mm plate using Lipofectamine 2000 (Invitrogen) as described (34). The pCH110 β-galactosidase expression plasmid was co-transfected to normalize for transfection efficiency. Cells were harvested 48 h after transfection and lysed in 200 μl of 1× Reporter Lysis Buffer (Promega). The luciferase and β-galactosidase assays were performed according to the manufacturer's protocols (Promega) as described (35).
Primer Extension Assay
A primer extension assay was performed to determine the transcription initiation site. Total neuronal RNA was extracted from SH-SY5Y cells using TRI-Reagent (Sigma). Yeast tRNA was used as a control. A reverse primer, corresponding to positions +46 to +28 bp, 5′-cacaagctttgtcagcagtctcccagc, was synthesized and radioactively end-labeled with [γ-32P]ATP (Amersham Biosciences). Primer extension and sequencing were performed as described (34).
Gel Shift Assay
Oligonucleotide RCAN1−272−237bp (5′-cagctgtcagaaaagcggaactggggacgaggactt) and its antisense strand (5′-aagtcctcgtccccagttccgcttttctgacagctg) were synthesized to generate a double-stranded RCAN1-GRE oligonucleotide probe. The GRE consensus oligonucleotide was generated by annealing 5′-agaggatcTGTACAggaTGTTCTagat with 5′- atctAGAACAtccTGTACAgatcctct (core sequence is italic and uppercase). The RCAN1-GRE probe was end-labeled with [γ-32P]ATP by T4 polynucleotide kinase. Gel shift assays were performed as described (36). The samples were analyzed by a 4% nondenaturing PAGE gel.
Virus Infection
The RCAN1 expression plasmid pcDNA3.1-RCAN1mycHis was generated as described (37). RCAN1-1 cDNA was cloned into a Semliki Forest virus vector pSFV to generate pSFV-RCAN1, and a green fluorescent protein cDNA was cloned into pSFV to generate pSFV-GFP. To generate the virus, pSFV1-EGFP, pSFV1-RCAN1myc, pSFV-RCAN1GFP, and helper virus pHelper-2 DNA were linearized by SpeI or SapI. Viral particles were generated according to the manufacturer's instructions (Invitrogen). For infection, the viral particles activated with chymotrypsin (Sigma) were added to neuronal cultures at a 1:10 dilution with culture medium. Cells were infected for 1 h at 37 °C, followed by replacement of the infection media with conditioned culture medium and incubated for 12–18 h prior to use. Then the infected neurons were monitored for expression of GFP or immunostained for detection of Myc tag by 9E10 antibody to visualize RCAN1 expression in SFV-RCAN1myc-infected neurons.
Antibodies and Immunoblotting
The rabbit anti-RCAN1 polyclonal antibody was raised against the C terminus of RCAN1 protein (RPKPKIIQTRRPEYTPIHLS). The antibody was characterized as being able to specifically detect RCAN1. Brain tissues from AD patients were obtained from the Department of Pathology, Columbia University (New York). Brain tissues of DS abortuses (17–21 gestational weeks) were obtained from University of Maryland Brain and Tissue Bank for Developmental Disorders. The brain tissues or cells were lysed in radioimmune precipitation assay lysis buffer (1% Triton X-100, 1% sodium deoxycholate and 4% SDS, 0.15 m NaCl, 0.05 m Tris-HCl, pH 7.2) supplemented with protease inhibitors (Complete, Roche Applied Science). The lysates were resolved by 12% SDS-PAGE for detecting RCAN1 and 16% Tris-Tricine PAGE for caspase-3 and caspase-9. The immunoblotting was performed as described (33). Rabbit anti-RCAN1 polyclonal antibody (1:1000 dilution) was used to detect RCAN1 expression. Caspase-3 and its cleaved form P20 were detected with a rabbit polyclonal antibody against amino acids 29–43 of caspase-3 (Sigma, catalog no. C9598, 1:1000). Caspase-3 cleavage was further confirmed with an anti-cleaved caspase-3 antibody (catalog no. 9661) from Cell Signaling. Internal control β-actin was analyzed using monoclonal anti-β-actin antibody AC-15 (Sigma, 1:1000). Caspase-9 was detected with anti-caspase-9 from Stressgen (catalog no. AAP-149).
RT-PCR
RNA was isolated from cells by TRI-Reagent (Sigma). PowerScriptTM reverse transcriptase (Invitrogen) was used to synthesize the first strand cDNA from an equal amount of the RNA sample following the manufacturer's instructions. The newly synthesized cDNA templates were further amplified by Platinum® TaqDNA polymerase (Invitrogen) in a 25-μl reaction. 20–35 cycles of PCR were used to cover the linear range of the PCR amplification. Two pairs of primers were used to specifically amplify RCAN1 isoform 1 (RCAN1-1) and isoform 4 (RCAN1-4), respectively. Primers for isoform 1 are 5′-ggaattcgccaccatggaggaggtggacctg and 5′-cgcgtcgactggctgaggtggatcggcgtg. Isoform 4 was amplified by using 5′-gggtctgtagcgctttcac and 5′-cgcgtcgactggctgaggtggatcggcgtg. A pair of gene-specific primers, 5′-tctggatcctcaccaccatggagaaggc and 5′-atactcgaggcagggatgatgttctg, was used to amplify a 324-bp fragment of GAPDH as an internal control. The samples were further analyzed on a 1.5% agarose gel. Eastman Kodak Co. Image Station 1000 software (PerkinElmer Life Sciences) was used to analyze the data.
MTT Assay
The MTT (Sigma) was added into culture medium of primary neurons and incubated for 4 h in a 37 °C incubator containing 5% CO2. The medium was then removed by a syringe, and 2-propanol-HCl was added to dissolve the dye. Lysates were analyzed with a spectrophotometer at a wavelength of 540 nm (Multiskan Ascent, ThermoLab System).
Apoptotic Cell Staining
Rat primary neurons were fixed, permeabilized, and stained with Hoechst 33258 (1 μg/ml, Sigma) at room temperature for 15 min. Cells were rinsed twice with PBS and sealed in mount medium (Vector Laboratories). Apoptotic cells were analyzed by a fluorescent microscope (Axiovert 200, Carl Zeiss Inc.). TUNEL staining was performed according to the manufacturer's instructions (Promega and Roche Applied Science). Results were analyzed by fluorescence microscopy.
Activities of Caspase-3/7 and Caspase-9 Assays
Activities of caspase-3/7 and caspase-9 were measured with Caspase-Glo® 3/7 (catalog no. G8090) and Caspase-Glo® 9 (catalog no. G8210) assay systems from Promega following the manufacturer's instructions. The luminescence was measured with a TD-20/20 luminometer (Turner Design).
Mitochondrial Fractionation
90% confluent HEK293 cells were harvested and washed three times with cold 1× PBS. The cells were then suspended in mitochondrial isolation buffer (250 mm sucrose, 10 mm Hepes, 10 mm KCl, 1.5 mm MgCl2, 1 mm EDTA) supplemented with Complete Protease Inhibitors (Roche Applied Science)) and incubated on ice for 30 min. Cells were lysed with a glass homogenizer (15–25 strokes) on ice. The homogenate was centrifuged twice at 1000 × g for 5 min in order to get rid of the nuclei and remaining intact cells. The supernatant was removed and centrifuged at 140,000 × g for 15 min. The resultant pellet containing mitochondria was washed with mitochondrial isolation buffer. The supernatant was transferred to a new tube and centrifuged for an additional time in order to remove any remaining mitochondria.
RESULTS
RCAN1 Expression Is Elevated in the Brains of DS Fetus and AD Patients
Previous studies have shown that RCAN1 mRNA is elevated in the brains of DS and AD patients (17, 38), indicating that RCAN1 gene expression is up-regulated at the transcriptional level. To further examine whether RCAN1 is also elevated at the protein level, an antibody specific to RCAN1, DCT3, was used to detect RCAN1 proteins in the brain tissue lysates from AD patients and DS abortuses as well as their age-matched controls. A synthetic peptide DCT with sequence PRKPKIIQTRRPEYTPIHLS corresponding to the C terminus of the human RCAN1 protein was used to immunize a rabbit, and a polyclonal antibody DCT3 was raised against the human RCAN1 protein C terminus. To characterize this RCAN1 antibody, HEK293 cells were transfected with empty vector or pRCAN1-mycHis plasmid. Fig. 1A shows that Myc-tagged RCAN1 proteins were detected by both mouse monoclonal 9E10 anti-Myc antibody and rabbit polyclonal antibody DCT3. Overexpressed RCAN1 protein could not be detected with the preimmunization serum pre-DCT. Furthermore, preincubation of DCT3 with excess DCT peptides resulted in clearance of RCAN1-specific antibody, and the precleared antibody could not detect RCAN1 protein (Fig. 1A). These data clearly demonstrate that our DCT3 antibody is able to specifically detect RCAN1 protein.
FIGURE 1.
RCAN1 protein is elevated in brains from AD and DS patients. A, characterization of RCAN1 antibody DCT3. Anti-RCAN1 antibody DCT3 targeting the last 20 amino acids of the human RCAN1 C terminus was raised in a rabbit. Cell lysates from HEK293 cells transfected with plasmid constructs pRCAN1-mycHis or its empty vector pcDNA3.1mycHis(c) were separated on a 12% glycine SDS-polyacrylamide gel. RCAN1 was immunoblotted with DCT3 at a dilution of 1:1000 (lanes 3 and 4). 9E10 antibody targeting the Myc tag fused with RCAN1 was used as positive control (lanes 1 and 2). Lanes 5 and 6 show immunoblots with the preimmune serum drawn from the rabbit before immunization. Lanes 7 and 8 were blotted with DCT3 preincubated with the antigen DCT. B, RCAN1 is enriched in the neuronal layers in human hippocampus and cortex. a, immunohistochemical staining with DCT3 antibody showed that RCAN1 is localized in the granule cell layer in human hippocampus (100×). Higher magnification shows its localization in both cytosol and nucleus in neurons. b, RCAN1 is enriched in pyramidal neuronal layers in cortex (100×). Higher magnification (400×) shows that the characteristic morphology of pyramidal neurons was stained by DCT3 antibody targeting the RCAN1 C terminus. C, representative gels showing Western blots of RCAN1 in tissues from the cortex of DS. Cortical tissues from gestational week 17–21 DS abortuses and their age-matched controls were lysed in 4% SDS radioimmune precipitation assay lysis buffer. 150 μg of total protein were separated on a 12% Tris-glycine SDS-polyacrylamide gel. Rabbit polyclonal anti-RCAN1 antibody DCT3 (1:1000 dilution) was used to detect RCAN1. β-Actin was detected by a monoclonal anti-β-actin antibody (AC15 from Sigma) and was used as loading control. D, quantification of Western blots showed that RCAN1 was increased in DS brains. Values represent mean ± S.E. (error bars) (n = 8). *, p = 0.0372 by Student's t test. E, representative gels showing Western blots of RCAN1 in tissues from the cortex of AD patients. F, quantification of Western blots showed that RCAN1 was markedly increased in AD brains. Values represent mean ± S.E. (n = 10). *, p = 0.0121 by Student's t test. Con, control.
To examine the localization of RCAN1 protein expression in human brain, human hippocampal and cortical slices were immunostained with DCT3 antibody specifically targeting RCAN1. Immunohistochemistry studies show that DCT3 antibody specifically stains the granule cell layer in the hippocampus and the pyramidal neuronal layers in the cortex (Fig. 1B). RCAN1 expression is detected in both the nucleus and cytosol of neurons. Higher magnification reveals that RCAN1 is preferentially localized in nucleus of pyramidal neurons (Fig. 1B). These results show that RCAN1 is highly and specifically expressed in neurons in the hippocampus and cortex. Because neurons of the hippocampus and cortex are specifically affected in Alzheimer disease, the high and specific expression of RCAN1 in neurons of the hippocampus and cortex suggests that RCAN1 may be involved in AD pathogenesis.
RCAN1 proteins were examined in the brain tissue lysates from AD patients and DS abortuses and patients as well as their age-matched controls. Western blot analysis using DCT3 antibody showed that levels of RCAN1 protein are significantly elevated in cortical tissue from DS abortuses (156.7 ± 17.04% relative to control, p < 0.05) (Fig. 1, C and D). RCAN1 protein levels are also significantly increased in the cortex of AD patients (173.4 ± 18.84% relative to control, p < 0.05) (Fig. 1, E and F). Our data show that RCAN1 is overexpressed in AD and DS brains. The result is consistent with previous reports (39, 40). Because nearly all DS patients will develop AD after their 30s, up-regulation of RCAN1 in both DS and AD suggests that RCAN1 may be involved in AD pathogenesis. Increased RCAN1 levels in DS may be attributed to its triplication on chromosome 21. However, the mechanism of RCAN1 up-regulation in AD is unknown.
Cloning and Functional Analysis of the Human RCAN1 Gene Promoter
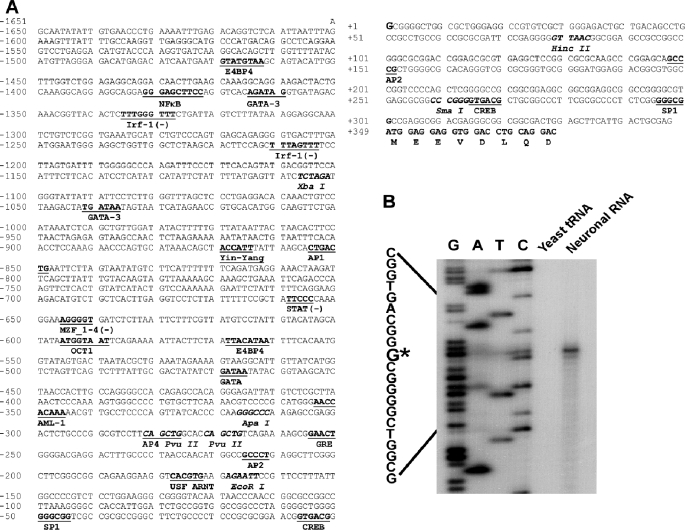
To investigate the molecular mechanism by which RCAN1 gene expression is up-regulated in AD pathogenesis, we cloned and functionally analyzed a 2000-bp fragment of the 5′-flanking region of the first exon of the human RCAN1 gene. The sequence was deposited to GenBankTM under accession number EF577083 (Fig. 2A). To determine the transcription start site of the human RCAN1 gene, a primer extension assay was performed. A 32P-labeled antisense primer (5′-tgtcagcagtctcccagc) located 302 bp upstream of the translational start site AUG was used to hybridize with neuronal RNA. The primer extension assay yielded a 45-bp major cDNA product. DNA sequencing gel analysis indicates that the major transcription start site is located at 348 bp upstream from the translation start site. Transcription starts with guanine, and this site was designated as +1 (Fig. 2B). A computer-based transcription factor binding site search revealed that this 2.0-kb 5′-flanking region contains several putative regulatory elements, such as AP1, AP2, GATA, OCT1, and USF (Fig. 2A).
FIGURE 2.
Characterization of the human RCAN1 gene promoter. A, the nucleotide sequence of the human RCAN1 gene promoter. A 2000-bp fragment of the 5′-flanking region and the first exon of the human RCAN1 gene was isolated from the human genomic library and sequenced. The guanidine at +1 represents the transcription start site. The positions of some of the unique and common restriction enzymes are indicated in italic type. The putative transcription factor binding sites are underlined in boldface type. The codon of the first exon is also indicated. B, a primer extension assay was performed to map the RCAN1 gene transcription start site. Neuronal RNA was extracted by TRI-Reagent, and yeast tRNA was used as a control. A 32P-labeled primer complementary to the region of +46 to +29 bp was used for both primer extension and the sequencing reaction. The plasmid pRCANluc-D, containing a DNA fragment of −311 to +46 bp, was used as a sequencing template. The samples were analyzed on 6% denaturing polyacrylamide gel electrophoresis. *, major transcription start site.
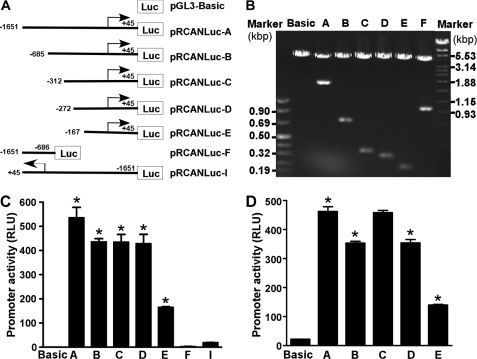
To determine if the 5′-flanking fragment obtained from genomic DNA contains the promoter of the RCAN1 gene, we cloned the fragment into a promoterless plasmid vector, pGL3-Basic (Fig. 3A). The pGL3-Basic vector lacks eukaryotic promoter and enhancer sequences upstream of a reporter luciferase gene. Luciferase activity in cells transfected with this plasmid depends on insertion and proper orientation of a functional promoter upstream from the luciferase gene. Luciferase activity is indicative of promoter activity. pRCANluc-A plasmid was constructed to contain a 1.70-kb 5′-flanking region from −1651 to +45 bp of the RCAN1 gene upstream of the luciferase reporter gene. Plasmid DNA was transfected into HEK293 cells and SH-SY5Y cells, and luciferase activity was measured by a luminometer to reflect promoter activity. Compared with an empty vector control, pRCANluc-A-transfected cells had significant luciferase activity at 554.2 ± 44.48 relative luciferase units (RLU) in HEK293 and 461.6 ± 16.82 RLU in SH-SY5Y cells (p < 0.001 relative to controls) (Fig. 3, C and D, lane A and Basic). However, plasmid pRCANlucR-I containing the RCAN1 promoter sequence from −1651 to +45 bp in the reverse orientation (Fig. 3C, lane F) and plasmid pRCANluc-F containing the sequence from −1651 to −686 lacking the transcription initiation site (Fig. 3C, lane I) have little or no luciferase activity. These results indicate that the 1.70-kb fragment contains the functional promoter of the human RCAN1 gene.
FIGURE 3.
Characterization of the RCAN1 promoter. A, schematic diagram of the promoter constructs. RCAN1 promoter deletion constructs were cloned into a promoterless vector, pGL3-Basic, in front of the reporter luciferase gene (Luc). The arrow denotes the direction of transcription. The numbers represent the end points of each construct. B, the corresponding deletion plasmids were confirmed by sequencing and restriction enzyme digestion. Digested samples were analyzed on a 1.0% agarose gel. The size of the vector is 4.7 kb, and the RCAN1 gene 5′-flanking fragment insert sizes range from 0.2 to 2.0 kb. C and D, luciferase assay of the deletion promoter constructs. The constructed plasmids were cotransfected into HEK293 cells (C) or SH-SY5Y cells (D) with pCH110. Luciferase activity was measured 48 h after transfection by a luminometer and expressed in RLU. β-Galactosidase activity was used to normalize for transfection efficiency. The values represent mean ± S.E. (error bars) (n = 4). *, p < 0.001 by analysis of variance with post hoc Newmann-Keuls test.
To analyze the transcriptional activity of the RCAN1 promoter, a series of luciferase reporter gene plasmids containing different upstream deletions of the RCAN1 promoter were constructed and transfected into HEK293 and SH-SY5Y cells. The inserts of pRCANluc-B, -C, -D, and -E are the fragments of the RCAN1 promoter from −684, −311, −271, and −167 bp to +45 bp, respectively (Fig. 3A). Analysis of deletion plasmids indicates that pRCANluc-A, pRCANluc-B, pRCANluc-C, and pRCANluc-D have strong promoter activity in HEK293 and SH-SY5Y cells. Deletion of 969 bp from −1651 to −685 resulted in significant reduction of RCAN1 promoter activity from 554.2 ± 44.48 to 450.8 ± 13.48 RLU in HEK293 cells (p < 0.05) (Fig. 3C, lanes A and B) and from 461.6 ± 16.82 to 352.2 ± 7.10 RLU in SH-SY5Y cells (p < 0.005) (Fig. 3D, lanes A and B). Deletion of an additional 413 bp from −685 had no significant effect on the promoter activity in HEK293 cells. pRCANluc-C and pRCANluc-D had similar promoter activity to pRCANluc-B in HEK293 cells, 449.1 ± 33.45 and 442.5 ± 40.73 RLU, respectively (p > 0.05) (Fig. 3C, lane C and D). However, deletion of 373 bp from −685 resulted in an increased promoter activity in SH-SY5Y cells from 352.2 ± 7.10 to 457.4 ± 8.75 RLU (p < 0.005) (Fig. 4D, lanes B and C). The promoter activities of pRCANLuc-C and pRCANLuc-A were not significantly different in SH-SY5Y cells (p > 0.05) (Fig. 3D). Further deletion of 40 bp from −312 reduced promoter activity to 353.0 ± 12.49 RLU in SH-SY5Y cells (p < 0.005, Fig. 3D lane D versus lane C). These data suggested that the RCAN1 promoter has cell-specific transcriptional activity.
FIGURE 4.
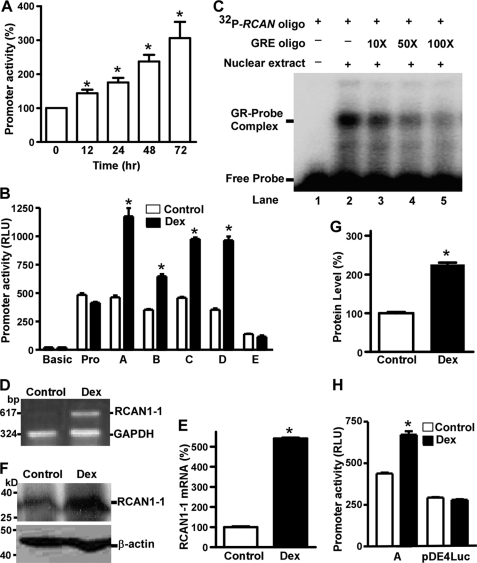
The human RCAN1 gene promoter contains a GRE. A, dexamethasone increased RCAN1 promoter activity in a time-dependent fashion. pRCANluc-A plasmid was cotransfected with GR expression plasmid and pCH11O into HEK293 cells. Cells were treated with 100 nm dexamethasone for 0, 12, 24, 48, and 72 h prior to the luciferase assay. The luciferase activity was measured by a luminometer and expressed in RLU. The x axis represents the time, and the y axis represents the percentage increase relative to the non-treatment control. Values represent mean ± S.E. (error bars) (n = 3–6). *, p < 0.05 by analysis of variance with post hoc Newmann-Keuls test. B, mapping of GRE in the RCAN1 promoter. The various deletion plasmid constructs were cotransfected with the GR expression plasmid and pCH11O in SH-SY5Y cells. Cells were treated with 100 nm dexamethasone for 48 h before harvest. Values represent mean ± S.E. (n = 4). *, p < 0.01 by Student's t test. C, gel shift assay. The gel shift assay was performed as described under “Experimental Procedures” using a 32P-labeled double-stranded oligonucleotide probe, RCAN1−272−237bp. Lane 1, labeled probe alone without nuclear extract. Incubation of 32P-labeled RCAN1−272−237bp with HeLa nuclear extracts retarded the migration rate of the labeled probe, which formed a new shifted DNA-protein complex band (lane 2). Competition assays were performed by further adding different concentrations of molar excess of unlabeled GRE competition consensus oligonucleotide (lanes 3–5). D, dexamethasone increased the RCAN1-1 mRNA level. RT-PCR was used to amplify RCAN1-1 from SH-SY5Y cells transfected with GR expression plasmid treated with or without 100 nm dexamethasone for 48 h. GAPDH was amplified as the internal control. E, quantification of RT-PCR shows that RCAN1-1 mRNA was elevated ∼5-fold after dexamethasone treatment. Values represent mean ± S.E. (n = 3). *, p = 0.0003 by Student's t test. F, dexamethasone increased RCAN1 protein expression. Western blot of RCAN1 in SH-SY5Y cells, which were transfected with GR expression plasmid and treated with or without 100 nm dexamethasone for 48 h. 150 μg of cell lysates were separated in a 12% Tris-glycine polyacrylamide gel. RCAN1 was detected with anti-RCAN1 antibody DCT3. β-Actin was detected by anti-β-actin and served as the internal control. G, quantification of E shows that RCAN1 protein was increased ∼2-fold following dexamethasone treatment. The values represent mean ± S.E. (n = 3). *, p = 0.0006 by Student's t test. H, differential activation of RCAN1 alternative promoters in SH-SY5Y cells by dexamethasone. The pRCANluc-A and pDE4Luc plasmids were cotransfected with GR expression plasmid and pCH11O into SH-SY5Y cells. pGL3-Basic was used as a negative control. Cells were treated with or without 100 nm dexamethasone for 48 h before the luciferase assay. The isoform 1 promoter plasmid pRCANluc-A has a higher activity in the neuronal cell line than the isoform 4 promoter plasmid pDE4Luc. *, p = 0.0002 by Student's t test. Dexamethasone increases the promoter activity of isoform 1 promoter pRCANluc-A (*, p = 0.0120 by Student's t test), whereas the drug has no effect on RCAN1 isoform 4 promoter (p > 0.05 by Student's t test). Values represent mean ± S.E., n = 3. Dex, dexamethasone.
To identify the minimal promoter region required for RCAN1 gene expression, additional deletion plasmids were constructed. Further deletion of 95 bp from −272 bp in pRCANluc-D to −167 bp in pRCANluc-E significantly reduced luciferase activity from 442.5 ± 40.73 to 170.7 ± 2.68 RLU in HEK293 cells (p < 0.0005) and from 353.0 ± 12.49 to 138.8 ± 3.61 RLU in SH-SY5Y cells (p < 0.0001) (Fig. 3, C and D). These results indicate that a 317-bp fragment from −272 to +45 bp contains the minimal human RCAN1 gene promoter.
The RCAN1 Promoter Contains a Glucocorticoid Response Element (GRE)
It was previously reported that RCAN1 mRNA was markedly increased by glucocorticoid induction in pre-B human leukemia cells (41). Neurons in hippocampus and cortex express high levels of glucocorticoid receptor (GR) (42). To investigate if glucocorticoids also activate the RCAN1 gene promoter, a GR expression plasmid was cotransfected with pRCANluc-A into HEK293 cells, and the cells were subjected to 100 nm dexamethasone treatment for 0, 12, 24, 48, and 72 h. A luciferase assay showed that the RCAN1 gene promoter activity was significantly up-regulated in a time-dependent manner by dexamethasone, with 143.6 ± 10.63, 175.4 ± 13.38, 237.0 ± 19.95, and 306.1 ± 47.53% increases at 12, 24, 48, and 72 h (p < 0.0001) (Fig. 4A). Similar results were also observed in SH-SY5Y cells. Up-regulation of RCAN1 promoter activity by dexamethasone suggests that the RCAN1 promoter may contain a GRE.
To identify the GRE site in the RCAN1 promoter, we examined the effect of dexamethasone on the deletion constructs containing various RCAN1 promoter regions, including pRCANluc-A, -B, -C, -D, and -E. pGL3-Basic and pGL3-Promoter, which do not contain any GRE sites, were used as negative controls. Luciferase activity in the cells transfected with pGL3-Basic and pGL3-Promoter plasmids was not affected by dexamethasone treatment (p > 0.05), whereas luciferase activity in cells transfected with pRCANluc-A, -B, -C, and -D plasmids was markedly increased (Fig. 4B). Dexamethasone increased promoter activity by 254.45 ± 27.32, 183.48 ± 9.89, 213.32 ± 4.85, and 273.09 ± 16.28% in cells transfected with pRCANluc-A, -B, -C, and -D plasmids, respectively (p < 0.001). However, further deletion of 95 bp from −272 bp in pRCANluc-D to −167 bp in pRCANluc-E abolished the up-regulatory effect of dexamethasone on RCAN1 promoter activity, and dexamethasone had no effect on RCAN1 promoter activity of pRCANluc-E, 83.13 ± 14.81% of control (p > 0.05) (Fig. 4B, lane E). Thus, the results suggest that a GRE is located in the region of −272 to −167 bp in the RCAN1 gene promoter.
To further confirm that the −272 to −167 bp region of the RCAN1 promoter contains a GRE, gel shift assays were performed. Double-stranded oligonucleotide probes corresponding to −272 to −237 bp, −251 to −217 bp, −231 to −197 bp, and −211 to −166 bp of the RCAN1 promoter were synthesized and end-labeled with 32P radioactive isotope. A shifted DNA-protein complex band was detected after hybridizing the 32P-labeled RCAN1−272−237bp probe with HeLa nuclear extract (Fig. 4C, lane 2) but not with other labeled oligonucleotide probes. These data suggest that the GRE site may be located in the −272 to −237 bp region of the RCAN1 promoter. To determine the binding specificity, GRE consensus oligonucleotides (5′-agaggatctgtacaggatgttctaga-3′) were added to compete for GR binding with the labeled RCAN1−272−237bp probe. The binding intensity of this shifted band was partially reduced after adding a 10- or 50-fold molar excess of unlabeled GRE consensus competition oligonucleotides, and the shifted band was abolished by the addition of a 100-fold excess of GRE consensus oligonucleotides (Fig. 4C, lanes 3–5). These results confirm that the shifted band represents the complex of RCAN1−272−237bp oligonucleotide bound with GR, and the human RCAN1 promoter contains a GRE in the region of −272 to −237 bp.
To investigate if this element plays an important role in transcription of the human RCAN1 gene, the effect of dexamethasone on endogenous RCAN1 gene expression was examined. Quantitative RT-PCR was performed to amplify RCAN1-1 mRNA by a pair of gene-specific primers (5′-gccaccatggaggaggtggacctg and 5′-tggctgaggtggatcggcgtg). Dexamethasone treatment increased the RCAN1-1 mRNA level more than 5-fold in GR-transfected SH-SY5Y cells (p < 0.0001) (Fig. 4, D and E). To further confirm the regulatory effect of GRs on RCAN1 gene expression, endogenous levels of RCAN1 protein were examined with anti-RCAN1 antibody DCT3. Consistent with our data showing that dexamethasone up-regulated RCAN1-1 expression at the transcription level, endogenous RCAN1-1 protein level was also markedly elevated by dexamethasone treatment (225.49 ± 5.78% relative to control, p < 0.0001) (Fig. 4, F and G). Taken together, these results demonstrate that the RCAN1-1 gene promoter contains a functional GRE in the region of −272 to −237 bp, through which glucocorticoids exert an up-regulatory effect on RCAN1 gene expression.
RCAN1 has a tissue-specific expression pattern. The RCAN1 gene spans ∼45 kb of genomic DNA and contains seven exons and six introns. Two major isoforms, RCAN1-1 and isoform 4 (RCAN1-4), are generated by alternative splicing of the first four exons. RCAN1-1 has a large form with 252 amino acids (RCAN1-1L) and a short form, RCAN1-1S. RCAN1-1S and RCAN1-4 both consist of 197 amino acids. The isoforms differ only at their N terminus, and the last 168 amino acids of the C terminus, encoded by exons 5–7, are the same in all forms of the isoforms. RCAN1-1 is highly expressed in the central nervous system (CNS), whereas RCAN1-4 is mostly expressed in heart muscle and fetal kidney (13, 43). It was reported that the calcineurin-dependent isoform 4 is also highly expressed in areas of the brain in which calcineurin is highly expressed (44, 45). We have cloned a 1.7-kb region upstream of the first exon of the human RCAN1 gene containing a functional promoter that controls the transcription of RCAN1-1 (Fig. 2). It has been reported that there is an alternative promoter upstream of exon 4 of the RCAN1 gene, which responds to the calcineurin-NFAT signaling pathway (30, 46). Therefore, the RCAN1 gene contains two promoters and two translation initiation codons: one in the 5′-UTR of exon 1 and the other in the 5′-UTR of exon 4, which are responsible for transcriptional control of RCAN1-1 and RCAN1-4, respectively. To examine whether the effect of glucocorticoids is specific for the RCAN1 exon 1 promoter, a 1200-bp DNA fragment upstream of RCAN1 exon 4 was cloned into promoterless luciferase reporter plasmid pGL3-Basic to generate pDE4Luc. Dexamethasone markedly increased isoform 1 promoter activity (from 175.33 ± 3.74 to 268.86 ± 14.2 RLU, p < 0.001), whereas it had no effect on isoform 4 promoter activity (from 117.47 ± 2.51 to 112.17 ± 2.39 RLU, p > 0.05) (Fig. 4H). The data suggest that glucocorticoids specifically up-regulate RCAN1-1 expression but not RCAN1-4 expression.
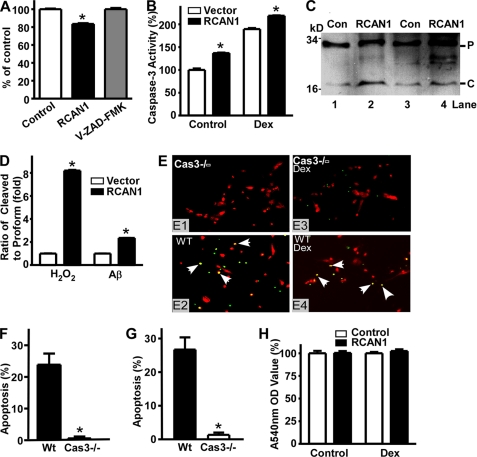
Overexpression of RCAN1 Induces Neuronal Apoptosis
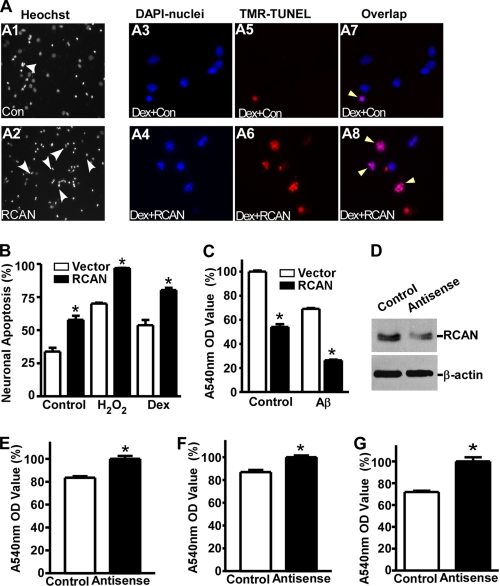
Our results thus far have shown that RCAN1 expression can be activated by dexamethasone, which has been shown to induce apoptosis in primary neurons. Previous studies have shown that overexpression of RCAN1 deters melanoma tumor growth and suppresses metastasis in mice (29, 47, 48). Although muscle-specific overexpression of RCAN1 does not always result in embryonic lethality (49), there was a report that RCAN1 overexpression could lead to early embryonic fatality in transgenic mice (50), suggesting that RCAN1 may be proapoptotic. RCAN1 is preferentially localized in neurons of the hippocampus and cortex, where there is a marked neuronal loss in the brains of AD patients (38). To investigate whether RCAN1 overexpression could induce apoptosis or exacerbate neuronal death induced by apoptotic inducers, cDNA encoding for the RCAN1 isoform 1 was cloned into a Semliki Forest virus vector, pSFV, to generate pSFV-RCAN1. A green fluorescent protein vector, pSFV-GFP, was used as control. Rat E18 primary neurons were infected with SFV-RCAN1 or SFV-GFP. The infection efficiency is about 30–40%. A colorimetric MTT assay and Hoechst and TUNEL staining were performed to determine the cell viability of primary neurons. Hoechst staining revealed that there is more nuclear condensation and fragmentation in RCAN1-1-overexpressing neurons exposed to H2O2 (Fig. 5A, compare A2 with A1). The apoptotic features of RCAN1-overexpressing neurons with dexamethasone treatment were confirmed by TUNEL staining (Fig. 5A, A3–A8). Compared with SFV-GFP control, overexpression of RCAN1-1 renders more neuronal death, a 57.90 ± 5.31% apoptosis ratio relative to 33.87 ± 4.86% in control by Hoechst staining (p < 0.005) (Fig. 5B, lane 2 versus lane 1). RCAN1-1 overexpression also exacerbated the toxicity of H2O2 on neurons, 97.02 ± 0.20% compared with 70.18 ± 0.93% in control (p < 0.005) (Fig. 5B, lane 4 versus lane 3), and significantly increased the dexamethasone-induced neuronal apoptosis (80.37 ± 2.80%, compared with 53.81 ± 5.80% in control, p < 0.001) (Fig. 5B, lane 6 versus lane 5). The proapoptotic effect of RCAN1 was further confirmed by an MTT assay. RCAN1 overexpression resulted in a 54.01 ± 5.17% reduction in A540 nm relative to control (p < 0.0001) (Fig. 5C). To examine whether RCAN1 overexpression could further exacerbate neuronal death induced by Aβ, a 10 μm concentration of aggregated Aβ peptides was added to SFV-infected neurons for 12 h. Consistent with a previous report (51), Aβ induced neuronal apoptosis, as indicated by A540 nm (69.11 ± 1.45% of control) in the Aβ-treated neuronal culture (p < 0.001) (Fig. 5C, lane 3 versus lane 1). The addition of Aβ to the RCAN1-1-overexpressing neurons further increased neuronal apoptosis, with the MTT assay showing a reduction to 26.39 ± 1.14% of control (p < 0.0001) (Fig. 5C, lane 4 versus lane 3). These data clearly indicate that RCAN1-1 facilitates neuronal apoptosis.
FIGURE 5.
Overexpression of RCAN1-1 induces neuronal apoptosis. A, apoptotic cells shown by Hoechst (A1 and A2) and TUNEL staining (A3–A8). Shown is Hoechst staining of rat primary neurons infected with SFV-GFP (A1) and SFV-RCAN1-1 (A2) for 14 h and further treated with 200 μm H2O2 for 6 h. SFV-GFP served as control. Results were analyzed by a fluorescent microscope with ×200 magnification. SFV-GFP- and SFV-RCAN1-expressing RCAN1-1-infected primary neurons were treated with 1 μm dexamethasone and stained with TUNEL (A5 and A6). Nuclei of neurons were counterstained with DAPI (A3 and A4). Apoptotic cells were indicated by a magenta color (A7 and A8), which corresponds to an overlap of red TUNEL and blue DAPI staining. The results were analyzed by a fluorescent microscope with ×400 magnification. White arrows indicate apoptotic cells. B, Hoechst staining showed more apoptotic cells in SFV-RCAN1-infected neurons compared with SFV-GFP controls. Lanes 1 and 2, primary neurons were infected with SFV-GFP and SFV-RCAN1 for 14 h before Hoechst staining. Lanes 3 and 4, SFV-GFP- and SFV-RCAN1-infected neurons were further treated with 200 μm H2O2 for 4 h. Lanes 5 and 6, SFV-GFP- and SFV-RCAN1-infected neurons were further treated with 5 μm dexamethasone for 12 h. The values represent mean ± S.E. (error bars) (n = 3). *, p < 0.001 by Student's t test. C, MTT showed that overexpression of RCAN1 induced neuronal death and rendered neurons more vulnerable to Aβ. Lanes 1 and 2, primary neurons were infected with SFV-GFP and SFV-RCAN1 for 16 h. Lanes 3 and 4, SFV-GFP- and SFV-RCAN1-infected neurons were further treated with 10 μm aged Aβ for 12 h. Values represent mean ± S.E. (n = 5). *, p < 0.0001 by Student's t test. D, inhibition of RCAN1 expression by RCAN1 antisense oligonucleotides. RCAN1 antisense oligonucleotides and control were cotransfected with the RCAN1-1 expression plasmid pcDNA3.1-RCAN1mycHis into HEK293 cells. Cell lysates were separated on a 12% SDS-polyacrylamide gel. RCAN1 was detected with 9E10 antibody targeting the Myc tag. β-Actin was detected by using a monoclonal anti-β-actin antibody (AC15 from Sigma) and served as a loading control. E, transfection of RCAN1 antisense oligonucleotides reduced the neuronal apoptosis induced by dexamethasone. The values represent mean ± S.E. (n = 4). *, p = 0.0005 by Student's t test. F, RCAN1 antisense oligonucleotides reduced Aβ-induced neuronal apoptosis. The values represent mean ± S.E. (n = 4). *, p < 0.001 by Student's t test. G, primary neurons were transfected with RCAN1 antisense oligonucleotides or control oligonucleotides using Oligofectamine 2000 (Invitrogen) and treated with 100 μm H2O2 for 4 h. Neuronal apoptosis was measured by an MTT assay. Values represent mean ± S.E. (n = 4). *, p = 0.0123 by Student's t test. The OD value of RCAN1 antisense was set to 100% in E–G.
Previous studies reported that dexamethasone can induce cortical neuronal death, an effect rescued by a caspase inhibitor (52). Our results also showed that exposure of RCAN1-1-overexpressing neurons to 1 μm dexamethasone rendered more neuronal death, as indicated by the MTT assay (Fig. 5A). To further determine if RCAN1 mediated apoptosis, RCAN1 expression was inhibited by using RCAN1-specific antisense oligonucleotides (5′- ezeftgtccttgtcfzfog) (Fig. 5D). Knockdown of endogenous RCAN1 expression with the antisense oligonucleotides inhibited dexamethasone-induced neurotoxicity (100 ± 11.02% versus 83.50 ± 5.84% in controls, p < 0.0001) (Fig. 5E). Furthermore, endogenous RCAN1 knockdown was also able to rescue neurons from Aβ-induced neuronal death (100 ± 3.41% versus 86.90 ± 4.29% in controls, p < 0.01) (Fig. 5F) and H2O2-induced neuronal death (100 ± 8.73% versus 71.87 ± 2.76% in controls, p < 0.05) (Fig. 5G). These results demonstrate that RCAN1 exacerbates the neuronal apoptosis induced by dexamethasone, Aβ, and H2O2.
RCAN1 Activates the Caspase-3 Apoptotic Pathway
The mechanism by which RCAN1 overexpression induces neuronal apoptosis is unknown. The caspase family of proteins is the principal molecular machinery that executes apoptosis (53–56). Altered expression of apoptosis-related proteins, such as Par-4, Bak, Bad, Bax, Bcl-2, p53, caspase-3, and Fas, has been reported in AD brains (57–61). To investigate if the neuronal death induced by RCAN1 overexpression is mediated by the caspase signaling pathway, the caspase inhibitor benzyloxycarbonyl-VAD-fluoromethyl ketone was added to primary neuronal cultures. The caspase inhibitor benzyloxycarbonyl-VAD-fluoromethyl ketone blocked the neuronal apoptosis induced by SFV-RCAN1 (100.0 ± 2.30%, compared with 83.44 ± 1.60%, p < 0.0001) (Fig. 6A), suggesting that the caspase pathway is indeed involved in neuronal death induced by RCAN1 overexpression. Caspase-3 is a major executor of the caspase signaling pathway (62). To investigate whether caspase-3 activation is involved in the proapoptotic effect of RCAN1, the activities of caspase-3/7 in SFV-RCAN1- or SFV-GFP-infected primary neurons were measured by the Caspase-Glo® 3/7 assay. Overexpression of RCAN1 increased caspase-3/7 activity by 136.63 ± 6.4% relative to vector control (p < 0.0001); exposure to dexamethasone further up-regulated caspase-3/7 activity from 189.28 ± 10.12 to 218.41 ± 3.11% (p < 0.0001) (Fig. 6B). The results also indicate that dexamethasone could activate caspase-3/7 (189.28 ± 10.12% of control, p < 0.0001) (Fig. 6B, lane 3 versus lane 1). Activation of caspase-3 results in its cleavage from the 32-kDa pro form to the P17 and P12 fragments. To further confirm that caspase-3 is activated by RCAN1 isoform 1 overexpression, Western blot assays were performed to detect the cleavage of caspase-3 in primary neurons infected by SFV-RCAN1 or SFV-GFP in the presence of Aβ and H2O2 treatment (Fig. 6B). Generation of the cleaved caspase-3 fragment P17 was markedly increased in RCAN1-1-overexpressing primary neurons compared with controls by 2.34 ± 0.01-fold in Aβ-treated neurons (p < 0.0001) and 8.20 ± 0.10-fold in H2O2-treated neurons (p < 0.0001) (Fig. 6C). In the absence of insults, the caspase-3 cleavage was not detectable by Western blot, probably due to the low activity of the caspase pathway (data not shown). These data demonstrate that RCAN1 overexpression activates the caspase-3 signaling pathway, thereby inducing neuronal apoptosis.
FIGURE 6.
Overexpression of RCAN1 activates caspase-3 in primary neurons. A, the pancaspase inhibitor benzyloxycarbonyl-VAD-fluoromethyl ketone (Z-VAD-FMK) blocked neuronal death induced by overexpression of RCAN1-1 Values represent mean ± S.E. (n = 3). *, p < 0.0001 by Student's t test. B, RCAN1-1 expression increased caspase-3/7 activity. Primary neurons were infected with SFV-GFP and SFV-RCAN1 for 16 h and assayed by the Caspase-Glo® 3/7 assay system (from Promega, catalog no. G8090). Luciferase activity was measured with a luminometer. Lanes 1 and 2, SFV-GFP and SFV-RCAN1 virus alone. Lanes 3 and 4, GFP- and RCAN1-infected neurons were further treated with 1 μm dexamethasone for 12 h. Values represent mean ± S.E. (n = 6). *, p < 0.0001 by Student's t test. C, cleaved form of caspase-3 was markedly increased by RCAN1-1 overexpression. Whole cell lysates from neurons infected with SFV-RCAN1 and SFV-GFP were separated on a 16% Tris-Tricine gel and blotted with anti-caspase-3 antibody (from Sigma). Lanes 1 and 2, SFV-GFP- and SFV-RCAN1-infected neurons were treated with 200 μm H2O2 for 6 h. Lanes 3 and 4, SFV-GFP- and SFV-RCAN1-infected neurons were treated with 10 μm aged Aβ for 12 h. P and C indicate the full-length pro form caspase-3 (32 kDa) and the cleaved form caspase-3 (17 kDa). D, quantification of Western blot (C) shows RCAN1-1 overexpression increases caspase-3 cleavage. Values represent mean ± S.E. (error bars) (n = 3). *, p < 0.0001 by Student's t test. E, primary neurons derived from caspase-3−/− (Cas3−/−) newborn mice (E1 and E3) as well as control neurons from wild type mice (E2 and E4) were infected with SFV-RCAN1 for 14 h. TUNEL staining revealed less neuronal death in caspase-3−/− neurons (E1) than in wild type neurons (E2). Less neuronal death occurred in caspase-3−/− neurons treated with 10 μm dexamethasone (E3) than in wild type neurons (E4). Yellow color merged from green TUNEL and red propidium iodide staining indicates apoptotic cells (white arrow). F, quantification showed that neuronal death was significantly reduced in caspase-3−/− neurons compared with wild type neurons. Values represent mean ± S.E. (n = 6). *, p < 0.0001 by Student's t test. G, quantification of TUNEL staining showed that neuronal death induced by dexamethasone treatment was also significantly reduced in caspase-3−/− neurons compared with wild type neurons. Values represent mean ± S.E. (n = 6). *, p < 0.0001 by Student's t test. H, MTT showed that knock-out of caspase-3 blocked neuronal death induced by RCAN1 overexpression in neurons. Primary neurons derived from caspase-3−/− newborn mice were infected with SFV-GFP and SFV-RCAN1 for 16 h. Values represent mean ± S.E. (n = 4). p > 0.05 by Student's t test.
To further confirm the involvement of caspase-3 in RCAN1-mediated neuronal apoptosis, primary neuron cultures were derived from caspase-3 knock-out (caspase-3−/−) and wild type newborn mice (32). Caspase-3−/− and wild type neurons were infected with SFV-RCAN1 and further subjected to dexamethasone treatments. TUNEL staining showed markedly reduced neuronal apoptosis in neurons derived from the caspase-3−/− mice compared with neurons derived from wild type mice (0.60 ± 0.59% apoptosis ratio in caspase-3−/− mice compared with 23.91 ± 3.48% in wild type mice (p < 0.0001) (Fig. 6, E (E1 and E2) and F)). Similar results were observed in the presence of dexamethasone treatments (1.36 ± 0.67% versus 27.99 ± 2.39%, p < 0.0001) (Fig. 6, E (E3 and E4) and G). Furthermore, an MTT assay showed that the neuronal death induced by RCAN1 isoform 1 overexpression was abolished in caspase-3−/− primary neurons with or without dexamethasone treatment (Fig. 6H). These data demonstrate that disruption of the caspase-3 gene blocks dexamethasone-induced and RCAN1-mediated neuronal apoptosis in caspase-3−/− mice, and furthermore, caspase-3 is required for dexamethasone-induced and RCAN1-mediated neuronal apoptosis.
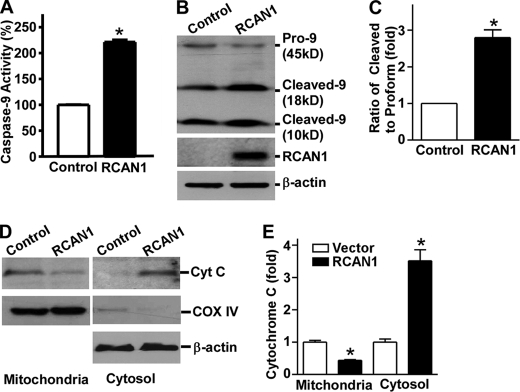
RCAN1 Overexpression Activates Caspase-9
Our data suggested that overexpression of RCAN1 isoform 1 can activate caspase-3, so we then performed the following experiments to determine the upstream signaling molecules by which caspase-3 is activated. Upstream initiators of caspase-3 include caspase-8, -10, and -9. The cytochrome c-mediated pathway is essential in CNS development and stress-induced cell death (63). Upon exposure to intracellular proapoptotic agents, cytochrome c is relocated from the mitochondria to the cytoplasm, where it binds to Apaf-1 to form the so-called apoptosome, which subsequently recruits procaspase-9 and facilitates autoactivation of caspase-9. The activated caspase-9 then cleaves the downstream effector caspases, such as caspase-3. To investigate whether caspase-9 initiates RCAN1 overexpression-induced caspase-3 activation, caspase-9 activity was measured using the Caspase-Glo® 9 assay. Caspase-9 activity was significantly increased by 2.18 ± 0.1-fold in RCAN1-1-overexpressing neurons (p < 0.005) (Fig. 7A). Furthermore, a Western blot assay revealed that there were higher levels of cleaved forms of caspase-9 and reduced levels of procaspase-9 in RCAN1-1-overexpressing HEK293 cells (Fig. 7B). The ratio of the cleaved form to the pro form was increased to 2.80 ± 0.22-fold by RCAN1-1 overexpression (p < 0.05) (Fig. 7C). The results indicate that RCAN1-1 overexpression in cells activates caspase-9, which then recruits and activates its downstream executor, caspase-3, which subsequently cleaves its substrates and initiates the neuronal apoptotic pathway.
FIGURE 7.
Activation of caspase-9 by RCAN1 overexpression. A, activity of caspase-9 was elevated due to overexpression of RCAN1 in primary neurons. Primary neurons were infected with SFV-RCAN1 or SFV-GFP. Values represent mean ± S.E. (error bars) (n = 4). *, p < 0.0001 by Student's t test. B, increased cleavage of caspase-9 in RCAN1-overexpressing HEK293 cells. Different forms of caspase-9 were separated with a 16% Tris-Tricine SDS-polyacrylamide gel and detected with anti-caspase-9 antibody, which detected procaspase-9 as well as the cleaved forms of caspase-9. RCAN1 was detected with 9E10 antibody. β-Actin detected by anti-β-actin served as a loading control. C, quantification of caspase-9 cleavages. Values indicate the ratio of cleaved form (18 kDa) to pro form. Values represent mean ± S.E. (n = 3). *, p = 0.0142 by Student's t test. D, RCAN1-1 expression increased translocation of cytochrome c from mitochondria to cytosol in HEK293 cells. Mitochondrial and cytosolic fractions were separated on a 16% Tris-Tricine SDS-polyacrylamide gel. Cytochrome c was detected by the anti-cytochrome c antibody (Cell Signaling). Anti-COX IV antibody (Cell Signaling) was used to detect COX IV in mitochondria. E, quantification of cytochrome c. In the mitochondrial fraction, cytochrome c was normalized with COX IV. In the cytosolic fraction, cytochrome c was normalized to β-actin. Values represent mean ± S.E. (n = 3). *, p < 0.01 by Student's t test.
Caspase-11 is another upstream initiator of caspase-3 and mainly associated with inflammation in murine (64, 65). Caspase-11 and caspase-3 activation have been observed in the spinal cord of mice with amyotrophic lateral sclerosis (66). To test if the activation of caspase-9 and caspase-3 by RCAN1 has a specific effect or if it has a panactivation effect on all of the caspase families, caspase-11 was examined in rat glial C6 cells transduced with SFV-GFP and SFV-RCAN1. Spleen tissue lysates derived from mice treated with LPS (40 mg/kg) were used as caspase-11 protein markers (67). A monoclonal anti-caspase-11 antibody was used to detect caspase-11, and our study showed no significant difference in SFV-GFP- and SFV-RCAN1-transduced C6 cells (data not shown). These results suggest that overexpression of RCAN1 can specifically activate caspase-9 and caspase-3 but not caspase-11.
Activation of caspase-9 requires cytochrome c release from the mitochondria. To investigate whether RCAN1 overexpression affects cytochrome c release, a Western blot assay was used to examine cytochrome c levels in cytosolic and mitochondrial fractions (Fig. 7D). RCAN1-1 overexpression in HEK293 cells resulted in significantly reduced levels of cytochrome c in the mitochondria and markedly increased levels in the cytosol (p < 0.01) (Fig. 7E). Taken together, our data suggest that the cytochrome c, caspase-9, and caspase-3 signaling pathway is activated by RCAN1-1 overexpression and mediates neuronal apoptosis induced by RCAN1-1 overexpression.
DISCUSSION
Prominent neuronal death, neuritic plaques, and neurofibrillary tangles are neuropathological hallmarks in AD brains (68). The neuronal loss is closely correlated to memory impairment in AD patients. Most studies in AD have been focused on the initial steps leading to neuritic plaque and neurofibrillary tangle formation in AD brains, whereas the mechanism underlying neuronal death still remains elusive. RCAN1 has been shown to play an important role in memory and learning. Although disruption of RCAN1 in mice does not show overt abnormalities in vivo (69), both disruption and overexpression of nebula, a RCAN1 ortholog in Drosophila, lead to severe learning and memory deficits (70). RCAN1 is highly expressed in neurons in the brain and is overexpressed in the brains of AD and DS patients (38, 40, 43). Recently, we reported that degradation of (RCAN1 is mediated by both the chaperone-mediated autophagy and ubiquitin proteasome pathways, and alteration of the autophagy and ubiquitin proteasome pathways could result in dysfunction of RCAN1 signaling (71). Knock-out of RCAN1 in mice resulted in increased enzymatic calcineurin activity and cleaved calcineurin fragments and caused learning and memory deficits and impaired late phase long term potentiation (45). The effect of RCAN1 on AD and DS pathogenesis is unknown.
Epidemiological studies have revealed that risk factors for AD, including aging, atherosclerosis, stroke, and diabetes, will induce stress and increase oxidative free radicals in the brain, resulting in neuronal death in patients. Furthermore, hippocampal atrophy and severity of cognitive impairment in AD patients are correlated with dysfunctions of the HPA axis (72). Corticosteroids can facilitate neuronal apoptosis in the hippocampus as well as memory impairment (73–76). In this report, we found that RCAN1 transcription is tightly regulated, and the stress hormone dexamethasone can specifically up-regulate RCAN1 gene expression by activating the neuronal tissue-enriched isoform 1 promoter of the RCAN1 gene. Neurons in the hippocampus and cortex express a high level of GRs. HPA hyperactivity may result in elevated glucocorticoids, which in turn can up-regulate RCAN1 gene expression by binding to GRE on the RCAN1-1 gene promoter. Indeed, our results showed that RCAN1 protein expression was significantly increased in cortical tissues from DS and AD patients. We showed that the caspase-9 and caspase-3 apoptotic pathways were activated by overexpression of RCAN1-1 in neurons. Previous studies also suggest that activation of caspase-3 increases Aβ production (77, 78). Moreover, Aβ42 can directly stimulate RCAN1 expression in a U-373 cell line (38). Our studies showed that RCAN1-1 overexpression activated caspase-3. Such results may suggest a vicious cycle of Aβ-RCAN1-caspase-Aβ and a link between Aβ accumulation and neuronal death in AD pathogenesis. RCAN1-1 protein was shown to inhibit calcineurin and protects against acute calcium-mediated stress damage, including transient oxidative stress, using hamster fibroblast HA-1 cells (44). It is also reported that disruption of RCAN1 in neurons protects neurons against oxidative stress (79), which is consistent with our findings using primary neuronal cultures. Our study implies that inhibition of RCAN1 may prevent neuronal death and benefit AD patients.
DS patients show AD-associated neuropathological changes in their 30s, which is 20–30 years earlier than sporadic AD cases (80). We previously reported that Aβ is abnormally elevated in brains of DS patients (81). In addition to overproduction and accumulation of Aβ, our current study implies that RCAN1, a gene localized on chromosome 21 and overexpressed in DS, may further contribute to the early onset of AD pathogenesis in DS due to its proapoptotic effect. Abnormal autophagy lysosome and ubiquitin proteasome pathways, resulting in protein accumulation, have been implicated in AD pathogenesis, and our recent report showed that RCAN1 degradation is mediated by both chaperone-mediated autophagy and ubiquitin proteasome pathways (71). Overexpression of RCAN1, due to its triplication in DS, can render neurons more vulnerable to stress and neurotoxic insults leading to hippocampal and cortical neuronal loss. We have demonstrated that Aβ further increases neuronal apoptosis in RCAN1-1-overexpressing neurons. AD pathology is characterized by a specific demise of hippocampal and cortical neurons; however, how the specific population of cells is affected remains elusive. Our immunohistochemistry data showed that RCAN1 is specifically expressed in cortical and hippocampal neurons. The specific expression pattern of RCAN1 may explain the particular population of neuronal death in AD because RCAN1 overexpression in neurons leads to neuronal death. Our study provides a novel mechanism by which RCAN1 functions as a mediator of stress and Aβ-induced neuronal death, and overexpression of RCAN1 due to an extra copy of the RCAN1 gene on chromosome 21 contributes to AD pathogenesis in DS. Our study suggests that inhibition of RCAN1 signaling may have pharmaceutical potential for reducing neuronal loss and treating cognitive impairments, thereby benefiting AD and DS patients.
It has been reported that the RCAN1 isoform 4 promoter can be regulated by the calcineurin-NFAT signaling pathway, thus forming a negative feedback loop in RCAN1 isoform 4 gene regulation because RCAN1 can inhibit calcineurin activity (14). The negative feedback loop in RCAN1 gene regulation implies that tight regulation of RCAN1 is vital to cell survival. The negative loop may also serve a self-protective function in cells by way of preventing RCAN1 overexpression, leading to cell demise. Previous studies showed that RCAN1 physically binds to calcineurin and inhibits its function (15, 17, 24, 82). Activation of caspase-3 by RCAN1 is probably not due to this known function because previous studies show that calcineurin overexpression induces caspase-3 activation and apoptosis (83, 84). Calcineurin promotes apoptosis via BAD dephosphorylation, and there is also evidence indicating that calcineurin inhibitors such as FK506 are neuroprotective (85, 86). This suggests that the proapoptotic effect of RCAN1 is independent of its function of inhibiting calcineurin and that RCAN1 is a multifunctional protein. A previous report showed that nebula, the RCAN1 homolog in Drosophila, is crucial for mitochondrial integrity and function (87). Our fractionation and confocal microscopy imaging data indicate that RCAN1 is localized in cytosol, mitochondria, and nucleus, suggesting that RCAN1 is a multifunctional protein. The mitochondrial localization of RCAN1 may contribute to its proapoptotic effect. The underlying mechanism by which RCAN1 overexpression triggers cytochrome c release and initiates cell death remains unknown. Future studies will elucidate the involvement of RCAN1 in the mitochondrial apoptotic pathway.
Acknowledgments
We thank the University of Maryland Brain and Tissue Bank for Developmental Disorders and the New York Brain Bank at Columbia University for Trisomy-21, AD, and control brain tissues. We thank Fang Cai, Kelley Bromley, and Odysseus Zis for technical assistance and helpful discussion.
This work was supported by the Canadian Institutes of Health Research, the Jack Brown and Family Alzheimer's Research Foundation, the Michael Smith Foundation for Health Research, and the National Natural Science Foundation of China (Distinguished Young Scholars (Oversea) Fund Grant 30528015) (to W. S.).
- DS
- Down syndrome
- AD
- Alzheimer disease
- NFAT
- nuclear factor of activated T cells
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- RLU
- relative luciferase units
- GR
- glucocorticoid receptor
- GRE
- glucocorticoid response element.
REFERENCES
- 1. Down J. L. (1866) London Hospital Reports 3, 259–262 [Google Scholar]
- 2. Glenner G. G., Wong C. W. (1984) Biochem. Biophys. Res. Commun. 122, 1131–1135 [DOI] [PubMed] [Google Scholar]
- 3. Glenner G. G., Wong C. W. (1984) Biochem. Biophys. Res. Commun. 120, 885–890 [DOI] [PubMed] [Google Scholar]
- 4. Hardy J., Selkoe D. J. (2002) Science 297, 353–356 [DOI] [PubMed] [Google Scholar]
- 5. Mattson M. P. (2004) Nature 430, 631–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Busciglio J., Yankner B. A. (1995) Nature 378, 776–779 [DOI] [PubMed] [Google Scholar]
- 7. Stadelmann C., Deckwerth T. L., Srinivasan A., Bancher C., Brück W., Jellinger K., Lassmann H. (1999) Am. J. Pathol. 155, 1459–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eckert A., Marques C. A., Keil U., Schüssel K., Müller W. E. (2003) Ann. N.Y. Acad. Sci. 1010, 604–609 [DOI] [PubMed] [Google Scholar]
- 9. Friedlander R. M. (2003) N. Engl. J. Med. 348, 1365–1375 [DOI] [PubMed] [Google Scholar]
- 10. Jacobs P. A., Baikie A. G., Court Brown W. M., Strong J. A. (1959) Lancet 1, 710. [DOI] [PubMed] [Google Scholar]
- 11. Lejeune J., Turpin R., Gautier M. (1959) Bull. Acad. Natl. Med. 143, 256–265 [PubMed] [Google Scholar]
- 12. Reeves R. H., Baxter L. L., Richtsmeier J. T. (2001) Trends Genet. 17, 83–88 [DOI] [PubMed] [Google Scholar]
- 13. Fuentes J. J., Pritchard M. A., Planas A. M., Bosch A., Ferrer I., Estivill X. (1995) Hum. Mol. Genet. 4, 1935–1944 [DOI] [PubMed] [Google Scholar]
- 14. Arron J. R., Winslow M. M., Polleri A., Chang C. P., Wu H., Gao X., Neilson J. R., Chen L., Heit J. J., Kim S. K., Yamasaki N., Miyakawa T., Francke U., Graef I. A., Crabtree G. R. (2006) Nature 441, 595–600 [DOI] [PubMed] [Google Scholar]
- 15. Chan B., Greenan G., McKeon F., Ellenberger T. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13075–13080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rothermel B. A., McKinsey T. A., Vega R. B., Nicol R. L., Mammen P., Yang J., Antos C. L., Shelton J. M., Bassel-Duby R., Olson E. N., Williams R. S. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3328–3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fuentes J. J., Genescà L., Kingsbury T. J., Cunningham K. W., Pérez-Riba M., Estivill X., de la Luna S. (2000) Hum. Mol. Genet. 9, 1681–1690 [DOI] [PubMed] [Google Scholar]
- 18. Rothermel B., Vega R. B., Yang J., Wu H., Bassel-Duby R., Williams R. S. (2000) J. Biol. Chem. 275, 8719–8725 [DOI] [PubMed] [Google Scholar]
- 19. van Rooij E., Doevendans P. A., Crijns H. J., Heeneman S., Lips D. J., van Bilsen M., Williams R. S., Olson E. N., Bassel-Duby R., Rothermel B. A., De Windt L. J. (2004) Circ. Res. 94, e18–26 [DOI] [PubMed] [Google Scholar]
- 20. Davies K. J., Ermak G., Rothermel B. A., Pritchard M., Heitman J., Ahnn J., Henrique-Silva F., Crawford D., Canaider S., Strippoli P., Carinci P., Min K. T., Fox D. S., Cunningham K. W., Bassel-Duby R., Olson E. N., Zhang Z., Williams R. S., Gerber H. P., Pérez-Riba M., Seo H., Cao X., Klee C. B., Redondo J. M., Maltais L. J., Bruford E. A., Povey S., Molkentin J. D., McKeon F. D., Duh E. J., Crabtree G. R., Cyert M. S., de la Luna S., Estivill X. (2007) FASEB J. 21, 3023–3028 [DOI] [PubMed] [Google Scholar]
- 21. Abbasi S., Lee J. D., Su B., Chen X., Alcon J. L., Yang J., Kellems R. E., Xia Y. (2006) J. Biol. Chem. 281, 7717–7726 [DOI] [PubMed] [Google Scholar]
- 22. Abbasi S., Su B., Kellems R. E., Yang J., Xia Y. (2005) J. Biol. Chem. 280, 36737–36746 [DOI] [PubMed] [Google Scholar]
- 23. Hilioti Z., Gallagher D. A., Low-Nam S. T., Ramaswamy P., Gajer P., Kingsbury T. J., Birchwood C. J., Levchenko A., Cunningham K. W. (2004) Genes Dev. 18, 35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vega R. B., Yang J., Rothermel B. A., Bassel-Duby R., Williams R. S. (2002) J. Biol. Chem. 277, 30401–30407 [DOI] [PubMed] [Google Scholar]
- 25. Genescà L., Aubareda A., Fuentes J. J., Estivill X., De La Luna S., Pérez-Riba M. (2003) Biochem. J. 374, 567–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lin H. Y., Michtalik H. J., Zhang S., Andersen T. T., Van Riper D. A., Davies K. K., Ermak G., Petti L. M., Nachod S., Narayan A. V., Bhatt N., Crawford D. R. (2003) Free Radic. Biol. Med. 35, 528–539 [DOI] [PubMed] [Google Scholar]
- 27. Lee E. J., Seo S. R., Um J. W., Park J., Oh Y., Chung K. C. (2008) J. Biol. Chem. 283, 3392–3400 [DOI] [PubMed] [Google Scholar]
- 28. Liu Q., Busby J. C., Molkentin J. D. (2009) Nat. Cell Biol. 11, 154–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stathatos N., Bourdeau I., Espinosa A. V., Saji M., Vasko V. V., Burman K. D., Stratakis C. A., Ringel M. D. (2005) J. Clin. Endocrinol. Metab. 90, 5432–5440 [DOI] [PubMed] [Google Scholar]
- 30. Minami T., Horiuchi K., Miura M., Abid M. R., Takabe W., Noguchi N., Kohro T., Ge X., Aburatani H., Hamakubo T., Kodama T., Aird W. C. (2004) J. Biol. Chem. 279, 50537–50554 [DOI] [PubMed] [Google Scholar]
- 31. Cano E., Canellada A., Minami T., Iglesias T., Redondo J. M. (2005) J. Biol. Chem. 280, 29435–29443 [DOI] [PubMed] [Google Scholar]
- 32. Kuida K., Zheng T. S., Na S., Kuan C., Yang D., Karasuyama H., Rakic P., Flavell R. A. (1996) Nature 384, 368–372 [DOI] [PubMed] [Google Scholar]
- 33. Sun X., He G., Song W. (2006) FASEB J. 20, 1369–1376 [DOI] [PubMed] [Google Scholar]
- 34. Sun X., Wang Y., Qing H., Christensen M. A., Liu Y., Zhou W., Tong Y., Xiao C., Huang Y., Zhang S., Liu X., Song W. (2005) FASEB J. 19, 739–749 [DOI] [PubMed] [Google Scholar]
- 35. Christensen M. A., Zhou W., Qing H., Lehman A., Philipsen S., Song W. (2004) Mol. Cell. Biol. 24, 865–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sun X., He G., Qing H., Zhou W., Dobie F., Cai F., Staufenbiel M., Huang L. E., Song W. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18727–18732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ma H., Xiong H., Liu T., Zhang L., Godzik A., Zhang Z. (2004) J. Neurochem. 88, 1485–1496 [DOI] [PubMed] [Google Scholar]
- 38. Ermak G., Morgan T. E., Davies K. J. (2001) J. Biol. Chem. 276, 38787–38794 [DOI] [PubMed] [Google Scholar]
- 39. Cook C. N., Hejna M. J., Magnuson D. J., Lee J. M. (2005) J. Alzheimers Dis. 8, 63–73 [DOI] [PubMed] [Google Scholar]
- 40. Harris C. D., Ermak G., Davies K. J. (2007) FEBS J. 274, 1715–1724 [DOI] [PubMed] [Google Scholar]
- 41. Yoshida N. L., Miyashita T. U. M., Yamada M., Reed J. C., Sugita Y., Oshida T. (2002) Biochem. Biophys. Res. Commun. 293, 1254–1261 [DOI] [PubMed] [Google Scholar]
- 42. Kawata M., Yuri K., Ozawa H., Nishi M., Ito T., Hu Z., Lu H., Yoshida M. (1998) J. Steroid Biochem. Mol. Biol. 65, 273–280 [DOI] [PubMed] [Google Scholar]
- 43. Fuentes J. J., Pritchard M. A., Estivill X. (1997) Genomics 44, 358–361 [DOI] [PubMed] [Google Scholar]
- 44. Ermak G., Harris C. D., Davies K. J. (2002) FASEB J. 16, 814–824 [DOI] [PubMed] [Google Scholar]
- 45. Hoeffer C. A., Dey A., Sachan N., Wong H., Patterson R. J., Shelton J. M., Richardson J. A., Klann E., Rothermel B. A. (2007) J. Neurosci. 27, 13161–13172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang J., Rothermel B., Vega R. B., Frey N., McKinsey T. A., Olson E. N., Bassel-Duby R., Williams R. S. (2000) Circ. Res. 87, E61–E68 [DOI] [PubMed] [Google Scholar]
- 47. Baek K. H., Zaslavsky A., Lynch R. C., Britt C., Okada Y., Siarey R. J., Lensch M. W., Park I. H., Yoon S. S., Minami T., Korenberg J. R., Folkman J., Daley G. Q., Aird W. C., Galdzicki Z., Ryeom S. (2009) Nature 459, 1126–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hampton T. (2005) JAMA 293, 284–285 [DOI] [PubMed] [Google Scholar]
- 49. Oh M., Rybkin I. I., Copeland V., Czubryt M. P., Shelton J. M., van Rooij E., Richardson J. A., Hill J. A., De Windt L. J., Bassel-Duby R., Olson E. N., Rothermel B. A. (2005) Mol. Cell. Biol. 25, 6629–6638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kluetzman K. S., Perez A. V., Crawford D. R. (2005) Biochem. Biophys. Res. Commun. 337, 595–601 [DOI] [PubMed] [Google Scholar]
- 51. Li Y. P., Bushnell A. F., Lee C. M., Perlmutter L. S., Wong S. K. (1996) Brain Res. 738, 196–204 [DOI] [PubMed] [Google Scholar]
- 52. Sousa N., Paula-Barbosa M. M., Almeida O. F. (1999) Neuroscience 89, 1079–1087 [DOI] [PubMed] [Google Scholar]
- 53. Shi Y. (2002) Mol. Cell 9, 459–470 [DOI] [PubMed] [Google Scholar]
- 54. Thornberry N. A., Lazebnik Y. (1998) Science 281, 1312–1316 [DOI] [PubMed] [Google Scholar]
- 55. Yuan J., Lipinski M., Degterev A. (2003) Neuron 40, 401–413 [DOI] [PubMed] [Google Scholar]
- 56. Shi Y. (2004) Cell 117, 855–858 [DOI] [PubMed] [Google Scholar]
- 57. Guo Q., Fu W., Xie J., Luo H., Sells S. F., Geddes J. W., Bondada V., Rangnekar V. M., Mattson M. P. (1998) Nat. Med. 4, 957–962 [DOI] [PubMed] [Google Scholar]
- 58. Su J. H., Deng G., Cotman C. W. (1997) J. Neuropathol. Exp. Neurol. 56, 86–93 [DOI] [PubMed] [Google Scholar]
- 59. Kitamura Y., Shimohama S., Kamoshima W., Matsuoka Y., Nomura Y., Taniguchi T. (1997) Biochem. Biophys. Res. Commun. 232, 418–421 [DOI] [PubMed] [Google Scholar]
- 60. Nishimura T., Akiyama H., Yonehara S., Kondo H., Ikeda K., Kato M., Iseki E., Kosaka K. (1995) Brain Res. 695, 137–145 [DOI] [PubMed] [Google Scholar]
- 61. Kitamura Y., Shimohama S., Kamoshima W., Ota T., Matsuoka Y., Nomura Y., Smith M. A., Perry G., Whitehouse P. J., Taniguchi T. (1998) Brain Res. 780, 260–269 [DOI] [PubMed] [Google Scholar]
- 62. Lakhani S. A., Masud A., Kuida K., Porter G. A., Jr., Booth C. J., Mehal W. Z., Inayat I., Flavell R. A. (2006) Science 311, 847–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jiang X., Wang X. (2004) Annu. Rev. Biochem. 73, 87–106 [DOI] [PubMed] [Google Scholar]
- 64. Kang S. J., Wang S., Kuida K., Yuan J. (2002) Cell Death Differ. 9, 1115–1125 [DOI] [PubMed] [Google Scholar]
- 65. Wang S., Miura M., Jung Y. K., Zhu H., Li E., Yuan J. (1998) Cell 92, 501–509 [DOI] [PubMed] [Google Scholar]
- 66. Kang S. J., Sanchez I., Jing N., Yuan J. (2003) J. Neurosci. 23, 5455–5460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang S., Miura M., Jung Y., Zhu H., Gagliardini V., Shi L., Greenberg A. H., Yuan J. (1996) J. Biol. Chem. 271, 20580–20587 [DOI] [PubMed] [Google Scholar]
- 68. Alzheimer A. (1906) Neurologisches Centralblatt 23, 1129–1136 [Google Scholar]
- 69. Rothermel B. A., Vega R. B., Williams R. S. (2003) Trends Cardiovasc. Med. 13, 15–21 [DOI] [PubMed] [Google Scholar]
- 70. Chang K. T., Shi Y. J., Min K. T. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15794–15799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Liu H., Wang P., Song W., Sun X. (2009) FASEB J. 23, 3383–3392 [DOI] [PubMed] [Google Scholar]
- 72. Pomara N., Greenberg W. M., Branford M. D., Doraiswamy P. M. (2003) Psychopharmacol. Bull. 37, 120–134 [PubMed] [Google Scholar]
- 73. de Quervain D. J., Henke K., Aerni A., Treyer V., McGaugh J. L., Berthold T., Nitsch R. M., Buck A., Roozendaal B., Hock C. (2003) Eur. J. Neurosci. 17, 1296–1302 [DOI] [PubMed] [Google Scholar]
- 74. Reagan L. P., McEwen B. S. (1997) J. Chem. Neuroanat. 13, 149–167 [DOI] [PubMed] [Google Scholar]
- 75. Newcomer J. W., Selke G., Melson A. K., Hershey T., Craft S., Richards K., Alderson A. L. (1999) Arch. Gen. Psychiatry 56, 527–533 [DOI] [PubMed] [Google Scholar]
- 76. Newcomer J. W., Craft S., Askins K., Hershey T., Bardgett M. E., Csernansky J. G., Gagliardi A. E., Vogler G. (1998) Psychoneuroendocrinology 23, 65–72 [DOI] [PubMed] [Google Scholar]
- 77. Tesco G., Koh Y. H., Tanzi R. E. (2003) J. Biol. Chem. 278, 46074–46080 [DOI] [PubMed] [Google Scholar]
- 78. Gervais F. G., Xu D., Robertson G. S., Vaillancourt J. P., Zhu Y., Huang J., LeBlanc A., Smith D., Rigby M., Shearman M. S., Clarke E. E., Zheng H., Van Der Ploeg L. H., Ruffolo S. C., Thornberry N. A., Xanthoudakis S., Zamboni R. J., Roy S., Nicholson D. W. (1999) Cell 97, 395–406 [DOI] [PubMed] [Google Scholar]
- 79. Porta S., Serra S. A., Huch M., Valverde M. A., Llorens F., Estivill X., Arbonés M. L., Martí E. (2007) Hum. Mol. Genet. 16, 1039–1050 [DOI] [PubMed] [Google Scholar]
- 80. Wisniewski K. E., Wisniewski H. M., Wen G. Y. (1985) Ann. Neurol. 17, 278–282 [DOI] [PubMed] [Google Scholar]
- 81. Sun X., Tong Y., Qing H., Chen C. H., Song W. (2006) FASEB J. 20, 1361–1368 [DOI] [PubMed] [Google Scholar]
- 82. Görlach J., Fox D. S., Cutler N. S., Cox G. M., Perfect J. R., Heitman J. (2000) EMBO J. 19, 3618–3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Asai A., Qiu J., Narita Y., Chi S., Saito N., Shinoura N., Hamada H., Kuchino Y., Kirino T. (1999) J. Biol. Chem. 274, 34450–34458 [DOI] [PubMed] [Google Scholar]
- 84. Shibasaki F., McKeon F. (1995) J. Cell Biol. 131, 735–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wang H. G., Pathan N., Ethell I. M., Krajewski S., Yamaguchi Y., Shibasaki F., McKeon F., Bobo T., Franke T. F., Reed J. C. (1999) Science 284, 339–343 [DOI] [PubMed] [Google Scholar]
- 86. Sharkey J., Butcher S. P. (1994) Nature 371, 336–339 [DOI] [PubMed] [Google Scholar]
- 87. Chang K. T., Min K. T. (2005) Nat. Neurosci. 8, 1577–1585 [DOI] [PubMed] [Google Scholar]