Abstract
The white rot fungus Phanerochaete chrysosporium, a saprophytic basidiomycete, possesses a large number of cytosolic glutathione transferases, eight of them showing similarity to the Omega class. PcGSTO1 (subclass I, the bacterial homologs of which were recently proposed, based on their enzymatic function, to constitute a new class of glutathione transferase named S-glutathionyl-(chloro)hydroquinone reductases) and PcGSTO3 (subclass II related to mammalian homologs) have been investigated in this study. Biochemical investigations demonstrate that both enzymes are able to catalyze deglutathionylation reactions thanks to the presence of a catalytic cysteinyl residue. This reaction leads to the formation of a disulfide bridge between the conserved cysteine and the removed glutathione from their substrate. The substrate specificity of each isoform differs. In particular PcGSTO1, in contrast to PcGSTO3, was found to catalyze deglutathionylation of S-glutathionyl-p-hydroquinone substrates. The three-dimensional structure of PcGSTO1 presented here confirms the hypothesis that it belongs not only to a new biological class but also to a new structural class that we propose to name GST xi. Indeed, it shows specific features, the most striking ones being a new dimerization mode and a catalytic site that is buried due to the presence of long loops and that contains the catalytic cysteine.
Keywords: Crystal Structure, Disulfide, Enzymes, Glutathione, Protein Structure
Introduction
Phanerochaete chrysosporium is considered as the model organism to study the physiology of white rot fungi, the only known microorganisms able to completely break down lignin to carbon dioxide and water (1–3). Analysis of the genome of this basidiomycete revealed a complex superfamily of cytochrome P450 monooxygenases representing around 1% of this genome (1, 4). The large size of this gene family is correlated with the extraordinary ability of this fungus to degrade and metabolize numerous recalcitrant compounds such as pesticides, polyaromatic hydrocarbons, halogenated aromatics, and textile dyes. Besides extracellular oxidative systems, including lignin and manganese peroxidases, cytochrome P450 monooxygenases are thought to be involved in the phase I of detoxication processes catalyzing the oxidation of various substrates (5–7). The oxidation of recalcitrant compounds is often followed by glutathione conjugation catalyzed by glutathione transferases (GSTs). The glutathione conjugates formed are less toxic and lipophilic than the parent compounds (8).
The distribution of GSTs in fungi has been recently investigated (9), highlighting that P. chrysosporium exhibits a large subfamily of cytosolic GSTs (at least 27 isoforms) in comparison with Saccharomyces cerevisiae for instance (11 isoforms). Based on primary structure similarity, these cytosolic GSTs belong to seven different classes as follows: Ure2p-like, GTT1, GTT2, EFBγ, MAK16, etherases, and Omega (GSTO)3 (9). Among the cytosolic GSTs, P. chrysosporium exhibits eight putative proteins showing homology with GSTs of the Omega class, named PcGSTO1 to PcGSTO8. In a previous phylogenetic analysis of fungal GSTs, we found that GSTOs fall into three different subclasses. Subclass 1 encloses PcGSTO1 and PcGSTO2, these latter being related to uncharacterized plant GST Lambda and bacterial GSTs (9). Members of subclasses II and III are related to the well characterized human HsGSTO1-1 (10, 11).
A major feature of GSTOs is the presence of a cysteinyl residue in the catalytic site present in the glutathione binding domain (G-site), explaining the deglutathionylation activities exhibited by these proteins. For instance, HsGSTO1-1 has been shown to remove glutathione from S-(phenacyl)glutathione (12), as this activity requires the presence of the catalytic Cys32 (13). In addition, GSTOs have been shown to be involved in the reduction of methyl and dimethyl arsonate, an intermediate in the arsenic biotransformation (14, 15). Bacterial GSTOs, which belong to subclass I, have been shown to be involved in pentachlorophenol catabolism acting as S-glutathionyl-(chloro)hydroquinone reductase; this activity was also related to the presence of a catalytic cysteine (16, 17). In fungi, few studies have been devoted to this GST class except in S. cerevisiae (ScGSTO). The three ScGSTOs belong to the subclass I and have been partially characterized particularly at the protein level. The three proteins exhibit glutaredoxin-like activities because they are able to reduce both hydroxyethyldisulfide and dehydroascorbate in the presence of glutathione. In addition, the transcription of the three genes is induced by oxidative stress (18, 19). Moreover, ScGSTO1 has been shown to be involved in sulfur metabolism and located in peroxisomes. PcGSTO1 and PcGSTO2, nevertheless, differ from their yeast counterparts particularly at the H-site (the domain thought to interact with the hydrophobic substrate).
From phylogenetic analysis and biochemical analysis, Xun et al. (17) recently proposed that the Omega subclass I members constitute a new class of GSTs named S-glutathionyl-(chloro)hydroquinone reductase (GHR) due to their activity with S-glutathionyl-hydroquinones.
The aim of this study was the investigation of the PcGSTOs, a class of GSTs that is over-represented in P. chrysosporium. Recombinant proteins of isoforms that belong to the GSTO subclasses I and II have been biochemically characterized, exhibiting substrate specificity. Furthermore, the three-dimensional structure of PcGSTO1 has been solved, allowing the first description of a S-glutathionyl-p-hydroquinone reductase at the structural level. From these biochemical and structural data, the potential functions of the different isoforms are discussed.
EXPERIMENTAL PROCEDURES
Materials
Hydroxyethyldisulfide (HED) and 5,5′-dithiobis-2-nitrobenzoic acid were from Aldrich and Pierce, respectively. All other reagents were from Sigma.
Cloning and Construction of Pcgsto1 and Pcgsto3 Mutants by Site-directed Mutagenesis
The open reading frame sequences encoding P. chrysosporium PcGSTO1 and PcGSTO3 were amplified from P. chrysosporium cDNA using Pcgsto1 and Pcgsto3 forward and reverse primers (see supplemental Table 1) and cloned into the NcoI and BamHI restriction sites (underlined in the primers) of pET3d (Novagen). For PcGSTO3, the amplified sequence encoded a protein in which an alanine has been added during cloning. Using two complementary mutagenic primers, the two cysteines of PcGSTO3 were individually substituted into serines, and the primers are listed in supplemental Table 1. For PcGSTO1, due to the presence of the signal peptide, a first construction has been made, and the resulting protein starts with the N-terminal sequence MATTICLRH. Because we obtained a mixture of different proteins after production and purification (see under “Results”), a new construction has been made. The resulting protein with an additional alanine started thus with the N-terminal sequence MASFTTGST (see supplemental Fig. 1). PcGSTO1 used for structure determination consists also of the 352 last residues of the full-length protein.
Expression and Purification of the Recombinant Proteins
For protein production, the Escherichia coli BL21(DE3) strain that contained the pSBET plasmid was co-transformed with the different recombinant plasmids (20). Cultures were progressively amplified up to 2.4 liters in LB medium supplemented with ampicillin and kanamycin at 37 °C. Protein expression was induced at exponential phase by adding 100 μm isopropyl β-d-thiogalactopyranoside for 4 h at 37 °C. The cultures were centrifuged for 15 min at 4400 × g. The pellets were resuspended in 30 ml of TE NaCl buffer (30 mm Tris-HCl, pH 8.0, 1 mm EDTA, 200 mm NaCl), and the suspension was stored at −20 °C.
Cell lysis was performed by sonication (three times for 1 min with intervals of 1 min), and the soluble and insoluble fractions were separated by centrifugation for 30 min at 27,000 × g. The soluble part was then fractionated with ammonium sulfate in two steps, and the protein fraction precipitated between 40 and 80% of the saturation contained the recombinant protein, as estimated by 15% SDS-PAGE. The protein was purified by size exclusion chromatography after loading on an ACA44 (5 × 75 cm) column equilibrated in TE NaCl buffer. The fractions that contained the protein were pooled, dialyzed by ultrafiltration to remove NaCl, and loaded onto a DEAE-cellulose column (Sigma) in TE buffer (30 mm Tris-HCl, pH 8.0, 1 mm EDTA). The proteins were eluted using a 0–0.4 m NaCl gradient. Finally, the fractions of interest were pooled, dialyzed, concentrated by ultrafiltration under nitrogen pressure (YM10 membrane; Amicon), and stored in TE buffer at −20 °C. Purity was checked by SDS-PAGE. Protein concentrations were determined spectrophotometrically using a molar extinction coefficient at 280 nm of 64,860 m−1·cm−1 for the PcGSTO1 WT, C86S, and 51340 m−1·cm−1 for PcGSTO3 WT and C37S.
E. coli strain B121 containing the pET-Pcgsto1 plasmid was cultured as described previously (21) and was used as a source of selenomethionine-substituted form of PcGSTO1 (SeMet-PcGSTO1). The purification of SeMet-PcGSTO1 was performed following a procedure similar to that described for PcGSTO1.
Mass Spectrometry Analysis
PcGSTOs WT and mutated were analyzed by quadrupole-TOF MS as described by Koh et al. (22).
Fluorescence Properties of Wild-type and Mutated PcGSTOs
The fluorescence characteristics of PcGSTOs in the reduced and oxidized forms were recorded with a spectrofluorometer (Cary Eclipse; VARIAN) in TE buffer at a protein concentration of 10 μm.
Determination of Free Thiol Groups
The number of free thiol groups in either untreated, denatured, or reduced conditions was determined spectrophotometrically with 5,5′-dithiobis-2-nitrobenzoic, as described by Koh et al. (22). All thiol titrations were performed on enzymes either as purified or after SDS treatment or after dithiothreitol (DTT) reduction. In the latter case, proteins were first reduced by incubation with 10 mm DTT for 30 min at room temperature, then precipitated with 1 volume of 20% (w/v) trichloroacetic acid (TCA), and finally stored on ice for 30 min. The mixture was then centrifuged for 10 min at 13,000 × g, and the pellets were washed twice with 2% (w/v) TCA. The precipitate was resuspended in 30 mm Tris-HCl, pH 7.0, 1 mm EDTA, and 1% (w/v) SDS. The concentration was estimated by UV spectrophotometry. For denatured proteins, samples were incubated in the presence of 2% SDS treatment during 30 min before 5,5′-dithiobis-2-nitrobenzoic addition.
Activity Measurements
The activity measurements of WT or mutant PcGSTOs in the HED assay or for reduction of dehydroascorbate (DHA) were performed as described by Couturier et al. (23). Dimethylarsinate reductase and glutathionyl-phenylacetophenone assay were performed as described previously (12, 13). The GST activity was measured with 0.2 mm CDNB as the substrate in 30 mm Tris buffer, pH 8.0, and the continuous absorbance at 340 nm was monitored. S-Glutathionyl-p-hydroquinone reductase activity was monitored as follows: 1 mm 1,4-benzoquinone in 30 mm Tris-HCl, pH 8, was used as base line for an absorbance spectrum (230–400 nm). After 2 min, 1 mm reduced glutathione (GSH) was added to the reaction mixture, and the spectra were monitored every minute during 5 min. At last, purified recombinant proteins were added in the reaction mixture, and spectra were recorded every minute. In complementary experiments, 1 mm 1,4-benzoquinone in 30 mm Tris-HCl, pH 8, was incubated in the presence of 1 mm GSH, 200 μm NADPH, 0.5 IU glutathione reductase before the addition of the protein of interest. The activity was followed by monitoring the decrease in absorbance arising from NADPH oxidation in this coupled enzyme assay system that showed the formation of glutathione disulfide (GSSG).
In complementary experiments, S-(phenacylacetophenone)glutathione (PAP-SG) and 2-methyl-S-glutathionyl-naphthoquinone (thiodione) were prepared as described previously (24, 25), and the purity and the nature of the products were verified by mass spectrometry. Activities of the WT and variant enzymes were assayed by following the appearance of deglutathionylated menadione (at 425 nm) and phenylacetophenone (at 290 nm). The reactions were performed in 30 mm Tris-HCl, pH 8, containing GSH (5 mm) and substrates (from 15 to 2 mm) in a final volume of 650 μl and initiated by adding WT or mutant enzyme. The concentration of enzyme added was adjusted based on preliminary activity experiments, ranging from 0.1 and 1 μm. The reactions were stopped by adding 350 μl of ethanol, then strongly mixed by vortex, then centrifuged at 14,500 rpm during 15 min, and analyzed by HPLC using a Gemini 5-μm C18 column (150 mm long × 4.6 mm inner diameter) at 22 °C.
To visualize the deglutathionylation reactions, PAP-SG or thiodone (15 μm) was added in a 650-μl reaction mixture containing 30 mm Tris-HCl, pH 8, and 15 μm enzyme. After stopping the reaction by adding ethanol, samples were analyzed by HPLC as described above.
Size Exclusion Chromatography
Size exclusion chromatography experiments have been performed using a Superose 12HR column connected to a fast protein liquid chromatography (FPLC) system. The column was equilibrated with 30 mm Tris-HCl buffer, pH 8, containing 0.15 m NaCl, and the proteins were eluted in the same buffer at a flow rate of 0.25 ml·min−1.
Crystallization and X-ray Diffraction Data Collection
Prior to crystallization, protein samples were concentrated to 40 mg·ml−1 PcGSTO1, in TE buffer. Crystals of the SeMet-PcGSTO1 were obtained by using the microbatch method. 1.5 μl of the PcGSTO1 solution at various concentrations and 1.5 μl of various crystallization solutions were deposited in a 72-well Microbatch plate (Hampton Research) pre-filled with paraffin oil and stored at 4 °C. Preliminary results have been obtained from two commercial screening solutions as follows: the condition B2 from the JBScreen classic kit 2 (Jena Bioscience) and the Crystal Screen solution 6 from Hampton Research. Further trials aimed to improve reproducibility and crystal quality gave the best results for PcGSTO1 solutions at 30–40 mg·ml−1 and crystallization solutions composed of 20–30% polyethylene glycol 4000 (PEG 4000), 0 to 0.2 m magnesium or calcium chloride, and 0.1 m Tris-HCl, pH 8.5. However, polycrystals grew frequently so that only rare portions of them were suitable for x-ray data collection. No crystal of nonselenomethionylated PcGSTO1 suitable for data collection has been obtained.
The diffraction data were collected at 100 K on the synchrotron beamline FIP-BM30A (26) at ESRF (France) with a wavelength of 0.9805 Å corresponding to the peak of the Se K-edge, from a crystal grown with 40 mg·ml−1 PcGSTO1, 25% PEG 4000, and no salt, and flash-frozen after a quick immersion in the crystallization solution mixed with 20% glycerol. The data sets were processed with XDS (27) and scaled with SCALA from the CCP4 Package (28). Statistics are summarized in Table 1.
TABLE 1.
Statistics of X-ray diffraction data collection and model refinement
| Data collection | |
| Space group | C2 |
| Cell dimensions | a, 166.8 Å, b, 70.3 Å, c, 72.6 Å; β, 98.8° |
| Resolution | 46.66 to 2.30 Å (2.43 to 2.30 Å)a |
| Rmerge | 3.7% (18.8%) |
| I/σI | 19.8 (4.8) |
| Completeness | 98.9% (96.0%) |
| Redundancy | 4.3 |
| Refinement | |
| Resolution | 46.66 to 2.30 Å |
| No. of reflections | 36,622 |
| Rall/Rfreeb | 19.2/22.8 |
| No. of atoms | |
| Protein | 2572 (monomer A) + 2451 (monomer B) |
| Ligand | 1 glutathione (20 atoms) in monomer A |
| Water | 223 |
| B-factors | |
| Protein | 37.9 (monomer A); 41.6 (monomer B) |
| Ligand | 49.4 |
| Water | 38.6 |
| Ramachandran statistics | |
| Residues in preferred regions | 95.7% |
| Residues in allowed regions | 4.3% |
| Outlier residues | 0.0% |
| Root mean square deviations | |
| Bond lengths | 0.011 Å |
| Bond angles | 1.25° |
a Values in parentheses are for highest resolution shell.
b Rall is determined from all the reflections (working set + test set), and Rfree corresponds to a subset of 5% of reflections (test set).
Structure Solution and Quality of the Model
The PcGSTO1 structure was solved using the single wavelength anomalous dispersion protocol of Auto-Rickshaw at the EMBL-Hamburg automated crystal structure determination platform (29). The input diffraction data were prepared and converted for use in Auto-Rickshaw using programs of the CCP4 suite (28). Nine heavy atoms among the 10 requested were found using the program SHELXD (30). Initial phases were calculated after density modification using the program SHELXE (31), at 3.0 Å resolution. The initial phases were improved using density modification and phase extension to 2.30 Å resolution using the program DM (32). 70% of the model was built by ARP/wARP (33, 34). It was progressively completed and refined by using iteratively COOT (35) and REFMAC5 (36), including diffraction data up to 2.3 Å resolution. An electron density further attributed to one GSH bound in the active site appeared in monomer A during refinement, although no GSH was added to crystallize PcGSTO1. On the contrary, the corresponding area of monomer B only displayed a few poor spherical densities that more probably corresponded to water molecules. Final model statistics are shown in Table 1.
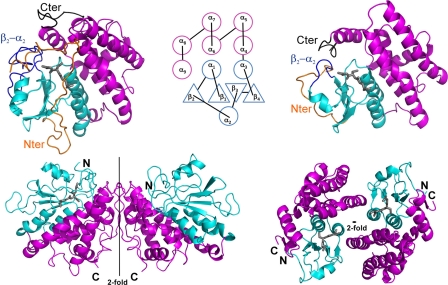
Most residues corresponded to well defined electron density, except for the N-terminal part of both monomers that appeared too disordered to be observed. Thus, models of monomers A and B contained residues Glu38–Asp351 and residues Phe54–Asp351, respectively. Furthermore, the residues Glu38–Ser53 only observed in monomer A displayed a weaker electron density and higher B factors with respect to the rest of the structure. Figs. 4 and 5 were prepared by using PyMOL.
FIGURE 4.
Schematic drawing of the PcGSTO1 (left) and HsGSTO1-1 (10) (right) structures. The monomers are displayed at the top separated by their common topology diagram mentioning the secondary structure numbering; the dimers are shown at the bottom, with orientations chosen to render their assembling at most. In each case, the thioredoxin-like domain is colored in cyan, and the α-helix domain is in purple. In monomers, the other colors emphasize the features that distinguish PcGSTO1 from HsGSTO1-1 as follows: the N-terminal coil in orange, the loop that connects the strand β2 to the α-helix α2 in blue, and the C-terminal coil in black. The gray sticks correspond to glutathione, only present in one monomer of PcGSTO1 and in the two monomers of HsGSTO1-1.
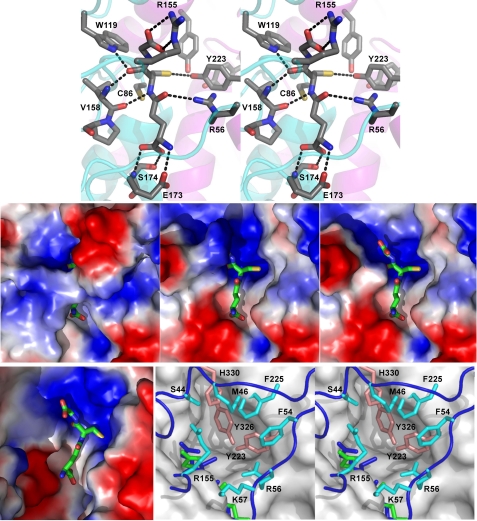
FIGURE 5.
Substrate-binding sites of PcGSTO1 and HsGSTO1-1 (10). Top, stereo view of the hydrogen bond network formed by the active site residues around GSH in PcGSTO1. The schematic trace of the polypeptide main chain is suggested in cyan (thioredoxin-like domain) and purple (α-helix domain). Middle, PcGSTO1 electrostatic surface (blue, positive charge; red, negative charge) of PcGSTO1 in the region of GSH (shown as sticks). Left panel, calculated with all the residues observed for monomer A in the crystallographic model (Glu38–Asp351); middle panel, calculated without the N-terminal coil (i.e. with residues Tyr78–Asp351); right panel, same as previous but without Arg155. Bottom, HsGSTO1-1 active site compared with PcGSTO1. Left panel, electrostatic surface calculated for the HsGSTO1-1dimer, in the region of GSH (shown as sticks). Right panel, stereo view of the accessible surface of HsGSTO1-1 (gray) in the region of GSH (green), superimposed to the PcGSTO1 active site containing the GSH (blue sticks). Residues Tyr223, Tyr326, and His330 fill the pocket defined by Board et al. (10) as the HsGSTO1-1 active site. Residues in cyan and in orange form a second and a third layers of residues that almost close the PcGSTO1 active site.
RESULTS
PcGSTO1 belongs to the first subclass of GSTs Omega displaying a CPWATR potential active site compatible with the CP(W/F)(A/T)(H/Q)R characteristic of the members of this subclass (Fig. 1, top and bottom) (9). According to the prediction softwares, PcGSTO1 possesses a signal peptide targeting to mitochondria. Different attempts to produce heterologously the full-length protein in E. coli resulted in a mixture of truncated proteins and suggested that this N-terminal extension contained putative cleavage sites recognized by the E. coli machinery or an intrinsic instability of the precursor. These data were in accordance with the presence of a signal peptide in the N-terminal part of the protein as shown previously for different other redox proteins (37, 38). To obtain a homogeneous protein preparation, a shorter form of PcGSTO1 was produced and purified removing the N-terminal extension of the protein (46 amino acids). Nevertheless, the measured enzymatic activities and specificities (full-length and without signal peptide) described in this study were similar for both forms (data not shown).
FIGURE 1.
Alignment of PcGSTO1 (top) and PcGSTO3 (bottom) with homologs. Amino acids involved in the catalytic site are labeled. NCBI accession numbers are as follows: PcGSTO1 (EU791894), PcGSTO3 (EU 791893), PcGSTO4 (JGI accession number 7168), Sphingobium chlorophenolicum PcpF (AAM 96671), Escherichia coli YqjG (NP_417573), and Homo sapiens GSTO1–1 NP_004823. For PcGSTO1, the putative signal peptide is in italic.
PcGSTO3 belongs to the subclass II of the GSTs Omega with an FCPFVQ active site quite similar to the FCPFAE of the HsGSTO1-1 (Fig. 1, bottom). A second cysteinyl residue is present at the same position into both proteins (Cys94, PcGSTO3 numbering). Different attempts have been made to produce heterologously the mutated protein PcGSTO3 C94S, but they remained unsuccessful because the resulting protein was fully insoluble.
Thiol Content Determination
To investigate the putative role of the conserved cysteines in the catalytic mechanism of both proteins, site-directed mutagenesis has been used to produce the recombinant proteins in E. coli, PcGSTO1 C86S and PcGSTO3 C37S. The thiol content of these proteins has been determined in different conditions as follows: native oxidized corresponding to nonreducing and nondenaturing conditions; denatured oxidized corresponding to a titration in the presence of SDS; reduced and denatured corresponding to a reduction followed by a protein precipitation (see “Experimental Procedures”). A summary of these data is shown in Table 2. Nearly three and two thiols per protein were titrated for the reduced and denatured PcGSTO1 WT and PcGSTO3 WT proteins, respectively, in accordance with the expected thiol content. Under denaturing conditions and after reduction, the obtained values for PcGSTO1 C86S (around two thiols per protein) and PcGSTO3 C37S (around one thiol per protein) were in agreement with the theoretical values. Comparison of values obtained for denatured PcGSTO1 WT and C86S suggested that the sulfur of Cys86 was involved at least partially in a DTT-reducible link. For PcGSTO3 WT, one cysteinyl residue was probably involved in a disulfide bridge, whereas for PcGSTO3 C37S, one additional thiol group seemed to be noncovalently bound in the protein.
TABLE 2.
Number of free thiols in WT and mutated PcGSTOs under various redox conditions
The native column is indicative of the protein thiol content measured after purification. Proteins were denatured using SDS treatment or reduced by DTT and subsequently precipitated using TCA. The thiol content per protein was quantified by 5,5-dithiobis(nitrobenzoic acid). Data are represented as mean ± S.D. (n ± 4).
| Native | Denatured | Reduced and denatured | |
|---|---|---|---|
| PcGSTO1 | 0.46 ± 0.12 | 2.50 ± 0.25 | 3.0 ± 0.2 |
| PcGSTO1 C86S | 0.15 ± 0.03 | 2.0 ± 0.1 | 2.04 ± 0.12 |
| PcGSTO3 | 0.15 ± 0.03 | 1.00 ± 0.03 | 1.80 ± 0.13 |
| PcGSTO3 C37S | 0.92 ± 0.09 | 1.78 ± 0.12 | 1.00 ± 0.04 |
Fluorescence Spectrometry and Mass Analysis
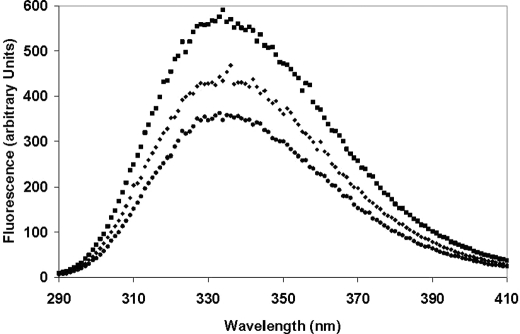
PcGSTO3 contains two tryptophans adjacent to the active site. Given that proximity, we have investigated whether the intrinsic fluorescence of PcGSTO3 could change under reducing (DTT) or oxidizing (GSSG or diamide) conditions (Fig. 2). The native protein displayed an emission spectrum with a maximum at 350 nm characteristic of a fluorescence signal strongly dominated by Trp. Adding 400 μm DTT to native PcGSTO3 immediately led to a strong increase of the fluorescence signal (Fig. 2). The addition of GSSG (10 mm) on previously DTT-reduced PcGSTO3 strongly decreased the fluorescence signal, whereas addition of diamide (10 mm) did not change the reduced PcGSTO3 fluorescence spectrum (data not shown). Similar experiments have been performed with PcGSTO3 C37S; however, in this case no changes in the fluorescence spectrum have been obtained whatever the reductant and oxidant used. In accordance with thiol titrations, these data suggested that the Cys37 of the native PcGSTO3 WT was glutathionylated and that the tryptophan environment strongly changed after glutathionylation of this active site cysteine.
FIGURE 2.
Fluorescence spectra of PcGSTO3 under different redox states. Emission spectra of native (♦), DTT-reduced (■), and subsequently GSSG-oxidized (●) PcGSTO3 (excitation at 290 nm) were recorded with 10 μm protein at 25 °C in TE buffer, pH 8.0,. The reduced PcGSTO3 was obtained by a treatment of 10 μm protein with 400 μm DTT. PcGSTO3 was oxidized using 10 mm GSSG after DTT reduction.
To confirm this hypothesis, PcGSTO3 WT and PcGSTO3 C37S were analyzed by mass spectrometry. A Q-TOF analysis of the DTT-reduced PcGSTO3 revealed a single protein with a molecular mass of 29,615 Da, which was consistent with a protein where the N-terminal methionine was cleaved. After treatment with GSSG, the mass of the protein increased by around 305 Da, a feature consistent with the formation of one glutathione adduct. This obtained value of 29,921 Da also corresponded to the mass of the native protein, and thus confirmed this latter also possessed a glutathione adduct on Cys37. For the mutant PcGSTO3 C37S, the analysis revealed a single protein with a molecular mass of 29,599 Da, which was consistent with a protein without the first methionine and which harbored the Cys to Ser mutation. This result combined with the thiol titration was in agreement with the noncovalent binding of one reduced glutathione to PcGSTO C37S.
Similar experiments (fluorescence and mass spectrometry) have been conducted with PcGSTO1 and PcGSTO1 C86S. Even when a tryptophan residue was present near the putative catalytic cysteine, the fluorescence spectrum remained unchanged whatever the reductant (GSH/DTT) or the oxidant (GSSG/diamide) used. A Q-TOF analysis of the DTT-reduced protein revealed a single protein with a molecular mass of 40,187 Da, which was consistent with a protein where the methionine was cleaved. After treatment with GSSG, the mass of the reduced protein increased by around 306 Da, a feature consistent with the formation of one glutathione adduct. This obtained value of 40,493 Da corresponded also to the mass of the native protein and confirmed this latter also possessed a glutathione adduct. These data combined with thiol titration results were consistent with the potential glutathionylation of Cys86, because no glutathione adduct was observed on PcGSTO1 C86S.
Substrate Specificity between PcGSTO1 and PcGSTO3
Both PcGSTOs were found to be active in the two classical glutaredoxin assays as follows: the reduction of DHA and HED assays. The activity of the PcGSTOs in these two assays increased linearly with increasing protein concentrations in the 0–750 and 0–100 nm concentration ranges, respectively. As expected, these observed “thiol transferase” activities are related to the presence of a cysteine in their catalytic site, because the mutants PcGSTO1 C86S and PcGSTO3 C37S are fully inactive in these assays (data not shown). The kinetic analyses revealed catalytic efficiency values (kcat/Km) similar between both PcGSTOs against DHA, whereas PcGSTO1 exhibited a slightly stronger activity against HED than PcGSTO3 (Table 3). Interestingly, whereas PcGSTO1 C86S remained inactive in all tested assays, PcGSTO3 C37S exhibited a classical glutathione transferase activity detected using CDNB as the substrate. Both PcGSTOs WT were fully inactive against CDNB.
TABLE 3.
Kinetic parameters of PcGSTO1 and PcGSTO3 in HED, DHA, and CDNB activity assays
The apparent Km value for GSH in the DHA assay was determined using a GSH concentration range of 0.2–4 mm in the presence of 2 mm DHA for PcGSTO1 and PcGSTO3. The apparent Km value for GSH was determined for PcGSTO3 C37S in the CDNB assay using a GSH concentration range of 0.1–2 mm in the presence of 2 mm CDNB. The apparent Km value for DHA was determined using a concentration range of 0.1–4 mm in the presence of 3 mm GSH. The apparent turnover values (kcat) for HED were determined in the presence of 4 mm GSH and 4 mm HED. The apparent Km value for CDNB was determined using a concentration range of 0.1–2 mm in the presence of 2 mm GSH. Concerning PAP-SG and thiodione, the apparent Km values were determined using a substrate concentration range of 0.015–4 mm. The apparent Km and kcat values were calculated by nonlinear regression using the Michaelis-Menten equation. Data are represented as mean ± S.D. (n ± 3). ND means not detected. The detection limit was estimated at 0.5 mIU for spectrophotometric tests, whereas 0.5 nmol of either menadione or phenacylacetophenone was detected using analytical HPLC.
| GSH | HED | DHA | CDNB | PAP-SG | Thiodione | |
|---|---|---|---|---|---|---|
| Km (mm) | ||||||
| PcGSTO1 | 1.65 ± 0.33 | 1.84 ± 0.40 | ND | ND | 2.00 ± 0.62 | |
| PcGSTO3 | 1.44 ± 0.25 | 0.33 ± 0.07 | ND | 0.47 ± 0.19 | ND | |
| PcGSTO3 C37S | 0.18 ± 0.04 | 0.97 ± 0.27 | ND | ND | ||
| kcat (s−1) | ||||||
| PcGSTO1 | 2.67 ± 0.01 | 1.81 ± 0.17 | ND | 19.75 ± 2.31 | ||
| PcGSTO3 | 15.75 ± 0.26 | 0.97 ± 0.06 | 19.01 ± 2.89 | ND | ||
| PcGSTO3 C37S | ND | ND | 18.43 ± 1.90 | ND | ND | |
The HsGSTO1.1, ortholog of PcGSTO3 has been shown to possess activity against various substrates, including dimethylarsinate and 2-bromo-4′-phenylacetophenone (12, 13). In both cases, a glutathione adduct occurred followed by a deglutathionylation reaction catalyzed by the enzyme, preventing access to the initial substrate concentrations and also to kinetic parameters. As expected, PcGSTO3 was active in these tests in the presence of glutathione, whereas PcGSTO3 C37S, PcGSTO1 WT, and PcGSTO1 C86S were fully inactive (data not shown). To determine the catalytic parameters, PAP-SG was synthesized and purified, and the activity against this substrate was followed by analytical HPLC in the presence of GSH (Table 3). In these conditions, only PcGSTO3 is active with a catalytic efficiency of around 4·105 m−1 s−1, a value greater than that obtained for HsGSTO1-1 (104 m−1 s−1) (39). The incubation of previously reduced PcGSTO3 in the presence of PAP-SG without addition of GSH led to the formation of phenacylacetophone detected by analytical HPLC (supplemental Fig. 2A). This deglutathionylated product is not observed either in the presence of native purified protein (i.e. with a glutathione adduct) or in the presence of PcGSTO3 C37S. These results are in accordance with the involvement of Cys37 in the deglutathionylation activity leading to the formation of a disulfide bridge with the glutathione removed from PAP-SG. This hypothesis was confirmed by mass spectrometry analysis of the resulting protein showing the presence of a glutathione adduct (data not shown).
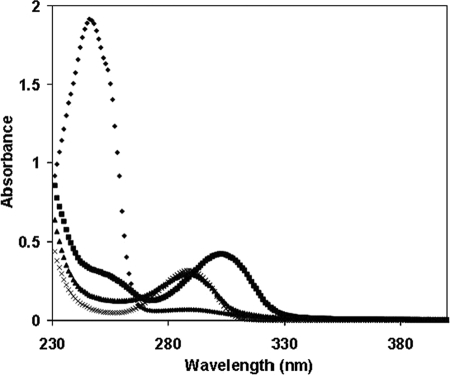
In additional experiments, we tested quinones previously incubated with reduced glutathione as substrates for PcGSTOs. In these conditions, Michael addition occurred leading to the formation of glutathione adducts corresponding to S-glutathionyl-p-hydroquinones (40, 41). As shown in Fig. 3, addition of reduced glutathione strongly modified the absorbance spectrum of 1,4-benzoquinone with the appearance of a peak maximum at 305 nm in accordance with the formation of S-glutathionyl-p-benzohydroquinone as described previously (42). Addition of PcGSTO1 led to a strong modification of the absorbance spectrum, with the disappearance of the 305-nm peak and the formation of a peak at 290 nm corresponding to p-benzohydroquinone. In contrast, no effect was observed in the presence of PcGSTO1 C86S and PcGSTO3 (data not shown). All these data were consistent with a deglutathionylation reaction of S-glutathionyl-p-benzohydroquinone. As expected, the activity of PcGSTO1 against S-glutathionyl-p-benzohydroquinone led to the production of GSSG as revealed by a coupled enzymatic test performed using glutathione reductase (data not shown). Similar data were obtained using menadione (2-methyl-1,4-naphthoquinone), the glutathionylated product (S-glutathionyl-naphthoquinone) being characterized by an absorbance peak at 425 nm (data not shown) (43). The obtained results suggested a preference of PcGSTO1 for aromatic substrates in contrast to PcGSTO3. To obtain catalytic parameters, S-glutathionyl-naphthoquinone, also called thiodione, was synthesized and purified, and the activity against this substrate was followed by analytical HPLC (Table 3). The activity of PcGSTO1 against thiodione leads to the formation of a product that elutes with the same retention time as menadione (data not shown). The kcat value with this substrate is around 10-fold higher than the values obtained with HED and DHA. No activity against thiodione was detected using PcGSTO1 C86S, confirming the importance of the cysteinyl residue in the catalysis of the deglutathionylation reaction. In additional experiments, no glutathione was added in a mixture containing PcGSTO1 and thiodone, the results of the reaction being analyzed by HPLC (supplemental Fig. 2B). In these conditions, addition of reduced PcGSTO1 led to the formation of menadione. Analysis by mass spectrometry of the resulting protein showed the presence of a glutathione adduct (data not shown). No menadione formation was observed when PcGSTO1 C86S was used.
FIGURE 3.
PcGSTO1 activity against S-glutathionyl-p-benzohydroquinone. Absorption spectra have been recorded adding successively in 1 ml at 25 °C in TE buffer, pH 8.0, 1 μm p-benzoquinone (♦), 5 μm reduced GSH (■), and 0.28 μm PcGSTO1 (♦). In a separate experiment, as control, absorption spectrum of 1 μm p-benzohydroquinone was recorded at 25 °C in TE buffer, pH 8.0.
Dimerization
Size exclusion chromatography and dynamic light scattering were used to determine the oligomeric nature of the native forms of PcGSTO1 and PcGSTO3. The retention times of the size exclusion chromatography allowed estimation of the following native molecular masses: ∼85,000 Da for PcGSTO1 and ∼62,000 Da for PcGSTO3. Similar results have been obtained using dynamic light scattering suggesting that both enzymes were dimers.
Crystal Structure of PcGSTO1
The structure of PcGSTO1 has been solved to 2.3 Å resolution by single wavelength anomalous dispersion using crystals of SeMet-PcGSTO1 (Table 1). The crystals belonged to the space group C2, and the asymmetric unit consisted of two polypeptide chains (monomer A, residues Glu38–Asp351; monomer B, residues Phe54–Asp351; root mean square deviation, 0.348 Å for 298 superimposed Cα atoms), one GSH molecule not covalently bound and 223 water molecules. Crystal packing analysis using PISA (44) suggested that the asymmetric unit content corresponded to a dimer. The two protomers are related to each other by a noncrystallographic 2-fold symmetry axis.
PcGSTO1 adopts the canonical GST fold composed of an N-terminal thioredoxin-like domain and a C-terminal α-helix domain. It also possesses several additional features as follows: a long N-terminal coil of 77 residues; a loop that connects β2 to α2 elongated by 20 residues compared with typical GSTs; and a C-terminal extension corresponding to a ninth α-helix followed by a coil of 20 residues (Fig. 4 and supplemental Fig. 1).
This helix α9 (Phe319 to Ile333) is the only secondary structure among these supplementary features. It adds to the α-helix domain, near the C-terminal end of helix α6, so that it also interacts with the thioredoxin-like domain via the short loop that connects the strand β1 and the α-helix α1 and via the long loop that goes from β2 to α2. Helix α9 is a structural characteristic of GST Omega (10) and is also present in GST Tau (45) and GST Delta (46). In these GSTs, the few residues that follow α9 (when they exist) go toward the thioredoxin-like domain to interact with the loops β1-α1 and β2-α2. In PcGSTO1, the long loop observed between β2 and α2 hinders this interaction, so the 20 residues that follow α9 in PcGSTO1 form a coil that runs at the opposite side of the thioredoxin-like domain, antiparallel to α9.
This C-terminal coil allows the formation of a dimer that completely differs from the usual GST dimer, where the two PcGSTO1 monomers associate exclusively via their α-helix domains (Fig. 4), with a large buried area (3100 Å2) as determined by PISA (44). After α9, the 20 C-terminal residues of one monomer mainly interact with residues of the helix α5 N-terminal end (H-bonds to the side chains of Gln231 and Tyr234), next with a residue of the α6 C-terminal end (Thr283), and then with a residue of the loop that follows (Asn288), in the other monomer. These direct hydrogen bonds only concern the main chain atoms of the C-terminal end and add to several hydrophobic contacts. Is this dimer a crystal artifact or a physiological entity? The large surface of contact between the protomers and the involvement of the C-terminal coil, specific to PcGSTO1 and related proteins, in the inter-monomer interactions argue for a physiological dimer, as do the dynamic light scattering and size exclusion chromatography results. Furthermore, this dimer, different from the canonical GST dimer, is similar to the dimer assigned by Cuff et al. (Midwest Center for Structural Genomics) in the crystal structure of a putative glutathione S-transferase from Corynebacterium glutamicum recently released by the Protein Data Dank (PDB code 3M1G).4 The similar dimerization mode appears clearly related to a monomer structure found by DALI (47) to be the closest to the PcGSTO1 structure (root mean square deviation, 1.092 Å for 233 superimposed residues).
PcGSTO1 also possesses an N-terminal extension with respect to other GSTs, by far much longer than the 20 residues observed in GSTOs, because 77 residues precede the entrance to β1. No electron density was observed for the 37 first residues, which were thus supposed to be highly disordered. Residues Glu38 to Ser53 were only observed in monomer A (see under “Experimental Procedures”). The main chain of residues 38–53 first runs between the long loop that connects β2 to α2 and the loop that goes from α2 to β3 in the N-terminal thioredoxin-like domain. Then it interacts with the short loop between α4 and α5 in the α-helix domain, where it forms a turn before it enters the region Phe54–Arg77 observed in both monomers. This segment adopts an extended conformation to return toward the N-terminal domain. There it interacts with the loops that connect α2 to β3 and β4 to α3, before it runs all along α3, parallel to it. Then it enters β1. The conformation of the PcGSTO1 N-terminal end confers it with the original property to form part of the active site, composed of the G-site for GSH binding and the H-site, which receives the hydrophobic part of the glutathionylated substrate.
The G-site
Only monomer A displayed an electron density consistent with modeling of a GSH molecule (see under “Experimental Procedures”). However, no major structural difference between monomers A and B was observed that could account for an asymmetric binding of GSH. Nevertheless, in this structure only residues that belong to monomer A contributed to GSH binding.
No disulfide bridge exists between the GSH cysteine side chain and the catalytic residue Cys86 situated at the N-terminal end of helix α1 (Fig. 5). The sulfur to sulfur distance is 3.9 Å; the electron density is unambiguous, and careful analysis of the diffraction data reveals no radiation damage that could have disrupted a pre-existing disulfide bond. Superposition of the structures of PcGSTO1 and HsGSTO1-1 (where a disulfide bridge exists) shows a conserved orientation of the catalytic cysteine side chain. On the contrary, a displacement of the GSH Sγ2 atom is observed in PcGSTO1, which results from slight differences in the rotation angles around the bond between Cγ1 and Cδ1 in the GSH glutamyl residue and around the bond between Cα2 and Cβ2 in the GSH cysteinyl side chain. The latter points toward three tyrosine residues (Fig. 5), the hydroxyl groups of which form a hydrogen bond network; in particular, the GSH Sγ2 atom lies at 3.25 Å from the Tyr223 OH group tightly hydrogen-bonded to the Tyr326 OH.
In addition to the usual interactions with the main chain of a residue (Val158) that precedes the conserved cis-proline at the entrance of the strand β3, the GSH cysteine main chain forms an additional hydrogen bond between its CO group and the Nϵ1 atom of Trp119, situated in the long loop between β2 and α2 absent in most GSTs (Fig. 5). In the same way, besides the usual stabilization of the GSH glutamyl residue (main chain by residues Glu173 and Ser174 from the loop between β4 and α3 and aliphatic part of the side chain by hydrophobic contact with Trp88 from the N-terminal end of α1), an additional hydrogen bond was observed between the GSH glutamyl atom Oϵ1 and the Nη1 atom of Arg56 in the long N-terminal extension of PcGSTO1. Finally, the oxygen atoms of the carboxylic group of the GSH glycyl residue form two hydrogen bonds with the side chain of Arg155 situated in the loop between α2 and β3 slightly longer than in the other GSTs (Fig. 5), although usually only one bond is observed with a basic residue (48).
To summarize, some of the additional features that PcGSTO1 displays with respect to other GSTs (the long N-terminal end, the long loop between α2 and β2, and the few supplementary residues between α2 and β3) contribute to GSH binding. They squeeze GSH so much that only 45 Å2 of its surface is accessible to solvent (Fig. 5). As a comparison, this value is 148 Å2 in HsGSTO1.1. The active site seems so enclosed that it raises the question of its accessibility not only to the GSH part of the glutathionylated substrate but also to its hydrophobic part.
The Putative H-site
The lack of a structure of PcGSTO1 with a bound S-glutathionylated substrate forbids direct experimental delineation of the H-site. The PcGSTO1 surface shows no clear accessible cavity that could accommodate the hydrophobic part of the aromatic S-glutathionylated substrate (Fig. 5). Comparison with HsGSTO1-1 emphasizes the absence of a large binding pocket; the bottom of the HsGSTO1-1 putative H-site (formed by Phe31, Cys32, Pro33, Arg183, Trp222, and nearby residues (10)) is filled, in PcGSTO1, by Tyr223 and Tyr326, which surround the GSH cysteinyl side chain, and by Arg56, Cys86, and His325 (Figs. 1 and 5). Ser44, Met46, Phe54, Lys57, Arg155, Phe227, and His330 form a narrow, deep cylindrical hole, incompatible with access and binding of a bulky substrate.
Searching in the PDB for X-ray structures of GSTs in complex with S-glutathionylated substrates, 1-(S-glutathionyl)-(2,4-dinitrobenzene) was found to be the closest analog to S-glutathionyl-p-benzohydroquinone. It has been used as a substrate by the GSTs of the classes Mu from rat (PDB code 5GST (49)) and human (PDB code 1XWK (50)), Pi from human (PDB code 18GS (51), Sigma from squid (PDB code 1GSQ (52)), and Alpha from chicken (PDB code 1VF3).5 In each of these structures, 1-(S-glutathionyl)-(2,4-dinitrobenzene) leans its benzene ring against the hydrophobic side chains of the residues that belong to the α4 C-terminal end, and it often interacts with the first N-terminal residue of α1 (using PcGSTO1 numbering of secondary structures). Comparable interactions could be obtained using Trp88, Tyr223, and Phe227, but they would require a slightly different positioning of the 1-(S-glutathionyl)-(2,4-dinitrobenzene) glutathionyl part with respect to the current position of GSH in PcGSTO1. Indeed, in the structure, GSH appears too shifted toward the α-helix domain, with its cysteinyl sulfur atom exactly aligned on the α1 axis. The current position of the long N-terminal loop shows that residues such as Met46, Phe54, or Arg56 could also be involved, but once again, the main question concerns access of the substrate to its binding site, which would likely require a movement of the N-terminal loop.
DISCUSSION
Compared with other organisms, the GSTO class of enzymes is considerably larger in saprotrophic fungi and particularly in P. chrysosporium. From primary structure homology, this class could be divided in three different subclasses in fungi (9), one related to plant Lambda GSTs and bacterial GSTs, whereas the second and the third subclasses are related to animal isoforms. In accordance with our first phylogenetic analysis (9), Xun et al. (17) have recently proposed from a study of the bacterial homologs of PcpF that the proteins belonging to the Omega subclass I constitute a new class of GSTs named S-glutathionyl-(chloro)hydroquinone reductases.
The present x-ray structure of PcGSTO1 confirms that it should be considered as the member of a new class of GSTs. Indeed, the GST canonical fold (a thioredoxin-like domain followed by a α-helix domain) is complemented by additional specific features as follows: the long N-terminal coil (77 residues) that covers the active site and participates in glutathione binding, as does the long loop (about 30 residues) between the strand β2 and the α-helix α2, and finally the C-terminal coil (20 residues after the ninth α-helix) that allows an original dimerization mode where the monomers interact via their α-helix domain (also observed for a putative GST from C. glutamicum.4 All these features clearly contrast with the HsGSTO1-1 structure (10).
As shown for members of the Omega class, the main characteristic of GHRs is the presence of a conserved cysteine in their catalytic site. From a biochemical point of view, as shown previously (10) and in this study, GHRs and GSTOs share the ability to reduce various substrates such as HED and DHA using glutathione. Concerning PcGSTO1 and PcGSTO3, we propose from fluorescence experiments (for PcGSTO3) and free thiol and mass measurements that the catalytic cysteine is involved in the deglutathionylation activity leading to the formation of a disulfide bridge with the removed glutathione from their substrate. This hypothesis is in accordance with experiments performed using thiodione or PAP-SG as substrates, because the appearance of the deglutathionylated products is observed in the absence of added glutathione, a glutathione-adduct being detected in these conditions on the resulting proteins. Furthermore, these activities require the presence of the catalytic cysteine. This common mechanism has been described at least for monothiol glutaredoxin (23), human GSTOs (10), and bacterial GHRs (16). In PcGSTO3, as shown previously for HsGSTO1-1, a change of the catalytic cysteine to a serine allows reversal of the activity of the protein allowing the mutated protein to catalyze glutathione transfer, and it also confirms the presence of a disulfide bridge between Cys-37 and glutathione in PcGSTO3 WT. For PcGSTO1, biochemical data are also in accordance with the formation of a disulfide bridge between the catalytic Cys-86 and glutathione. Similar results have been obtained for the bacterial ortholog PcpF (16). However, this disulfide bridge is not present in the crystal structure of PcGSTO1. X-rays could have disrupted the bond, but this should have taken place very rapidly at the beginning of the radiation exposure because no residual electron density corresponding to this bond was observed and the sulfur to sulfur distance is large (3.9 Å), contrary to documented cases where such a phenomenon has been described. At the moment, the reasons for the discrepancy between the PcGSTO1 x-ray structure and the results of thiol titrations concerning the presence of a disulfide bond remain to be elucidated.
As defined by Xun et al. (17), the main biochemical characteristic of GHRs is the ability to remove glutathione from S-glutathionyl-(chloro)hydroquinone substrates. In this study, we have shown that in contrast to PcGSTO3, PcGSTO1 is active against S-glutathionyl-p-hydroquinone. Binding of S-glutathionyl-p-hydroquinone can be modeled from the PcGSTO1 structure, one monomer of which was observed in complex with GSH. Glutathione adopts a conformation very close to that observed in HsGSTO1-1. However, on the contrary to the latter, GSH is more buried in PcGSTO1. It results from new interactions with residues that belong to the features characteristic of PcGSTO1 as follows: Arg56 in the N-terminal coil, Trp119 in the loop β2-α2, and Arg155 in the loop α2-β3. The additional coils close the cleft usually present between the Trx-like domain and the α-helix domain, near the loop β1-α1 in which is the catalytic residue Cys86. Furthermore, the side chains of Tyr223 and Tyr326 here shut up the pocket present near the catalytic cysteine in HsGSTO1-1. Even if conformational adaptation occurs upon substrate binding, it would hardly concern these residues because they belong to α-helices. Hence, a substrate-binding site different from the putative pocket in HsGSTO1-1 has to be considered for the hydroquinone adduct. It could involve Trp88, Tyr223, and Phe227. All the PcGSTO1 residues involved either in the G-site or the putative H-site are conserved in the bacterial homologs (ScPcpF and EcYqjG, see Fig. 1), although they are different or absent in other GSTs (see supplemental Fig. 1). Once again, comparison of the PcGSTO1 and HsGSTO1-1 structures in the light of their substrate-binding site emphasizes the need to consider GHRs as having a new GST structure, different from the Omega class.
In agreement with our previous phylogenetic analysis (9), the biochemical and structural data presented in this study and the recent proposition of Xun et al. (17), we propose to change the name of PcGSTO1 in PcGHR1 with regard to the enzymatic function and to define a new structural class of GSTs named GST xi, characterized by their canonical GST fold complemented by specific features and their new dimerization mode. Thus, this paper describes this newly defined S-glutathionyl-(chloro)hydroquinone reductase from P. chrysosporium as a member of the new structural class xi of GSTs, which would also contain the structure of the putative GST from C. glutamicum.
As for PcpF (16), PcGHR1 in P. chrysosporium could be involved in the catabolism of pentachlorophenol. In this fungus, the reductive dehalogenation of pentachlorophenol requires indeed at least two enzymes, a glutathione transferase for glutathionylation and an enzyme called glutathione conjugate reductase for deglutathionylation of the resulting S-glutathionyl-p-hydroquinones (53–55). From the biochemical data presented in this study, we postulated that PcGHR1 or the second isoform PcGHR2 (formerly named PcGSTO2 (9)) is responsible for this latter reaction. The wide presence of GHRs in bacteria, fungi, and plants suggests an additional physiological role beyond the degradation of the recently introduced pentachlorophenol. Concerning the white rot fungi, their main physiological feature is their ability to mineralize wood components and in particular lignin. It has been suggested recently that chlorination of lignin is a phenomenon occurring through, at least in part, the activity of chloroperoxidase (56, 57). As a result, soils and decayed plant litter contain significant quantities of chlorinated aromatic polymers, which could lead after oxidation via various oxidative systems to the formation of chlorinated quinones. GHRs could play a central role in the intracellular detoxication pathways of such molecules. On the other hand, the significance of the Omega class extension in saprotrophic fungi remains unclear, and the function of the different GSTO isoforms in white rot fungi remains to be elucidated.
Supplementary Material
Acknowledgments
We thank the staff of the BM30A beamline at ESRF for their kind assistance during data collection, Arnaud Gruez for numerous discussions, and Jean-Michel Girardet for technical assistance.
This work was supported by Agence Nationale de la Recherche Research Grants ANR-06-BLAN-0386 and ANR-09-BLAN-0012, Ministère de l'Enseignement Supérieur de la Recherche et de la Technologie, Institut National de la Recherche Agronomique, and CNRS.
This work is dedicated to the memory of the late Dr. André Aubry.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1, Figs. 1 and 2, and additional references.
The atomic coordinates and structure factors (code 3PPU) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
M. E. Cuff, N. Marshall, G. Cobb, and A. Joachimiak, unpublished results.
S. C. Lin, Y. C. Lo, M. F. Tam, and Y. C. Law, unpublished results.
- GSTO
- glutathione transferase Omega
- DHA
- dehydroascorbate
- GHR
- S-glutathionyl-(chloro)hydroquinone reductase
- HED
- hydroxyethyldisulfide
- PDB
- Protein Data Bank
- PAP-SG
- S-(phenacylacetophenone)glutathione
- Se
- selenium
- CDNB
- 1-chloro-2,4-dinitrobenzene.
REFERENCES
- 1. Martinez D., Larrondo L. F., Putnam N., Gelpke M. D., Huang K., Chapman J., Helfenbein K. G., Ramaiya P., Detter J. C., Larimer F., Coutinho P. M., Henrissat B., Berka R., Cullen D., Rokhsar D. (2004) Nat. Biotechnol. 22, 695–700 [DOI] [PubMed] [Google Scholar]
- 2. Morel M., Kohler A., Martin F., Gelhaye E., Rouhier N. (2008) New Phytol. 180, 391–407 [DOI] [PubMed] [Google Scholar]
- 3. Morel M., Ngadin A. A., Jacquot J. P., Gelhaye E. (2009) Adv. Bot. Res. 52, 153–186 [Google Scholar]
- 4. Doddapaneni H., Yadav J. S. (2005) Mol. Genet. Genomics 274, 454–466 [DOI] [PubMed] [Google Scholar]
- 5. Kasai N., Ikushiro S., Hirosue S., Arisawa A., Ichinose H., Wariishi H., Ohta M., Sakaki T. (2009) Biochem. Biophys. Res. Commun. 387, 103–108 [DOI] [PubMed] [Google Scholar]
- 6. Kasai N., Ikushiro S., Shinkyo R., Yasuda K., Hirosue S., Arisawa A., Ichinose H., Wariishi H., Sakaki T. (2010) Appl. Microbiol. Biotechnol. 86, 773–780 [DOI] [PubMed] [Google Scholar]
- 7. Matsuzaki F., Wariishi H. (2004) Biochem. Biophys. Res. Commun. 324, 387–393 [DOI] [PubMed] [Google Scholar]
- 8. Schröder P., Scheer C. E., Diekmann F., Stampfl A. (2007) Environ. Sci. Pollut. Res. Int. 14, 114–122 [DOI] [PubMed] [Google Scholar]
- 9. Morel M., Ngadin A. A., Droux M., Jacquot J. P., Gelhaye E. (2009) Cell. Mol. Life Sci. 66, 3711–3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Board P. G., Coggan M., Chelvanayagam G., Easteal S., Jermiin L. S., Schulte G. K., Danley D. E., Hoth L. R., Griffor M. C., Kamath A. V., Rosner M. H., Chrunyk B. A., Perregaux D. E., Gabel C. A., Geoghegan K. F., Pandit J. (2000) J. Biol. Chem. 275, 24798–24806 [DOI] [PubMed] [Google Scholar]
- 11. Mannervik B., Board P. G., Hayes J. D., Listowsky I., Pearson W. R. (2005) Methods Enzymol. 401, 1–8 [DOI] [PubMed] [Google Scholar]
- 12. Board P. G., Coggan M., Cappello J., Zhou H., Oakley A. J., Anders M. W. (2008) Anal. Biochem. 374, 25–30 [DOI] [PubMed] [Google Scholar]
- 13. Whitbread A. K., Masoumi A., Tetlow N., Schmuck E., Coggan M., Board P. G. (2005) Methods Enzymol. 401, 78–99 [DOI] [PubMed] [Google Scholar]
- 14. Burmeister C., Lüersen K., Heinick A., Hussein A., Domagalski M., Walter R. D., Liebau E. (2008) FASEB J. 22, 343–354 [DOI] [PubMed] [Google Scholar]
- 15. Zakharyan R. A., Sampayo-Reyes A., Healy S. M., Tsaprailis G., Board P. G., Liebler D. C., Aposhian H. V. (2001) Chem. Res. Toxicol. 14, 1051–1057 [DOI] [PubMed] [Google Scholar]
- 16. Huang Y., Xun R., Chen G., Xun L. (2008) J. Bacteriol. 190, 7595–7600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xun L., Belchik S. M., Xun R., Huang Y., Zhou H., Sanchez E., Kang C., Board P. G. (2010) Biochem. J. 428, 419–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garcerá A., Barreto L., Piedrafita L., Tamarit J., Herrero E. (2006) Biochem. J. 398, 187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Herrero E., Ros J., Tamarit J., Bellí G. (2006) Photosynth. Res. 89, 127–140 [DOI] [PubMed] [Google Scholar]
- 20. Schenk P. M., Baumann S., Mattes R., Steinbiss H. H. (1995) BioTechniques 19, 196–198, 200 [PubMed] [Google Scholar]
- 21. Ramakrishnan V., Finch J. T., Graziano V., Lee P. L., Sweet R. M. (1993) Nature 362, 219–223 [DOI] [PubMed] [Google Scholar]
- 22. Koh C. S., Navrot N., Didierjean C., Rouhier N., Hirasawa M., Knaff D. B., Wingsle G., Samian R., Jacquot J. P., Corbier C., Gelhaye E. (2008) J. Biol. Chem. 283, 23062–23072 [DOI] [PubMed] [Google Scholar]
- 23. Couturier J., Koh C. S., Zaffagnini M., Winger A. M., Gualberto J. M., Corbier C., Decottignies P., Jacquot J. P., Lemaire S. D., Didierjean C., Rouhier N. (2009) J. Biol. Chem. 284, 9299–9310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nickerson W. J., Falcone G., Strauss G. (1963) Biochemistry 2, 537–543 [DOI] [PubMed] [Google Scholar]
- 25. Vince R., Daluge S., Wadd W. B. (1971) J. Med. Chem. 14, 402–404 [DOI] [PubMed] [Google Scholar]
- 26. Roth M., Carpentier P., Kaïkati O., Joly J., Charrault P., Pirocchi M., Kahn R., Fanchon E., Jacquamet L., Borel F., Bertoni A., Israel-Gouy P., Ferrer J. L. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 805–814 [DOI] [PubMed] [Google Scholar]
- 27. Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 28. Collaborative Computational Project No. 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 29. Panjikar S., Parthasarathy V., Lamzin V. S., Weiss M. S., Tucker P. A. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 449–457 [DOI] [PubMed] [Google Scholar]
- 30. Schneider T. R., Sheldrick G. M. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 31. Sheldrick G. M. (2002) Z. Kristallogr. 217, 644–650 [Google Scholar]
- 32. Cowtan K. (1994) Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31, 34–38 [Google Scholar]
- 33. Morris R. J., Zwart P. H., Cohen S., Fernandez F. J., Kakaris M., Kirillova O., Vonrhein C., Perrakis A., Lamzin V. S. (2004) J. Synchr. Rad. 11, 56–59 [DOI] [PubMed] [Google Scholar]
- 34. Perrakis A., Morris R., Lamzin V. S. (1999) Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 35. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 36. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 37. Gelhaye E., Rouhier N., Gérard J., Jolivet Y., Gualberto J., Navrot N., Ohlsson P. I., Wingsle G., Hirasawa M., Knaff D. B., Wang H., Dizengremel P., Meyer Y., Jacquot J. P. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 14545–14550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gelhaye E., Rouhier N., Laurent P., Sautière P. E., Martin F., Jacquot J. P. (2002) Physiol. Plant. 114, 165–171 [DOI] [PubMed] [Google Scholar]
- 39. Board P. G., Anders M. W. (2007) Chem. Res. Toxicol. 20, 149–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rao D. N., Takahashi N., Mason R. P. (1988) J. Biol. Chem. 263, 17981–17986 [PubMed] [Google Scholar]
- 41. Song Y., Wagner B. A., Witmer J. R., Lehmler H. J., Buettner G. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9725–9730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brunmark A., Cadenas E. (1988) Chem. Biol. Interact. 68, 273–298 [DOI] [PubMed] [Google Scholar]
- 43. Mauzeroll J., Bard A. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7862–7867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 45. Thom R., Cummins I., Dixon D. P., Edwards R., Cole D. J., Lapthorn A. J. (2002) Biochemistry 41, 7008–7020 [DOI] [PubMed] [Google Scholar]
- 46. Oakley A. J., Harnnoi T., Udomsinprasert R., Jirajaroenrat K., Ketterman A. J., Wilce M. C. (2001) Protein Sci. 10, 2176–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Holm L., Rosenström P. (2010) Nucleic Acids Res. 38, Suppl W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vararattanavech A., Ketterman A. J. (2007) Biochem. J. 406, 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ji X., Armstrong R. N., Gilliland G. L. (1993) Biochemistry 32, 12949–12954 [DOI] [PubMed] [Google Scholar]
- 50. Patskovsky Y., Patskovska L., Almo S. C., Listowsky I. (2006) Biochemistry 45, 3852–3862 [DOI] [PubMed] [Google Scholar]
- 51. Oakley A. J., Lo Bello M., Nuccetelli M., Mazzetti A. P., Parker M. W. (1999) J. Mol. Biol. 291, 913–926 [DOI] [PubMed] [Google Scholar]
- 52. Ji X., von Rosenvinge E. C., Johnson W. W., Tomarev S. I., Piatigorsky J., Armstrong R. N., Gilliland G. L. (1995) Biochemistry 34, 5317–5328 [DOI] [PubMed] [Google Scholar]
- 53. McCarthy D. L., Navarrete S., Willett W. S., Babbitt P. C., Copley S. D. (1996) Biochemistry 35, 14634–14642 [DOI] [PubMed] [Google Scholar]
- 54. Reddy G. V., Gold M. H. (1999) Biochem. Biophys. Res. Commun. 257, 901–905 [DOI] [PubMed] [Google Scholar]
- 55. Reddy G. V., Gold M. H. (2001) Arch. Biochem. Biophys. 391, 271–277 [DOI] [PubMed] [Google Scholar]
- 56. Ortiz-Bermúdez P., Hirth K. C., Srebotnik E., Hammel K. E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3895–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ortiz-Bermúdez P., Srebotnik E., Hammel K. E. (2003) Appl. Environ. Microbiol. 69, 5015–5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.