Abstract
Enhancer of zeste homolog 2 (EZH2) regulates stem cells renewal, maintenance, and differentiation into different cell lineages including neuron. Changes in intracellular Ca2+ concentration play a critical role in the differentiation of neurons. However, whether EZH2 modulates intracellular Ca2+ signaling in regulating neuronal differentiation from human mesenchymal stem cells (hMSCs) still remains unclear. When hMSCs were treated with a Ca2+ chelator or a PLC inhibitor to block IP3-mediated Ca2+ signaling, neuronal differentiation was disrupted. EZH2 bound to the promoter region of PIP5K1C to suppress its transcription in proliferating hMSCs. Interestingly, knockdown of EZH2 enhanced the expression of PIP5K1C, which in turn increased the amount of PI(4,5)P2, a precursor of IP3, and resulted in increasing the intracellular Ca2+ level, suggesting that EZH2 negatively regulates intracellular Ca2+ through suppression of PIP5K1C. Knockdown of EZH2 also enhanced hMSCs differentiation into functional neuron both in vitro and in vivo. In contrast, knockdown of PIP5K1C significantly reduced PI(4,5)P2 contents and intracellular Ca2+ release in EZH2-silenced cells and resulted in the disruption of neuronal differentiation from hMSCs. Here, we provide the first evidence to demonstrate that after induction to neuronal differentiation, decreased EZH2 activates the expression of PIP5K1C to evoke intracellular Ca2+ signaling, which leads hMSCs to differentiate into functional neuron lineage. Activation of intracellular Ca2+ signaling by repressing or knocking down EZH2 might be a potential strategy to promote neuronal differentiation from hMSCs for application to neurological dysfunction diseases.
Keywords: Calcium, Differentiation, Gene Expression, Gene Regulation, Neuron, EZH2, PIP5K1C, Intracellular Calcium, Mesenchymal Stem Cells, Neuronal Differentiation
Introduction
Human mesenchymal stem cells (hMSCs)4 derived from bone marrow are easily obtained (1), safely expanded in vitro and are not susceptible to malignant transformation; thus, hMSCs are suitable for therapeutic applications (2). hMSCs can be induced to differentiate into multiple lineages, including bone, fat, cartilage, as well as neuron in vitro (1, 3–5). Transplanted MSCs are able to differentiate into neuronal lineage and improve the functions of central nervous system (CNS) with ischemia (6), Parkinson disease (7), Alzheimer disease (8), spinal cord injury (9, 10), and other neurodegenerative disorders (11, 12) modeled in rodents. These findings demonstrate the potential of hMSCs for cell therapy in human CNS.
Spontaneous and transient elevation in intracellular Ca2+ concentration during an early period is required and critical for neuronal differentiation both in vitro and in embryonic neuron development (13). However, whether intracellular Ca2+ signaling is required for neuronal differentiation from hMSCs remains unclear. The expression of transient receptor potential (TRP) proteins, TRPC1 and TRPC3, is elevated to activate store-operated calcium entry (SOCE) after differentiation of H19–7 hippocampal neuronal cells (14). It is also known that inositol 1,4,5-trisphosphate (IP3) stimulates a ligand-gated channel, IP3 receptor (IP3R), to release Ca2+ from intracellular stores in neurons (15). IP3 is generated by phospholipase C (PLC)-mediated hydrolysis of phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2], which is synthesized by type I phosphatidylinositol-4-phosphate 5-kinase (PIP5K1) (16). PIP5K1 comprises three isoforms namely α (PIP5K1A), β (PIP5K1B), and γ (PIP5K1C) (17). Unlike ubiquitously distributed PIP5K1A and PIP5K1B (18, 19), PIP5K1C is highly expressed in brain, suggesting that PIP5K1C may play a distinctive role in neuron functions (20, 21). Moreover, it has been shown that PIP5K1C is critical in the regulation of IP3-mediated Ca2+ signaling after stimulation of G protein-coupled receptor by histamine (22) and is required for cardiovascular and neuronal development (23). Nonetheless, the regulation of PIP5K1C and intracellular Ca2+ transient during neuronal differentiation from hMSCs is still unclear.
The polycomb group (PcG) proteins consist of two functionally distinct multimeric polycomb repressive complexes (PRCs) referred to as PRC1 and PRC2 (24). PRC2 binds to target genes and initiates tri-methylation at lysine 27 of histone H3 (H3K27me3); PRC1 recognizes the H3K27me3 through its chromodomain to mediate ubiquitylation of H2AK119 and maintain gene repression (25). The core components of PRC2 include suppressor of zeste-12 (SUZ12) and embryonic ectoderm development (EED) for complex stability and enhancer of zeste homolog 2 (EZH2) for the methyltransferase activity (26). EZH2 not only catalyzes histone H3 tri-methylation at lysine 27 (27) but also recruits DNA methyltransferases to silence gene expression (28). The PcG proteins play a critical role in the regulation of gene expressions in stem cells maintenance and lineage specification (29). For example, EZH2 is highly expressed in embryonic stem cells (ESCs) and is required for the derivation of pluripotent ESCs (30). EZH2 is also involved in differentiation into different cell lineages in mice, including skeletal muscle (31, 32), hepatocytes (33), epidermis (34), as well as neuron (35). However, whether and how EZH2 regulates differentiation of hMSCs into neuronal lineage remain to be explored.
In the current study, we demonstrate that EZH2 targets PIP5K1C promoter to suppress its expression and negatively regulates intracellular Ca2+ concentration to maintain homeostasis in proliferating hMSCs. After induction to neuronal differentiation, dissociation of EZH2 protein from PIP5K1C promoter induces the expression of PIP5K1C to elevate intracellular Ca2+ contents and promote neuronal differentiation. This is the first evidence to show that PIP5K1C-mediated Ca2+ signaling in neuronal differentiation from hMSCs is regulated by EZH2.
EXPERIMENTAL PROCEDURES
Materials
Chemicals used in induction of neuronal differentiation, dexamethasone, ascorbic acid-2-phosphate, indomethacin, insulin, and 3-isobutyl-1-methyl-xanthine, an intracellular Ca2+ chelator, 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid tetrakis-acetoxymethyl ester (BAPTA-AM), polybrene, and puromycin were purchased from Sigma. The cell-permeant Ca2+ indicator, Fluo-4-acetoxymethyl ester (Fluo4-AM) and Fura-2-acetoxymethyl ester (Fura-2 AM) were obtained from Molecular Probes. The following antibodies were purchased from commercial companies: anti-CD105, anti-MAP2, AlexFluor 488 conjugated anti-MAP2A and AlexFluor 647 conjugated anti-β-tubulin III (BD); anti-β-tubulin III (Covance); anti-Neu-N, clone A60 (Chemicon); anti-β-actin and anti-α-tubulin (Sigma); anti-EZH2, clone AC22, and anti-PIP5K1C (Cell Signaling).
Cell Culture and Induction to Differentiation into Neuronal Lineage
Human mesenchymal stem cells (hMSCs), size-sieved stem cells from human bone marrow, were isolated and characterized previously, and the hMSCs had the capacity for multi-lineage potential to form bone, fat, cartilage (1), as well as electrically active neural cells (3). The hMSCs were immortalized without neoplastic transformation by transduction with HPV16 E6/E7 genes (5). The E6/E7-immortalized hMSC derivative named 3A6 contains the human telomerase reverse transcriptase (hTERT) gene for more stem-like properties (36). The 3A6 cells were grown in Dulbecco's modified Eagle's medium-low glucose (DMEM-LG) (HyClone) with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin in a humidified incubator with 5% CO2 at 37 °C. To induce neuronal differentiation, cells were seeded at a density of 4000 cells/cm2 in the regular medium the day before experiment, and then treated with serum-free DMEM-high glucose (DMEM-HG) (HyClone) neuronal induction medium (NIM) containing 10−7 m dexamethasone, 50 μg/ml ascorbic acid-2-phosphate, 50 μm indomethacin, 10 μg/ml insulin, and 0.45 mm 3-isobutyl-1-methyl-xanthine for 1–5 days, and the neuronal induction medium was changed per 3 days (37).
Western Blot Analysis
Cells were washed twice with phosphate-buffered saline (PBS, containing 137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 2 mm KH2PO4), and then lysed in RIPA buffer (50 mm Tris at pH 7.5, 150 mm NaCl, 1 mm EDTA, 0.25% Na-deoxycholate, 1% Nonidet P-40, 1 mm NaF, 1 mm Na3VO4, 1 mm PMSF, 1 μg/ml aprotinin) by sonication. The soluble extraction was collected from the supernatant after centrifugation at 15,000 × g for 10 min at 4 °C. The extract was boiled at 100 °C for 5 min, separated by SDS-PAGE, and transferred to a PVDF membrane. Subsequently, the membrane was blocked with 5% skim milk in PBST buffer (PBS containing 0.1% Tween-20) for 1 h at room temperature, and then hybridized with primary antibody with gentle agitation overnight at 4 °C. After washing with PBST, the membrane was incubated with HRP-conjugated secondary antibody (Chemicon) for 1 h at room temperature. The immunoreactive band was visualized by the enhanced chemiluminescence (ECL) detection reagent (GE Healthcare, Amersham Biosciences Place).
RNA Extraction and Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
Total RNA was extracted with TRIzol reagent (Invitrogen) according to the manufacturer's instruction. The complementary DNA (cDNA) was synthesized from 5 μg of total RNA in a reaction mixture containing 2.5 μm oligo (dT) primer, 0.5 mm dNTP mixture, 200 units of SuperScript III reverse transcriptase, 40 units of RNaseOUT, an RNase inhibitor (all from Invitrogen). After incubation at 50 °C for 50 min, the reaction mixture was heat inactivated at 85 °C for 5 min and then treated with 2 units of RNase H at 37 °C for 20 min. The quantitative PCR (q-PCR) was performed by detection of the hydrolyzed fluorescent probes from Universal ProbeLibrary (UPL; Roche) using the LightCycler 480 equipment (Roche). Primers and UPL probes were designed using the ProbeFinder software. The primer sets and the matched UPL probe numbers are as following: PIP5K1C: 5′-CGCCACCGACATCTACTTTC-3′ (forward), 5′-ATAGTGGAGCGGGGAGTACA-3′ (reverse), and UPL probe 17; β-actin (ACTB): 5′-ATTGGCAATGAGCGGTTC-3′ (forward), 5′-GGATGCCACAGGACTCCAT-3′ (reverse), and UPL probe 11. The q-PCR was examined by incubating the cDNA in a reaction mixture containing, 0.5 μm of each primer, 0.1 μm UPL probe, and 1-fold concentration of Probes Master reagent (Roche). The amplification conditions were initial denaturation at 95 °C for 10 min, followed by 45 cycles of 95 °C for 10 s, 55 °C for 30 s, and 72 °C for 1 s. The fluorescent signal was detected at 72 °C step of each cycle. The relative quantification of the gene of interest was normalized by β-actin and calculated by the value of cross-point (CP) in each fluorescence curve of each gene. For traditional PCR, the reaction mixture containing 2 μl of cDNA, 0.2 mm dNTP mixture, 2 μm of each primers, 1 unit of TaqDNA polymerase, and 1-fold concentration of ThermalPol Buffer (New England BioLabs) was started by denatured 95 °C for 5 min, followed by amplification of indicated cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s. The numbers of cycles for amplification of NSE, PITX-3, NURR1, and EZH2 were all 35, and that of β-actin was 25. The specific primer sequences for these genes are as follows: NSE: 5′-CATCGACAAGGCTGGCTACACG-3′ (forward), 5′-GACAGTTGCAGGCCTTTTCTTC-3′ (reverse); PITX-3: 5′-GTCTATCGGGACCCGTGTAA-3′ (forward), 5′-CCAGTCAAAATGACCCCAGT-3′ (reverse); NURR1: 5′-CAATGCGTTCGTGGCT-3′ (forward), 5′-GGGTACGAAGTTCTGGG-3′ (reverse); EZH2: 5′-CAGTAAAAATGTGTCCTGCAAGAA-3′ (forward), 5′-TCAAGGGATTTCCATTTCTCTTTCGA-3′ (reverse); and β-actin: 5′-GCACTCTTCCAGCCTTCCTTCC-3′ (forward), 5′-TCACCTTCACCGTTCCAGTTTTT-3′ (reverse).
Ca2+ Influx ([Ca2+]i) Measurement
Ca2+ influx measurement was carried out as previously described (3). The hMSCs were seeded on coverslips. After treatment, cells were incubated with 5 μm Fura-2 AM for 1 h and then washed out the excess Fura-2 AM. The coverslip was mounted on an inverted microscope (Olympus IX-70) under ×40 magnification. The excitation peak for Fura-2 shifts from 380 nm for the calcium-free chelator to about 340 nm for the calcium-saturated form. The excitation lights (340 nm and 380 nm) were provided by T.I.L.L. Polychrome IV spectrophotometer (T.I.L.L. Photonics), and the emission light (510 nm) was recorded by Princeton charged-coupled device camera CCD-130 (Roper Scientific). The [Ca2+]i was represented as a ratio (R340/380) between emission intensities at 510 nm induced by 340 nm and 380 nm. For stimulation, loading buffer contained glutamate (25 μm) or high K+ (the NaCl was replaced with an equimolar KCl) was puffed from a glass micropipette placed right beside the cell. The opening of the micropipette was ∼1.5 μm. The puff pressure was controlled by Eppendorf Microinjector 5242 (Eppendorf) with a P2 setting of 200 hPa.
In addition, we also used another method to measure [Ca2+]i (38). Cells were seeded on the Biocoat poly-d-lysine 96-well black/clear plate (BD) for 2 days, and then induced to neuronal differentiation for 5 days. Cells were pre-incubated with 5 μm Fluo4-AM for 1 h. After washout of Fluo4-AM, cells were stimulated with 100 μm glutamate (Sigma) in a calcium buffer solution containing 5 mm EGTA, 100 mm KCl, and 30 mm MOPS with free 0.15 μm Ca2+ (Sigma). The plates were measured immediately at excitation (485 nm) and emission (528 nm) every 40 s interval using Synergy 2 microplate reader (BioTek Instruments, Inc.).
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay was performed with the EZ ChIP kit (Upstate). In brief, chromatin and proteins from approximate 2 × 106 cells were crosslinked with 1% formaldehyde for 10 min at room temperature. These cells were collected, lysed, and sonicated on ice to shear the chromatin DNA to a length between 200 bp and 1000 bp using Sonicator 3000 (Misonix) equipped with a microtip (setting: output level 4, 5 times of pulse for 10 s, and pause for 1 min). The sonicated chromatin lysate was immunoprecipitated with anti-EZH2 antibody, and collected with protein A/G-agarose beads (Pierce). The protein/DNA crosslinks of the immunoprecipitated complexes were reversed by incubation in 0.2 m NaCl at 65 °C for 4–5 h, and then the DNA was purified and applied to PCR as described above to determine the binding ability of EZH2 to its potential target gene, PIP5K1C. The sequences of the primers specific to the promoter of PIP5K1C are 5′-GACCTACACAGCACATGCCA-3′ (forward), and 5′-GCATGTATTGTGCATATCCG-3′ (reverse).
Gene Knockdown by Short-hairpin RNA (shRNA)
Knockdown of genes were performed with the specific shRNAs delivered by the lentiviral system from National RNAi Core Facility (Academia Sinica, Taipei, Taiwan) according to the instruction manual. Briefly, to generate the lentivirus containing specific shRNA, 293T cells were cotransfected with 2.25 μg of pCMV-ΔR8.91 plasmid harboring Gag and Pol genes, 0.25 μg of pMD.G plasmid containing VSV-G gene for expression of envelope glycoprotein, and 2.5 μg of pLKO.1 plasmid bearing the specific shRNA for 16 h, and cells were then cultured in growth medium containing 1% BSA for another 24 h. The cultured medium containing lentivirus was collected and stored at −80 °C as aliquots for further use. To deliver the specific shRNA construct, approximate 80% confluent cells were infected with the lentivirus bearing specific shRNA in growth medium containing 8 μg/ml polybrene and incubated at 37 °C for 24 h. Afterward, cells were subcultured and selected with 2 μg/ml puromycin. The shRNA constructs targeting the interested genes are as follows: PIP5K1C: TRCN0000037666 corresponding to the sequences, 5′-CGTGGTCAAGATGCACCTCAA-3′; EZH2: TRCN0000040076 referring to the sequence, 5′-CGGAAATCTTAAACCAAGAAT-3′. The shRNA construct against luciferase (shLuc), TRCN0000072244 referring to the sequence, 5′-ATCACAGAATCGTCGTATGCA-3′ was used as negative control.
Detection of Intracellular Ca2+ by Flow Cytometer
Fluo4-AM dissolved in DMSO was used to detect intracellular Ca2+ (39), which bound to cytoplasmic-free Ca2+ and emitted a green fluorescence (peak at 516 nm). Cells were seeded on 35-mm dishes at a density of 104 cells/dish the day before experiment and incubated with 2 μm Fluo4-AM at 37 °C for 30 min in the dark. Cells were then washed with PBS and incubated at 37 °C for further 1 h to allow for complete deesterification of the dye by cytosolic esterases inside cells. Subsequently, cells were collected and applied to detect the fluorescence of Ca2+ bound to Fluo4 within cells using FL1 of the FACSCalibur flow cytometry (BD Biosciences). Simultaneously, cells were treated with DMSO in parallel as a negative control in each treatment. The intensity of the fluorescence from Fluo4-AM treated cells was normalized by the background from DMSO-treated cells, and the geographic mean value (Geo Mean) of fluorescence in each experiment was calculated as the value of normalized Geo Mean using CellQuest program (BD Biosciences).
PI(4,5)P2 Measurement
PI(4,5)P2 mass was determined by the PI(4,5)P2 mass ELISA kit (Echelon Biosciences Inc.) according to the manual instruction (40). In brief, ∼2 × 106 cells were collected, and lipids were extracted. The extracted lipids were incubated with a PI(4,5)P2 detector protein at room temperature for 1 h, and then transferred to the PI(4,5)P2-coated plate for competitive binding, and incubated at room temperature for another 1 h. After removing the reaction mixture and washing three times with PBST, the peroxidase-linked secondary detection reagent was added into the wells, and the plate was incubated at room temperature for 1 h to detect the PI(4,5)P2 detector protein bound to the plate. Finally, after discarding the reaction mixture from the plate, the 3,3′,5,5′-tetramethylbenzidine (TMB) substrate was added and incubated for 5–30 min to allow the color development, and the absorbance at 450 nm was measured by Synergy 2 microplate reader (BioTek Instruments, Inc.). The colorimetric signal is inversely proportional to the amounts of PI(4,5)P2. The amounts of PI(4,5)P2 in each sample was calculated according to the standard curve from serious diluents of pure PI(4,5)P2.
Neuronal Differentiation from hMSCs in Animal Brains
Adult male Sprague-Dawley (SD) rats (weight > 300 × g; age, 7–8 weeks) were used in this study. All animal experiments were approved by the Institutional Review Board of Animal Experiments, China Medical University Hospital. Prior to implantation, hMSCs with or without knockdown of EZH2 were incubated with 1 μg/ml bis-benzimide Hoechst 33342 (Sigma) to label nuclei with blue fluorescence for 5 h at 37 °C. After washing three times with PBS, the labeled hMSCs were counted using a cytometer to ensure an adequate cell number for implantation. The SD rats were anesthetized with chloral hydrate (0.4 g/kg, intraperitoneal) and then were injected stereotaxically with approximate 1 × 106 cells in 3–5 μl of DMEM medium through a 26-gauge Hamilton syringe into 3 cortical areas adjacent to the right middle cerebral artery, 3.0 to 5.0 mm below the dura mater. Cyclosporin A, an immunosuppressant drug, (CsA; 1 mg/kg/d, intraperitoneal; Novartis) injections were given daily to each experimental rat for 3 weeks (41).
Laser Scanning Confocal Microscopy for Immunofluorescence Colocalization Analysis
To determine the neuronal differentiation of hMSCs in rat brain, tissue sections from hMSCs implanted rat brains were stained with specific primary antibodies (1:300) against EZH2 and a neuronal specific marker, MAP2. Subsequently, the tissue sections were stained with Cy5- and Cy3-conjugated secondary antibodies (1:500, Jackson Immunoresearch). The immunofluorescent colocalization study with three-dimensional images was performed to test for the expression of hMSC (blue fluorescence), EZH2 (red fluorescence), and MAP2 (green fluorescence). The three-dimensional images were observed under a Carl Zeiss LSM510 laser-scanning confocal microscope.
RESULTS
Characterization of Neuronal Differentiation from 3A6 hMSCs
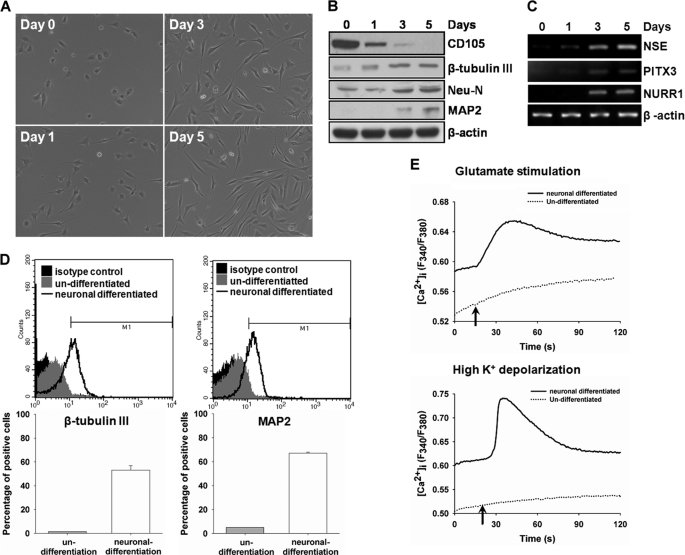
The immortalized hMSC line, KP-hMSCs, possesses the characteristics for differentiation into several cell types, including fat, cartilage, and neuron (5). We further validated the capacity of its derivative line, 3A6, containing the human telomerase reverse transcriptase (hTERT) gene for more stem-like properties (36) and induction into neuronal lineage. Extension of neurite was observed at 1 day after induction in the neuronal induction medium (NIM), and the length of neurite outgrowth was increased after 3–5 days of treatment (Fig. 1A). During the induction of neuronal differentiation, protein expression of neuron markers, β-tubulin III, Neu-N, and MAP2, and mRNA expression of NSE, PITX3, and NURR1 increased with time (Fig. 1, B and C). However, the expression of MSC marker, CD105, was simultaneously reduced (Fig. 1B). The flow cytometry results showed that the percentage of cells expressing β-tubulin III and MAP2 dramatically increased to 55∼65% after induction of differentiation for 5 days (Fig. 1D). To further confirm that hMSCs possess the potential to differentiate into functional neuronal cell, neuron-like function was validated by measuring the [Ca2+]i change after stimulation with glutamate or high K+. As shown in Fig. 1E, elevated [Ca2+]i stimulated by glutamate or high K+ was observed significantly in differentiated hMSCs. These results demonstrated that the 3A6 hMSC line could be successfully induced to differentiate into functional neuron lineage.
FIGURE 1.
Characterization of neuronal differentiation from hMSCs. 3A6 hMSCs were seeded at a density of 4000 cells/cm2 the day before induction of neuronal differentiation, and then treated with NIM for 1–5 days as described under “Experimental Procedures.” A, cell morphology at indicated time interval was observed at ×100 magnification under an inverted phase microscope. B, protein expressions of a MSC marker, CD105, and neuron markers, β-tubulin III, Neu-N, and MAP2 were examined. C, mRNA expressions of neuron markers, NSE, PITX3, and NURRI were determined as well. β-Actin was used as an internal control. D, expressions of neuronal markers, β-tubulin III and MAP2, were labeled by AlexFluor 647- and AlexFluor 488-conjugated antibodies, respectively, and then detected by flow cytometry. The β-tubulin III- and MAP2-positive cells in comparison with the cells labeled with isotype IgG were margined as M1 region. The bottom bar plots show the percentage of cells in the M1 region in un-differentiated and neuronal differentiated hMSCs. E, hMSCs were treated with (solid line) or without (dotted line) NIM for 5 days to induce differentiation into the neuron lineage. Change of [Ca2+]i after stimulated with glutamate (upper panel) or high K+ buffer (lower panel) was used to determine the neuron-like function. The arrow indicates the time point of stimulation.
EZH2 Negatively Regulates Intracellular Ca2+ Required for hMSCs Neuronal Differentiation
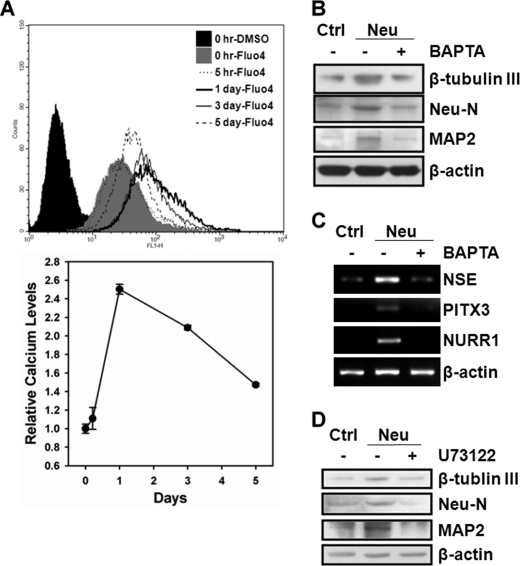
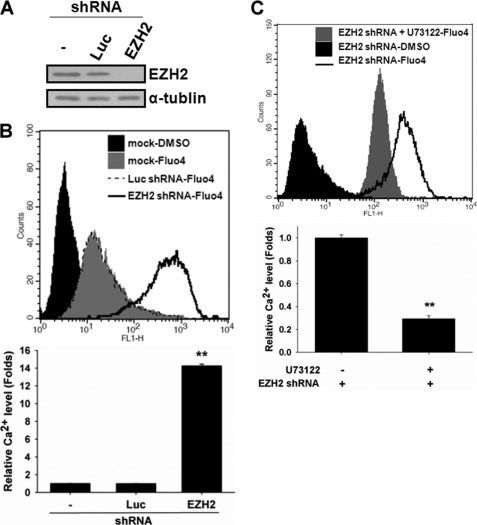
To investigate whether intracellular Ca2+ change is required for neuronal differentiation from hMSCs, we measured the intracellular Ca2+ contents with Fluo4-AM during differentiation and examined the effect of an intracellular Ca2+ chelator, BAPTA-AM, on neuronal differentiation. As shown in Fig. 2A, intracellular Ca2+ contents were transiently elevated during neuronal differentiation, which peaked at day 1 post-induction and declined afterward. hMSCs treated with BAPTA-AM significantly decreased the expression of neuron markers after induction to neuronal differentiation as demonstrated by both Western blot (Fig. 2B) and RT-PCR (Fig. 2C). To further show that hMSCs neuronal differentiation involves IP3-mediated Ca2+ signaling, we used a PLC inhibitor, U73122, to disrupt PI(4,5)P2 hydrolysis into IP3 and diacylglycerol (DAG) and examined its effect on neuronal differentiation. Cells that were treated with U73122 suppressed the expression of neuron markers after induction to neuronal differentiation (Fig. 2D), supporting that IP3-mediated Ca2+ signaling is required for inducing differentiation from hMSCs into neuronal lineage. We silenced the EZH2 gene by specific short hairpin RNA (shRNA) (Fig. 3A) to further understand the role of EZH2 in the regulation of intracellular Ca2+ contents. Specifically, knockdown of EZH2 dramatically increased the intracellular Ca2+ content by 14-fold, indicating that EZH2 might be a negative regulator of intracellular Ca2+ content in proliferating undifferentiated hMSCs (Fig. 3B). In addition, treating EZH2-silenced hMSCs with U73122 inhibited about 70% of intracellular Ca2+ contents compared with untreated cells (Fig. 3C), suggesting that EZH2 diminished the intracellular Ca2+ content that is likely through repression of IP3-mediated Ca2+ release.
FIGURE 2.
hMSCs neuronal differentiation requires IP3-mediated intracellular Ca2+ signaling. A, intracellular Ca2+ contents during neuronal differentiation from 3A6 hMSCs was measured with Fluo4-AM by flow cytometer. The bottom plot is the relative intracellular Ca2+ level comparing to that at the initial induction time (0 h) during neuronal differentiation. B and C, 3A6 hMSCs were pretreated with or without 10 μm intracellular Ca2+ chelator, BAPTA-AM, for 1 h, and then induced to neuronal differentiation for 5 days. The protein (B) and mRNA (C) expression of the indicated neuron markers in undifferentiated (lane 1) and neuron-differentiated (lanes 2 and 3) were determined. D, 3A6 hMSCs were pretreated with or without 20 μm PLC inhibitor, U73122, for 30 min, and then induced to neuronal differentiation for 5 days. The expression of neuron markers were examined after each treatment.
FIGURE 3.
Negative regulation of IP3-mediated intracellular Ca2+ contents by EZH2 in hMSCs. A, 3A6 hMSCs were infected without (lane 1) or with lentivirus carrying shRNAs against luciferase (lane 2) or EZH2 (lane 3) gene. The shRNA against luciferase was used as a negative control of shRNA. The expressions of EZH2 and α-tubulin were examined. B, effect of EZH2 knockdown on intracellular Ca2+ contents was determined by flow cytometer. The bottom plot showed the ratio of intracellular Ca2+ contents from shRNA-containing cells over mock control cells. Bars represented mean ± S.D., and the symbol, **, indicated p value < 0.01 by t test. C, EZH2-silenced hMSCs were treated with or without 20 μm U73122 for 30 min, and then the intracellular Ca2+ contents were determined. The bottom plot is the relative intracellular Ca2+ level normalized by that from untreated cells.
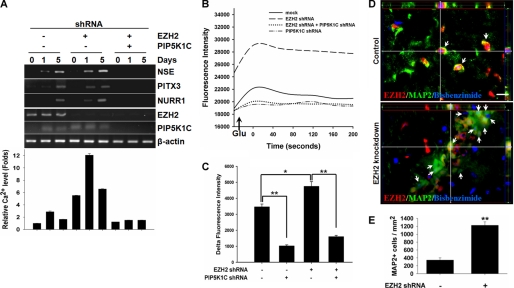
EZH2 Negatively Regulates Intracellular Ca2+ through Suppression of PIP5K1C
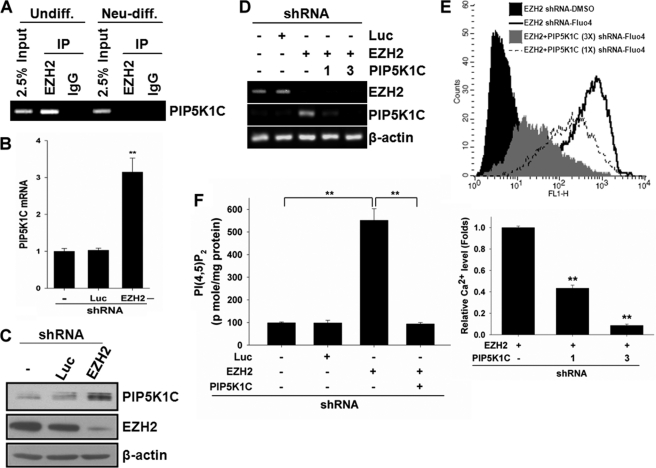
Because IP3-mediated Ca2+ signaling is required for neuronal differentiation from hMSCs (Fig. 2D), and PIP5K1C synthesizes a precursor of IP3, PI(4,5)P2, which is critical in the process (22) and highly expressed in brain (20, 21), we questioned whether EZH2 affects intracellular Ca2+ contents through regulation of PIP5K1C gene expression. To address whether EZH2 targets PIP5K1C gene promoter, we performed ChIP with anti-EZH2 antibody followed by amplification of the promoter region of PIP5K1C. EZH2 bound to the promoter of PIP5K1C in undifferentiated hMSCs but not neuron-differentiated hMSCs. However, after induction to neuronal differentiation, binding of EZH2 on PIP5K1C promoter was significantly reduced (Fig. 4A). Knockdown of EZH2 dramatically increased the expression of PIP5K1C (Fig. 4, B and C), indicating that EZH2 targets the promoter of PIP5K1C and represses its expression in proliferating undifferentiated hMSCs. To further support our hypothesis that EZH2 negatively regulates intracellular Ca2+ signaling through PIP5K1C, we knocked down PIP5K1C in EZH2-silenced cells (Fig. 4D) and determined the effect on intracellular Ca2+ contents. Infection of increasing amount of lentivirus carrying shRNA against PIP5K1C (1- versus 3-fold; middle panel, Fig. 4D) in EZH2-silenced hMSCs resulted in significant reduction of intracellular Ca2+ contents caused by knockdown of EZH2 in a dose-dependent manner (up to 90% of inhibition) (Fig. 4E). In addition, knockdown of EZH2 increased the amount of the product of PIP5K1C, PI(4,5)P2, which was abolished by further knockdown of PIP5K1C (Fig. 4F). Taken together, these results showed that EZH2 negatively regulates intracellular Ca2+ contents by suppressing PIP5K1C expression.
FIGURE 4.
The role of PIP5K1C in EZH2-mediated calcium signaling. A, undifferentiated and neuron-differentiated 3A6 hMSCs were collected and applied to ChIP assay with anti-EZH2 antibody or immunoglobulin (IgG, negative control). The immunoprecipitated chromatin DNA was amplified by PCR with primers specific to the promoter region of PIP5K1C gene. B and C, effects of EZH2 knockdown on the expression of PIP5K1C was determined by quantitative RT-PCR (bar3 in B) and Western blotting (lane 3 in C). D, 3A6 hMSCs were infected with or without lentivirus containing shRNAs against luciferase or EZH2, and EZH2-silenced cells were infected with increased amounts of lentivirus harboring shRNA to PIP5K1C (1- and 3-fold amounts). Total RNA was extracted and the expressions of EZH2, PIP5K1C, and β-actin were determined by RT-PCR. E, effect of PIP5K1C knockdown in EZH2-silencing cells on intracellular Ca2+ contents was examined by flow cytometer. The bottom plot was the relative quantity of intracellular Ca2+ contents. The bars represented mean ± S.D., and the symbol, **, indicated p value < 0.01 by t test. F, 3A6 hMSCs were infected with or without lentivirus containing shRNAs against luciferase or EZH2, and PIP5K1C was further knocked down in EZH2-silenced cells. The PI(4,5)P2 mass after each treatment was determined as described under “Experimental Procedures.”
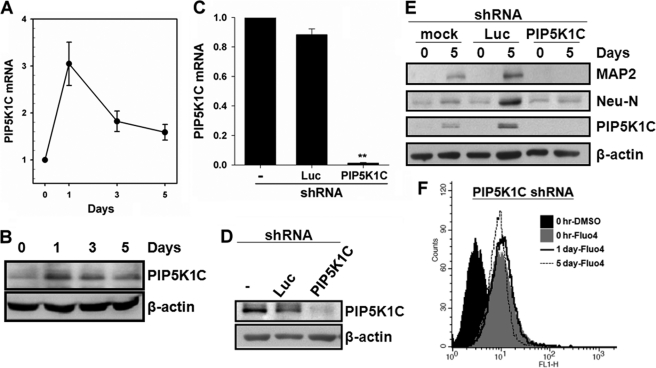
PIP5K1C Is Required for hMSCs Neuronal Differentiation
To explore the role of the EZH2 target gene, PIP5K1C, in neuronal differentiation from hMSCs, we silenced PIP5K1C gene expression by shRNA to determine its effect on neuronal differentiation. As shown in Fig. 5, A and B, the expression of PIP5K1C increased after inducing hMSCs to neuronal differentiation, which peaked at day 1 and declined at days 3 and 5 post-induction. The shRNAs against PIP5K1C showed 98% inhibition of its mRNA expression (Fig. 5C). Likewise, we also observed a significant decrease in PIP5K1C protein expression by Western blot (Fig. 5D). To clarify the influence of PIP5K1C on neuronal differentiation from hMSCs, we compared the expression of neuron markers in 3A6 hMSCs transfectants, which had been infected with or without shRNA against Luc (as negative control) or PIP5K1C, at day 0 and day 5 post-induction to neuronal differentiation. The expression of PIP5K1C as well as neuron markers, Neu-N and MAP2, was significantly increased in mock-treated and Luc-silenced cells but not in PIP5K1C-silenced cells (Fig. 5E), and the intracellular Ca2+ content was not significantly changed in PIP5K1C-silenced cells (Fig. 5F), indicating that PIP5K1C is required for intracellular Ca2+ transient and neuronal differentiation from hMSCs.
FIGURE 5.
Neuronal differentiation from hMSCs requires PIP5K1C. A and B, 3A6 hMSCs were induced to differentiation into neuronal lineage. The expression of PIP5K1C at each time interval was determined by quantitative RT-PCR (A) and Western blotting (B). C and D, cells were infected with or without lentivirus containing shRNAs against luciferase or PIP5K1C. The effect of these shRNAs on PIP5K1C expression was examined by detecting its mRNA (C) and protein (D) level. E, shRNA containing cells were induced to neuronal differentiation. The expressions of indicated neuron markers were determined at day 0 and day 5 after induction. β-Actin was used as an internal control. F, intracellular Ca2+ contents in hMSCs with PIP5K1C knockdown during induction of neuronal differentiation at indicated time interval were measured by flow cytometry.
Knockdown of EZH2 Enhances Neuronal Differentiation from hMSCs in Vitro and in Vivo
To address the potential of activation of Ca2+ signaling to enhance hMSCs differentiation into functional neuron, we compared the capabilities of neuronal differentiation from hMSCs with or without knockdown of EZH2. As shown in Fig. 6A, knockdown of EZH2 increased the expression of neuron markers (NSE, PITX3, and NURR1) and PIP5K1C (upper panel), and the intracellular Ca2+ content was also elevated (lower panel). Further knockdown of PIP5K1C in EZH2-silenced cells disrupted neuronal differentiation as indicated by loss of neuron marker expression and attenuated the elevation of Ca2+ transient. The neuron-like function measured by change of [Ca2+]i after glutamate stimulation was significantly higher in EZH2-silenced hMSCs than that in parental hMSCs, and knockdown of PIP5K1C in both parental and EZH2-silenced hMSCs disturbed the neuron-like function (Fig. 6, B and C). These results suggest that knockdown of EZH2 enhanced hMSCs differentiation into functional neuron, and this is likely due to the activation of PIP5K1C-mediated Ca2+ signaling. To further investigate the effect of EZH2 knockdown in neuronal differentiation in vivo, we implanted hMSCs with or without knockdown of EZH2 to the brains of SD rats for 3 weeks and examined the expression of MAP2 in their tissue sections. Images from the colocalization study (three-dimensional image) showing exogenous implanted hMSCs and MAP2 (green fluorescence) positive cells in implanted hMSCs (blue fluorescence) as indicated by the arrows. Quantitative analysis of the implanted MAP2-positive cells in EZH2-silenced hMSCs was significantly more than that in the mock-treated hMSCs (Fig. 6E), suggesting that knockdown of EZH2 in hMSCs might be a potential strategy to enhance neuronal differentiation in vivo.
FIGURE 6.
Knockdown of EZH2 enhances neuronal differentiation from hMSCs in vitro and in vivo. A, mRNA expression profiles of neuron markers (NSE, PITX3, and NURR1), EZH2, PIP5K1C, as well as β-actin, were determined by RT-PCR at indicated time intervals post-induction to neuronal differentiation in parental and shRNA-containing hMSCs. The bottom plot shows the relative intracellular Ca2+ contents at indicated time point during neuronal differentiation of each hMSC transfectant. The original flow cytometry profiles were shown in supplemental Fig. S2. B, neuron-like function in each hMSC transfectant after induction of differentiation for 5 days was determined by change of Ca2+ influx after stimulation of 100 μm glutamate. The plot shows the kinetic profile of glutamate stimulated [Ca2+]i. C, difference of fluorescence intensity between the basal (before stimulation) and maximum (during stimulation) values (mean ± S.D.) from plot B. The symbols, * and ** indicate p value <0.05 and < 0.01, respectively by t test. D, representative three-dimensional image of bisbenzimide-labeled hMSCs (blue fluorescence) implantation in rat brain, significant down-regulated expression of EZH2 (red fluorescence) was found in EZH2 knockdown hMSCs-treated rats compared with that of control mock hMSCs-treated rats. The white arrows indicate the implanted MAP2 (green fluorescence)-positive hMSCs in rat brains. Scale bar: 50 μm. E, quantitative analysis of the implanted MAP2-positive cells numbers in both the EZH2 knockdown hMSCs-treated rats and control mock hMSCs-treated rats.
DISCUSSION
Although the involvement of Ca2+ in neuronal differentiation has been reported from 1990s (42), it remains unclear whether intracellular Ca2+ transient is required for differentiation from hMSCs into neuronal lineage. In the current study, we demonstrated that intracellular Ca2+ transient peaks at day 1 post-induction (Fig. 2A) and is required for neuronal differentiation from hMSCs (Fig. 2, B and C). We also showed that the effect of intracellular Ca2+ on neuronal differentiation from hMSCs, at least in part, is through IP3-mediated calcium signaling (Figs. 2D and 3C). Recently, a study reported that a ryanodine-sensitive receptor (RyR) agonist, caffeine, stimulates Ca2+ response that increases throughout neuronal differentiation in embryonic P19 carcinoma stem cells (CSCs) and adult murine MSCs (43). Intracellular Ca2+ concentration has been demonstrated to be controlled by multiple mechanisms, including well characterized Ca2+ influx through voltage-gated channels, ligand-gated channels (44, 45), and non-voltage-gated channels (14), as well as Ca2+ releasing from the endoplasmic reticulum (ER) via intracellular RyR and IP3R (15). Here, we uncovered for the first time that the intracellular Ca2+ contents could be regulated by a PcG protein, EZH2, through modulating the gene expression of PIP5K1C in addition to above mentioned calcium channels (Figs. 3 and 4). The expression profile of PKP5K1C (Fig. 5, A and B) is positively correlated to that of intracellular Ca2+ contents (Fig. 2A) during induction of neuronal differentiation from hMSCs.
PcG proteins epigenetically repress transcription and their target genes have been genome-wide-mapped in murine ESCs (46) and human embryonic lung fibroblast TIG3 cell line (47), revealing that numerous development regulators and differentiation-related genes are repressed by binding of PcG proteins. Cells deficient in PRC2 component, EED, de-repress PcG target genes and activate neuronal differentiation in murine ESCs (46). Inactivation of PcG protein by knock-out of EED or EZH2 promotes neurogenesis of neural precursor cells (NPCs, or neural stem cells, NSCs), (48). Our study showed that EZH2 targets the promoter of PIP5K1C gene in proliferating hMSCs, and the binding could not be detected in neuron-differentiated hMSCs. Knockdown of EZH2 enhances PIP5K1C expression, PI(4,5)P2 generation, elevates intracellular Ca2+ contents (Fig. 4), and promotes hMSCs differentiating into functional neuron lineage (Fig. 6, A–C). Furthermore, knockdown of EZH2 in hMSCs also enhanced neuronal differentiation in the rat brain (Fig. 6, D and E). Although the mRNA expression of EZH2 was almost no different during induction to neuronal differentiation in hMSCs (Fig. 6A), the protein level of EZH2 was decreased after day 1 (supplemental Fig. S1). This is likely the cause of increased amount of PIP5K1C mRNA after day 1 post-induction to neuronal differentiation. Likewise, decreased level of EZH2 has been reported in ESCs differentiation (49). In addition, a previous report demonstrated that EZH2 protein is highly expressed in proliferating NSCs from embryonic mice and decreased after NSCs differentiating into neurons and astrocytes (35).
It has been well characterized that PIP5K1s catalyze the production of PI(4,5)P2 in membrane lipid metabolism (50). In terms of their cellular functions, PIP5K1s are also involved in the regulation of actin reorganization and focal adhesion dynamics, which are critical in cell migration (51) and neurite outgrowth (52, 53). Among all PIP5K1s, PIP5K1C is mainly expressed in brain and required for cardiovascular and neuronal development. The PIP5K1C-null embryos result in extensive prenatal lethality (23). A common regulatory mechanism of PIP5K1s is phosphorylation at specific site to alter the association with binding partners. For instance, phosphorylation at Tyr-649 of human PIP5K1C by Src increases the affinity of its C-terminal for talin, and phosphorylation at Ser-650 by cyclin B1/Cdk1 blocks this interaction in focal adhesion (54, 55). Here, we provide evidence to establish that PIP5K1C is transcriptionally regulated by EZH2 (Fig. 4, A–C), and knockdown of PIP5K1C disrupts neuronal differentiation from hMSCs (Fig. 5E) and EZH2-silenced hMSCs (Fig. 6, A–C), indicating that PIP5K1C is essential for neuronal differentiation from hMSCs, and silencing EZH2 enhanced neuronal differentiation might be mediated via activation of PIP5K1C.
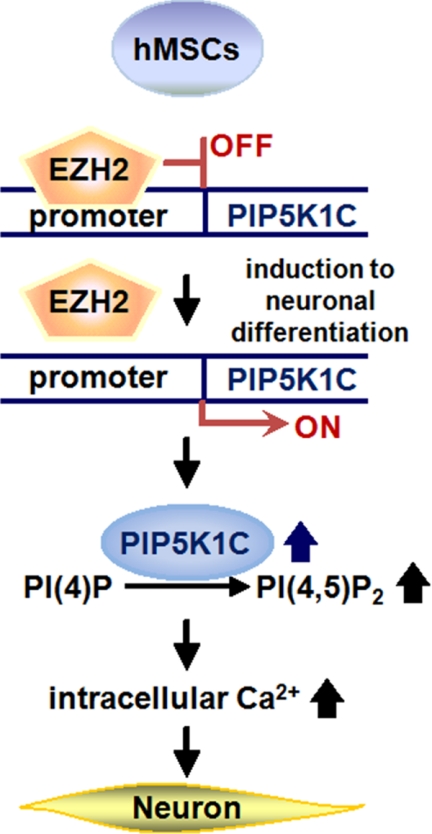
In summary, the current study demonstrates a novel regulatory mechanism of intracellular Ca2+ signaling by EZH2 in hMSCs. A proposed model is shown in Fig. 7. After induction of neuronal differentiation, disassembly of EZH2 protein from the promoter of PIP5K1C gene increases the expression of PIP5K1C and generation of PI(4,5)P2, leading to elevated intracellular calcium signaling and advance neuronal differentiation from hMSCs. To the best of our knowledge, this is the first report that demonstrates intracellular Ca2+ signaling could be modulated by a PcG protein, EZH2, through transcriptional regulation of PIP5K1C. Our study provides a new insight to the role of EZH2 in neuronal differentiation from hMSCs. Activation of intracellular Ca2+ signaling, which is suppressed by EZH2, might be a potential strategy to promote neuronal differentiation for an application to cure neurodegenerative diseases or spinal cord injury.
FIGURE 7.
A proposed model of EZH2-mediated PIP5K1C-dependent neuronal differentiation from hMSCs. In proliferating undifferentiated hMSCs, EZH2 protein binds to the promoter of PIP5K1C gene to repress its transcription to maintain the homeostasis of intracellular Ca2+ contents in relative low level. After induction to neuronal differentiation, EZH2 protein is dissociated from the promoter of PIP5K1C to enhance its gene expression and results in increase of PI(4,5)P2 generation, leading to activation of IP3-mediated Ca2+ signaling to promote neuronal differentiation from hMSCs.
Supplementary Material
Acknowledgment
We thank the National RNAi Core Facility (Academia Sinica, Taipei, Taiwan) for providing the shRNAs.
This study was supported, in whole or in part, by National Institutes of Health Grants R01 CA109311 and P01 CA099031. This work was also supported by The University of Texas M.D. Anderson-China Medical University and Hospital Sister Institution Fund, Cancer Center Research of Excellence DOH-99-TDC-111-05 (to M. C. H.); NSC-2632-B-039-001-MY3 (to M. C. H.); NSC-2320-B-039-032-MY3 (to Y. L. Y.); and NSC-3111-B-039 (to M. C. H., and Y. L. Y.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- hMSC
- human mesenchymal stem cell
- EZH2
- enhancer of zeste homolog 2
- PIP5K1C
- type I phosphatidylinositol-4-phosphate 5-kinase isoform C
- PLC
- phospholipase C
- PI(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- IP3
- inositol 1,4,5-trisphosphate
- DAG
- diacylglycerol
- ESCs
- embryonic stem cells
- CSCs
- carcinoma stem cells
- CNS
- central nervous system
- TRP
- transient receptor potential
- SOCE
- store-operated calcium entry
- PcG
- polycomb group
- PRCs
- polycomb repressive complexes
- SUZ12
- suppressor of zeste-12
- EED
- embryonic ectoderm development
- H3K27me3
- histone H3 tri-methylation at lysine 27
- H2AK119
- histone 2A at lysine 119
- ChIP
- chromatin immunoprecipitation
- shRNA
- short-hairpin RNA
- IP3R
- IP3 receptor
- RyR
- ryanodine-sensitive receptor.
REFERENCES
- 1. Hung S. C., Chen N. J., Hsieh S. L., Li H., Ma H. L., Lo W. H. (2002) Stem Cells 20, 249–258 [DOI] [PubMed] [Google Scholar]
- 2. Bernardo M. E., Zaffaroni N., Novara F., Cometa A. M., Avanzini M. A., Moretta A., Montagna D., Maccario R., Villa R., Daidone M. G., Zuffardi O., Locatelli F. (2007) Cancer Res. 67, 9142–9149 [DOI] [PubMed] [Google Scholar]
- 3. Hung S. C., Cheng H., Pan C. Y., Tsai M. J., Kao L. S., Ma H. L. (2002) Stem Cells 20, 522–529 [DOI] [PubMed] [Google Scholar]
- 4. Hung S. C., Lu C. Y., Shyue S. K., Liu H. C., Ho L. L. (2004) Stem Cells 22, 1321–1329 [DOI] [PubMed] [Google Scholar]
- 5. Hung S. C., Yang D. M., Chang C. F., Lin R. J., Wang J. S., Low-Tone Ho L., Yang W. K. (2004) Int. J. Cancer 110, 313–319 [DOI] [PubMed] [Google Scholar]
- 6. Zhao L. R., Duan W. M., Reyes M., Keene C. D., Verfaillie C. M., Low W. C. (2002) Exp. Neurol. 174, 11–20 [DOI] [PubMed] [Google Scholar]
- 7. Dezawa M., Kanno H., Hoshino M., Cho H., Matsumoto N., Itokazu Y., Tajima N., Yamada H., Sawada H., Ishikawa H., Mimura T., Kitada M., Suzuki Y., Ide C. (2004) J. Clin. Invest. 113, 1701–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee J. K., Jin H. K., Endo S., Schuchman E. H., Carter J. E., Bae J. S. (2010) Stem Cells 28, 329–343 [DOI] [PubMed] [Google Scholar]
- 9. Cho S. R., Kim Y. R., Kang H. S., Yim S. H., Park C. I., Min Y. H., Lee B. H., Shin J. C., Lim J. B. (2009) Cell Transplant 18, 1359–1368 [DOI] [PubMed] [Google Scholar]
- 10. Gu W., Zhang F., Xue Q., Ma Z., Lu P., Yu B. (2010) Neuropathology 30, 205–217 [DOI] [PubMed] [Google Scholar]
- 11. Jin H. K., Carter J. E., Huntley G. W., Schuchman E. H. (2002) J. Clin. Invest. 109, 1183–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bae J. S., Han H. S., Youn D. H., Carter J. E., Modo M., Schuchman E. H., Jin H. K. (2007) Stem Cells 25, 1307–1316 [DOI] [PubMed] [Google Scholar]
- 13. Spitzer N. C., Lautermilch N. J., Smith R. D., Gomez T. M. (2000) Bioessays 22, 811–817 [DOI] [PubMed] [Google Scholar]
- 14. Wu X., Zagranichnaya T. K., Gurda G. T., Eves E. M., Villereal M. L. (2004) J. Biol. Chem. 279, 43392–43402 [DOI] [PubMed] [Google Scholar]
- 15. Banerjee S., Hasan G. (2005) Bioessays 27, 1035–1047 [DOI] [PubMed] [Google Scholar]
- 16. van den Bout I., Divecha N. (2009) J. Cell Sci. 122, 3837–3850 [DOI] [PubMed] [Google Scholar]
- 17. Kanaho Y., Kobayashi-Nakano A., Yokozeki T. (2007) Biol. Pharm. Bull. 30, 1605–1609 [DOI] [PubMed] [Google Scholar]
- 18. Ishihara H., Shibasaki Y., Kizuki N., Katagiri H., Yazaki Y., Asano T., Oka Y. (1996) J. Biol. Chem. 271, 23611–23614 [DOI] [PubMed] [Google Scholar]
- 19. Loijens J. C., Anderson R. A. (1996) J. Biol. Chem. 271, 32937–32943 [DOI] [PubMed] [Google Scholar]
- 20. Ishihara H., Shibasaki Y., Kizuki N., Wada T., Yazaki Y., Asano T., Oka Y. (1998) J. Biol. Chem. 273, 8741–8748 [DOI] [PubMed] [Google Scholar]
- 21. Giudici M. L., Emson P. C., Irvine R. F. (2004) Biochem. J. 379, 489–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Y. J., Li W. H., Wang J., Xu K., Dong P., Luo X., Yin H. L. (2004) J. Cell Biol. 167, 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Y., Lian L., Golden J. A., Morrisey E. E., Abrams C. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11748–11753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schwartz Y. B., Pirrotta V. (2008) Curr. Opin. Cell Biol. 20, 266–273 [DOI] [PubMed] [Google Scholar]
- 25. Sparmann A., van Lohuizen M. (2006) Nat. Rev. Cancer 6, 846–856 [DOI] [PubMed] [Google Scholar]
- 26. Pasini D., Bracken A. P., Jensen M. R., Lazzerini Denchi E., Helin K. (2004) EMBO J. 23, 4061–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R. S., Zhang Y. (2002) Science 298, 1039–1043 [DOI] [PubMed] [Google Scholar]
- 28. Viré E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J. M., Bollen M., Esteller M., Di Croce L., de Launoit Y., Fuks F. (2006) Nature 439, 871–874 [DOI] [PubMed] [Google Scholar]
- 29. Rajasekhar V. K., Begemann M. (2007) Stem Cells 25, 2498–2510 [DOI] [PubMed] [Google Scholar]
- 30. O'Carroll D., Erhardt S., Pagani M., Barton S. C., Surani M. A., Jenuwein T. (2001) Mol. Cell. Biol. 21, 4330–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Caretti G., Di Padova M., Micales B., Lyons G. E., Sartorelli V. (2004) Genes Dev. 18, 2627–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Juan A. H., Kumar R. M., Marx J. G., Young R. A., Sartorelli V. (2009) Mol. Cell 36, 61–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aoki R., Chiba T., Miyagi S., Negishi M., Konuma T., Taniguchi H., Ogawa M., Yokosuka O., Iwama A. (2010) J. Hepatol. 52, 854–863 [DOI] [PubMed] [Google Scholar]
- 34. Ezhkova E., Pasolli H. A., Parker J. S., Stokes N., Su I. H., Hannon G., Tarakhovsky A., Fuchs E. (2009) Cell 136, 1122–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sher F., Rössler R., Brouwer N., Balasubramaniyan V., Boddeke E., Copray S. (2008) Stem Cells 26, 2875–2883 [DOI] [PubMed] [Google Scholar]
- 36. Tsai C. C., Chen C. L., Liu H. C., Lee Y. T., Wang H. W., Hou L. T., Hung S. C. (2010) J. Biomed. Sci. 17, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chu M. S., Chang C. F., Yang C. C., Bau Y. C., Ho L. L., Hung S. C. (2006) Cell Signal. 18, 519–530 [DOI] [PubMed] [Google Scholar]
- 38. Tondreau T., Dejeneffe M., Meuleman N., Stamatopoulos B., Delforge A., Martiat P., Bron D., Lagneaux L. (2008) BMC Genomics 9, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gee K. R., Brown K. A., Chen W. N., Bishop-Stewart J., Gray D., Johnson I. (2000) Cell Calcium. 27, 97–106 [DOI] [PubMed] [Google Scholar]
- 40. Gray A., Olsson H., Batty I. H., Priganica L., Peter Downes C. (2003) Anal. Biochem. 313, 234–245 [DOI] [PubMed] [Google Scholar]
- 41. Shyu W. C., Liu D. D., Lin S. Z., Li W. W., Su C. Y., Chang Y. C., Wang H. J., Wang H. W., Tsai C. H., Li H. (2008) J. Clin. Invest. 118, 2482–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Holliday J., Spitzer N. C. (1990) Dev. Biol. 141, 13–23 [DOI] [PubMed] [Google Scholar]
- 43. Resende R. R., da Costa J. L., Kihara A. H., Adhikari A., Lorençon E. (2010) Stem Cells Dev. 19, 379–394 [DOI] [PubMed] [Google Scholar]
- 44. Tsien R. W., Lipscombe D., Madison D., Bley K., Fox A. (1995) Trends Neurosci. 18, 52–54 [PubMed] [Google Scholar]
- 45. Jackson M. B. (1999) Adv. Neurol. 79, 511–524 [PubMed] [Google Scholar]
- 46. Boyer L. A., Plath K., Zeitlinger J., Brambrink T., Medeiros L. A., Lee T. I., Levine S. S., Wernig M., Tajonar A., Ray M. K., Bell G. W., Otte A. P., Vidal M., Gifford D. K., Young R. A., Jaenisch R. (2006) Nature 441, 349–353 [DOI] [PubMed] [Google Scholar]
- 47. Bracken A. P., Dietrich N., Pasini D., Hansen K. H., Helin K. (2006) Genes Dev. 20, 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hirabayashi Y., Suzki N., Tsuboi M., Endo T. A., Toyoda T., Shinga J., Koseki H., Vidal M., Gotoh Y. (2009) Neuron 63, 600–613 [DOI] [PubMed] [Google Scholar]
- 49. de la Cruz C. C., Fang J., Plath K., Worringer K. A., Nusinow D. A., Zhang Y., Panning B. (2005) Chromosoma 114, 183–192 [DOI] [PubMed] [Google Scholar]
- 50. Heck J. N., Mellman D. L., Ling K., Sun Y., Wagoner M. P., Schill N. J., Anderson R. A. (2007) Crit. Rev. Biochem. Mol. Biol. 42, 15–39 [DOI] [PubMed] [Google Scholar]
- 51. Ling K., Schill N. J., Wagoner M. P., Sun Y., Anderson R. A. (2006) Trends Cell Biol. 16, 276–284 [DOI] [PubMed] [Google Scholar]
- 52. van Horck F. P., Lavazais E., Eickholt B. J., Moolenaar W. H., Divecha N. (2002) Curr. Biol. 12, 241–245 [DOI] [PubMed] [Google Scholar]
- 53. Yamazaki M., Miyazaki H., Watanabe H., Sasaki T., Maehama T., Frohman M. A., Kanaho Y. (2002) J. Biol. Chem. 277, 17226–17230 [DOI] [PubMed] [Google Scholar]
- 54. Ling K., Doughman R. L., Iyer V. V., Firestone A. J., Bairstow S. F., Mosher D. F., Schaller M. D., Anderson R. A. (2003) J. Cell Biol. 163, 1339–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lee S. Y., Voronov S., Letinic K., Nairn A. C., Di Paolo G., De Camilli P. (2005) J. Cell Biol. 168, 789–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.