Abstract
Bacteria are equipped with two-component systems to cope with environmental changes, and auxiliary proteins provide response to additional stimuli. The Cpx two-component system is the global modulator of cell envelope stress in Gram-negative bacteria that integrates very different signals and consists of the kinase CpxA, the regulator CpxR, and the dual function auxiliary protein CpxP. CpxP both inhibits activation of CpxA and is indispensable for the quality control system of P pili that are crucial for uropathogenic Escherichia coli during kidney colonization. How these two essential biological functions of CpxP are linked is not known. Here, we report the crystal structure of CpxP at 1.45 Å resolution with two monomers being interdigitated like “left hands” forming a cap-shaped dimer. Our combined structural and functional studies suggest that CpxP inhibits the kinase CpxA through direct interaction between its concave polar surface and the negatively charged sensor domain on CpxA. Moreover, an extended hydrophobic cleft on the convex surface suggests a potent substrate recognition site for misfolded pilus subunits. Altogether, the structural details of CpxP provide a first insight how a periplasmic two-component system inhibitor blocks its cognate kinase and is released from it.
Keywords: Antibiotics, Bacterial Signal Transduction, Peptide Arrays, Protein Structure, Protein-Protein Interactions, CpxP, Two-component Signaling, Auxiliary Proteins
Introduction
Two-component signal transduction systems are the predominant mechanisms used by bacteria for coupling response to changes in the environment (1). The core structures of two-component signal transduction systems are a sensor kinase perceiving the stimulus and a cognate response regulator mediating the output response. Stimulus perception results in the autophosphorylation of the sensor kinase by ATP at a conserved histidine residue. Subsequently, the phosphoryl group is transferred to an aspartate residue on the response regulator, and the phosphorylated response regulator functions in general as a transcription factor for target genes. The target genes of a particular two-component signal transduction system are customized to the specific signal to which the particular two-component signal transduction system corresponds (2, 3). This specificity is also reflected by the high specificity of sensor kinase and response regulator pairs (4, 5). However, contrary to the growing knowledge on signal transmission between conserved cytosolic domains in two-component signal transduction system proteins, signal integration is only poorly understood (2, 3).
In contrast to other two-component signal transduction systems, and therefore underlying its impressive role for biological processes in Gram-negative bacteria, the Cpx pathway composed of the sensor kinase CpxA and the response regulator CpxR integrates very different signals. These include physical (osmolarity), chemical (ethanol, pH, indol), and biological (adhesion, lipids) stresses as well as misfolded proteins (adhesin subunits, β-barrel outer membrane proteins, and misfolded variants of the maltose-binding protein) (6–9). In addition, the Cpx pathway is negatively modulated by the periplasmic auxiliary protein CpxP (10). Molecular biological and biochemical analysis of several Cpx signals supports the notion that most signals are specific (11). This highlights the Cpx pathway as a model system to determine the mechanisms involved in signal transduction by a two-component signal transduction system, ranging from signal integration by the sensor kinase (CpxA) to the output response by the response regulator (CpxR).
Mechanistic details about the precise action of any Cpx-modulating signal remain unknown (11). Only for the inhibition by CpxP and the activation by misfolded proteins have some details been clarified (11–13). Thereby, cpxP was identified as a CpxR target gene that combats expression of toxic envelope proteins including misfolded pilus subunits of P pili that are crucial for uropathogenic Escherichia coli during kidney colonization (11, 14–16). The biogenesis of P pili depends on the specific pilus chaperone PapD; without PapD, pilus subunits misfold (17). Misfolded forms of the pilus subunit PapE and the adhesin PapG induce the Cpx envelope stress response and are toxic for wild-type cells (15). Because the overexpression of CpxP leads to degradation of misfolded PapE and PapG by the DegP protease, CpxP was proposed as an adaptor for DegP (11, 18). However, it is not clear how the two important biological functions of CpxP are linked (10).
Here, we performed structural and functional studies to understand how CpxP is able to support degradation of misfolded pilus subunits and to inhibit CpxA. We now report the structure of CpxP at 1.45 Å resolution. Our data provide for the first time structural and biochemical evidence for the notion that Cpx pathway inhibition results from direct protein-protein interaction between the CpxA sensor domain and CpxP. Interaction of misfolded pilus subunits with an elongated hydrophobic cleft on the convex surface of CpxP induces the release of CpxP from CpxA. This activation of the Cpx pathway results in the expression of Cpx targets including all players essential for the biogenesis and quality control of pili and consequently in adhesion. Accordingly, our results suggest the protein-protein interaction between CpxP and pilus subunits as a novel target for a new class of antimicrobials.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
CpxP and CpxA were amplified from E. coli MG1655 (19), and PapE was amplified from uropathogenic strain CFT073 (20) by PCR. For overproduction and purification of CpxP without its signal peptide, pRF06 plasmid was used (12). CpxPΔ151, which is in addition deleted for the C-terminal residues Lys151–Gln166, is a derivative of pRF06Δ151. pSHE100 and pTcpxP-His encode CpxP without and with a C-terminal His6 fusion, respectively, on pTrc99A. To investigate CpxP-dependent degradation of the PapE adhesin, papE was cloned with PCR-fused Strep-tag of the vector pASK-IBA3 (IBA GmbH) into pTrc99A, resulting in pSHE101. pSHE102 and pSHE102Δ151 encode CpxP and CpxPΔ151, respectively, with the Shine-Dalgarno sequence of pBAD24 on pBAD33 (21). The nucleotide sequence of the periplasmic signaling domain of CpxA encoding residues Pro28–Pro164 was cloned into pET15b, resulting in pZXH03. Mutagenesis was performed with the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. All constructs were confirmed by sequencing. A list of all constructs is given in supplemental Table 1.
Purification of CpxP and CpxPΔ151 followed established protocols (12) with additional purification by a Sephacryl-S-200 column (GE Healthcare). For functional studies, the His6 fusion was cleaved off using the thrombin CleanCleave kit essentially as described (Sigma). Purified protein was buffered into crystallization buffer P3 (10 mm Tris/HCl, pH 7.5, 100 mm NaCl, 5% glycerol (v/v)). Protein labeling with selenomethionine (Se-Met)7 was conducted as described (22), and protein was purified as described for the unlabeled protein without thrombin cleavage.
[35S]Methionine labeling of CpxA28–164 and CpxP followed an established protocol (23). [35S]Methionine-labeled protein was purified with the Protino kit (Macherey-Nagel) according to the manufacturer's instructions.
Steady-state Analysis by Immunological Determination
CpxP-dependent degradation of the PapE adhesin was determined according to an established protocol (11) with minor modifications. In brief, E. coli strains TG1 and TG1cpxP::kan (24) transformed with pSHE101 and pSHE102 or pSHE102Δ151, respectively, were subcultured from fresh overnight cultures to A600 = 0.1 in 10 ml of LB supplemented with 0.02% arabinose at 37 °C. Expression of PapE-Strep was induced at an A600 = 0.5 by the addition of 1 mm isopropyl-1-thio-β-d-galactopyranoside, and cells were grown for one further hour. Cells were harvested by centrifugation, and cell pellets were normalized to the same A600 and resuspended in TS buffer (20 mm Tris/HCl, pH 7.5; 0.5 m sucrose). Cells were mixed with SDS-PAGE loading buffer and lysed by boiling for 5 min. 20 μl of each sample were separated on a 15% SDS-polyacrylamide gel. Proteins were electroblotted, and immunoblots were probed with antiserum to CpxP (produced against purified His6-CpxP by Pineda, Berlin, Germany), with peroxidase conjugated anti-Strep antiserum (IBA GmbH) or with antiserum to MalE.
Preparation of Proteoliposomes and Kinase Assay
Preparation of proteoliposomes and measurement of CpxA autophosphorylation activity were performed essentially as described (12). Three determinations of independently prepared proteoliposomes were averaged to obtain the indicated values.
Crystallization of Full-length CpxP and CpxPΔ151
Se-Met-labeled full-length CpxP and CpxPΔ151 were used for crystallization at concentrations up to 8 mg/ml. Crystallization screens by the sparse matrix method were carried out by the sitting drop vapor diffusion method testing more than 2,000 crystallization conditions (25) at 295 K using 24-well Linbro plates. Each sitting drop was prepared on a siliconized crystallization bridge by mixing equal volumes (5 μl each) of CpxPΔ151 and reservoir solution (16–20% polyethylene glycol 3350, 5% glycerol, 100 mm NaCl, 10 mm CaCl2 and 10 mm Tris/HCl, pH 7.0). Se-Met-labeled crystals appeared within 8–12 days and continued to grow for 14 days. Crystals were frozen in reservoir buffer supplemented with 10% polyethylene glycol 400 as cryoprotectant.
Data Collection and Structure Determination
Diffraction data of native and Se-Met-labeled crystals were collected at synchrotron beamline BL 14.2 at Bessy-MX/Helmholtz Zentrum Berlin für Materialien und Energie with a MAR-225CCD detector. All diffraction images were indexed, integrated, and scaled using HKL2000 (26), showing that native and Se-Met-labeled CpxPΔ151 crystals belong to either hexagonal space group P61 or hexagonal space group P65 (a = b = 61.46 Å, c = 130.08 Å, α = β = 90°, γ = 120°). Three multiwavelength anomalous dispersion datasets at different wavelengths (inflection, peak, and remote) were collected to 2.0, 2.0, and 1.45 Å resolutions, respectively. Initial phases were determined to 2.0 Å resolution based on all multiwavelength anomalous dispersion datasets by using SHELXC/D/E (27) via the graphical user interface HKL2MAP (28) to solve the selenium-atom substructure. Substructure solution with SHELXD was successful for space group P65, yielding 14 selenium sites (with occupancies greater than 0.62). This solution gave a mean figure-of-merit of 0.724 after phasing with SHELXE that increased to 0.770 after density modification with the program DM (29). The electron density map was readily interpretable, and the phases were input for initial automated model building using ARP/wARP 7.0.1 (30). The initial model consisting of 208 residues had an agreement factor Roverall of 24.5%. In subsequent steps, this model was subjected to torsion angle molecular dynamics simulated annealing using a slow cooling protocol and a maximum likelihood target function, energy minimization, and B-factor refinement against the remote Se-Met dataset in the extended resolution range 53.22–1.45 Å by the program CNS (31). Restrained individual B-factors were refined, and the crystal structure was finalized by REFMAC5 and other programs in CCP4 (32). The final model had agreement factors Rfree and Rcryst of 18.9 and 16.8%, respectively. Manual rebuilding of the CpxPΔ151 model and electron density interpretation were performed after each refinement cycle using the program COOT (33). Structure validation was performed with the programs PROCHECK (34) and WHAT_CHECK (35). Potential hydrogen bonds involved in inter- and intramolecular contacts (supplemental Table 1) were analyzed using the programs HBPLUS (36) and LIGPLOT (37). The CpxP structure was analyzed for salt bridges utilizing the WHAT IF server (38). Electrostatic surface potentials were calculated through solution of the Poisson-Boltzmann equation using the program APBS (39). The solvent-accessible area was calculated using the PISA server (40). Fold recognition for CpxP was carried out utilizing the DALI server (41). All molecular graphics representations were created using PyMOL (31).
Size-exclusion Chromatography
Size-exclusion chromatography was carried out on a Superdex 200 (HR10/30) column (GE Healthcare) equilibrated in 50 mm Tris/HCl, pH 7.5, containing 150 mm NaCl and 0.5% glycerol (v/v) at 4 °C. Protein samples (100 μl at 0.7 mg ml−1) were applied to the column at a flow rate of 0.5 ml min−1. The following standard proteins were used for calibration (supplemental Fig. S3A): ferritin (Mr = 440,000; Stokes radius, RS = 6.15 nm), aldolase (Mr = 158,000, RS = 4.70 nm), bovine serum albumin (Mr = 66,399, RS = 3.48 nm), and carbonic anhydrase (Mr = 29,000, RS = 2.01 nm). There is a relationship between Kav and the Stokes radius of a protein (42, 43). Kav is defined as
where Ve is the elution volume of the protein, V0 is the void volume of the column, and Vt is the total volume of the column. The void volume was determined by elution of blue dextran 2000. A calibration curve of the column was obtained by plotting −(log Kav)½ versus Stokes radii.
Sucrose Gradient Sedimentation Analysis
Linear gradients (10 ml) of 5–20% sucrose in 50 mm Tris-HCl, pH 7.5 were prepared. 400 μg of purified CpxPΔ151 protein in 300 μl of crystallization buffer P3 were layered on the top and centrifuged at 100,000 × g at 14 °C for 18 h. 500-μl fractions were collected from the bottom of the tube and analyzed by SDS-PAGE. The gradient was standardized by sedimenting 500 μg of the following standard proteins of known sedimentation coefficients (supplemental Fig. S3B): thyroglobulin (Mr = 669,000; S = 18.32 × 10−13 s), apoferritin (Mr = 443,000; S = 16.84 × 10−13 s), alcohol dehydrogenase (Mr = 150,000; S = 7.61 × 10−13 s), bovine serum albumin (Mr = 669,000; S = 4.3 × 10−13 s), and carbonic anhydrase (Mr = 669,000; S = 2.89 × 10−13 s). After determination of the sedimentation coefficient of CpxPΔ151, the following equation was used to calculate the molecular mass (43)
where η is the viscosity of the medium (value used = 1), N is Avogadro's number, a is the Stokes radius, S is the sedimentation coefficient, υ the partial specific volume (value used = 0.735 cm3 g−1), and ρ is the density of the medium (value used = 1 g cm−3).
β-Galactosidase Activity Analysis and Determination of CpxP Expression Levels
β-Galactosidase assays were performed in strain SP594 (16) as described (44) with four biological replicates each with technical triplicates. Cells from β-galactosidase activity analysis were fractionated by spheroplast preparation as described (8). Periplasmic fractions were analyzed by immune blotting with antiserum to CpxP (Pineda) or with antiserum to MalE and detected with an ECL kit (GE Healthcare). Total protein bands of one sample were visualized and quantified by phospho imaging using Molecular Imager FX and associated software Quantity One (Bio-Rad).
Chaperone Assays
CpxP effects on thermal aggregation at 43 °C and on renaturation of citrate synthase were analyzed by determining citrate synthase activity as described (45). For refolding, citrate synthase was denatured in 6 m guanidinium hydrochloride, 50 mm Tris/HCl, pH 8.0, 20 mm dithiothreitol at room temperature for at least 2 h.
Peptide Synthesis on Cellulose Membranes (SPOT Synthesis) and Screening of Peptide Arrays
Cellulose-bound peptide libraries were prepared by semiautomatic SPOT synthesis on modified Whatman 50 cellulose membranes as described (46). Sequence files and array design were generated with the in-house software LISA. Peptides derived from E. coli MG1655 CpxA (GenBankTM GeneID: 948405) and CpxP (GenBank GeneID: 2847688) were used for pepscan analyses. Peptide arrays consisting of 15-, 20-, or 25-mer peptides, overlapping by 12, 17, and 22 amino acid residues, respectively, were synthesized. Complete substitutional and length analyses of the interacting 25-mer peptide were generated using the software LISA and subsequently synthesized as described (46, 47).
Dried membranes were washed in ethanol for 10 min and in TBS (50 mm Tris/HCl, pH 8, 137 mm NaCl, 27 mm KCl) for 3 × 10 min. Blocking was performed in TBS, supplemented with 5% blocking buffer (Sigma) and 5% (w/v) sucrose, at room temperature for 3 h. After washing with TTBS (TBS with 10% (v/v) Tween 20) for 10 min, peptide arrays were incubated with 35S-labeled protein (105 cpm) in blocking buffer, supplemented with 20% glycerol with gentle shaking at 4 °C over night. Unbound protein was removed with TBS, and peptide-bound protein was visualized and quantified by phospho imaging using Molecular Imager FX and associated software Quantity One (Bio-Rad).
RESULTS
Overall Structure of CpxPΔ151
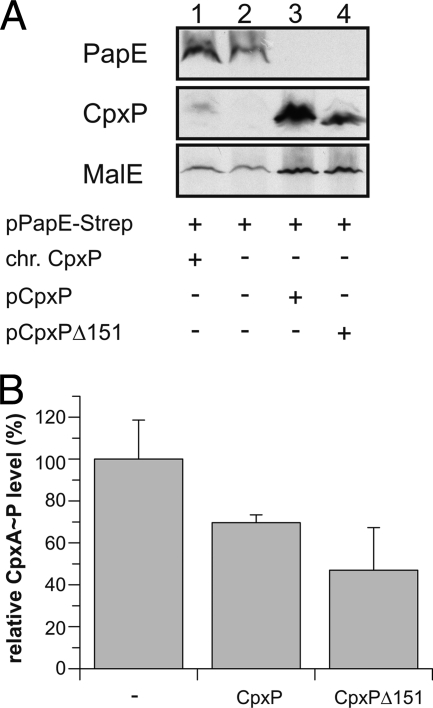
To gain insight into the dual functions of CpxP, i.e. sensor kinase inhibition and supporting the degradation of misfolded pilus subunits, it was of interest to determine the structure of CpxP. We expressed and purified the E. coli CpxP protein (CpxP21–167) without its signal sequence from the cytosol (12). Crystals prepared from this truncated CpxP protein diffracted only to moderate a 5 Å resolution. We therefore created a more protease-resistant variant CpxPΔ151 lacking the 16 C-terminal residues that were predicted to be unfolded (CpxP21–151) (supplemental Figs. S1 and S2). This variant was indistinguishable from the full-length protein with respect to both known functions, i.e. supporting the degradation of misfolded pilus subunits (Fig. 1A), and prevented activation of the CpxA sensor kinase (Fig. 1B).
FIGURE 1.
CpxPΔ151 CpxP has two functions. A, steady-state analysis of PapE by immunological determination according to an established protocol (11) with two modifications as described in detail under “Experimental Procedures.” Cells expressing PapE-Strep and indicated CpxP variants were subjected to immunological determination using antiserum to the Strep-tag, the CpxP protein, and the MalE protein (loading control), respectively. B, CpxA autophosphorylation activity was measured by incubating proteoliposomes containing purified CpxA-His6 (1 μm) in phosphorylation buffer containing [γ-32P]ATP for 20 min. To test the influence of CpxP variants on CpxA autophosphorylation, the experiment was performed as described (12) with wild-type CpxP or CpxPΔ151-loaded proteoliposomes. Samples were separated by SDS-PAGE and, analyzed as phosphorimages, and the amounts of [32P]ATP were quantified by phospho imaging using [γ-32P]ATP as a standard. Shown are averages ± S.E. from three different experiments (t test).
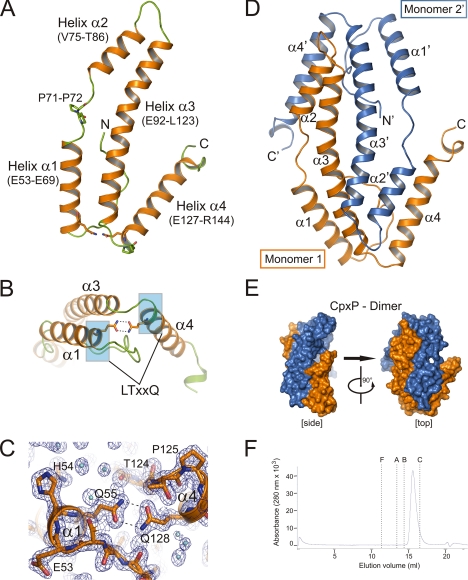
We crystallized CpxPΔ151 in the hexagonal space group P65 with two monomers in the asymmetric unit that form dimers of non-crystallographic C2 symmetry. The structure of CpxPΔ151 was solved at 1.45 Å resolution by multiwavelength anomalous dispersion using selenomethionine-labeled crystals (Table 1). The CpxPΔ151 final structural model comprises 111 out of 131 residues of the truncated CpxPΔ151 monomer (Fig. 2A) and shares homologies with the structure of the spheroplast protein Spy (2. 7 Å resolution), a protein with unknown function (48). The CpxPΔ151 monomer can be divided into three distinct regions (Fig. 2A): 1) a partially unfolded N-terminal region of which only residues Ser40 to Thr52 were visible in the electron density map; 2) a curved laminar region divided into two antiparallel α-helical segments, the first consisting of helix α1 (Glu53–Glu69) and helix α2 (Val75–Thr86) connected by a linker that contains two consecutive prolines (Gln70–Asn74) and the second being the elongated helix α3 (Glu92–Leu123); and 3) a region consisting of helix α4 (Glu127–Arg144), which projects at a 35° tilt angle from α3, and a short C-terminal segment (Asp145–Gln150). Remarkably, the V-shaped structure formed by helices α3 and α4 in a CpxPΔ151 monomer is stabilized by a strong intramolecular interaction consisting of a double hydrogen bond between the side-chain amide groups of Gln128 in helix α4 and Gln55 in helix α1 (Fig. 2, B and C). Gln55 and Gln128 are key residues of two highly conserved LTXXQ motifs that can be used to identify CpxP homologues (supplemental Fig. S1). Mutations at these two glutamine residues affect both inhibitory function and protein stability (49). Thus, the structure of CpxPΔ151 elucidates the importance of the conserved glutamines and suggests an essential role of the LTXXQ motifs for the fold of CpxP.
TABLE 1.
Data collection and refinement statistics for CpxPΔ151
| Se-Meta CpxPΔ151 (λremote = 0.9184Å) | Se-Meta (λpeak = 0.9797Å) | Se-Meta (λinflection = 0.9799Å) | |
|---|---|---|---|
| Data collection | |||
| Space group | P65 | ||
| Cell dimensions | |||
| a, b, c (Å) | 61.46, 61.46, 130.08 | ||
| α, β, γ (°) | 90.0, 90.0, 120.0 | ||
| Resolution (Å) | 120.0-1.45 (1.50-1.45)b | 50.0-2.00 (2.03-2.00)b | 50.0-2.00 (2.03-2.00)b |
| Rmerge | 0.084 (0.567) | 0.048 (0.117) | 0.049 (0.158) |
| 〈Ι/σΙ〉 | 18.9 (2.4) | 16.6 (6.1) | 16.0 (4.2) |
| Completeness (%) | 100.0 (100.0) | 99.8 (99.9) | 99.8 (99.9) |
| Redundancy | 5.7 (5.3) | 1.9 (1.9) | 1.9 (1.9) |
| Refinement statistics | |||
| Resolution (Å) | 53.22-1.45 | ||
| No. of reflections | 46,652 | ||
| Rwork/Rfree | 16.8/18.9 | ||
| No. of atoms/residues(2 monomers per asymmetric unit) | |||
| Protein (CpxPΔ151) | 1,922/220 | ||
| Water | 348/348 | ||
| Mean B-factor (all atoms; Å2) | 16.02 | ||
| r.m.s.c deviations | |||
| Bond lengths (Å) | 0.013 | ||
| Bond angles (°) | 1.416 | ||
| Ramachandran plotd,e (%) | |||
| Most favored | 95.1d/100e | ||
| Allowed | 4.9d/0e | ||
| Generously allowed and disallowed | 0.0d/0e | ||
FIGURE 2.
Crystal structure of CpxPΔ151. A, ribbon representation of a CpxPΔ151 monomer containing four helices α1 to α4 (helices, orange; loops, green). Two characteristic glutamine residues of the conserved LTXXQ repeat motifs (Leu51–Gln55 and Leu124–Gln128) and a PP motif (Pro71–Pro72) are shown as sticks. B, the monomer conformation with a V-shaped structure formed by helices α3 and α4 is stabilized by a double hydrogen bond between the conserved LTXXQ motifs (highlighted in blue) in helices α1 and α4. C, close-up view of the region surrounding the double hydrogen bond between Gln55 and Gln128 shown with 2Fo − Fc as electron density map (blue mesh) contoured at 1.2σ. D, the asymmetric unit contains a CpxPΔ151 dimer with two monomers that are interdigitated like left hands. Distinct perspectives of the CpxPΔ151 dimer are shown as surface representation (CpxPΔ151 monomer 1, blue; CpxPΔ151 monomer 2′, orange). E, ribbon representation of the CpxPΔ151 dimer using the same color scheme as in D. F, size-exclusion chromatography of CpxP was carried out on a Superdex 200 (HR10/30) column (GE Healthcare) equilibrated in 50 mm Tris/HCl, pH 7.5, containing 150 mm NaCl and 0.5% glycerol (v/v) at 4 °C. Protein samples (100 μl at 0.7 mg ml−1) were applied to the column at a flow rate of 0.5 ml min−1. The elution volumes for the molecular weight standards are shown by dashed lines (F = ferritin, A = aldolase, B = bovine serum albumin, C = carbonic anhydrase). See supplemental Fig. S3 for molecular mass determination of CpxPΔ151 by the combination of size-exclusion chromatography and sucrose-gradient sedimentation analysis.
CpxP Functions as a Dimer
Two CpxP monomers are interdigitated like “left hands” forming a cap-shaped dimer with helices α4 and α4′ being the “thumbs” (Fig. 2, D and E). We characterized the oligomeric state of CpxPΔ151 further by size-exclusion chromatography and sucrose gradient sedimentation (Fig. 2F and supplemental Fig. S3). According to these, the Stokes radius and the sedimentation coefficient were determined to be 2.28 nm and 3.223 × 10−13 s, respectively. Using Equation 2, we calculated a molecular mass of 31.0 kDa, which fits very well with the theoretical molecular mass for the CpxPΔ151 dimer of 31.7 kDa.
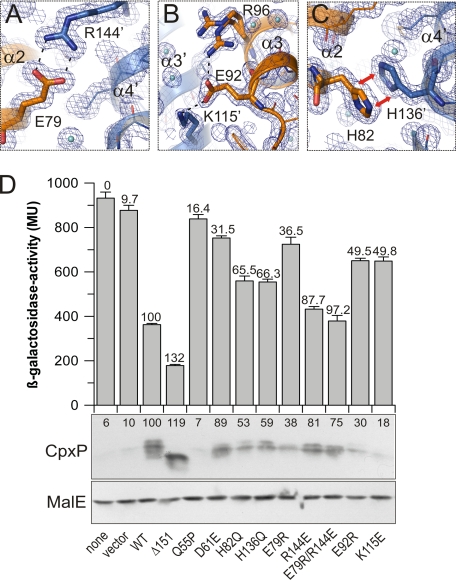
The CpxP dimer interface exhibits two remarkable salt bridges and a striking intermolecular π-π-stacking interaction as key features locking the two monomers in a stable dimer conformation. The first salt bridge is formed between Glu79 and Arg144 in helices α2 and α4′, respectively (Fig. 3A and supplemental Table 2). The second salt bridge involves Glu92 and Lys115 and connects helices α3 and α3′, respectively (Fig. 3B and supplemental Table 2). The electrostatic component of this interaction is additionally screened by Arg96, which adopts two conformations in the crystal structure. These solvent-exposed salt bridges are suggested to be strong as they require only low desolvation energies, and weakening of the electrostatic interactions by solvent screening may be more than compensated by very strong hydrogen bonds of favorable geometry (50). The π-π-stacking interaction is formed between His82 in helix α2 and His136 of helix α4′, respectively (Fig. 3C and supplemental Table 2), and His82 interacts via a further hydrogen bond with Asn133 in helix α4′. The dimer interface is also stabilized by a hydrogen bond between Val85 and Asn118 of helices α2 and α3′, respectively, and several hydrophobic interactions involving helices α2, α3, and α4 as shown by a list of van der Waals contacts (supplemental Table 2).
FIGURE 3.
CpxPΔ151 dimer stability is accomplished by intermolecular contacts. A–C, salt bridges between Glu79 and Arg144 (A) and Glu92 and Lys115 (B) and a π-π-stacking between His82 and His136 (C) stabilize the CpxPΔ151 dimer. D, CpxP-mediated Cpx pathway inhibition was investigated by β-galactosidase activity measurement (upper panel). Shown are averages ± S.E. from four independent determinations each with three replicates (t test). CpxP levels in periplasmic fractions were analyzed by immunoblotting (middle panel). Numbers above bars give the relative inhibition of β-galactosidase activity (given in Miller Units MU) or the relative protein amount, both normalized for wild-type CpxP. The CpxP loss-of-function variants Q55P and D61E were used as functional controls (49) (supplemental Fig. S4), and MalE was used as loading control (lower panel).
We verified the functional and structural importance of the prominent two salt bridges and the π-π-stacking interaction sites for the CpxP dimer by analyzing the inhibitory effect of overproduced CpxP variants, which carried mutations of the amino acid residues involved in these interactions on the Cpx pathway (Fig. 3D). All mutants were impaired in their inhibitory capacity, although to different extents. Moreover, the reduction of inhibition correlated overall with protein levels, indicating an effect on protein stability. In contrast to expression in the periplasm, all CpxP mutants were purified as stable dimers from the cytosol (supplemental Fig. S3). Furthermore, when compared with native CpxP, higher protein levels of CpxPΔ151 as well as of a C-terminally tagged CpxP were found in the periplasm (supplemental Fig. S4), indicating a role of the C-terminal peptide for the recognition of CpxP by the quality control system.
CpxP Exhibits a Chaperone-like Activity
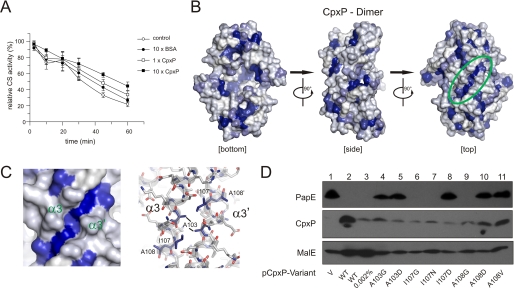
It has been proposed that Cpx pathway activation is caused by titrating CpxP away from CpxA (11, 51). A prerequisite of this scenario is the detection of unfolded proteins by CpxP, which might result from a chaperone-like activity (52, 53) but has never been shown. We used two classical chaperone activity assays (45) and found that CpxP significantly prevents the thermal aggregation of citrate synthase at a 10-fold molar excess (Fig. 4A) but was unable to restore citrate synthase activity (data not shown). We propose this moderate chaperone-like activity to be sufficient to allow titration of CpxP away from CpxA and to reflect preferential binding of CpxP to proteins that have started to unfold as well as high substrate specificity, as described for the periplasmic chaperone SurA (54).
FIGURE 4.
Chaperone-like function of CpxP. A, chaperone function of CpxP was investigated by determining the activity of citrate synthase (CS) during thermal stress in the absence of additional protein (control) or in the presence of BSA or CpxP at the indicated concentrations. Shown are averages ± S.E. from three independent determinations (t test). B, hydrophobic surface representation of the CpxPΔ151 dimer. The coloring scheme reflects the hydropathic index at a surface position as calculated by the method of Kyte and Doolittle (61) to analyze the hydrophobic character of the surface of the protein. Regions with a hydropathy index above 0 are hydrophobic in character and depicted in blue. Hydrophilic regions are shown in gray. The hydrophobic cleft on the convex surface (green) of the CpxPΔ151 dimer is highlighted. C, detailed views of the hydrophobic cleft between the two symmetry-related long helices α3 and α3′ as hydrophobic surface and stick representations. D, steady-state analysis of PapE by immunological determination was performed as described for Fig. 1. For control purposes, we used the empty vector (V, lane 1) and wild-type CpxP with 10-fold reduced arabinose concentration (lane 3).
The hallmark for classical chaperones, an expanded hydrophobic patch that binds unfolded proteins (53), is absent at the surface of CpxP (Fig. 4B). However, the concave surface of the cap-shaped dimer includes two potential hydrophobic recognition sites related by the non-crystallographic C2 symmetry and located in the intermolecular contact region of the dimer between helices α2 and α4′ and helices α2′ and α4, respectively, including the hydrophobic amino acid residues Pro72, Val73, Val76, and Leu78 on helices α2 and α2′ and residues Met140, Leu143, and Trp149 on helices α4 and α4′ (Fig. 4B).
Even more prominent is an elongated hydrophobic cleft on the convex surface (Fig. 4B). This cleft is situated in the intermolecular contact region between the two symmetry-related, antiparallel oriented long helices α3 and α3′ and is formed by pairs of symmetry-related hydrophobic amino acid residues Ala103, Ile107, and Ala108 (Fig. 4C). To corroborate the functionality of the hydrophobic cleft, we analyzed the capacities of the CpxP single-site A103G, A103D, I107G, I107D, I107N, A108G, A108D, and A108V substitutions to promote PapE degradation (Fig. 4D). All substitutions impaired protein levels, indicating a significant contribution of the intermolecular contact region between helices α3 and α3′ to dimer assembly. However, all substitutions resulted in protein levels that at least correlate to the protein level observed for native CpxP after only slight induction (Fig. 4D, lane 3). Substitutions A103G, A103D, I107D, A108D, and A108V significantly impaired the degradation of PapE, supporting the relevance of the hydrophobic cleft for interaction between CpxP and PapE. Accordingly, we suggest that CpxP is involved in pilus sensing.
Direct Protein-Protein Interaction between CpxP and CpxA
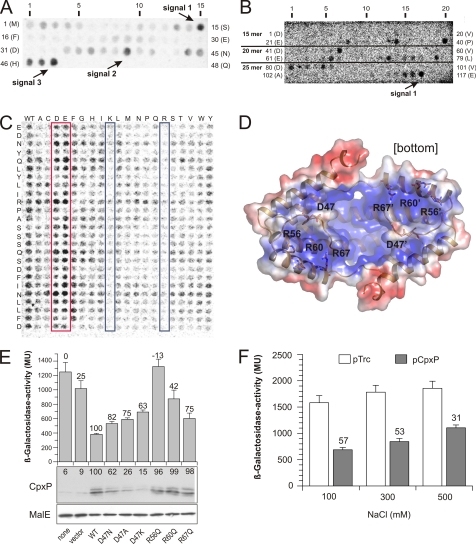
It is hypothesized that direct protein-protein interaction between CpxP and the periplasmic sensor domain of CpxA is essential for CpxP inhibitor function (6, 12). To test this hypothesis, we screened cellulose-bound peptide arrays, representing the complete CpxP sequence (Met1–Gln166), for interaction with the purified, soluble periplasmic CpxA sensor domain. Three regions interacting with the CpxA sensor domain were detected on CpxP, one covering helix α1 and two covering the entire C terminus (Fig. 5A and supplemental Fig. S1). Reversely, only the C-terminal region of the CpxA sensor domain was clearly accessible for purified CpxP (Fig. 5B and supplemental Fig. S5). Surprisingly, a complete substitution analysis of this CpxA region did not identify any specific epitope essential for CpxP binding (Fig. 5C). Amino acid substitutions that introduced an additional negative charge enhanced binding, whereas substitutions that added a positive charge abolished binding. Analyzing the polarity of the CpxP dimer, we noted a positively charged polar patch on the concave surface (Fig. 5D). This polarity is mainly contributed by residues Asp47, Arg56, Arg60, and Arg67. Of these, an R60Q substitution was described as a stable loss-of-function mutant (49). We further determined the inhibitory capacity of the CpxP single-site D47N, D47A, D47K, R56Q, R60Q, and R67Q substitutions on the Cpx pathway activity (Fig. 5E). Substitution of Asp47 results in unstable variants, likely due to its role in stabilizing the conformation of the N-terminal loop. However, substitutions of Arg56, Arg60, and Arg67 on helix α1 of CpxP resulted in stable variants that were unable to inhibit Cpx pathway activation (Fig. 5E). Moreover, the inhibitory capacity of CpxP decreased with increased salt concentration in the medium (Fig. 5F). Altogether, it is very likely that the cap-shaped CpxP dimer interacts via its concave polar surface with the negatively charged sensor domain on CpxA.
FIGURE 5.
Protein-protein interaction between CpxP and CpxA. A, a peptide array derived from the sequence of CpxP was screened for binding of the 35S-labeled CpxA sensor domain (CpxA28–164-His6) as described under “Experimental Procedures.” Rows are labeled for peptide numbers and the N-terminal peptide residues. Those spots exhibiting the strongest signal within a serial of increasing signals are highlighted. B, reverse experiment to A. Peptide arrays (15-mer, 20-mer, and 25-mer) derived from the sequence of the CpxA sensor domain were prepared and incubated with 35S-labeled CpxP-His6. One spot exhibiting the strongest signal within a serial of increasing signals of the 25-mer peptide array that covers sequences of signals of the 15-mer and 20-mer peptide arrays is highlighted. C, complete substitutional and length analysis of the interacting 25-mer peptide of the CpxA-derived peptide recognized by CpxP was generated using the software LISA and subsequently synthesized as described under “Experimental Procedures.” Each amino acid of the peptides (25-mer) corresponding to signal 1 in B is substituted by all other 20 l-amino acids in alphabetical order (shown on top of the membrane) and tested for binding of 35S-labeled CpxP-His6. All spots in the left column comprise the wild-type sequence (WT) of the peptide. Those spots with increased negative or positive charged amino acids are highlighted by red and blue boxes, respectively. D, surface representation of CpxPΔ151 dimer. The electrostatic surface potentials were calculated using the program APBS (39) with the non-linear Poisson-Boltzmann equation and contoured at ± 3 k * T/e, where k is Boltzmann's constant; T, the temperature in K, and e, the charge of an electron. Negatively and positively charged surface areas are colored in red and blue, respectively. E, CpxP-mediated Cpx pathway inhibition was investigated as described for Fig. 3. F, the salt dependence of CpxP-mediated Cpx pathway inhibition was investigated by β-galactosidase activity measurement using the strain SP594 (16). CpxP was expressed from vector pTrc99A (Invitrogen). Averages ± S.E. from five independent determinations each with three replicates are shown. Numbers above the bars give the relative inhibition of β-galactosidase activity at the indicated NaCl concentrations.
DISCUSSION
Signal integration by sensor kinases is still only poorly understood (1, 3). This lack of knowledge is due to the high degree of variation in sequence and structure between the individual sensing domains of sensor kinases and reflects the diversity of signals detected by this class of sensors (1, 3). An important aspect in two-component system signaling is provided by auxiliary proteins (2, 10), which confer responsiveness of two-component systems to additional signals. Accordingly, structural and functional studies on auxiliary proteins allow a deeper insight into signal perception and regulation of sensor kinase activities. The crystal structure of CpxP presented here is the first of a dual function auxiliary protein.
The structure of CpxP is special for several reasons. First, in the CpxP monomer, the V-shaped structure formed by two α-helices is stabilized by an essential intramolecular double hydrogen bond between the glutamine residues of two highly conserved LTXXQ motifs (Fig. 2, B and C). The importance of this intramolecular interaction is supported by a finding of the Raivio group (49) that mutation of each of the glutamine residues results in unstable proteins. Second, CpxP forms a stable cap-shaped dimer. The stability of the dimer is promoted by two salt bridges and a π-π-stacking interaction (Fig. 3, A–C). The significance of this finding is that only the CpxP dimer is able to promote both functions of the protein, i.e. inhibition of a sensor kinase and escorting of misfolded pilus subunits to degradation. The structure presented here supports the idea that the dual functions of the CpxP dimer are accomplished by two specific surface features: a hydrophobic cleft on its concave surface (Fig. 4C) and a positively charged polar patch on its convex surface (Fig. 5D).
Our data provide for the first time structural and biochemical evidence for the notion that Cpx pathway inhibition results from direct protein-protein interaction between the CpxA sensor domain and helix α1 of the positively charged polar patch on the convex surface of CpxP (Fig. 5D). We suggest that this interaction prevents not only signaling pathway activation by an inhibitory interaction keeping the kinase in an “off” mode (12) but also by positioning the CpxP dimer as a patch to shield the CpxA sensor domain from inducing signals and/or to prevent dimerization of CpxA essential for the autophosphorylation activity of any sensor kinase. In addition, we prove here for the first time a chaperone-like function for CpxP (Fig. 4A) that is likely to be promoted by the hydrophobic cleft (Fig. 4, C and D). Thus, our results not only support a model that misfolded proteins titrate CpxP away from CpxA (11) but also indicate that sensing pilus subunits by interaction with the hydrophobic cleft on the convex surface of CpxP induces the release of CpxP from CpxA (Fig. 6). This activation of the Cpx pathway results in the expression of Cpx targets including all players essential for the biogenesis and quality control of pili. Meanwhile, CpxP escorts the misfolded pilus subunits for degradation.
FIGURE 6.
A model of signal integration by CpxP. Direct interaction of the cap-shaped CpxP dimer (ribbon representation) via its concave polar surface with the negatively charged sensor domain of CpxA keeps the kinase in an off mode. Direct interaction of misfolded pilus subunits with the hydrophobic cleft on the convex surface on CpxP results in the release of CpxP from CpxA and switches the kinase in an “on” mode. The release of CpxP from CpxA results in CpxR activation, which acts as inducer (arrow) or repressor (bar) for target gene expression.
CONCLUSION
The mechanism how CpxP regulates CpxA activities differs substantially from known or suggested regulatory mechanisms of other auxiliary proteins (reviewed in 10). In contrast to other auxiliary proteins, the regulatory function of CpxP is based on two protein-protein interactions: one with the sensor kinase CpxA and the other with a misfolded pilus subunit as substrate. Furthermore, interaction with CpxA is not permanent, as shown for the autoinducer-binding protein LuxP and the bioluminescence controlling LuxQ sensor kinase in Vibrio harveyi (55), but rather substrate-regulated, as reported for the sugar-binding protein ChvE that induces VirA-mediated tumor formation of dicotyledonous plants by Agrobacterium tumefaciens (56). However, binding of the sugar-ChvE complex to the VirA HK does not inhibit but activates virulence gene expression (56).
Due to their central role in bacterial virulence regulation and their absence in animals including man, two-component signal transduction systems have been suggested as targets for antimicrobials (57, 58). Cpx targets include important virulence factors of different pathogenic bacteria as well as proteins essential for the secretion (59), biogenesis, and quality control of different surface structures including pili (51, 60). We suggest that blocking protein-protein interaction between CpxP and misfolded pilus subunits would prevent activation of the Cpx pathway and therefore suppress the formation of the essential biogenesis and quality control system of pili and consequently inhibit adhesion. Thus, the structural details from this study highlight CpxP as a suitable target for antimicrobials against a two-component signal transduction system acting from the exterior of the bacterial cell.
Supplementary Material
Acknowledgments
We thank E. Schneider, C. M. Spahn and K. P. Hofmann for continuous advisory support, K. Tedin for critical reading of the manuscript, and H. Landmesser for technical support. Strain TG1cpxP::kan was kindly provided by J.-M. Ghigo, strain SP594 was provided by T. J. Silhavy, and antiserum to MalE was provided by E. Schneider. We are grateful to the European Synchrotron Radiation Facility (ESRF, Grenoble, France), U. Müller, and the scientific staff of the BESSY-MX/Helmholtz Zentrum Berlin für Materialien und Energie at beamlines BL 14.1 and BL 14.2, where the data were collected, for continuous support.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (Hu1011/1-3) and the Yousef Jameel Foundation.
The atomic coordinates and structure factors (code 3ITF) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. S1–S5.
- Se-Met
- selenomethionine.
REFERENCES
- 1. Gao R., Stock A. M. (2009) Annu. Rev. Microbiol. 63, 133–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Szurmant H., White R. A., Hoch J. A. (2007) Curr. Opin. Struct. Biol. 17, 706–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cheung J., Hendrickson W. A. (2010) Curr. Opin. Microbiol. 13, 116–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Skerker J. M., Perchuk B. S., Siryaporn A., Lubin E. A., Ashenberg O., Goulian M., Laub M. T. (2008) Cell 133, 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Casino P., Rubio V., Marina A. (2009) Cell 139, 325–336 [DOI] [PubMed] [Google Scholar]
- 6. MacRitchie D. M., Buelow D. R., Price N. L., Raivio T. L. (2008) Adv. Exp. Med. Biol. 631, 80–110 [DOI] [PubMed] [Google Scholar]
- 7. Bury-Moné S., Nomane Y., Reymond N., Barbet R., Jacquet E., Imbeaud S., Jacq A., Bouloc P. (2009) PLoS Genet. 5, e1000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hunke S., Betton J. M. (2003) Mol. Microbiol. 50, 1579–1589 [DOI] [PubMed] [Google Scholar]
- 9. Gerken H., Leiser O. P., Bennion D., Misra R. (2010) Mol. Microbiol. 75, 1033–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Buelow D. R., Raivio T. L. (2010) Mol. Microbiol. 75, 547–566 [DOI] [PubMed] [Google Scholar]
- 11. Isaac D. D., Pinkner J. S., Hultgren S. J., Silhavy T. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17775–17779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fleischer R., Heermann R., Jung K., Hunke S. (2007) J. Biol. Chem. 282, 8583–8593 [DOI] [PubMed] [Google Scholar]
- 13. Keller R. F., Hunke S. (2009) Res. Microbiol. 160, 396–400 [DOI] [PubMed] [Google Scholar]
- 14. Hung D. L., Raivio T. L., Jones C. H., Silhavy T. J., Hultgren S. J. (2001) EMBO J. 20, 1508–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jones C. H., Danese P. N., Pinkner J. S., Silhavy T. J., Hultgren S. J. (1997) EMBO J. 16, 6394–6406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Danese P. N., Silhavy T. J. (1998) J. Bacteriol. 180, 831–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sauer F. G., Pinkner J. S., Waksman G., Hultgren S. J. (2002) Cell 111, 543–551 [DOI] [PubMed] [Google Scholar]
- 18. Krojer T., Sawa J., Schäfer E., Saibil H. R., Ehrmann M., Clausen T. (2008) Nature 453, 885–890 [DOI] [PubMed] [Google Scholar]
- 19. Blattner F. R., Plunkett G., 3rd, Bloch C. A., Perna N. T., Burland V., Riley M., Collado-Vides J., Glasner J. D., Rode C. K., Mayhew G. F., Gregor J., Davis N. W., Kirkpatrick H. A., Goeden M. A., Rose D. J., Mau B., Shao Y. (1997) Science 277, 1453–1462 [DOI] [PubMed] [Google Scholar]
- 20. Welch R. A., Burland V., Plunkett G., 3rd, Redford P., Roesch P., Rasko D., Buckles E. L., Liou S. R., Boutin A., Hackett J., Stroud D., Mayhew G. F., Rose D. J., Zhou S., Schwartz D. C., Perna N. T., Mobley H. L., Donnenberg M. S., Blattner F. R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 17020–17024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995) J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hendrickson W. A., Horton J. R., LeMaster D. M. (1990) EMBO J. 9, 1665–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Swidersky U. E., Rienhöfer-Schweer A., Werner P. K., Ernst F., Benson S. A., Hoffschulte H. K., Müller M. (1992) Eur. J. Biochem. 207, 803–811 [DOI] [PubMed] [Google Scholar]
- 24. Beloin C., Valle J., Latour-Lambert P., Faure P., Kzreminski M., Balestrino D., Haagensen J. A., Molin S., Prensier G., Arbeille B., Ghigo J. M. (2004) Mol. Microbiol. 51, 659–674 [DOI] [PubMed] [Google Scholar]
- 25. Jancarik J., Kim S. H. (1991) J. Appl. Crystallogr. 24, 409–411 [Google Scholar]
- 26. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 27. Sheldrick G. M. (2008) Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 28. Pape T., Schneider T. R. (2004) J. Appl. Crystallogr. 37, 843–844 [Google Scholar]
- 29. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 32. Collaborative Computational Project, Number Four (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 33. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 35. Hooft R. W., Vriend G., Sander C., Abola E. E. (1996) Nature 381, 272. [DOI] [PubMed] [Google Scholar]
- 36. McDonald I. K., Thornton J. M. (1994) J. Mol. Biol. 238, 777–793 [DOI] [PubMed] [Google Scholar]
- 37. Wallace A. C., Laskowski R. A., Thornton J. M. (1995) Protein Eng. 8, 127–134 [DOI] [PubMed] [Google Scholar]
- 38. Rodriguez R., Chinea G., Lopez N., Pons T., Vriend G. (1998) Bioinformatics 14, 523–528 [DOI] [PubMed] [Google Scholar]
- 39. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 41. Holm L., Sander C. (1997) Nucleic Acids Res. 25, 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sablonniere B., Lefebvre P., Formstecher P., Dautrevaux M. (1987) J. Chromatogr. 403, 183–196 [DOI] [PubMed] [Google Scholar]
- 43. Siegel L. M., Monty K. J. (1966) Biochim. Biophys. Acta 112, 346–362 [DOI] [PubMed] [Google Scholar]
- 44. Miller J. (1992) A Short Course in Bacterial Genetics 2: Escherichia coli and Related Bacteria, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 45. Buchner J., Grallert H., Jakob U., George H. L., Thomas O. B. (1998) Methods Enzymol. 290, 323–338 [DOI] [PubMed] [Google Scholar]
- 46. Wenschuh H., Volkmer-Engert R., Schmidt M., Schulz M., Schneider-Mergener J., Reineke U. (2000) Peptide Science 55, 188–206 [DOI] [PubMed] [Google Scholar]
- 47. Volkmer-Engert R., Ehrhard B., Höhne W., Kramer A., Hellwig J., Schneider-Mergener J. (1994) Lett. Peptide Sci. 1, 243–253 [Google Scholar]
- 48. Kwon E., Kim D. Y., Gross C. A., Gross J. D., Kim K. K. (2010) Protein Sci. 19, 2252–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Buelow D. R., Raivio T. L. (2005) J. Bacteriol. 187, 6622–6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kumar S., Nussinov R. (1999) J. Mol. Biol. 293, 1241–1255 [DOI] [PubMed] [Google Scholar]
- 51. Rowley G., Spector M., Kormanec J., Roberts M. (2006) Nat. Rev. Microbiol. 4, 383–394 [DOI] [PubMed] [Google Scholar]
- 52. DiGiuseppe P. A., Silhavy T. J. (2003) J. Bacteriol. 185, 2432–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hartl F. U., Hayer-Hartl M. (2002) Science 295, 1852–1858 [DOI] [PubMed] [Google Scholar]
- 54. Behrens S., Maier R., de Cock H., Schmid F. X., Gross C. A. (2001) EMBO J. 20, 285–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Neiditch M. B., Federle M. J., Pompeani A. J., Kelly R. C., Swem D. L., Jeffrey P. D., Bassler B. L., Hughson F. M. (2006) Cell 126, 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. He F., Nair G. R., Soto C. S., Chang Y., Hsu L., Ronzone E., DeGrado W. F., Binns A. N. (2009) J. Bacteriol. 191, 5802–5813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stephenson K., Hoch J. A. (2002) Curr. Opin. Pharmacol. 2, 507–512 [DOI] [PubMed] [Google Scholar]
- 58. Cegelski L., Marshall G. R., Eldridge G. R., Hultgren S. J. (2008) Nat. Rev. Microbiol. 6, 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. van Stelten J., Silva F., Belin D., Silhavy T. J. (2009) Science 325, 753–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Raivio T. L. (2005) Mol. Microbiol. 56, 1119–1128 [DOI] [PubMed] [Google Scholar]
- 61. Kyte J., Doolittle R. F. (1982) J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.