Abstract
For nuclear export of proteins, the formation of a ternary export complex composed of the export substrate, a cellular export factor and Ran-GTP is crucial. CRM1 is a cellular export factor for proteins containing leucine-rich nuclear export signals (NESs). Although the NES sequence is crucial for nuclear export, its exact role in the formation of the ternary export complex is controversial. Here we demonstrate an interaction between human CRM1 (hCRM1) and influenza A virus NS2 protein, which contains an NES motif in its N-terminal region. Replacement of the hydrophobic amino acids in the NES motif did not abolish NS2’s interaction with hCRM1. Using our recently established systems for the generation of influenza virus or virus-like particles from cloned cDNAs, we found that NS2 is essential for nuclear export of influenza virus ribonucleoprotein (RNP) complexes, and that alteration of the NS2-NES abrogated this event and influenza virus generation. These findings suggest that the NS2-NES is not crucial for the interaction of this protein with hCRM1, but is for the formation of the ternary export complex with Ran-GTP.
Keywords: CRM1/influenza/A virus/NS2 protein/nuclear export
Introduction
The transport of proteins or RNAs across the nuclear envelope is an active, energy-dependent process (reviewed in Mattaj and Englmeier, 1998) that requires specific targeting sequences within the substrates to be imported or exported. For the import of proteins, several types of nuclear localization signals (NLSs) that interact with cellular import receptors (e.g. karyopherin α and β) have been identified. Nuclear export is controlled by export receptors [e.g. the highly conserved CRM1 (chromosome region maintenance 1 protein)] that mediate the nuclear export of nuclear export signal (NES)-containing substrates. CRM1 is a nuclear export receptor for proteins containing leucine-rich NES characterized by 3–5 hydrophobic amino acids with characteristic spacing (Table I) (Fukuda et al., 1996; Fornerod et al., 1997a,b; Stade et al., 1997).
Table I. Alignment of previously identified leucine-rich NESs with the leucine-rich motif of influenza A virus NS2 protein.
| NES sequence | Reference | |
|---|---|---|
| Consensus | ψnψxxψxψ | |
| HIV-Rev | LxLxxLxxLxL | Fischer et al. (1995) |
| PKI-α | LxLxLxxLxI | Wen et al. (1995) |
| Influenza A NS2 | 12-ILMxMxxMxL-21 | O’Neill et al. (1998) |
The consensus sequence for leucine-rich NESs is shown at the top. ψ indicates hydrophobic residues, including leucine, isoleucine, methionine and valine. x indicates any amino acid. Hydrophobic residues are shown in bold. Numbers indicate the amino acid position of A/WSN/33 NS2 protein.
Most nucleocytoplasmic transport processes studied so far require the Ran-GTPase protein that shuttles between the nucleus and the cytoplasm (reviewed in Mattaj and Englmeier, 1998; Gorlich and Kutay, 1999; Moroianu, 1999). The low intrinsic GTPase activity of Ran is stimulated by two cytoplasmic proteins, the Ran GTPase-activating protein (RanGAP1) (Bischoff et al., 1994, 1995) and the Ran-binding protein 1 (RanBP1) (Coutavas et al., 1993), resulting in GDP-bound Ran in the cytoplasm. Conversely, the replacement of GDP with GTP is stimulated by the nuclear nucleotide exchange factor RCC1 (Ohtsubo et al., 1989; Bischoff and Ponstingl, 1991), leading to high concentrations of Ran-GTP in the nucleus. The asymmetric distribution of Ran-GDP and Ran-GTP is believed to determine the directionality of transport processes. For protein transport across the nuclear membrane, import receptors bind their substrates in the cytoplasm and translocate them to the nucleus where Ran-GTP induces the release of the imported substrate from the import receptor. In contrast, export receptors such as CRM1 bind their substrates only in the presence of Ran-GTP (Fornerod et al., 1997a; Fukuda et al., 1997; Stade et al., 1997), forming a ternary complex with the substrate that is released in the cytoplasm after the stimulation of GTP hydrolysis by RanGAP and RanBP1. The leucine-rich NES is crucial for nuclear export of the respective substrate; however, its exact role in the formation of the ternary export complex is controversial. Whereas most studies suggest that the leucine-rich NES is critical for direct interaction with CRM1 (Fornerod et al., 1997a; Fukuda et al., 1997; Ossareh-Nazari et al., 1997; Bogerd et al., 1998; Ossareh-Nazari and Dargemont, 1999), one study indicates that it is dispensable for CRM1 binding but required for the formation of the ternary export complex (Askjaer et al., 1998).
Influenza virus, unlike most other RNA viruses, replicates in the nucleus of infected cells (reviewed in Lamb and Krug, 1996). After receptor-mediated endocytosis, viral ribonucleoprotein (vRNP) complexes, composed of negative-strand viral RNA (vRNA), nucleoprotein (NP) and three polymerase proteins (PA, PB1 and PB2), dissociate from the matrix (M1) protein and enter the nucleus (Martin and Helenius, 1991a,b), where vRNAs are replicated and transcribed. The newly synthesized NP, M1 and polymerase proteins are transported into the nucleus and assembled into vRNPs. Little is known about the mechanism by which vRNPs are exported from the nucleus late in infection. O’Neill et al. (1998) proposed a leucine-rich NES in the N-terminal region of the NS2 protein (Table I) because it interacts with nucleoporins in the yeast two-hybrid system. Alteration of the NES abolished this interaction. Furthermore, the N-terminal region of NS2 replaced the functional domain of HIV-Rev, which promotes the nuclear export of HIV RNA, and mediated nuclear export when fused to a reporter protein (O’Neill et al., 1998). These findings suggested a role for NS2 in vRNP nuclear export. In contrast, Bui et al. (2000) concluded that NS2 is not required for vRNP nuclear export. This conclusion was based on the findings that vRNPs were retained in the nucleus of virus-infected cells treated with the protein kinase inhibitor H7, which downregulates expression of the M1 and NS2 proteins; however, vRNP nuclear export was restored by providing M1 from an expression vector. Since, in this experiment, low amounts of NS2 may have been expressed even in the presence of H7, a contribution of this protein to vRNP nuclear export cannot be excluded.
Recently, we and others established a system for the generation of infectious influenza virus entirely from cloned cDNA (Fodor et al., 1999; Neumann et al., 1999). The transfection of plasmids that contain cDNAs encoding all eight vRNAs of A/WSN/33 (H1N1) virus, controlled by RNA polymerase I promoter and terminator sequences, results in vRNA synthesis by cellular RNA polymerase I. Cotransfection of cells with plasmids for the synthesis of all viral structural proteins yields >107 infectious viruses per milliliter of supernatant (Neumann et al., 1999). We also established a system for the production of virus-like particles (VLPs) by expressing all nine structural proteins and virus-like RNA from plasmids (Neumann et al., 2000). With these advances, any mutation can now be introduced into the genome of influenza virus, allowing one to address long-standing questions concerning the viral proteins and signals that regulate vRNP nucleocytoplasmic transport. In this study, we determined directly whether NS2 is required for vRNP nuclear export. Moreover, we provide evidence for an interaction between NS2 and CRM1, and address the significance of the putative NS2-NES for this interaction as well as for the influenza viral life cycle.
Results
NS2 interacts with the cellular export factor hCRM1
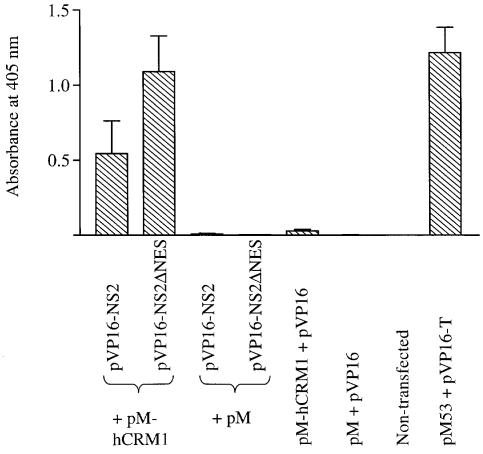
The influenza A virus NS2 protein contains a sequence resembling a leucine-rich NES, composed of M16, M19 and L21 (Table I). To determine whether NS2 interacts with human CRM1 (hCRM1), a cellular export factor for proteins with leucine-rich NESs (Fornerod et al., 1997a; Stade et al., 1997), we studied the interaction between these proteins in the mammalian two-hybrid system. 293T cells were cotransfected with a construct that encodes the GAL4 DNA-binding domain fused to hCRM1 (pM-hCRM1), a construct that encodes the herpes simplex virus (HSV) 16 activation domain fused to NS2 (pVP16-NS2), as well as a reporter gene construct (pG5CAT) expressing chloramphenicol acetyltransferase (CAT). At 48 h after transfection, the level of CAT expression in cell lysates was determined by ELISA. This analysis revealed an interaction between wild-type NS2 (pVP16-NS2) and hCRM1 (pM-hCRM1) (Figure 1). Negative control experiments included cotransfection of the reporter gene construct with pVP16 + pM-hCRM1, pVP16-NS2 + pM or pM + pVP16, as well as non-transfected cells. None of these experiments yielded significant levels of CAT expression (Figure 1). A strong reaction was observed in a positive control (Luo et al., 1997): 293T cells cotransfected with pM53 (encoding p53 fused to the GAL4 DNA-binding domain), pVP16-T (encoding SV40 T-antigen fused to the HSV 16 activation domain) and pG5CAT (Figure 1).
Fig. 1. Interaction of NS2 or its mutant with hCRM1 in the mammalian two-hybrid assay. 293T cells were cotransfected with the reporter gene construct pG5CAT, pM-hCRM1 (hCRM1 fused to the Gal4 DNA-binding domain) and wild-type or mutant NS2, fused to the HSV 16 activation domain (pVP16-NS2 or pVP16-NS2ΔNES). Negative controls included non-transfected cells, and cells transfected with the reporter gene and pVP16 + pM-hCRM1, pVP16-NS2 or pVP16-NS2ΔNES + pM, or pVP16 + pM. As a positive control, we determined the interaction between p53, fused to the Gal4-binding domain (pM53), and SV40 T-antigen, fused to the HSV 16 activation domain (pVP16-T). Forty-eight hours after transfection, CAT-ELISAs were performed. The bars denote the absorbance at 405 nm, which corresponds to the level of CAT expression and thus the level of interaction between hCRM1 and the protein of interest. The results are the mean ± SD for three experiments.
Replacement of the leucine-rich motif of NS2 does not abolish its interaction with hCRM1
Leucine-rich NESs are critical for the nuclear export of the NES-containing substrate; however, the exact role of the leucine-rich motif in the formation of the ternary export complex, composed of an export substrate (e.g. NS2), an export receptor (e.g. CRM1) and Ran-GTP, is controversial (Fornerod et al., 1997a; Fukuda et al., 1997; Ossareh-Nazari et al., 1997; Askjaer et al., 1998; Bogerd et al., 1998; Ossareh-Nazari and Dargemont, 1999). Therefore, we determined the significance of the putative NS2-NES for interaction with hCRM1 by replacing amino acids M16, M19 and L21 of NS2 with alanine. The plasmid expressing this mutant NS2 (pVP16-NS2ΔNES) was transfected into 293T cells together with pM-hCRM1 and pG5CAT. The alteration of the putative NS2-NES did not affect the binding of the mutated NS2 to hCRM1 (Figure 1). In fact, we consistently detected stronger interactions between these proteins. These data suggest that NS2 interacts with hCRM1 independently of the leucine-rich motif.
NS2 is essential for the viral life cycle
To establish whether the leucine-rich motif in the N-terminal region of NS2 forms a functional NES, we first determined whether NS2 is essential for influenza virus replication. Transfection of 293T cells with all eight RNA polymerase I plasmids for vRNA production, together with protein expression plasmids for all viral structural proteins, resulted in >107 infectious viruses per milliliter of supernatant, as described (Neumann et al., 1999). However, the omission of pPolI-WSN-NS, which drives the production of NS vRNA, abrogated the generation of virus (Table II). Thus, protein(s) encoded by segment 8 (NS1 and/or NS2) are crucial for the viral life cycle. Since influenza virus can survive in the absence of NS1 (Garcia-Sastre et al., 1998), our failure to generate viruses lacking NS vRNA was likely to be due to the absence of NS2.
Table II. Effects of NS gene products on NP localization and virus generationa.
| RNA polymerase I plasmid used | Protein(s) encoded by the RNA polymerase I plasmid | NP localizationb | Virus generationc |
|---|---|---|---|
| pPol-WSN-NS | NS1(wt), NS2(wt) | cytoplasm + nucleus | yes |
| No pPolI-WSN-NS | No NS1, No NS2 | nucleus | no |
| pPolI-WSN-NSΔSplice | NS1(wt), No NS2 | nucleus | no |
| pPolI-WSN-NS-STOP124 | NS1(aa1–124)d, NS2(wt) | cytoplasm + nucleus | yes |
| pPolI-WSN-NS-STOPΔNES | NS1(aa1–124), NS2ΔNESe | nucleus | no |
aResults are from three independent experiments.
b293T cells were transfected with protein expression plasmids for all viral structural proteins, the respective RNA polymerase I construct for NS vRNA synthesis and the remaining RNA polymerase I plasmids. Forty-eight hours after transfection of 293T cells, MDCK cells were infected with aliquots of supernatants from the former cells. Cells were fixed 24 h after infection and processed for indirect immunofluorescence assays, using antibodies against NP (see also Figure 4).
c293T cells were transfected with the same set of plasmids as described above. At 48 h after transfection, virus in the culture supernatant was titrated in MDCK cells. ‘No’ indicates no virus generation, whereas ‘yes’ indicates ≥104 TCID50/ml.
dNS1(aa1–124) indicates a truncated NS1 protein expressing only the N-terminal 124 amino acids.
eNS2ΔNES indicates an altered NS2-NES (16-AxxAxA-21).
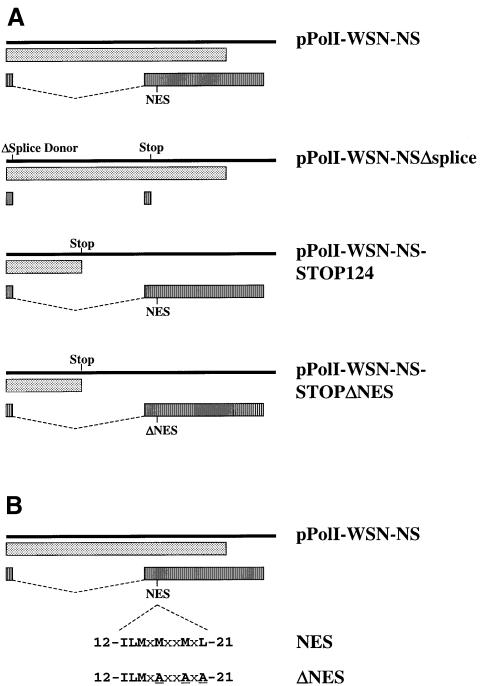
To test this prediction, we attempted to generate a virus encoding wild-type NS1, but not NS2, which is encoded by a spliced mRNA derived from the NS segment (Figure 2A). We therefore introduced a point mutation into the splice donor sequence (G56A, with reference to WSN-NS cRNA) and created a stop signal at codon 17 of NS2 (C548A) (Figure 2A). Cotransfection of the resulting plasmid (pPolI-WSN-NSΔsplice) with the remaining RNA polymerase I and protein expression plasmids did not yield virus (Table II). This failure cannot be attributed to the lack of particle production in transfected 293T cells expressing NS2 from a plasmid. Virus particles were produced and infected Madin–Darby canine kidney (MDCK) cells, as determined by NP expression (see below and Figure 4A), but no infectious progeny viruses were generated, presumably due to the lack of a functional NS2 coding region. These findings establish the importance of NS2 in the viral life cycle.
Fig. 2. (A) Schematic representation of RNA polymerase I constructs for NS vRNA synthesis. Solid bars represent NS vRNAs synthesized by RNA polymerase I. Translation products are shown as boxes (dotted boxes, NS1; striped boxes, NS2). pPolI-WSN-NSΔSplice: ‘ΔSplice Donor’ and ‘Stop’ indicate alteration of the splice donor sequence (G56A) and introduction of a stop signal at codon 17 in the NS2 reading frame, respectively. For all other constructs, ‘Stop’ indicates the introduction of two stop signals at codons 125 and 126 in the NS1 coding sequence. NES represents the leucine-rich motifs of NS2 (see below), whereas ΔNES indicates the altered export signal. (B) Amino acid sequences of NS2-NES and NS2ΔNES. To generate NS2ΔNES, we replaced the conserved hydrophobic amino acids with alanine, as indicated. The numbers indicate the amino acid position of A/WSN/33 NS2 protein.
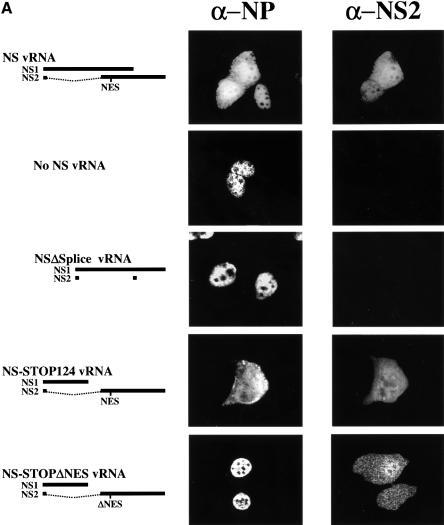
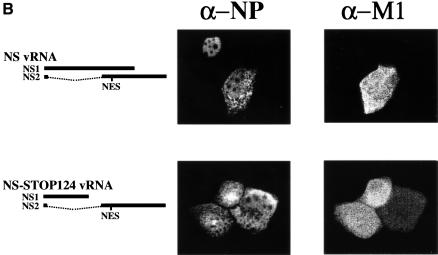
Fig. 4. Intracellular localization of NP and NS2 (A), or NP and M1 (B). MDCK cells were infected with VLPs, and immunofluorescence assays were performed 24 h p.i. NP localization was assessed with an anti-NP monoclonal antibody. NS2 and M1 were detected with rabbit antisera against these proteins.
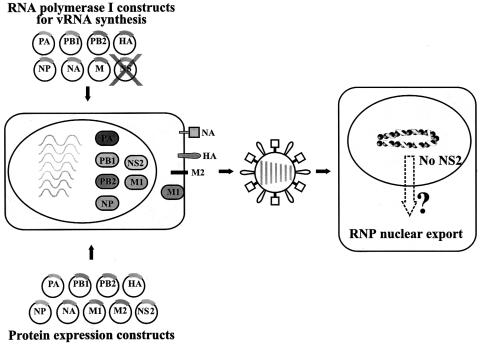
NS2 is essential for NP nuclear export
To determine whether the failure to generate virus in the absence of NS2 is due to vRNP nuclear retention, we studied NP localization [a generally accepted indicator of vRNP localization (Martin and Helenius, 1991a,b; Kemler et al., 1994; Bui et al., 1996; Whittaker et al., 1996)] in cells infected with VLPs lacking the coding capacity for NS2 (Figure 3). We produced VLPs by transfecting 293T cells with protein expression plasmids for all viral structural proteins together with seven RNA polymerase I constructs, omitting the RNA polymerase I NS construct, or with seven RNA polymerase I constructs for wild-type vRNAs and an RNA polymerase I construct encoding NS1 but not NS2. VLP-containing supernatant of 293T cells was collected 48 h after transfection and used to infect MDCK cells. Twenty-four hours later, cells were fixed and the intracellular localization of NP was determined by double immunofluorescence assays using antibodies against NP and NS2. In cells infected with artificially generated wild-type virus (as a positive control), NP localized to both the nucleus and the cytoplasm in cells that express NS2 (Figure 4A, top panel), but was confined to the nucleus in cells infected with virus particles lacking NS vRNA (Figure 4A, second panel) or encoding NS1 but not NS2 protein (Figure 4A, third panel). In the latter experiment, double immunofluorescence assays using antibodies against NP and NS1 confirmed the expression of NS1 and thus the presence of the altered NS vRNA (data not shown). Since the M1 protein has also been suggested to participate in vRNP nuclear export (Martin and Helenius, 1991a; Bui et al., 1996, 2000) we performed double immunofluorescence assays with antibodies against NP and M1. About 90% of the NP-positive MDCK cells were also positive for M1 in all samples. In cells infected with artificially generated wild-type influenza virus, NP localized to both the nucleus and the cytoplasm in cells expressing M1, but accumulated in the nucleus in cells lacking M1 expression (Figure 4B, top panel). Taken together, nuclear NP accumulation in cells infected with NS2-deficient particles was not due to the lack of M1 but to the lack of NS2, suggesting the essential role of the latter protein in vRNP nucleocytoplasmic transport.
Fig. 3. Schematic diagram of a system for the analysis of the role of NS2 in NP nuclear export, using VLPs. 293T cells were transfected with plasmids expressing all viral structural proteins and seven RNA polymerase I plasmids for vRNA synthesis, omitting pPolI-WSN-NS. Forty-eight hours after transfection, supernatants derived from transfected cells were used to infect MDCK cells. Cells were fixed at 24 h p.i. and processed for indirect immunofluorescence assays, using antibodies against NP and NS2 to evaluate the effects of NS2 on RNP nuclear export.
Amino acids M16, M19 and L21 of NS2 are crucial for viral replication
Others have identified a leucine-rich NES-like sequence comprised of M16, M19 and L21 of NS2 as a potential contributor to vRNP nuclear export (O’Neill et al., 1998), but did not provide direct evidence of an essential role for the proposed NES in virus-infected cells. To substantiate this putative role, we planned to replace conserved hydrophobic amino acids in the NES with alanine. However, since such replacements would alter the overlapping NS1 reading frame, we first introduced two consecutive stop signals downstream of codon 124 of NS1 (Figure 2A). This modification seemed logical, as influenza A and B viruses with C-terminal deletions of NS1 occur naturally (Norton et al., 1987; Tanaka et al., 1988; Tobita et al., 1990), and viruses encoding only the N-terminal 99 (Talon et al., 2000) or 124 amino acids of NS1 (Egorov et al., 1998) have been generated by conventional reverse genetics. Also, a virus lacking the entire NS1 gene has been generated (Garcia-Sastre et al., 1998). Cotransfection of the resulting construct (pPolI-WSN-NS-STOP124) with the remaining plasmids for virus generation yielded infectious virus (Table II). We then replaced M16, M19 and L21 of NS2 with alanine without affecting the truncated NS1 reading frame, thus creating pPolI-WSN-NS-STOPΔNES (Figure 2A and B). Experiments in which this plasmid was used for virus production did not yield infectious virus (Table II), demonstrating the lethality of the NES alteration.
M16, M19 and L21 of NS2 are critical for NP nucleocytoplasmic transport
Failure to generate virus encoding an altered putative export signal might be caused by nuclear retention of NP. To test this hypothesis, we produced virus particles encoding a mutant NS2, with alanine to replace M16, M19 and L21, and determined whether these amino acids are required for NP nucleocytoplasmic transport. Infection of MDCK cells with virus particles encoding NS2ΔNES (16-AxxAxA-21) resulted in the nuclear confinement of NP (Figure 4A, bottom panel), and the localization of NS2ΔNES to the nucleus and the cytoplasm indicated that NP nuclear retention was not caused by the absence of NS2. To rule out the possibility that the C-terminal deletion of NS1 affected NP nuclear export, we generated virus particles expressing truncated NS1 but wild-type NS2, using pPolI-WSN-NS-STOP124 (Figure 2A; Table II). The extent of NP cytoplasmic accumulation in MDCK cells infected with these virus particles was comparable to that in cells infected with wild-type virus (Figure 4A, fourth panel), demonstrating the lack of any contribution from the deletion of the C-terminal 108 amino acids of NS1 to the nuclear retention of NP. As in experiments described earlier, ∼90% of NP-expressing cells also expressed NS2 (Figure 4A, fourth panel) and M1 (Figure 4B, second panel). Taken together, these results show that alanine replacement of M16, M19 and L21 of NS2 leads to the retention of NP in the nucleus, providing direct evidence that the proposed NES in the N-terminal region of this protein functions critically in the nuclear export of NP.
Discussion
In this report, we provide evidence for an interaction between the cellular nuclear export factor hCRM1 and the influenza virus NS2 protein. Alteration of the putative NS2-NES did not abolish this interaction, demonstrating that the NS2-NES is not required for interaction with hCRM1. However, replacement of the putative NS2-NES abolished the nuclear export of NP and the generation of influenza virus, demonstrating its function as an NES.
Some discrepancy exists about the exact role of NESs for interaction with CRM1. Most studies demonstrated that the alteration of leucine-rich NESs abolished the binding of the NES-containing substrate to CRM1 (Fornerod et al., 1997a; Fukuda et al., 1997; Ossareh-Nazari et al., 1997; Bogerd et al., 1998; Ossareh-Nazari and Dargemont, 1999). In contrast, Askjaer et al. (1998) reported binding of CRM1 to HIV-Rev independent of its NES. Here, we also demonstrate that the replacement of an NES did not abolish the interaction of the NES-containing substrate with CRM1. How can this discrepancy be explained? Two structural features are likely to influence the interaction between an NES-containing substrate and CRM1: the nature of the NES and regions surrounding the NES. The affinity for CRM1 differs among NESs (Askjaer et al., 1999). The NES of HIV-Rev, which interacts with CRM1 in an NES-independent manner, has a lower affinity for CRM1 than others (Askjaer et al., 1999). Thus, for HIV-Rev and possibly NS2, the contribution of regions surrounding the NES may be relatively high so that the alteration of these NESs does not affect their CRM1 binding. Alternatively, the discrepancy may be due to experimental conditions. Studies demonstrating an interaction of CRM1 with its substrate in an NES-independent manner used full-length proteins (Askjaer et al., 1998; this report), while in those demonstrating NES-dependent interaction with CRM1, NES-peptides rather than full-length proteins were used (Fukuda et al., 1997; Ossareh-Nazari and Dargemont, 1999). Although NES-peptides contain sufficient information to mediate nuclear export of their fusion partners (Fukuda et al., 1997; Ossareh-Nazari and Dargemont, 1999), optimal interaction with CRM1 may require additional protein domains.
Why does the alteration of the NS2-NES abolish NP nuclear export without affecting the interaction with hCRM1? The hydrophobic residues forming the NES may be critical for the formation of the ternary export complex, as supported by the finding that the alteration of the HIV-Rev NES did not abolish its interaction with CRM1 but did abolish the formation of the ternary export complex (Askjaer et al., 1998). Thus, the NES may promote the formation of a stable export complex. In the yeast two-hybrid system, replacement of the NS2-NES abolished the protein’s (likely to be indirect) interaction with nucleoporins (O’Neill et al., 1998). This finding could be explained by our hypothesis that NESs are not crucial for the direct interaction of NS2 with the cellular export factor (i.e. CRM1), but are required for the formation of the ternary export complex that interacts with nucleoporins, as has been shown in other systems (Fornerod et al., 1997a).
Disruption of the NS2-NES resulted in a stronger interaction with hCRM1 compared with that of wild-type NS2. Nuclear export of proteins and RNAs is a dynamic process that involves the formation of the export complex, its translocation across the nuclear membrane, disassembly and release of the export substrate into the cytoplasm. Thus, at any given time, only a subpopulation of wild-type NS2 proteins are bound to hCRM1. NES-deficient NS2, although bound by hCRM1, is not exported but remains in the nucleus in its hCRM1-bound state. This discrepancy in binding patterns would explain the stronger indication of NS2–hCRM1 binding in the mammalian two-hybrid system.
By generating VLPs, which lack a particular viral gene, we found that NS2 is required for vRNP nuclear export (Table II; Figure 4). To execute this function, NS2 has to interact with vRNPs. M1 interacts with both NS2 (Yasuda et al., 1993; Ward et al., 1995) and vRNPs (Ye et al., 1987), and may therefore link vRNPs to the viral export factor NS2, and hence to the cellular export factor hCRM1. Alternatively, NS2 may interact with RNPs through an as yet unidentified mechanism.
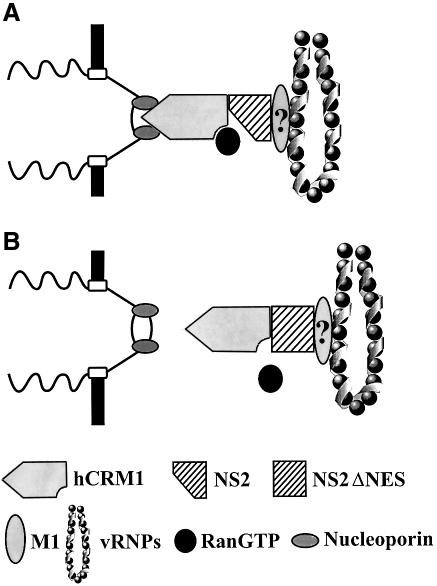
Taken together, our findings suggest the following model (Figure 5). vRNP nuclear export is mediated by NS2, which possibly interacts with vRNPs through M1 as well as with hCRM1 to form a functional export complex. Although the NS2-NES does not directly affect its interaction with hRCM1, its alteration is likely to abolish the formation of the ternary export complex with Ran-GTP, resulting in nuclear retention of vRNPs.
Fig. 5. A model for NS2-mediated vRNP nuclear export through the hCRM1 nuclear export pathway. (A) A ternary export complex is formed between hCRM1, Ran-GTP and NS2, which possibly interacts with vRNPs through M1. Interaction of the export complex with nucleoporins triggers nuclear export. (B) Replacement of the NS2-NES does not abrogate interaction with hCRM1, but does the formation of a stable ternary export complex and hence vRNP nuclear export, although experimental evidence is lacking.
Materials and methods
Cells
293T human embryonic kidney cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal calf serum. MDCK cells were grown in MEM supplemented with 5% newborn calf serum. Cells were maintained at 37°C and 5% CO2.
Construction of plasmids
The generation of RNA polymerase I constructs encoding wild-type vRNAs of A/WSN/33 (H1N1) virus was described previously (Neumann et al., 1999). The names of the plasmids indicate the vRNA segment transcribed from the respective constructs (e.g. pPolI-WSN-HA contains the A/WSN/33 HA gene controlled by RNA polymerase I promoter and terminator sequences).
To generate pPolI-WSN-NSΔSplice (Figure 2A), we used primers containing nucleotide replacements at positions 56 (G56A) and 548 (C548A) of the NS cRNA to amplify the NS gene, and then cloned the PCR product into the BamHI and PpuMI sites of pPolI-WSN-NS. Plasmid pPolI-WSN-NS-STOP124 was generated by PCR using primers that introduced stop signals at codons 125 and 126 of NS1. The putative NES of NS2 was altered by PCR using primers that replace M16, M19 and L21 with alanine. The resulting fragment was cloned into the HindIII and PpuMI sites of pPolI-WSN-NS, thus generating pPolI-WSN-NSΔNES. An SspI fragment derived from this construct was ligated with an SspI fragment derived from pPolI-WSN-NS-STOP124 to generate pPolI-WSN-NS-STOPΔNES. This construct encodes a mutated NS2-NES (16-AxxAxA-21) and a truncated NS1 protein (Figure 2A and B). The sequences of the primers will be provided upon request. All PCR-amplified regions were sequenced to ensure that the constructs did not contain unwanted mutations.
Protein expression plasmids for the synthesis of all viral structural proteins are described elsewhere (Neumann et al., 1999). Briefly, genes encoding the PB2, PB1 and PA proteins of A/PR/8/34 virus are controlled by the cytomegalovirus promoter (Perez and Donis, 1998). The chicken β-actin promoter controls expression of the HA, NP, NA and M1 genes (all derived from A/WSN/33 virus), as well as the M2 and NS2 genes (derived from A/PR/8/34 virus).
Generation of infectious influenza virus
Infectious virus was generated as described (Neumann et al., 1999). Forty-eight hours after transfection, virus in supernatants derived from transfected cells was titrated in MDCK cells.
Generation of VLPs with an altered genome
To generate VLPs either lacking the NS gene segment or containing mutations in this segment, we cotransfected protein expression plasmids with seven RNA polymerase I constructs (omitting pPolI-WSN-NS), or with seven wild-type RNA polymerase I constructs and an RNA polymerase I construct encoding mutant NS2 and/or NS1 protein. Forty-eight hours later, aliquots of the supernatant were used to infect MDCK cells. Cells were fixed at 24 h post-infection (p.i.) and processed for indirect immunofluorescence.
Immunofluorescence assays
Viral proteins were detected using monoclonal or polyclonal antibodies as follows. A monoclonal antibody (WSN 3/1; 1:500 dilution) was used for NP. The NS2 protein was detected with rabbit antiserum R5023 (1:100 dilution), which was produced against the A/PR/8/34 (H1N1) NS2 protein that contained a histidine tag and was expressed using the bacterial expression vector pET5a (Novagen, Madison, WI) and purified using Talon columns (Clontech, Palo Alto, CA). An antiserum to NS2 (Greenspan et al., 1985), kindly provided by Dr Peter Palese (Mount Sinai School of Medicine, New York, NY), was used to determine the authenticity of our anti-NS2 antiserum. Rabbit antiserum R74-95 (1:300 dilution) was used to detect M1. A mixture of anti-NS1 antibodies (9/1, 18/1, 19/1, 30/2, 31/3, 91/2, 1:500 dilution each; kindly provided by Dr Robert Webster, St Jude Children’s Research Hospital, Memphis, TN) allowed its detection. Fluorescein isothiocyanate- or Texas red-labeled goat anti-rabbit or anti-mouse IgG antibodies (1:200 dilution each) (Boehringer Mannheim, Germany, or JacksonImmuno, West Grove, PA) served as secondary antibodies.
Mammalian two-hybrid assay
In this assay, the proteins of interest are fused to the Gal4 DNA-binding domain (encoded by the vector pM; Clontech, Palo Alto, CA) or the HSV 16 activation domain (encoded by pVP16; Clontech, Palo Alto, CA). Their interaction brings the DNA-binding and activation domains into close proximity, initiating transcription of a reporter gene, in this instance, CAT.
To generate NS2 fusion proteins, we amplified wild-type NS2 or NS2ΔNES using PCR and a primer that spans the splice donor and acceptor sequences, as well as a primer that binds to the 3′ end of the coding region for NS2. The resulting PCR fragment, which encodes NS2 but not NS1, was cloned into the EcoRI and BamHI sites of pVP16, generating pVP16-NS2 and pVP16-NSΔNES. The coding region for hCRM1, derived from plasmid T7-hCRM1 kindly provided by M.Fornerod (EMBL, Heidelberg, Germany), was ligated to short synthetic DNA adaptors to provide BamHI and XhoI sites, which were filled in with Klenow polymerase to allow fusion to the Gal4 DNA-binding domain, yielding pM-hCRM1. Control plasmids pM53 and pVP16-T were also obtained from Clontech (Palo Alto, CA).
To analyze the interaction between hCRM1 and NS2, we transfected 293T cells with 2 µg of the pM and pVP16 fusion constructs and 0.4 µg of the reporter gene construct pG5CAT (Clontech, Palo Alto, CA). Forty-eight hours after transfection, CAT expression was detected with CAT-ELISA (Boehringer Mannheim, Germany). Briefly, 1:10 dilutions of cell lysates were added to plates coated with anti-CAT antibody, incubated for 1 h at 37°C, washed, and incubated with digoxigenin (DIG)-labeled antibody to CAT for 1 h at 37°C. Plates were washed again and incubated with anti-DIG horseradish peroxidase-labeled antibody for 1 h at 37°C. After a final washing step, the peroxidase substrate 2,2′-azino-bis[3-ethylbenzthiazoline-6-sulfonic acid] was added. The absorbance of the colored reaction product was quantified in an ELISA reader at 405 nm.
Acknowledgments
Acknowledgements
We thank Dr Maarten Fornerod (EMBL, Heidelberg, Germany) for T7-hCRM1, and Dr Robert Webster (St Jude Children’s Research Hospital, Memphis, TN) and Dr Peter Palese (Mount Sinai School of Medicine, New York, NY) for monoclonal antibodies against NS1 and antiserum against NS2, respectively. We also thank Yuko Kawaoka for illustrations, Martha McGregor and Krisna Wells for excellent technical assistance and John Gilbert for editing the manuscript. Automated sequencing was performed at the University of Wisconsin-Biotechnology Center. Support for this work came from National Institute of Allergy and Infectious Diseases Public Health Service research grants.
References
- Askjaer P., Jensen,T.H., Nilsson,J., Englmeier,L. and Kjems,J. (1998) The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J. Biol. Chem., 273, 33414–33422. [DOI] [PubMed] [Google Scholar]
- Askjaer P. et al. (1999) RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol. Cell. Biol., 19, 6276–6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F.R. and Ponstingl,H. (1991) Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature, 354, 80–82. [DOI] [PubMed] [Google Scholar]
- Bischoff F.R., Klebe,C., Kretschmer,J., Wittinghofer,A. and Ponstingl,H. (1994) RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc. Natl Acad. Sci. USA, 91, 2587–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F.R., Krebber,H., Kempf,T., Hermes,I. and Ponstingl,H. (1995) Human RanGTPase-activating protein RanGAP1 is a homologue of yeast Rna1p involved in mRNA processing and transport. Proc. Natl Acad. Sci. USA, 92, 1749–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd H.P., Echarri,A., Ross,T.M. and Cullen,B.R. (1998) Inhibition of human immunodeficiency virus Rev and human T-cell leukemia virus Rex function, but not Mason–Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1. J. Virol., 72, 8627–8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui M., Whittaker,G. and Helenius,A. (1996) Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J. Virol., 70, 8391–8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui M., Wills,E.G., Helenius,A. and Whittaker,G.R. (2000) Role of the influenza virus M1 protein in nuclear export of viral ribonucleoproteins. J. Virol., 74, 1781–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutavas E., Ren,M., Oppenheim,J.D., D’Eustachio,P. and Rush,M.G. (1993) Characterization of proteins that interact with the cell-cycle regulatory protein Ran/TC4. Nature, 366, 585–587. [DOI] [PubMed] [Google Scholar]
- Egorov A. et al. (1998) Transfectant influenza A viruses with long deletions in the NS1 protein grow efficiently in Vero cells. J. Virol., 72, 6437–6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U., Huber,H., Boelens,W.C., Mattaj,I. and Luhrmann,R. (1995) The HIV-Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell, 82, 475–483. [DOI] [PubMed] [Google Scholar]
- Fodor E., Devenish,L., Engelhardt,O.G., Palese,P., Brownlee,G.G. and Garcia-Sastre,A. (1999) Rescue of influenza A virus from recombinant DNA. J. Virol., 73, 9679–9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M., Ohno,M., Yoshida,M. and Mattaj,I.W. (1997a) CRM1 is an export receptor for leucine-rich nuclear export signals. Cell, 90, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Fornerod M., van Deursen,J., van Baal,S., Reynolds,A., Davis,D., Murti,K.G., Fransen,J. and Grosveld,G. (1997b) The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J., 16, 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M., Gotoh,I., Gotoh,Y. and Nishida,E. (1996) Cytoplasmic localization of mitogen-activated protein kinase kinase directed by its NH2-terminal, leucine-rich short amino acid sequence, which acts as a nuclear export signal. J. Biol. Chem., 271, 20024–20028. [DOI] [PubMed] [Google Scholar]
- Fukuda M., Asano,S., Nakamura,T., Adachi,M., Yoshida,M., Yanagida,M. and Nishida,E. (1997) CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature, 390, 308–311. [DOI] [PubMed] [Google Scholar]
- Garcia-Sastre A., Egorov,A., Matassov,D., Brandt,S., Levy,D.E., Durbin,J.E., Palese,P. and Muster,T. (1998) Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology, 252, 324–330. [DOI] [PubMed] [Google Scholar]
- Gorlich D. and Kutay,U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol., 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Greenspan D., Krystal,M., Nakada,S., Arnheiter,H., Lyles,D.S. and Palese,P. (1985) Expression of influenza virus NS2 nonstructural protein in bacteria and localization of NS2 in infected eucaryotic cells. J. Virol., 54, 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemler I., Whittaker,G. and Helenius,A. (1994) Nuclear import of microinjected influenza virus ribonucleoproteins. Virology, 202, 1028–1033. [DOI] [PubMed] [Google Scholar]
- Lamb R.A. and Krug,R.M. (1996) Orthomyxoviridae: the viruses and their replication. In Fields,B.N., Knipe,D.M. and Howley,P.M. (eds), Fields Virology. Lippincott-Raven, Philadelphia, PA, pp. 1353–1395. [Google Scholar]
- Luo Y., Batalao,A., Zhou,H. and Zhu,L. (1997) Mammalian two-hybrid system: a complementary approach to the yeast two-hybrid system. Biotechniques, 22, 350–352. [DOI] [PubMed] [Google Scholar]
- Martin K. and Helenius,A. (1991a) Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell, 67, 117–130. [DOI] [PubMed] [Google Scholar]
- Martin K. and Helenius,A. (1991b) Transport of incoming influenza virus nucleocapsids into the nucleus. J. Virol., 65, 232–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I.W. and Englmeier,L. (1998) Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem., 67, 265–306. [DOI] [PubMed] [Google Scholar]
- Moroianu J. (1999) Nuclear import and export pathways. J. Cell. Biochem. Suppl., 32–33, 76–83. [DOI] [PubMed] [Google Scholar]
- Neumann G. et al. (1999) Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl Acad. Sci. USA, 96, 9345–9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G., Watanabe,T. and Kawaoka,Y (2000) Plasmid-driven formation of influenza virus-like particles. J. Virol., 74, 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton G.P., Tanaka,T., Tobita,K., Nakada,S., Buonagurio,D.A., Greenspan,D., Krystal,M. and Palese,P. (1987) Infectious influenza A and B virus variants with long carboxyl terminal deletions in the NS1 polypeptides. Virology, 156, 204–213. [DOI] [PubMed] [Google Scholar]
- O’Neill R.E., Talon,J. and Palese,P. (1998) The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J., 17, 288–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo M., Okazaki,H. and Nishimoto,T. (1989) The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J. Cell Biol., 109, 1389–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossareh-Nazari B. and Dargemont,C. (1999) Domains of Crm1 involved in the formation of the Crm1, RanGTP and leucine-rich nuclear export sequences trimeric complex. Exp.Cell Res., 252, 236–241. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B., Bachelerie,F. and Dargemont,C. (1997) Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science, 278, 141–144. [DOI] [PubMed] [Google Scholar]
- Perez D.R. and Donis,R.O. (1998) The matrix 1 protein of influenza A virus inhibits the transcriptase activity of a model influenza reporter genome in vivo. Virology, 249, 52–61. [DOI] [PubMed] [Google Scholar]
- Stade K., Ford,C.S., Guthrie,C. and Weis,K. (1997) Exportin 1 (Crm1p) is an essential nuclear export factor. Cell, 90, 1041–1050. [DOI] [PubMed] [Google Scholar]
- Talon J., Salvatore,M., O’Neill,R.E., Nakaya,Y., Zheng,H., Muster,T., Garcia-Sastre,A. and Palese,P. (2000) Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl Acad. Sci. USA, 97, 4309–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Odagiri,T. and Tobita,K. (1988) Biological functions of the NS1 protein of an influenza B virus mutant which has a long carboxyl terminal deletion. Arch. Virol., 102, 173–185. [DOI] [PubMed] [Google Scholar]
- Tobita K., Tanaka,T., Odagiri,T., Tashiro,M. and Feng,S.-Y. (1990) Nucleotide sequence and some biological properties of the NS gene of a newly isolated influenza B virus mutant which has a long carboxyl terminal deletion in the NS1 protein. Virology, 174, 314–319. [DOI] [PubMed] [Google Scholar]
- Ward A.C., Castelli,L.A., Lucantoni,A.C., White,J.F., Azad,A.A. and Macreadie,I.G. (1995) Expression and analysis of the NS2 protein of influenza A virus. Arch. Virol., 140, 2067–2073. [DOI] [PubMed] [Google Scholar]
- Wen W., Meinkoth,J.L., Tsien,R.Y. and Taylor,S.S. (1995) Identification of a signal for rapid export of proteins from the nucleus. Cell, 82, 463–473. [DOI] [PubMed] [Google Scholar]
- Whittaker G., Bui,M. and Helenius,A. (1996) Nuclear trafficking of influenza virus ribonucleoproteins in heterokaryons. J. Virol., 70, 2743–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda J., Nakada,S., Kato,A., Toyoda,T. and Ishihama,A. (1993) Molecular assembly of influenza virus: association of the NS2 protein with virion matrix. Virology, 196, 249–255. [DOI] [PubMed] [Google Scholar]
- Ye Z.P., Pal,R., Fox,J.W. and Wagner,R.R. (1987) Functional and antigenic domains of the matrix (M1) protein of influenza A virus. J. Virol., 61, 239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]